Abstract
In previous studies, we reported that N-acetylglucosaminyltransferase III (GnT-III) activity and the enzyme product, bisected N-glycans, both were induced in cells cultured under dense conditions in an E-cadherin-dependent manner (Iijima, J., Zhao, Y., Isaji, T., Kameyama, A., Nakaya, S., Wang, X., Ihara, H., Cheng, X., Nakagawa, T., Miyoshi, E., Kondo, A., Narimatsu, H., Taniguchi, N., and Gu, J. (2006) J. Biol. Chem. 281, 13038–13046). Furthermore, we found that α-catenin, a component of the E-cadherin-catenin complex, was also required for this induction (Akama, R., Sato, Y., Kariya, Y., Isaji, T., Fukuda, T., Lu, L., Taniguchi, N., Ozawa, M., and Gu, J. (2008) Proteomics 8, 3221–3228). To further explore the molecular mechanism of this regulation, the roles of β-catenin, an essential molecule in both cadherin-mediated cell adhesion and canonical Wnt signaling, were investigated. Unexpectedly, shRNA knockdown of β-catenin resulted in a dramatic increase in GnT-III expression and its product, the bisected N-glycans, which was confirmed by RT-PCR and GnT-III activity and by E4-PHA lectin blot analysis. The induction of GnT-III expression increased bisecting GlcNAc residues on β1 integrin, which led to down-regulation of integrin-mediated cell adhesion and cell migration. Immunostaining showed that nuclear localization of β-catenin was greatly suppressed; intriguingly, the knockdown of β-catenin in the nuclei was more effective than that in cell-cell contacts in the knockdown cells, which was also confirmed by Western blot analysis. Stimulation of the Wnt signaling pathway by the addition of exogenous Wnt3a or BIO, a GSK-3β inhibitor, consistently and significantly inhibited GnT-III expression and its products. Conversely, the inhibition of β-catenin translocation into the nuclei increased GnT-III activation. Taken together, the results of the present study are the first to clearly demonstrate that GnT-III expression may be precisely regulated by the interplay of E-cadherin-catenin complex-mediated cell-cell adhesion and Wnt/β-catenin signaling, which are both crucial in the process of epithelial-mesenchymal transitions in physiological and pathological conditions.
Keywords: Carbohydrate, Carbohydrate Biosynthesis, Cell Adhesion, Cell Junctions, Glycosylation, Lectin, Glycosyltransferase, GnT-III, beta-Catenin
Introduction
Oligosaccharides (glycans) attached to proteins (glycosylation) are conserved in eukaryotes, and this is one of the most abundant posttranslational modification reactions (1). Glycosylation has been shown to play key roles in a variety of biological functions, such as bioactivity, folding, localization, and expression of the protein. In fact, changes in glycan structure are associated with many physiological and pathological events, including cell growth, migration, differentiation, and tumor invasion (2, 3). Production of glycoprotein glycans is catalyzed by various glycosyltransferases, and most of the cancer-associated changes of them are due to changes in glycosyltransferases (4). Functional glycomics, which uses sugar remodeling by glycosyltransferases, is known to be a promising tool for the understanding and characterization of glycan roles.
N-Acetylglucosaminyltransferase III (GnT-III)3 transfers N-acetylglucosamine (GlcNAc) from UDP-GlcNAc to a β1,4-mannose in N-glycans to form a “bisecting” GlcNAc linkage, as shown in Fig. 1A. Bisecting GlcNAc linkages are found in various hybrid and complex N-glycans. The addition of this bisecting GlcNAc residue alters not only the composition but also the conformation of the N-glycan (5). GnT-III is generally regarded as a key glycosyltransferase in N-glycan biosynthetic pathways. Introduction of the bisecting GlcNAc suppresses further processing and elongation of N-glycans catalyzed by GnT-V, which is strongly associated with cancer metastasis, because GnT-V cannot utilize the bisected oligosaccharide as a substrate (6–8). GnT-V activity and β1,6-branched N-glycans levels reportedly also are increased in highly metastatic tumor cell lines (9, 10). Consistently, cancer metastasis is greatly suppressed in GnT-V knock-out mice (11). GnT-III has, therefore, been proposed as an antagonist of GnT-V, thereby contributing to the suppression of cancer metastasis. In fact, overexpression of GnT-III in highly metastatic melanoma cells reduced β1,6 branching in cell surface N-glycans and increased bisected N-glycans (12). Cell-cell adhesion was enhanced due to prolonged turnover of E-cadherin on the cell surface in these GnT-III transfectants (13). These results further suggest that remodeling of glycosyltransferase-modified N-glycan structures modulates cell adhesion and cancer metastasis. It is, therefore, very important to clarify molecular mechanisms for the regulation of glycosyltransferase expression under physiological and pathological conditions.
FIGURE 1.
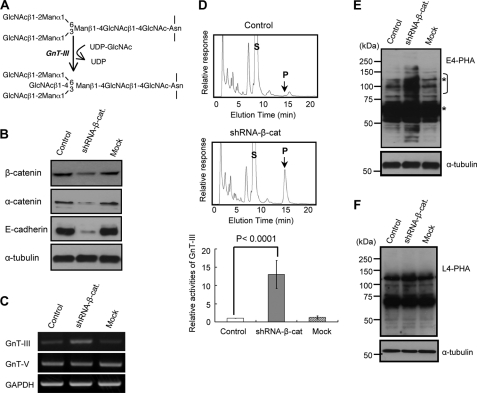
Effects of β-catenin knockdown on the GnT-III gene and its product expressions. A, reaction pathway for the synthesis of bisecting GlcNAc. Man, mannose. B, shRNA knockdown of β-catenin in a retroviral vector, as described under “Experimental Procedures.” Equal amounts of cell lysate protein were separated on 7.5% SDS-PAGE, and the membranes were probed with the indicated antibodies. α-Tubulin was used as a loading control. Control, without retroviral infection; Mock, retroviral infection with a random sequence. C, comparison of GnT-III gene expression levels among three cells. D, cells were then harvested under subconfluence. Equal amounts of cell lysate protein (20 μg) were used as the enzymatic source for GnT-III. Top and middle, a representative example of elution patterns of control and knockdown cells, respectively. S, substrate; P, product. The activity in control cells was set equal to 1. GnT-III activity is expressed as -fold increase relative to the activity in control cells (bottom). The shRNA knockdown of β-catenin resulted in a dramatic increase in the expression of bisected N-glycans but not β1,6-GlcNAc expression detected by E4-PHA (E) and L4-PHA (F), respectively. The asterisks indicate nonspecific staining of E4-PHA because those bands did not disappear after treatment with 100 mm acetic acid.
Cell-cell adherence junction formation and remodeling occur repeatedly throughout development. Epithelial cells are linked by adherence junctions that rely on the cadherin-catenin module. Cadherins, of which epithelial E-cadherin has been the most studied, are Ca2+-dependent transmembrane adhesion proteins forming homophilic and heterophilic bonds in trans between adjacent cells. The cytoplasmic carboxyl terminus of the E-cadherin is known to bind to p120-catenin and either of two closely related proteins, β-catenin or γ-catenin (plakoglobin), thereby linking the complex to α-catenin. Whereas p120-catenin acts to stabilize cadherins at the cell surface (14), β-catenin provides a link to α-catenin (15), which in turn has the ability to provide a functional link to the actin cytoskeleton, thus promoting junction protein clustering and stabilization of cellular adhesion. The ability of these junction core components to reorganize the actin cytoskeleton makes the assembly of cadherin-catenin adhesion complexes a highly dynamic process, which allows spatial reorganization of cells during normal development and cancer metastasis. In addition to their structural role in stabilizing adhesive contacts between the neighboring cells and directing actin cytoskeleton reorganization, components of the cadherin-catenin complex are tightly linked to several key signal transduction networks. The protein β-catenin plays a critical role in canonical Wnt signaling. The Wnt/β-catenin signaling pathway has a crucial role in the embryonic development of all animal species, in the regeneration of tissues in adult organisms, and in numerous other processes (16–18).
It is becoming clear that N-glycosylation can be regulated by cell-cell adhesion. We recently found that E-cadherin-mediated cell-cell interaction up-regulated GnT-III expression (19, 20). Significant up-regulation of GnT-III expression was observed only in epithelial cells that expressed basal levels of E-cadherin and GnT-III but not in E-cadherin-deficient cells, such as fibroblasts (20). The expression levels of GnT-III and its products, the bisected N-glycans, were up-regulated by cell-cell interaction via the E-cadherin-catenin-actin complex because disruption of actin polymerization or lack of α-catenin expression interfered with the regulation of GnT-III. The reintroduction of α-catenin into α-catenin-deficient cells rescued GnT-III expression enhanced under cell-cell adhesion (19), clearly indicating that the E-cadherin-catenin complex is essential for cell-cell adhesion-regulated GnT-III expression.
In the present study, the roles of β-catenin were investigated to further explore the detailed molecular mechanism for GnT-III regulation by E-cadherin-mediated cell adhesion. We found that β-catenin has both positive and negative effects on GnT-III expression, up-regulation by E-cadherin/β-catenin-mediated cell adhesion, and down-regulation by the Wnt/β-catenin pathway.
EXPERIMENTAL PROCEDURES
Cell Line and Cell Culture
Cells lacking α-catenin expression (DLD-1/Δα), a subclone of the human colon carcinoma DLD-1 cell line, were kindly provided by Dr. Shintaro T. Suzuki (Kwansei Gakuin University). The expression vector encoding the wild type α-catenin was transfected into DLD-1/Δα cells and selected using G418 to obtain a stable expression cell line (DLD-1/α-cat) (21, 22). DLD-1/α-cat cells used in this study were cultured in Dulbecco's modified Eagle's medium (DMEM) with high glucose (Sigma), supplemented with 10% fetal calf serum (FCS), 100 units/ml penicillin G, and 0.1 mg/ml streptomycin under a humidified atmosphere containing 5% CO2. Cells were plated at 5 × 106 and 5 × 105 on 150-mm dishes for dense culture and sparse culture, respectively, followed by incubation for 3 days (20). MCF-10A, a human non-tumorigenic immortalized breast epithelial cell line, was cultured in DMEM/F-12 medium supplemented with 5% horse serum, 20 ng/ml EGF, 10 μg/ml insulin, 0.5 μg/ml hydrocortisone, and 100 ng/ml cholera toxin under a humidified atmosphere containing 5% CO2 at 37 °C.
Western Blot, Immunoprecipitation, and Lectin Blot Analyses
Cells cultured under different conditions as indicated were washed with PBS and then lysed with lysis buffer (10 mm Tris-HCl, 1% Triton-X, 150 mm NaCl, aprotinin, leupeptin, and 1 mm phenylmethylsulfonyl fluoride). Insoluble materials were removed by centrifugation at 15,000 rpm for 10 min at 4 °C. Equal amounts of protein were separated using 7.5% SDS-PAGE, transferred to nitrocellulose, and probed with the appropriate antibodies, as indicated, or with biotinylated phytohemagglutinin-E4 (E4-PHA), phytohemagglutinin-L4 (L4-PHA), and Aleuria aurantia lectin (Seikagaku Kogyo Inc., Japan). Immunoreactive bands were visualized using a Vectastain ABC kit (Vector Laboratories, CA) and an ECL kit (Amersham Biosciences). Monoclonal antibodies against E-cadherin and β-catenin were purchased from BD Biosciences, and the anti-α-tubulin antibody was from Sigma. HRP-labeled anti-mouse IgG was obtained from Cell Signaling (Danvers, MA). For immunoprecipitation, the supernatant (2 mg of protein) was incubated with anti E-cadherin monoclonal antibody (3 μg/ml) (BD Biosciences) and anti-β1 integrin (P5D2), which was obtained from the Developmental Studies Hybridoma Bank, University of Iowa, for 1 h at 4 °C. Protein G beads (30 μl in 50% slurry) were then added, followed by incubation overnight at 4 °C with a rotator. After washing three times with lysis buffer, the immunoprecipitates were subjected to 8% SDS-PAGE, and the separated proteins were transferred to a nitrocellulose membrane. The membrane was incubated with a lectin for a lectin blot analysis or an antibody for immunoblot analysis.
GnT-III Activity Assay
After washing with PBS, the cultured cells were lysed by sonication. The cell lysate protein concentration was determined using a BCA protein assay kit (Pierce). Equal amounts of protein were used in the GnT-III activity assays, as described previously (23). The specific activity of GnT-III was determined using a substrate, 4-(2-pyridylamino)-butylamine-labeled GlcNAcβ1–2Manα1-6(GlcNAcβ1–2 Manα1–3)Manβ1–4GlcNAcβ1–4GlcNAc-Asn (24). Each assay used 5 mm substrate (in 10 μl of total reaction solution). The activity of endogenous GnT-III was measured by high performance liquid chromatography (HPLC), expressed as pmol of GlcNAc transferred/h/mg of proteins (20).
Microscopy and Cell Image
Cells were seeded on glass bottom dishes for 48 h before fixation. After washing two times with PBS, cells were fixed for 30 min in 3.7% paraformaldehyde solution at 37 °C. For permeabilization, the cells were treated with 0.2% (v/v) Triton X-100 in PBS. The fixed cells were blocked with 2% bovine serum albumin (BSA) in PBS for 1 h and were then incubated with anti-β-catenin and TO-PRO3 (Invitrogen) in blocking buffer for 1 h at room temperature. Following three washes in PBS, the cells were incubated with a 1:500 dilution of Alexa Fluor® 488 secondary antibody (Invitrogen) for 1 h at room temperature. After washing three times with PBS, the cells were analyzed using an Olympus fluorescence microscope (FV1000 system).
PCR for mRNA Expression Analysis
Total RNA was prepared with TRIzol (Invitrogen), and 2.0 μg of total RNA was reverse transcribed using the Superscript III RNase H reverse transcriptase kit (Invitrogen) according to the manufacturer's instructions. The following primers were used in PCR analysis for human GnT-III, GnT-V, and GAPDH mRNA expression: human GnT-III forward (5′-GCCGCGTCATCAACGCCATCAA-3′) and human GnT-III reverse (5′-CAGGTAGTCGTCGGCGATCCA-3′); human GnT-V forward (5′-GACCTGCAGTTCCTTCTTCG-3′) and human GnT-V reverse (5′-CCATGGCAGAAGTCCTGTTT-3′); human GAPDH forward (5′-AGCCACATCGCTCAGACA-3′) and human GAPDH reverse (5′-TGGACTCCACGACGTACT-3′). The GAPDH mRNA was used as a control in PCR runs, and the reaction products obtained were submitted to electrophoresis in 1.6% agarose gels containing ethidium bromide.
Construction of the shRNA Vector and Retroviral Infection
A retroviral vector carrying shRNA targeted to β-catenin (sense, GATCCAAGTCCTGTATGAGTGGGAACTTCAAGAGAGTTCCCACTCATACAGGACTTTTTTTTG; antisense, AATTCAAAAAAAAGTCCTGTATGAGTGGGAACTCTCTTGAAGTTCCCACTCATACAGGACTTG) and a random sequence (sense, GATCCAACAGTCGCGTTTGCGACTGGTTCAAGAGACCAGTCGCAAACGCGACTGTTTTTTTTG; antisense, AATTCAAAAAAAACAGTCGCGTTTGCGACTGGTCTCTTGAACCAGTCGCAAACGCGACTGTTG) were inserted in the sense and antisense directions into the pSINsi-mU6 cassette vector (Takara Bio). The retroviral supernatant was obtained by transfection of human embryonic kidney 293 cells using a Retrovirus Packaging Kit Eco (Takara Bio) according to the manufacturer's protocol. The recombinant retrovirus particles containing the target sequence or a random sequence as a control were infected into DLD-1/α-cat cells, and the puromycin-resistant clones were selected as a stable transfect. The expression of β-catenin and GnT-III activities were confirmed in the stable transfectants.
Extraction of Nuclear β-Catenin
The nuclear proteins were prepared using the Thermo Scientific NE-PER nuclear and cytoplasmic extraction kit (Rockford, IL). Briefly, 5 × 106 cells were harvested with trypsin-EDTA and then centrifuged at 500 × g for 5 min. After washing with PBS, the cells were transferred to 1.5 microcentrifuge tubes, leaving the cell pellets as dry as possible, and reagents were then added to nuclear proteins according to the manufacturer's instructions. The expression levels of β-catenin were examined by Western blot analysis. The staining of α-tubulin was used as a loading control.
Cell Adhesion Assay Using 96-well Plates
96-well plates (Corning Glass) were coated with 3 μg/ml fibronectin at 37 °C for 1 h and blocked with 1% BSA in DMEM at 37 °C for 1 h. The cells were detached with trypsin containing 1 mm EDTA and resuspended with 0.5 mg/ml trypsin inhibitor (Nacalai Tesque) in DMEM. The suspended cells were centrifuged at 1,000 rpm for 3 min and diluted to 8 × 105 cells/ml with assay medium and 0.1% BSA in DMEM. 100-μl aliquots of cell suspension were added to each well, and the plates were incubated at 37 °C for 30 min. After incubation, attached cells were fixed with 25% glutaraldehyde (Nacalai Tesque) and stained with 0.5% crystal violet. The absorbance at 590 nm was measured using an automated microtiter plate spectrometer, Powerscan® HT (Dainippon Sumitomo Pharma Co., Ltd., Osaka, Japan) operated with microplate data analysis software, KC4TM (BIO-TEC Instruments, Inc., Winooski, VT).
Wound-healing Assay
A confluent layer of cells was scraped/wounded using a yellow tip. The open gap was then inspected microscopically over time as the cells moved in and filled the damaged area. Micrographs were taken 0, 3, 9, and 18 h after wounding. The pictures show the cells as they began to migrate toward the center of the wound. Wound closure was measured by showing the distances between the sides of the wound at the indicated times.
Flow Cytometry
Flow cytometry was performed as previously described (25). Briefly, cells were detached by trypsinizing and incubated with biotinylated E4-PHA lectin, followed by streptavidin Alexa Fluor 488 conjugate (Invitrogen). Negative controls underwent the same procedure without E4-PHA lectin. The analyses were performed using a FACSCalibur instrument (BD Biosciences), equipped with CELLQuestPro software.
RESULTS
Knockdown of β-Catenin Resulted in a Dramatic Increase in GnT-III Expression
We have previously observed that GnT-III expression was markedly induced in cells cultured under dense conditions in an E-cadherin-α-catenin-actin-dependent manner in several cancer cell lines (19, 20). The mechanism was also confirmed in MCF-10A cells, a human non-tumorigenic immortalized breast epithelial cell line in the present study (data not shown), suggesting that it could be a universal observation. Here, we focused on β-catenin, which is a central player in E-cadherin-mediated cell-cell adhesion. Knockdown of β-catenin genes in the DLD-1/α-cat cells was used to confirm the necessity of β-catenin for GnT-III induction. The expression levels of β-catenin were effectively decreased to ∼20% of the levels in wild type or mock transfectants (Fig. 1B, top). It is worth noting that knockdown of β-catenin did decrease E-cadherin expression and α-catenin (Fig. 1B). It was previously found that newly synthesized E-cadherin associates with β-catenin, and the two proteins move together to the cell surface (26). Impairment of β-catenin binding leads to proteosomal destruction of cadherin (27). The catenin binding region of cadherins features a “PEST” sequence motif that is recognized by ubiquitin ligases but would be inaccessible in the complex with β-catenin (28). Thus, it appears that β-catenin prevents proteosomal destruction of cadherin, ensuring delivery of cadherin-β-catenin complexes to the cell surface.
Theoretically, it was thought that GnT-III expression was down-regulated in shRNA-β-cat cells because E-cadherin-catenin complex was essential for GnT-III induction by cell-cell adhesion. However, unexpectedly, the GnT-III expression in shRNA-β-cat cells, in all 10 of the clones selected by puromycin, dramatically increased by more than 10-fold, compared with those in wild type or mock transfectants, which was confirmed by RT-PCR and HPLC assays (Fig. 1, C and D). Furthermore, the GnT-III products, bisected N-glycans, were greatly enhanced in knockdown cells, which was confirmed by an E4-PHA lectin blot (which specifically recognizes bisecting GlcNAc) (Fig. 1E). On the other hand, the expression levels of GnT-V and α1,6-fucosyltransferase and their products confirmed by L4-PHA and A. aurantia lectin (data not shown), which selectively recognize β1,6-branching GlcNAc and α1,6-fucose, respectively, were not affected by the knockdown of β-catenin (Fig. 1F). In a similar fashion, the regulation of cell-cell interaction was also relatively specific to GnT-III but not other glycosyltransferases (20). Overall, these results suggest that β-catenin might specifically regulate the expression of GnT-III.
It remains unclear why the induction of GnT-III expression by β-catenin shRNA knockdown did not affect L4-PHA staining. Although an introduction of the bisecting GlcNAc usually suppresses the action of GnT-V in several cell lines and molecules, as we previously reported (29, 30), it is noteworthy that this is not always the case, which may be dependent on cells and molecules. In some cases, an introduction of the bisecting GlcNAc could enhance the products of GnT-V on some molecules, such as integrins and laminins.4 It could be argued that modification of the bisecting GlcNAc on certain sites can lead to molecular conformation changes, which may give GnT-V easy access to other N-glycosylation sites for β1,6-GlcNAc modification, because most glycoproteins contain multiple potential N-glycosylation sites. Recently, we found that GnT-III mainly modifies site 4 of the integrin α5 subunit, although it has 14 potential N-glycosylation sites (31). Thus, it may not be surprising that total L4-PHA staining is not affected by an increase in GnT-III activation, as shown in the present study.
Efficient shRNA Knockdown of β-Catenin in Nuclei
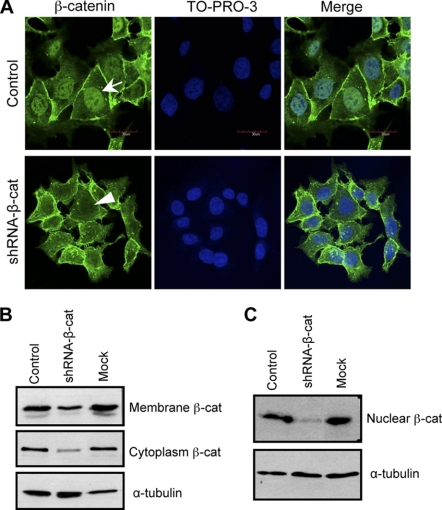
Based on the observation above, we hypothesized that there might be two pathways that participate in the regulation of GnT-III because it is known that β-catenin is a central player in not only E-cadherin-mediated cell-cell adhesion but also in the canonical Wnt signaling pathway. Experiments were thus carried out to examine the efficiency of the knockdown of β-catenin in cell-cell contact and the nuclei. Interestingly, the level of β-catenin in the nucleus was greatly decreased in shRNA-β-cat cells, compared with their corresponding control cells, in the immunofluorescence observation (Fig. 2A). To a lesser extent, decreased β-catenin staining was also observed in cell-cell contacts of knockdown cells. In order to confirm this phenomenon, nuclei were further isolated nuclei, and the expression levels of β-catenin were compared in membrane fraction and nuclear fraction by Western blot. The expression levels of β-catenin in membrane fractions and cytosol fractions were decreased in shRNA-β-cat cells, compared with control cells and mock cells (Fig. 2B). Consistent with the immunostaining data, β-catenin expression of nuclei in the shRNA-β-cat cells was substantially decreased as compared with those in control cells and mock cells (Fig. 2C).
FIGURE 2.
Localization of β-catenin in β-catenin knockdown cells. To visualize the effects of β-catenin knockdown, control and β-catenin knockdown cells were cultured for 48 h and stained with anti-β-catenin primary antibody and TO-PRO-3 and fluorescent secondary antibodies (A). The β-catenin protein expression levels were compared in membrane and cytoplasm fractions (B) and nuclear fractions (C). α-Tubulin was used as a load control.
Enhanced Expression of Bisected N-Glycans on Cell Surface Proteins
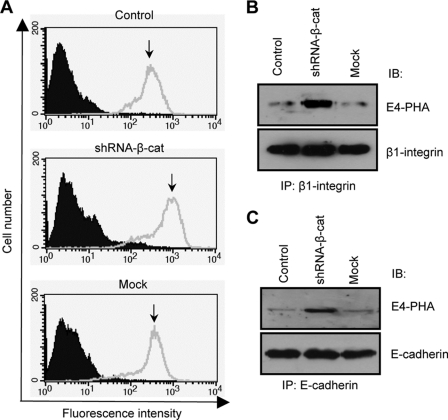
To examine whether increased GnT-III expression can affect the N-glycans expressed on the cell surface, a FACS analysis was performed using E4-PHA lectin. As shown in Fig. 3A, the E4-PHA reactivity was much stronger in shRNA-β-cat cells than in mock or control cells. Furthermore, the glycoproteins, such as β1 integrin and E-cadherin, were abundantly modified by GnT-III (Fig. 3, B and C). These results indicate that both β1 integrin and E-cadherin are targets of GnT-III induced by the knockdown of β-catenin.
FIGURE 3.
Enhanced expression levels of bisected N-glycans on cell surface and proteins. Cell surface expression levels of bisected N-glycans were examined using FACS analysis. Prior to analysis, cells were incubated with biotinylated E4-PHA lectin, followed by incubation with streptavidin Alexa Fluor 488 conjugate (A). Cell lysates from those three cells were immunoprecipitated using anti-β1 integrin (B) or anti-E-cadherin (C) antibodies. Immunoprecipitates were run on a 7.5% SDS-polyacrylamide gel and probed with the biotinylated E4-PHA lectin. IP, immunoprecipitation; IB, immunoblot.
Enhanced GnT-III Expression, Down-regulated Cell Adhesion, and Cell Migration
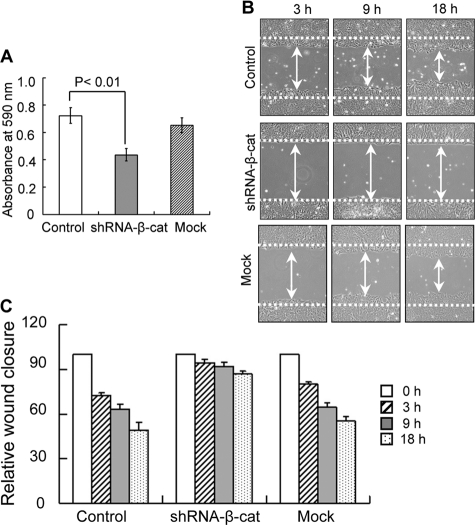
Previously, we reported that overexpression of GnT-III down-regulated integrin-mediated cell adhesion and cell migration. Here, cell adhesion and cell wound healing were examined. As expected, cell adhesion on fibronectin was significantly inhibited in β-catenin knockdown cells, compared with control or mock cells (Fig. 4A). In the wound-healing assay, the control cells efficiently moved in and filled the open gap at 3, 9, and 18 h after wounding. However, wound closure could not be observed in the knockdown cells (Fig. 4, B and C). These results suggest that the induction of GnT-III by shRNA-β-catenin functionally affects cell behavior. Of course, this does not exclude other reasons for the regulation of cell movement. For example, cadherin-mediated cell adhesion can activate small G proteins, such as Rho and Rac, which are also important for cell migration.
FIGURE 4.
Effects of β-catenin knockdown on fibronectin-mediated cell adhesion and migration. A, subconfluent cells were detached, and 40,000 cells were added to 96-well plates coated with 3 μg/ml fibronectin for the cell adhesion assay. The plates were incubated at 37 °C for 30 min and then washed twice with warmed PBS to remove non-adherent cells. The adherent cells were fixed with 25% glutaraldehydes and stained with 0.5% crystal violet, and then the absorbance at 590 nm was measured. Error bars, S.D. B, a confluent layer of cells was scraped/wounded using a yellow tip. The open gap was then inspected microscopically, and the distances between the sides of the wound were measured at the indicated times. C, quantitative data (mean value) for the cell migration from three independent experiments. The distance between the sides of the wound immediately following wounding (0 h) was set equal to 100. The relative wound closure is expressed as widths of the wound relative to the distance at 0 h.
Wnt/β-Catenin Signal Pathway Was Involved in Regulation of GnT-III Expression
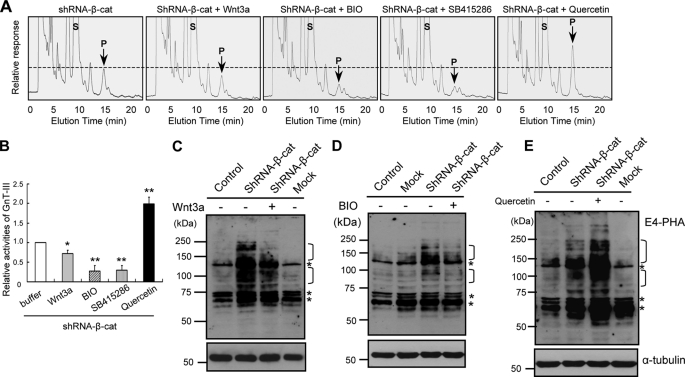
We previously reported that E-cadherin-catenin mediated cell adhesion positively regulates GnT-III expression. Logically, β-catenin knockdown should therefore decrease GnT-III expression. However, shRNA knockdown of β-catenin resulted in drastic up-regulation of GnT-III expression as described above. Because β-catenin is an essential molecule in both cadherin-mediated cell adhesion and canonical Wnt signaling, this finding prompted the hypothesis that Wnt signaling should inhibit GnT-III expression. To confirm this hypothesis, the effects of Wnt3a were examined. As expected, the addition of Wnt3a into the cultured medium significantly down-regulated GnT-III expression, confirmed by GnT-III activity assay as shown in Fig. 5A and 5B, and its products were confirmed by E4-PHA lectin blot (Fig. 5C). The addition of BIO or SB415286, an inhibitor of GSK-3β (glycogen synthase kinase-3β), which phosphorylates β-catenin for its proteasome degradation, consistently decreased GnT-III activities (Fig. 5, A and B) and E4-PHA staining to a great extent (Fig. 5D). By contrast, the inhibition of Wnt signaling by the presence of quercetin, a natural product that inhibits Wnt/β-catenin signaling without altering cytosolic β-catenin levels (32), significantly increased GnT-III activities (Fig. 5, A and B) and E4-PHA staining levels (Fig. 5E). Taken together, these results indicate that Wnt/β-catenin signaling inhibits GnT-III expression. The GnT-III activities were inhibited by treatment with GSK-3β inhibitors, Bio or SB415286, by ∼70%, a greater extent than those in cells treated with Wnt3a by ∼30% (Fig. 5, A and B). Considering Wnt binding also acts through β-catenin-independent, noncanonical pathways, the less inhibitory effects of Wnt3a on GnT-III activities could mean that the non-canonical Wnt pathway also affects GnT-III expression. Taken together, these results indicate that Wnt/β-catenin signaling inhibits GnT-III expression.
FIGURE 5.
Effects of Wnt signaling pathway on GnT-III expression. A, subconfluent β-catenin knockdown cells were incubated with or without Wnt3a (300 ng/ml), BIO (2 μm), SB415286 (50 μm), or quercetin (300 μm) for 48 h and then harvested for assay of GnT-III activities. Equal amounts of cell lysate protein (10 μg) were used as the enzymatic source for GnT-III. S, substrate; P, product. B, quantitative data for the relative activities of GnT-III from three independent experiments. Error bars, S.D. *, p < 0.01; **, p < 0.0001 compared with cells in the presence of buffer without these reagents. The subconfluent cells were cultured for 72 h in the absence or presence of Wnt3a (C), BIO (D), or quercetin (E) and then harvested and lysed for immunoblotting. Equal amounts of protein (20 μg) were separated on 7.5% SDS-PAGE under reducing conditions, and the membranes were probed with E4-PHA (top), and reprobed with anti-α-tubulin (bottom), which was used as a loading control. Control, without retroviral infection; Mock, retroviral infection with a random sequence. The asterisks indicate nonspecific staining of E4-PHA because those bands did not disappear after treatment with 100 mm acetic acid.
DISCUSSION
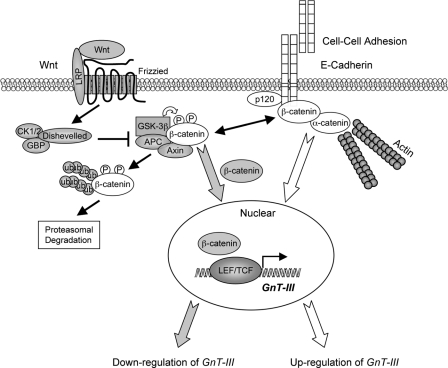
In the present study, it was found that GnT-III and its products, the bisected N-glycans, are up-regulated by knockdown of β-catenin or inhibition of Wnt/β-catenin signaling by treatment with quercetin. Conversely, stimulation of Wnt/β-catenin signaling by exogenous Wnt3a down-regulated GnT-III expression. β-catenin is an essential molecule both in cadherin-mediated cell adhesion and in canonical Wnt signaling, the loss of cadherin-mediated cell adhesion can promote β-catenin release and Wnt signaling, and then Wnt signaling in turn inhibits E-cadherin-catenin-mediated cell adhesion. Therefore, there is mutually exclusive cross-talk between β-catenin in two different compartments, the adhesion complex at the plasma membrane and a signaling complex in the nucleus. Given the previously described up-regulation by E-cadherin-mediated cell-cell adhesion, we postulate that there are positive and negative regulation pathways for GnT-III (i.e. E-cadherin-catenin-mediated cell adhesion signaling and Wnt/β-catenin signaling), and the intersection point is at β-catenin, as shown in Fig. 6. Thus, the expression levels of GnT-III can be closely regulated by the cellular distributions of β-catenin.
FIGURE 6.
A working model for reciprocal regulation of GnT-III expression by E-cadherin-catenin-mediated cell-cell adhesion and Wnt/β-catenin signaling. It was reported previously (19, 20) that GnT-III expression and bisected N-glycans were up-regulated by cell-cell adhesion in an E-cadherin-catenin-dependent manner. In the present study, GnT-III expression was down-regulated by Wnt/β-catenin signaling. The intersection point for the reciprocal regulation of GnT-III is at the β-catenin, which is a central player in both cadherin-mediated cell adhesion and canonical Wnt signaling. The diagram shows major proteins involved in the formation of E-cadherin plus α-, β-, and p120-catenin complexes at the plasma membranes of two juxtaposed cells. Wnt signaling is strongly implicated in the regulation of normal development, an intriguing connection with the E-cadherin-catenin complex. LEF/TCF, lymphoid enhancer factor-1/T-cell factor.
Cadherin-mediated cell-cell adhesion is highly dynamic and enables the reorganization and dispersal of cells (e.g. during the epithelial-to-mesenchymal transition in normal development and carcinogenesis) (33). In epithelium-derived tumors, the loss of cell-cell adhesion is correlated with down-regulation of E-cadherin as well as increased proliferation and tumor invasiveness (34). Ductal carcinoma, accounting for ∼80% of breast cancer, is associated with a reduction in both E-cadherin and α-catenin (34), and the loss of α-catenin is associated with advanced stages and poor patient survival (35). Taken together, these observations provide strong evidence that the regulation of E-cadherin and associated protein expression and localization are factors involved in carcinogenesis. On the other hand, E-cadherin can be N-glycosylated, and the N-glycosylation at Asn-633 is essential for E-cadherin expression, folding, and trafficking (36, 37). Our earlier study showed that E-cadherin-mediated cell-cell adhesion is regulated by post-transcriptional modification of N-glycans. Overexpression of GnT-III increased the retention of E-cadherin at the cell border, which resulted in an enhancement of E-cadherin-mediated homotypic adhesion. Thus, the expression of E-cadherin may be regulated not only by transcriptional factors but also by post-transcriptional processing, maturation, and modifications. It was recently found that GnT-III was conversely up-regulated by E-cadherin-catenin complex-mediated cell-cell adhesion. The regulation of GnT-III and E-cadherin expression may exist as a positive feedback loop (38). It is possible that up-regulation of GnT-III by cell-cell interaction might neutralize the signals responsible for maintenance of the cell differentiation phenotype.
Besides the roles of the cadherin-catenin complex in stabilizing adhesive contacts between neighboring cells and directing actin cytoskeleton reorganization, components of the complex are tightly linked to several key signal transduction networks. For example, β-catenin is a central player in the canonical Wnt signaling pathway, where it translocates to the nucleus and functions as a transcriptional cofactor (17, 39). One potential determinant of β-catenin's availability to participate in either the adhesion or transcriptional complexes may lie in its phosphorylation status. Indeed, β-catenin can be phosphorylated by a variety of serine/threonine and tyrosine kinases, and this can profoundly change the ability of β-catenin to interact with cadherin and α-catenin as well as its signaling function (40).
The Wnt/β-catenin signaling pathway has a crucial role in the embryonic development of all animal species, in the regeneration of tissues in adult organisms, and in numerous other processes. Mutations or deregulated expression of components of the canonical Wnt pathway can induce disease, most importantly cancer (41). The manifestation of cancer by aberrant Wnt signaling most likely results from inappropriate gene activation mediated by stabilized β-catenin. It is apparent that Wnt signaling causes cancer and that tumor promotion by this pathway can proceed through a number of different genetic defects. Here, we clearly showed that Wnt signaling sufficiently suppressed GnT-III expression, which usually relates to the inhibition of cancer metastasis. Thus, to a certain extent, the up-regulation of GnT-III by the knockdown of β-catenin could maintain cell differentiation rather than cell proliferation and invasion because cell adhesion and growth factor-mediated activation can be suppressed by GnT-III expression. Because β-catenin is a co-transcriptional factor, which binds to lymphoid enhancer factor-1/T-cell factor in order to turn on/off gene expression (42), we analyzed the promoter of the GnT-III gene using TESS (Transcription Element Search System) and found that there are three potential lymphoid enhancer factor-1/T-cell factor binding motifs: pyrimidine-rich elements (5′-PyCTTTG-3′ or complementary sequence 5′-PyGAAAC-3′) in the 5′-flanking 2000-bp region of the first exon of the GnT-III gene (43). Those potential sequences were identified at positions −1496, −1563, and −1652 (relative to the first base in the 5′-end of the first exon designated as +1 according to the published human GnT-III sequence) (44), suggesting that β-catenin may bind to transcription factor lymphoid enhancer factor-1/T-cell factor to turn off GnT-III gene expression. A detailed characterization of the promoter of the GnT-III gene is required for further studies.
Given the important biological functions of GnT-III described above, these results provide new insight into the molecular mechanism of relationships among cell-cell interaction and Wnt signaling during normal development, epithelial-to-mesenchymal transition, and cancer metastasis. A thorough understanding of these processes will require mechanistic explanations of how the signaling pathways implicated in epithelial-to-mesenchymal transitions trigger the rearrangements of cellular architecture and changes in cell behavior that permit this to happen through the regulation of N-glycosylation. It is also important to further examine whether the phenomena in vitro can also be observed in in vivo studies using animal models, such as GnT-III knock-out mice and epithelial tumor models, to elucidate the relationship between bisected N-glycosylation by GnT-III and β-catenin during cell adhesion, migration, and proliferation.
This work was partly supported by Core Research for Evolutional Science and Technology of the Japan Science and Technology Agency; the Academic Frontier Project for Private Universities of the Ministry of Education, Culture, Sports, Science, and Technology of Japan; and Grant-in-Aid for Scientific Research 21370059 from the Japan Society for the Promotion of Science.
T. Isaji, Y. Kariya, and J. Gu, unpublished data.
- GnT-III
- N-acetylglucosaminyltransferase III
- GnT-V
- N-acetylglucosaminyltransferase V
- E4-PHA
- phytohemagglutinin-E4
- L4-PHA
- phytohemagglutinin-L4.
REFERENCES
- 1. Apweiler R., Hermjakob H., Sharon N. (1999) Biochim. Biophys. Acta 1473, 4–8 [DOI] [PubMed] [Google Scholar]
- 2. Kobata A. (2000) Glycoconj. J. 17, 443–464 [DOI] [PubMed] [Google Scholar]
- 3. Dennis J. W., Granovsky M., Warren C. E. (1999) BioEssays 21, 412–421 [DOI] [PubMed] [Google Scholar]
- 4. Taniguchi N., Honke K., Fukuda M. (2001) Handbook of Glycosyltransferases and Related Genes, Springer-Verlag, Tokyo [Google Scholar]
- 5. Stanley P. (2002) Biochim. Biophys. Acta 1573, 363–368 [DOI] [PubMed] [Google Scholar]
- 6. Gu J., Nishikawa A., Tsuruoka N., Ohno M., Yamaguchi N., Kangawa K., Taniguchi N. (1993) J Biochem. 113, 614–619 [DOI] [PubMed] [Google Scholar]
- 7. Schachter H. (1986) Adv. Exp. Med. Biol. 205, 53–85 [DOI] [PubMed] [Google Scholar]
- 8. Schachter H., Narasimhan S., Gleeson P., Vella G. (1983) Can. J. Biochem. Cell Biol. 61, 1049–1066 [DOI] [PubMed] [Google Scholar]
- 9. Asada M., Furukawa K., Segawa K., Endo T., Kobata A. (1997) Cancer Res. 57, 1073–1080 [PubMed] [Google Scholar]
- 10. Pochec E., Litynska A., Amoresano A., Casbarra A. (2003) Biochim. Biophys. Acta 1643, 113–123 [DOI] [PubMed] [Google Scholar]
- 11. Granovsky M., Fata J., Pawling J., Muller W. J., Khokha R., Dennis J. W. (2000) Nat. Med. 6, 306–312 [DOI] [PubMed] [Google Scholar]
- 12. Yoshimura M., Nishikawa A., Ihara Y., Taniguchi S., Taniguchi N. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 8754–8758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yoshimura M., Ihara Y., Matsuzawa Y., Taniguchi N. (1996) J. Biol. Chem. 271, 13811–13815 [DOI] [PubMed] [Google Scholar]
- 14. Reynolds A. B., Carnahan R. H. (2004) Semin. Cell Dev. Biol. 15, 657–663 [DOI] [PubMed] [Google Scholar]
- 15. Aberle H., Butz S., Stappert J., Weissig H., Kemler R., Hoschuetzky H. (1994) J. Cell Sci. 107, 3655–3663 [DOI] [PubMed] [Google Scholar]
- 16. Klaus A., Birchmeier W. (2008) Nat. Rev. Cancer 8, 387–398 [DOI] [PubMed] [Google Scholar]
- 17. Clevers H. (2006) Cell 127, 469–480 [DOI] [PubMed] [Google Scholar]
- 18. Cadigan K. M., Nusse R. (1997) Genes Dev. 11, 3286–3305 [DOI] [PubMed] [Google Scholar]
- 19. Akama R., Sato Y., Kariya Y., Isaji T., Fukuda T., Lu L., Taniguchi N., Ozawa M., Gu J. (2008) Proteomics 8, 3221–3228 [DOI] [PubMed] [Google Scholar]
- 20. Iijima J., Zhao Y., Isaji T., Kameyama A., Nakaya S., Wang X., Ihara H., Cheng X., Nakagawa T., Miyoshi E., Kondo A., Narimatsu H., Taniguchi N., Gu J. (2006) J. Biol. Chem. 281, 13038–13046 [DOI] [PubMed] [Google Scholar]
- 21. Matsubara S., Ozawa M. (2001) J. Cell Biol. 154, 573–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ozawa M. (1998) J. Biol. Chem. 273, 29524–29529 [DOI] [PubMed] [Google Scholar]
- 23. Nishikawa A., Gu J., Fujii S., Taniguchi N. (1990) Biochim. Biophys. Acta 1035, 313–318 [DOI] [PubMed] [Google Scholar]
- 24. Uozumi N., Teshima T., Yamamoto T., Nishikawa A., Gao Y. E., Miyoshi E., Gao C. X., Noda K., Islam K. N., Ihara Y., Fujii S., Shiba T., Taniguchi N. (1996) J. Biochem. 120, 385–392 [DOI] [PubMed] [Google Scholar]
- 25. Isaji T., Sato Y., Zhao Y., Miyoshi E., Wada Y., Taniguchi N., Gu J. (2006) J. Biol. Chem. 281, 33258–33267 [DOI] [PubMed] [Google Scholar]
- 26. Hinck L., Näthke I. S., Papkoff J., Nelson W. J. (1994) J. Cell Biol. 125, 1327–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chen Y. T., Stewart D. B., Nelson W. J. (1999) J. Cell Biol. 144, 687–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Huber A. H., Weis W. I. (2001) Cell 105, 391–402 [DOI] [PubMed] [Google Scholar]
- 29. Gu J., Taniguchi N. (2008) Cell Adh. Migr. 2, 243–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gu J., Taniguchi N. (2004) Glycoconj. J. 21, 9–15 [DOI] [PubMed] [Google Scholar]
- 31. Sato Y., Isaji T., Tajiri M., Yoshida-Yamamoto S., Yoshinaka T., Somehara T., Fukuda T., Wada Y., Gu J. (2009) J. Biol. Chem. 284, 11873–11881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Park C. H., Chang J. Y., Hahm E. R., Park S., Kim H. K., Yang C. H. (2005) Biochem. Biophys. Res. Commun. 328, 227–234 [DOI] [PubMed] [Google Scholar]
- 33. Thiery J. P. (2003) Curr. Opin. Cell. Biol. 15, 740–746 [DOI] [PubMed] [Google Scholar]
- 34. Cowin P., Rowlands T. M., Hatsell S. J. (2005) Curr. Opin. Cell Biol. 17, 499–508 [DOI] [PubMed] [Google Scholar]
- 35. Nakopoulou L., Gakiopoulou-Givalou H., Karayiannakis A. J., Giannopoulou I., Keramopoulos A., Davaris P., Pignatelli M. (2002) Histopathology 40, 536–546 [DOI] [PubMed] [Google Scholar]
- 36. Liwosz A., Lei T., Kukuruzinska M. A. (2006) J. Biol. Chem. 281, 23138–23149 [DOI] [PubMed] [Google Scholar]
- 37. Zhou F., Su J., Fu L., Yang Y., Zhang L., Wang L., Zhao H., Zhang D., Li Z., Zha X. (2008) Glycoconj. J. 25, 727–740 [DOI] [PubMed] [Google Scholar]
- 38. Gu J., Sato Y., Kariya Y., Isaji T., Taniguchi N., Fukuda T. (2009) J. Proteome Res. 8, 431–435 [DOI] [PubMed] [Google Scholar]
- 39. Grigoryan T., Wend P., Klaus A., Birchmeier W. (2008) Genes Dev. 22, 2308–2341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Stepniak E., Radice G. L., Vasioukhin V. (2009)Cold Spring Harb. Perspect. Biol. 1, a002949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Moon R. T., Kohn A. D., De Ferrari G. V., Kaykas A. (2004) Nat. Rev. Genet. 5, 691–701 [DOI] [PubMed] [Google Scholar]
- 42. Willert K., Nusse R. (1998) Curr. Opin. Genet. Dev. 8, 95–102 [DOI] [PubMed] [Google Scholar]
- 43. Waterman M. L., Fischer W. H., Jones K. A. (1991) Genes Dev. 5, 656–669 [DOI] [PubMed] [Google Scholar]
- 44. Ihara Y., Nishikawa A., Tohma T., Soejima H., Niikawa N., Taniguchi N. (1993) J. Biochem. 113, 692–698 [DOI] [PubMed] [Google Scholar]