Abstract
Highly conserved G runs, G1M2 and ISE, regulate the proteolipid protein (PLP)/DM20 ratio. We have investigated recruitment of U1 small nuclear ribonuclear protein (snRNP) by G1M2 and ISE and examined the effect of splice site strength, distance, and context on G run function. G1M2 is necessary for initial recruitment of U1snRNP to the DM20 5′ splice site independent of the strength of the splice site. G1M2 regulates E complex formation and supports DM20 splicing when functional U1snRNP is reduced. By contrast, the ISE is not required for the initial recruitment of U1snRNP to the PLP 5′ splice site. However, in close proximity to either the DM20 or the PLP 5′ splice site, the ISE recruits U1snRNP to both splice sites. The ISE enhances DM20 splicing, whereas close to the PLP 5′ splice site, it inhibits PLP splicing. Splicing enhancement and inhibition are mediated by heterogeneous nuclear ribonuclear protein (hnRNP)H/F. The data show that recognition of the DM20 5′ splice site depends on G run-mediated recruitment of U1snRNA, whereas a complex interaction between the ISE G runs, context and position determines the functional outcome on splicing. The data suggest that different mechanisms underlie G run-mediated recognition of 5′ splice sites and that context and position play a critical role.
Keywords: RNA-binding Protein, RNA Processing, RNA-Protein Interaction, RNA Splicing, Spliceosome, Oligodendrocytes, Proteolipid Protein
Introduction
Alternative splicing is broadly utilized to generate multiple protein isoforms from a single transcript in a cell- and development-dependent fashion. Genome-wide analyses have shown that alternatively spliced sites are generally weak and that flanking regulatory sequences orchestrate their selection through the interaction with auxiliary splicing factors (for review, see Refs. 1 and 2).
Removal of introns from pre-mRNAs takes place within the spliceosome, a ribonuclear complex of RNA, and proteins whose assembly occurs in a stepwise fashion (reviewed in Refs. 3–5). The first step is the formation of the commitment complex, or E complex, that contains the U1snRNP bound to the 5′ splice site, U2AF bound to the 3′ acceptor site and at least one member of the SR (serine arginine) proteins. The second step is the formation of the prespliceosome, termed complex A, characterized by the ATP-dependent addition of U2 snRNP. Next, U1 snRNP is replaced by U5 and U6 snRNPs, which base pair with the 5′ splice site, and the splicing reaction is completed (6, 7).
U1snRNP is composed of the U1snRNA, U1-specific proteins (U1 70K, U1A, and U1C), and general Sm spliceosomal proteins that are also present in the U2, U5, and U6 snRNP complexes (8). The recognition of the 5′ splice site by the RNA moiety of the U1snRNP, through direct base pairing between the 5′ end of the U1snRNA and the 5′ splice site of the pre-RNA, commits the pre-RNA to splicing. However, in some genes, interactions other than direct base pairing of the U1snRNA with the 5′ splice site contribute significantly to the first step of splicing, and removal of the first 7 nucleotides of the U1snRNA does not prevent formation of the E complex (9–11). G runs, of which the G triplet is the basic functional unit, represent important regulatory sequences that have evolved to cluster preferentially, but not exclusively, downstream of weaker splice sites (12–20). G triplets function as intronic splicing enhancers (ISE),2 in most cases by binding hnRNPH/F (21–25). However, G triplets were also shown to recruit the U1snRNA through direct base pairing of nucleotides 8–11 of the U1snRNA with the G-rich sequences independently from nucleotides 2–7, which base pair with the 5′ splice site (11).
We have shown that highly conserved G runs, named G1M2 and ISE, located downstream of competing 5′ splice sites in the major myelin proteolipid protein (PLP) gene regulate the alternative exclusion/inclusion of exon 3B, generating DM20 and PLP, respectively (25–27). Tight regulation of the PLP/DM20 ratio is critical for brain development and function (26, 28, 29). Deletion of the ISE and mutations that strengthen the DM20 5′ splice site impair the developmental increase in the PLP/DM20 ratio in vivo, resulting in the reduction of the PLP product, which ultimately manifests itself as a neurological disorder in humans and mice (26, 30). We have previously shown that hnRNPH and hnRNPF regulate the PLP/DM20 ratio mostly by recruitment of U1snRNP through G1 and M2 located downstream of the DM20 5′ splice site. By contrast, hnRNPH/Fs have only a modest effect on the regulation of the PLP 5′ splice site through the ISE, although the G runs of the ISE are necessary for its splice enhancer activity and for binding hnRNPH and hnRNPF (25–27). Collectively, the data suggest that G1M2 and ISE regulate DM20 and PLP 5′ splice site selection through different mechanisms.
In the current study, we sought to gain additional insights into the mechanism of G1M2- and ISE-mediated regulation of 5′ splice site selection. We have examined whether G1M2 and ISE recruit U1snRNP to the DM20 and PLP 5′ splice sites by direct association with the U1snRNA and whether context, distance, and strength of the splice site influence the function of G-rich sequences to recruit U1snRNP, thereby regulating alternative splicing. We show that a direct interaction of U1snRNA with G1M2 plays a critical role in DM20 5′ splice site recognition and that this interaction does so independently from the strength of the splice site. G1M2 promotes E complex formation and supports DM20 splicing when functional U1snRNA is reduced. By contrast, the ISE is not required for the initial recruitment of U1snRNA to the PLP 5′ splice site, and its function in both U1snRNA recruitment and splicing is under complex regulation by gene context, position, and distance from the splice site. The data suggest that different mechanisms underlie G run-mediated recognition of 5′ splice sites and that context and position play a critical role.
EXPERIMENTAL PROCEDURES
RNase H Digestion Assays and Primer Extension
Oligonucleotide-mediated RNase H digestion was performed, as described previously (11) with some modifications. Briefly, 2 units of RNase H and 10 μg of an oligonucleotide complementary to nucleotides 1–11 of the U1snRNA (GAACYYACCUG) (see Fig. 2A) were added to HeLa cell splicing competent extracts (100 μg) and incubated at 30 °C. The subsequent complex formation was initiated by adding biotinylated RNA templates for streptavidin bead precipitations. Primer extension was carried out using an oligonucleotide complementary to nucleotides 64–75 in loop 2 of the U1snRNA (CAUUGCACUCCG) (see Fig. 2A). The products were separated in 6% acrylamide, 6 m urea denaturing gels and visualized by autoradiography. The bands were quantified using National Institutes of Health Image J software package.
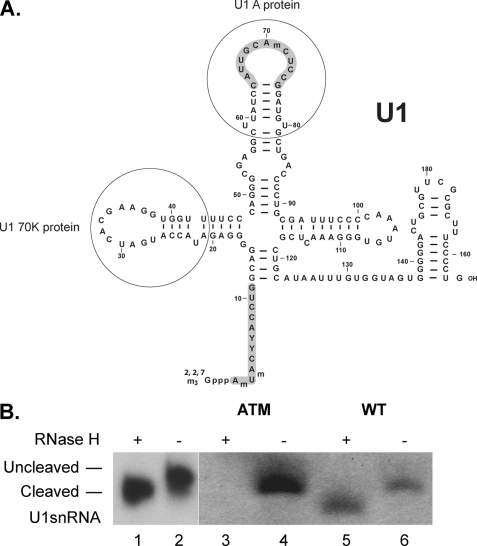
FIGURE 2.
Cleavage of the 5′ end of U1snRNA does not affect U1snRNA binding to the DM20 template. A, sequence of human U1snRNA (adapted from Ref. 44). The 5′ end of U1snRNA base paired with DNA oligonucleotide spanning nucleotides 1–11 (shaded) is cleaved by RNase H. Nucleotides 64–75 in loop 2 of the U1snRNA complementary to the oligonucleotide used for primer extension are shaded. The loops of U1snRNA that bind to U1A and U1 70K are shown. B, cleaved and uncleaved U1snRNA in HeLa extract with and without RNase H treatment. RNA was extracted from either untreated (lane 2) or RNase-treated (lane 1) HeLa nuclear extracts, annealed to a 32P-labeled oligonucleotide complementary to loop 2 of the U1snRNA and subjected to reverse transcription. The extension products were separated on 6%, 6 m urea denaturing acrylamide gels. The extension products from cleaved and uncleaved U1snRNA associated with ataxia telangectasia (ATM) and WT RNAs (Table 1) are shown. RNase H-treated (lanes 3 and 5) and untreated (lanes 4 and 6) HeLa nuclear extracts were incubated with the biotinylated RNA templates indicated (sequences in Table 1).
Splicing Extracts and RNA Affinity Precipitations
HeLa cell splicing competent nuclear extracts were a generous gift from Dr. McCullough. The sequences of the RNA templates used for RNA affinity precipitations and primer extension are shown in Table 1. RNA affinity precipitations were performed with biotinylated RNA templates, as described previously (27). All of the RNA templates were synthesized by Integrated DNA Technologies, Inc., with the exception of PLPISE, which was synthesized by Dharmacon. Natural or RNase-treated HeLa extracts (100 μg) were incubated with 500 pmol of the biotinylated RNA templates under splicing conditions at 30 °C for 30 min followed by streptavidin bead precipitation (27). RNA was extracted with phenol and chloroform, ethanol-precipitated, and subjected to primer extension. To ensure that binding reactions were carried out with excess template, we incubated 500 and 750 pmol of the ataxia telangectasia RNA template (see Table 1) (31, 32), used as control, with 100 μg of the HeLa extracts and performed primer extension. We show that 500 pmol of the template is in excess of the U1snRNA, and an extension product of similar intensity was detected with the higher concentration of template, indicating that differences in extension products quantitatively reflected the amount of U1snRNA bound to the template (supplemental Fig. S1A). For protein analysis, the RNA affinity precipitations were analyzed by Western blot with antibodies to hnRNPH (Bethyl Laboratories, Cambridge, MA), hnRNPF (rabbit polyclonal antibody, generous gift of Dr. Doug Black), U1A (Aviva Systems Biology, San Diego, CA), U170K (Abcam), and hnRNPL antibody (Abcam) diluted 1:2000 and reacted with ECL (Amersham Biosciences), as described previously (27). To ensure that the reactions were carried out with excess template, we incubated 500 and 750 pmol of PLPISE and DM20G1M2 with 50-μg HeLa extracts and performed Western blot with hnRNPL antibody (supplemental Fig. S1B).
TABLE 1.
Sequences of the RNAs
The sequences of the RNAs used for primer extension and RNA affinity precipitations are shown. The GU is italicized. The G1, M2, and ISE are underlined. The exonic sequences are in uppercase letters, and the intronic sequences are in lowercase letters. The names used in the text and figures for each template are shown to the left of the sequences.
| ATM | 5′-UGGCCAGGUAAGUGAUAUAU-3′ |
| DM20ss | 5′-GCAACGGUAACAGG-3′ |
| DM20G1M2 (WT) | 5′-GAGCGCAACGGUAACAGGGGGCCAGAAGGGGAGGGGUUCCAGAGG-3′ |
| MT | 5′-GAGCGCAACGGUAACAUUUUUCCAGAAUUUUUUUUUUUCCAGAGG-3 |
| G1M2 | 5′-CAGGGGGCCAGAAGGGGAGGGGUU-3′ |
| G1MT | 5′-CAUUUUUCCAGAAGGGGAGGGGUU-3′ |
| M2MT | 5′-CAGGGGGCCAGAAUUUUUUUUUUU-3′ |
| PLPss | 5′-UCCCGACAAGgugaucauccucaggauu-3′ |
| PLPISE (WT) | 5′-UCCCGACAAGgugaucauccucaggauuuuguggcaauaacaaggggugggggaaaauuggg-3′ |
| ISE | 5′-uaacaaggggugggggaaa-3′ |
| ISEdel | 5′-UCCCGACAAGgugaucauccucaggauuuuguggcaa.....auuggg-3′ |
| ISEMT3 | 5′-uugcaaaagguacaugaaa-3′ |
| C → Ass | 5′-GCAAAGGUAACAGG-3′ |
| C → A-MT | 5′-GAGCGCAAAGGUAACAUUUUUCCAGAAUUUUUUUUUUUCCAGAGG-3′ |
| AU-MT | 5′-GAGCGCAAAGGUAAGUUUUUUCCAGAAUUUUUUUUUUUCCAGAGG-3′ |
| AUss | 5′-GCAAAGGUAAGUGG-3′ |
| C → A | 5′-GAGCGCAAAGGUAACAGGGGGCCAGAAGGGGAGGGGUUCCAGAGG-3′ |
| AU | 5′-GAGCGCAAAGGUAAGUGGGGGCCAGAAGGGGAGGGGUUCCAGAGG-3′ |
| GLO | 5′-GAGCGUCAAGGUGAGCGGGGGCCAGAAGGGGAGGGGUUCCAGAGG-3′ |
| PLPISE+6 | 5′-UCCCGACAAGgugaucggggugggggaaaauuggg-3′ |
| PLPISE+12 | 5′-UCCCGACAAGgugaucauccucggggugggggaaaauuggg-3′ |
| G1-ISE | 5′-GAGCGCAACGGUAACAGGGGGCAACAAGGGGUGGGGGAAAAGAGG-3′ |
| G1MT-ISE | 5′-GAGCGCAACGGUAACAUUUUUCAACAAGGGGUGGGGGAAAAGAGG-3′ |
Plasmids
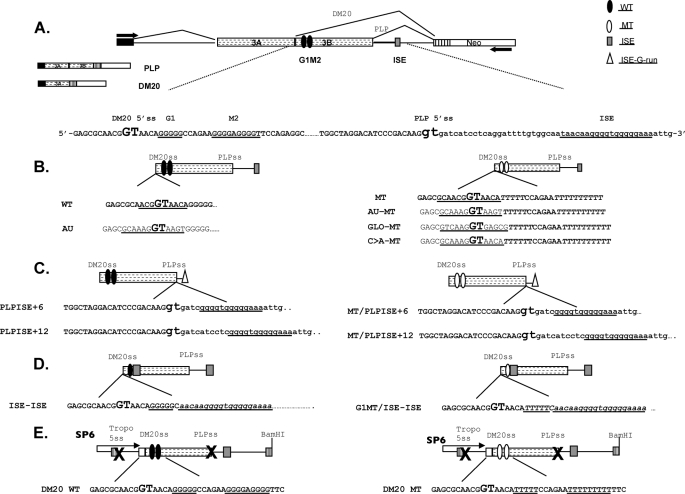
Mutant PLP-neo constructs were generated by site-directed mutagenesis using a QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). The sequences and schematics of all of the constructs are shown in Fig. 1. The minigene constructs used for in vitro splicing were generated in the single intron PY7NS minigene construct (kind gift of Dr. Buratti) (33, 34). We cloned the last 10 nucleotides of PLP exon 3A and exon 3B and the first 100 nucleotides of intron 3 into SmaI and NdeI restriction sites in the intron of the PY7NS construct. The primers used for PCR amplification of the PLP sequences in PLP-neo (WT) and G1M2MT (MT) (Fig. 1) (25) are: forward primer: 5′-AAAGCTCCCGGGGAGCGCAACGGTAACAG-3′ (underlined is the SmaI site); and reverse primer: 5′-AAAGCTCATATGCCCCTAGAGAGGACCCAGCCT-3′ (underlined is the NdeI site). To generate minigene splicing constructs containing only the DM20 5′ss (DM20 WT and DM20 MT), the tropomyosin 5′ss and the PLP 5′ss were abolished by site-directed mutagenesis using the QuikChange multisite-directed mutagenesis kit (Stratagene, La Jolla, CA). All of the constructs were verified by sequencing.
FIGURE 1.
PLP constructs. A, schematic of the PLP-neo splicing minigene construct. The arrows indicate the position of the PCR primers. The PLP and DM20 PCR products and partial sequences of PLP exon 3B (uppercase letters) and intron 3 (lowercase letters) in PLP-neo (WT) are shown; DM20 and PLP 5′ splice site are enlarged and in bold type; and G1, M2, and ISE are underlined. The filled ovals represent the wild type G1M2, empty ovals represent the G1M2 mutated to polyT, the shaded rectangle is the ISE, and the empty triangle represents the G runs of the ISE. B, schematics and sequences of the constructs in which the DM20 5′ss has been mutated to a stronger 5′ss: AU (canonical), GLO (α-globin), and C → A (disease-associated mutation (28). The sequences of natural DM20 5′ss and the strong 5′ss are underlined. C, schematics and sequences of the constructs in which the G runs of the ISE (empty triangle) are moved 6 nucleotides (PLPISE+6) and 12 nucleotides (PLPISE+12) downstream of the PLP 5′ss. The sequence of the ISE is underlined. D, schematics and sequences of the constructs in which the ISE replaces the M2. The G1, G1MT, and ISE sequences are underlined. E, schematics and sequences of the PY7 constructs generated for in vitro splicing and spliceosomal assembly (see “Experimental Procedures” for details). PLP exon 3A last 10 nucleotides, exon 3B, and intron 3 are cloned into the tropomyosin (Tropo) gene. The × indicates that the 5′ss of tropomyosin exon 2 and PLP 5′ss are mutated. DM20 WT contains the natural exon 3B sequences, whereas DM20 MT contains G1M2 mutated to poly Ts. SP6 refers to the promoter that drives in vitro transcription.
siRNAs, Cell Cultures, and Transfections
The custom-made double-stranded siRNA, siF/H (25), and Silencer® Negative Control 1 siRNA were purchased from Applied Biosystems (ABI, Foster City, CA). Oli-neu cells, an oligodendrocyte cell line, were cultured in SATO medium with 1% horse serum (25) and were co-transfected with plasmid DNAs (0.5 μg) and siRNAs (80 nm) using the siPORT amine transfection agent according to the reversed transfection protocol (ABI, Foster City, CA) (25).
RNA Extraction and RT-PCR
Total RNA was extracted from cultured cells using the RNeasy mini kit (Qiagen, Valencia, CA) and was treated with the DNA-free kit (Ambion, Austin, TX) according to the manufacturer's instructions. Reverse transcription was performed with 1 μg of total RNA using random hexamer primer mixture according to the manufacturer's instructions (ABI, Foster City, CA). The PCR products derived from the wild type, and mutated PLP-neo constructs were amplified using a primer set described previously (Fig. 1) (25, 29).
In Vitro Splicing and Spliceosomal Complex Assembly
In vitro splicing reactions were carried out in 30% nuclear extract containing SP6 generated 32P-labeled RNA transcripts as the template, 1.0 mm ATP, 20 mm creatine phosphate, 3.2 mm MgCl2. The reactions were incubated at 30 °C for a determined time course indicated in figure legends. Following incubation, the reactions were digested with proteinase K, phenol chloroform-extracted, ethanol-precipitated, resolved on 6% denaturing PAGE, and analyzed by Phosphor Imager analysis (Bio-Rad, Hercules, CA). The percentage spliced is defined as mol of spliced product/(mol unspliced product + mol spliced product). To derive kinetic rate constants, the percentage spliced/time was fit to a first order rate description for product appearance. The background was determined individually for each lane.
Reactions for in vitro complex formation were carried out as described (35), based on the protocol of Das and Reed (36) and incubated at 30 °C for a determined time course indicated in figure legends. All E complex reactions were fractionated on 1.75% low melt agarose gels using a 50 mm Tris, 50 mm glycine, pH 7.5 running buffer. Time points were loaded onto a running gel, causing a slight but apparent retardation of E complex at later time points. Analysis was carried out as described in the in vitro splicing section.
Ribonuclease H Digestion of U1 snRNA
Selective targeting of U1 snRNA was carried out via RNase H digestion with a U1 snRNA complementary DNA oligonucleotide. RNase H-mediated depletion of functional U1 snRNA was carried out in 1.5 mm ATP, 5 mm creatine phosphate, 1.6 mm MgCl2, 40 mm anti-snRNA DNA oligonucleotides, and 93% nuclear extract at 30 °C for 30 min. This treatment resulted in nuclear extract with significantly decreased splicing efficiency. The depleted extracts were then used to carry out splicing reactions at identical experimental conditions as described above. Control extract was also made by carrying out the same reaction with water in place of the DNA oligonucleotide. The sequence of DNA oligonucleotide used was 5′-GCCAGGTAAGTAT-3′ (against U1 snRNA).
RESULTS
Recruitment of U1snRNA to DM20 5′ Splice Site Depends on the U1snRNA Association with G1M2
To determine whether a direct interaction between G1M2 and the U1snRNA contributes to the recognition of the DM20 5′ splice site (DM20ss), we have incubated a biotinylated DM20G1M2 RNA template (WT) with splicing competent HeLa nuclear extracts either untreated or RNase H-treated for oligonucleotide-targeted removal of the 5′ end of the U1snRNA (nucleotide 1–11) (Fig. 2A). The WT substrate and associated U1snRNA were precipitated with streptavidin beads (27), and the U1snRNA was measured by primer extension using an oligonucleotide specific for nucleotides 64–75 in loop 2 of the U1snRNA. We first ascertained that the digestion of U1snRNA by RNase H was complete by measuring U1snRNA extracted from treated and untreated nuclear extracts (Fig. 2, A and B, lanes 1 and 2). A single extension product, shorter than the product from the untreated extracts, was detected from RNase H-treated extracts, thus indicating that the 5′ end of U1snRNA had been removed. Both cleaved and uncleaved U1snRNAs bound the WT RNA with similar intensities, indicating that the association of U1snRNA is largely independent of base pairing with the 5′ss (Fig. 2B, lanes 5 and 6). A strong U1snRNA product was detected in uncleaved extract incubated with ataxia telangectasia, used as control RNA (Table 1), which strongly base pairs with the U1snRNA 5′ end (32, 34, 37), whereas no product was detected in the cleaved extract (Fig. 2B, lanes 3 and 4).
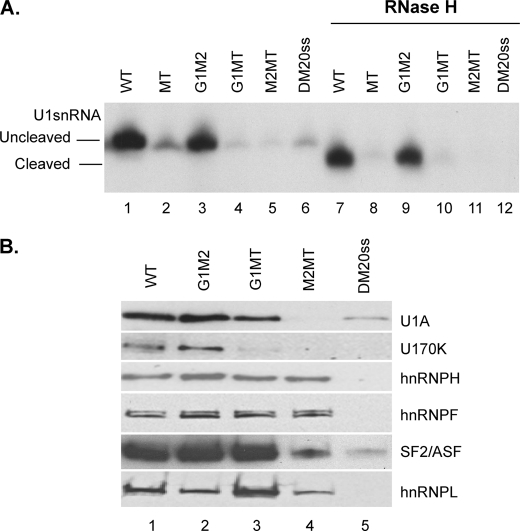
To determine whether G1M2 recruits U1snRNA to WT, we have measured the association of U1snRNA with the DM20 5′ss alone (DM20ss) and with an RNA, in which both G1 and M2 have been replaced by polyU (MT) (Table 1) (Fig. 3A) (27). Significantly less U1snRNA was associated with DM20ss and MT in the uncleaved extract compared with WT, 15 and 20%, respectively (Fig. 3A, compare lane 1 with lanes 2 and 6), whereas U1snRNA was barely detectable in cleaved extract (Fig. 3A, lanes 8 and 12). Cleaved and uncleaved U1snRNA were associated with G1M2 RNA, indicating that U1snRNA interacts with G1M2 independently of its 5′ end (Fig. 3A, lanes 3 and 9). The intensity of the extension product from G1M2 is slightly lower than that of WT, consistent with base pairing of the U1snRNA with the DM20 5′ss in WT (Fig. 3A, compare lane 3 with lane 1). Mutations of either G1 (G1MT) or M2 (M2MT) nearly abolished the association of cleaved and uncleaved U1snRNA with the RNA (Fig. 3A, compare lanes 4 and 5 with lane 3, and lanes 10 and 11 with lane 9). The fairly equal contribution of G1 and M2 to U1snRNA binding is in keeping with in vivo splicing results, which showed equal contribution of G1 and M2 to the regulation of the PLP/DM20 ratio (27).
FIGURE 3.
G1M2 recruits U1snRNA to the DM20 5′ splice site. A, representative primer extension analysis of U1snRNA bound to DM20 RNAs. Untreated (lanes 1–6) and RNase H-treated (lanes 7–12) HeLa splicing extracts were incubated with the biotinylated RNA templates indicated (sequences in Table 1). The RNA was extracted from the streptavidin bead precipitates and assayed by reverse transcription with a primer complementary to loop 2 of the U1snRNA. The extension products from cleaved and uncleaved U1snRNA are shown (n = 2). B, representative Western blot analysis of RNA affinity precipitates with the indicated RNA templates (sequences in Table 1) incubated with untreated HeLa nuclear extracts (n = 2). The precipitates were separated by SDS-PAGE, blotted, and probed with antibodies to U1A, U170K, SF2/ASF, hnRNPH, hnRNPF, and hnRNPL.
Next, we examined the assembly of hnRNPH/F and specific spliceosomal proteins onto G1MT, M2MT, and DM20ss compared with WT and G1M2 (Fig. 3B). U1A and U1 70K were detected in affinity precipitates with G1M2 similarly to WT (Fig. 3B, lanes 1 and 2), whereas they were hardly detected in precipitates with DM20ss and M2MT and were greatly reduced with G1MT (Fig. 3B, compare lane 5 with lane 1 and lanes 3 and 4 with lane 2), in keeping with the primer extension data. hnRNPH and hnRNPF were decreased by ∼50% in precipitates with G1MT and M2MT compared with G1M2, suggesting that hnRNPH/F interact with each G run with similar affinity. Interestingly, SF2/ASF was greatly reduced in precipitates with M2MT compared with G1MT and correlated with the near absence of U1A and U170K (Fig. 3B, compare lane 4 with lane 3). The data indicate that G1 and M2 are both required for recruitment of U1snRNA; however, M2 may have a greater impact on the assembly of U1-specific spliceosomal proteins, possibly through binding of SF2/ASF.
Relationship between G1M2 and 5′ Splice Site Strength
The G run class of enhancers is highly conserved downstream of 5′ splice sites, and their activity is highest for splice sites of intermediate strength (4–8 bits), suggesting functional conservation (19). Consistent with these findings, the DM20 and the PLP 5′ss are of intermediate strength, 5.4 and 6.8 bits, respectively (28). Importantly, PLP and DM20 5′ss strength contributes to the PLP/DM20 ratio in vivo, and a disease-associated mutation, predicted to strengthen the DM20 5′ss, increases DM20 splicing (28). Thus, we sought to determine whether increasing the strength of the DM20 5′ss reduces its dependence on G1M2. To test this, we have: 1) replaced the DM20 5′ss in WT with two strong 5′ss (>8 bits): the α-globin exon 2 (11) (GLO) and a canonical 5′ss (AU) (38) (Table 1) and 2) changed C → A at −2 in the DM20 5′ss (Table 1) (C → A), predicted to increase the strength of the splice site to 7.4 bits (28). U1snRNA bound to each template with similar intensity in uncleaved and cleaved extracts (Fig. 4A, compare lanes 1–3 with lanes 4–6), indicating that, in the presence of G1M2, recruitment of U1snRNA to the strengthened 5′ss is independent of U1snRNA base pairing with the splice site. The association of uncleaved U1snRNA to templates containing only the AUss and C → Ass or AU and C → A in the MT RNA (AU-MT and C → A-MT) (Table 1) (Fig. 4B, compare lane 2 with lane 1 and lane 4 with lane 3) was proportional to the predicted strength of the 5′ss. Cleaved U1snRNA was drastically reduced (Fig. 4B, lanes 5–8), consistent with base pairing of the 5′ end of U1snRNA with the 5′ss in these RNAs. Together, these data show that, in the presence of G1M2, recruitment of U1snRNA is largely independent of the 5′ss strength.
FIGURE 4.
Functional relationship of G1M2 with the strength of the 5′ splice site. A and B, representative primer extension analysis of U1snRNA bound by RNAs in which the DM20 5′ss is replaced by strong 5′ss. Untreated (A, lanes 1–3, and B, lanes 1–4) and RNase H-treated (A, lanes 4–6, and B, lanes 5–8) HeLa splicing extracts were incubated with the biotinylated RNA templates indicated (sequences in Table 1). The RNA was extracted from the streptavidin bead precipitates and assayed by reverse transcription with a primer complementary to loop 2 of the U1snRNA. The extension products from cleaved and uncleaved U1snRNA are shown (n = 2). C, schematic representation of the constructs. The sequences of the 5′ss: AU, C → A, and GLO that replace the natural DM20 5′ss are shown. The GT is indicated in bold type. Representative RT-PCR analysis of PLP and DM20 products amplified in RNA isolated from Oli-neu cells transfected with WT, AU, MT, AU-MT, GLO-MT, and C → A-MT (n = 2) (30 PCR cycles).
To directly test the functional relationship of G1M2 and the 5′ss strength in vivo, we have replaced the DM20 5′ss with the AUss (AU) (Figs. 1 and 4C) in the PLP-neo minigene. In addition, we have replaced the DM20 5′ss with AU, GLO, and C → A in the MT minigene construct, in which both G1M2 are replaced by polyT (27) (Figs. 1 and 4C). The PLP/DM20 ratio derived from the AU is much lower (<0.01) than from WT (0.34), consistent with the increased 5′ss strength (Fig. 4C). The C → A was previously shown to decrease the PLP/DM20 ratio, consistent with the predicted greater 5′ss strength (28). The PLP/DM20 ratio derived from AU-MT, GLO-MT, and C → A-MT is very low (<0.01) compared with MT (3.99 ± 0.98), reflecting the increased strength of the 5′ss (Fig. 4C). The data suggest that, although recruitment of the U1snRNA by G1M2 is largely independent of the 5′ splice site strength, DM20 splicing in vivo is strongly influenced by the 5′ss strength and is rescued by a strong 5′ss in the absence of G1M2, possibly through spliceosomal interactions at later stages of the splicing reaction rather than the initial recruitment of U1snRNA.
Spliceosomal Complex Assembly and in Vitro Splicing with DM20 WT and DM20 MT Pre-mRNAs
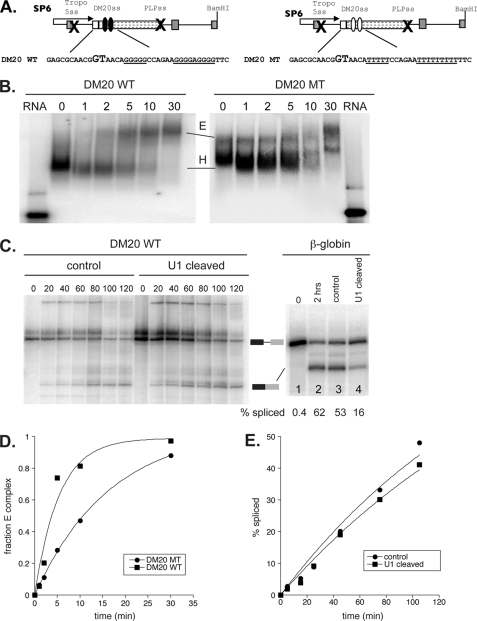
To better understand the mechanism by which G1M2 recruits U1snRNA to regulate DM20 splicing and to evaluate which step during spliceosomal assembly is targeted by the enhancer of the DM20 5′ss, we have analyzed spliceosomal complex assembly using in vitro splicing reactions. To this end, we have generated constructs in the PY7NS minigene (33–34), in which PLP sequences that span the last 10 nucleotides of PLP exon 3A, exon 3B, and the first 100 nucleotides of intron 3 are cloned between tropomyosin exon 2 and exon 3 (Figs. 1 and 5A). The 5′ss of tropomyosin exon 2 and the PLP 5′ss were mutated so that only one splicing event at the DM20 5′ss can take place. The minigene constructs carried either the natural G1M2 enhancer (DM20 WT) or the mutant G1M2-MT enhancer (DM20 MT) (Figs. 1 and 5A). Spliceosomal complex assembly was analyzed by native gel electrophoresis using radiolabeled transcripts. A prediction from our data was that, in the absence of G1M2, there would be a reduction in E complex formation. To this end, we have carried out reactions in which HeLa extract was depleted of ATP, which stalls spliceosomal assembly at E complex. Time-dependent E complex formation was then analyzed by separating H and E complexes using native gel analysis (Fig. 5B). E complex with the DM20 WT pre-mRNA is first detected at 2 min, increases at 5 and 10 min, and is complete at 30 min (Fig. 5B). By contrast, the formation of the E complex with the DM20 MT pre-mRNA is markedly reduced (greater than 3-fold), highlighted by the slowed accumulation of E complex at 2, 5, and 10 min (Fig. 5, B and D). The data show that the efficiency of E complex formation is reduced when the G1M2 sequences are mutated. As expected from these results, a delay in the ATP-dependent formation of subsequent spliceosomal complexes (A, B, and C complexes) was also observed for DM20 MT when compared with DM20 WT in assays using HeLa cell extracts in the presence of ATP (data not shown). We conclude that the G1M2 sequence enhances DM20 splicing by aiding initial splice site recognition.
FIGURE 5.
In vitro splicing and spliceosome complex assembly formation. A, schematics and sequence of the PY7 constructs generated for in vitro splicing and spliceosomal assembly (see “Experimental Procedures” for details). PLP exon 3A last 10 nucleotides, exon 3B, and intron 3 are cloned into the tropomyosin (Tropo) gene. The × indicates that the 5′ss of tropomyosin exon 2 and PLP 5′ss are mutated. DM20 WT contains the natural exon 3B sequences, whereas DM20 MT contains G1M2 mutated to poly Ts. SP6 refers to the promoter that drives in vitro transcription. B, representative autoradiogram comparing E complex formation efficiency between DM20 WT (left panel) and an isogenic mutant of the G1M2 enhancer (DM20 MT, right panel). E and H refer to E and H complexes, respectively. Time points are defined by the numbers above the gel. C, in vitro splicing of DM20 WT (left panel) and β-globin (right panel) in control or U1 snRNA depleted nuclear extracts. The numbers below the β-globin profile represent the percentage spliced. Time points are defined by the numbers above the gel. The migration of unspliced and spliced RNA species is indicated between the gels. D, quantitation of the data presented in B. The observed rates of complex formation are 0.2/min for DM20 WT and 0.06/min for DM20 MT. E, quantitation of the time course experiment in C. Within error, the observed rates of splicing are 0.3/h for the control and the U1 snRNA depleted extract conditions.
To take our primer extension data showing that the association of U1snRNA to the DM20 5′ss is largely independent of base pairing with the 5′ss one step further, we sought to demonstrate that the 5′ end of the U1snRNA is largely dispensable for DM20 splicing. To this end, we have performed in vitro splicing reactions with the DM20 WT pre-mRNA using HeLa cell nuclear extracts containing uncleaved and cleaved U1 snRNA and fractionated the products by denaturing PAGE (Fig. 5C). Splicing rates were determined through time course analysis as described under “Experimental Procedures” (Fig. 5, C and E). As a control, we have used the highly efficient β-globin splicing substrate, which depends on the base pairing of the U1snRNA for splicing (Fig. 5C). The rate constant of splicing of the DM20 WT pre-mRNA was nearly identical in uncleaved and U1snRNA-cleaved extracts and was calculated to be ∼0.3 mol/h (Fig. 5D). By contrast, the overall splicing kinetics of the β-globin pre-mRNA were severely reduced in the cleaved U1snRNA extract (Fig. 5C). These results clearly demonstrate that the 5′ end of U1snRNA is dispensable for DM20 splicing and further support the conclusion that recruitment of U1snRNA is mediated by G1M2 independently from base pairing with the DM20 5′ss.
Recruitment of U1snRNA to PLP 5′ss Does Not Depend on Its Association with the ISE
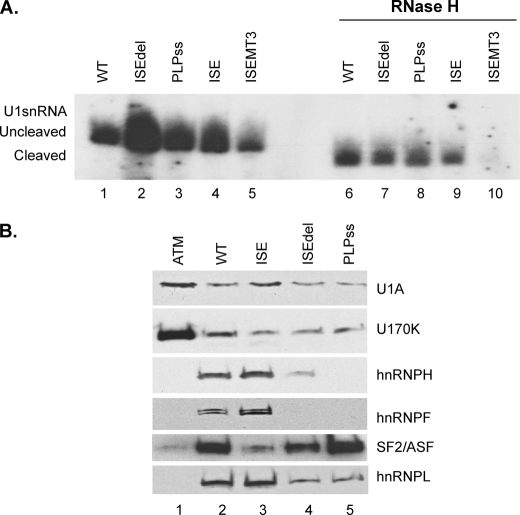
The ISE is a critical enhancer of PLP splicing and spans G runs that are essential for its enhancer function and are similar in configuration to M2 (26, 27, 30). Deletion of the ISE causes a neurological disorder in humans and in a knockin mouse (26, 30). To elucidate the ISE-mediated regulation of PLP and gain insight into disease mechanisms, we tested the role of the ISE in the recruitment of U1snRNA to the PLP 5′ss. Untreated and RNase-treated HeLa nuclear extracts were incubated with biotinylated PLPISE (WT), PLP splice site (PLPss), ISE and a PLP template in which the ISE is deleted (ISEdel) (Table 1) and measured the associated U1snRNA by primer extension. Uncleaved U1snRNA bound similarly to ISE, PLPss, and WT (Fig. 6A, lanes 1, 3, and 4), whereas more U1snRNA was associated with ISEdel (Fig. 6A, lane 2). Cleaved U1snRNA associated with WT, ISEdel, PLPss, and ISE was ∼50–70% lower than the uncleaved U1snRNA (Fig. 6A, compare lanes 6–9 with lanes 1–4). The data indicate that base pairing of the 5′ end of the U1snRNA contributes substantially to its association with both PLP 5′ss and ISE. In the absence of the ISE, we detected a greater association of U1snRNA to the PLP 5′ splice site, suggesting that the ISE and PLP 5′ss may compete for U1snRNA binding (see “Discussion”).
FIGURE 6.
The PLP U1snRNP-pre-mRNA complex does not require the ISE. A, representative primer extension analysis of U1snRNA bound to PLP RNAs. Untreated (lanes 1–5) and RNase H-treated (lanes 6–10). HeLa nuclear extracts were incubated with the biotinylated RNA templates indicated (sequences in Table 1). The RNA was extracted from the streptavidin bead precipitates and assayed by reverse transcription with a primer complementary to loop 2 of the U1sn RNA (n = 3). The extension products from cleaved and uncleaved U1snRNA are shown. B, representative Western blot analysis of RNA affinity precipitates of the indicated RNA templates (sequences in Table 1) incubated with untreated HeLa nuclear extracts (n = 2). The precipitates were separated by SDS-PAGE, blotted, and probed with antibodies to U1A, U170K, SF2/ASF, hnRNPH, hnRNPF, and hnRNPL.
The ISE is a complex enhancer, in which G runs and flanking sequences contribute to its activity (26, 27). Mutations that disrupt the G runs and the 5′-flanking sequences of the ISE (ISEMT3) drastically reduced PLP splicing in vivo (27). To examine the role of G runs and flanking sequences in recruiting U1snRNA, we have measured uncleaved and cleaved U1snRNA associated with ISEMT3 RNA (Table 1). Compared with the ISE, uncleaved U1snRNA bound to ISEMT3 is greatly reduced (>10-fold), demonstrating that these sequences are required for the association of U1snRNA with the ISE (Fig. 6A, compare lane 5 with lane 4). Cleaved U1snRNA did not bind to ISEMT3, indicating that the 5′ end of the U1 snRNA is required for the association with the ISEMT3 (Fig. 6A, lane 10).
Next, we characterized spliceosomal and nonspliceosomal proteins assembled onto WT, PLPss, ISEdel, and ISE templates by RNA affinity precipitations (Fig. 6B). Similar amounts of U1A, U1 70K were present in precipitates with WT, ISE, PLPss, and ISEdel RNAs (Fig. 6B, lanes 2–5). hnRNPH and hnRNPF were detected only on WT and ISE RNAs, in keeping with their binding to the G runs (Fig. 6B, lanes 2 and 3). SF2/ASF and hnRNPL were also present, in varying amounts, in the complexes assembled onto the various templates (Fig. 6B). hnRNPL was present in precipitates with WT and ISE but was greatly reduced in precipitates with ISEdel and PLPss, consistent with its association with the G runs of the ISE (25) (Fig. 6B). SF2/ASF was present in similar amount in precipitates with PLPss and WT, whereas it was greatly reduced with ISEdel and nearly absent with ISE (Fig. 6B). These data show that assembly of spliceosomal-specific proteins is similar with all templates in keeping with the primer extension data; however, there are differences among the nonspliceosomal proteins assembled in the U1snRNP complex. The significance of the latter observation is not yet clear.
Together, these data show that U1snRNA binds to the ISE as efficiently as to the PLP 5′ss and suggest that the ISE does not contribute significantly to the initial recruitment of U1snRNA to the PLP 5′ss. Furthermore, the interaction of the ISE with the U1snRNA is sensitive to the loss of U1snRNA 5′ end, and this may result from the presence of a 5′ss-like sequence in the ISE (see “Discussion”).
The Function of the ISE Is Dependent on Position and Distance from the 5′ Splice Site
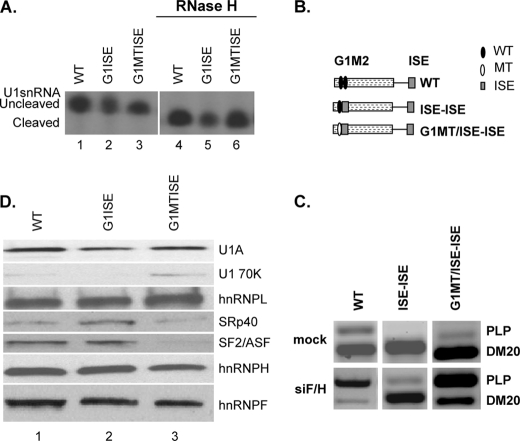
The absence of a role of the ISE in the initial recruitment of U1snRNA to the PLP 5′ss is in sharp contrast with the critical role that G1M2 has in recruiting U1snRNA to the DM20 5′ss. Because G1M2 is closer to the DM20 5′ss than the ISE is to the PLP 5′ss, we considered whether context sequences and distance of the ISE from the upstream 5′ss control the mechanism of U1snRNA recruitment by the ISE. To test this possibility, we have replaced M2 and flanking sequences with the ISE either in the presence of G1 (G1-ISE) or G1 mutated to polyU (G1MT-ISE) (Table 1). Cleaved and uncleaved U1snRNA associated with each template, indicating that the ISE-mediated recruitment of U1snRNA to the DM20 5′ss is not sensitive to the loss of the 5′ end of the U1snRNA similarly to the recruitment mediated by G1M2 (Fig. 7A). We consistently found that the amount of cleaved and uncleaved U1snRNA bound to G1ISE was lower than with the other RNAs (Fig. 7A). Interestingly, mutation of G1 did not affect recruitment of U1snRNA, in contrast to the severe decrease in U1snRNA recruitment with G1MT-M2 (compare Fig. 7A, lanes 2 and 3 with Fig. 3A, lanes 3 and 4), suggesting that, in this position, the ISE alone may be sufficient to recruit U1snRNA to the DM20 5′ss.
FIGURE 7.
The ISE recruits U1snRNA to the DM20 5′ splice site and is a strong enhancer of DM20 splicing. A, representative primer extension analysis of U1snRNA bound to the RNAs. Untreated (lanes 1–3) and RNase H-treated (lanes 4–6) HeLa nuclear extracts were incubated with the biotinylated RNA templates indicated (sequences in Table 1). The RNA was extracted from the streptavidin bead precipitates and assayed by reverse transcription with a primer complementary to loop 2 of the U1sn RNA (n = 3). The extension products from cleaved and uncleaved U1snRNA are shown. B, schematic representation of minigene constructs (see also Fig. 1). C, representative RT-PCR analysis of PLP and DM20 products amplified in RNA isolated from Oli-neu cells transfected with WT, ISE-ISE, G1MT/ISE-ISE, and treated with siF/H (n = 3) (30 PCR cycles). Mock are cells transfected with the same plasmids and treated with scrambled siRNA (30 PCR cycles). D, representative Western blot analysis of RNA affinity precipitates of WT (lane 1), G1ISE (lane 2), and G1MTISE (lane 3) RNAs (sequence is shown in Table 1) incubated with untreated HeLa nuclear extract (n = 2). The precipitates were separated by SDS-PAGE, blotted, and probed with antibodies to U1A, U170K, SF2/ASF, SRp40, hnRNPL, hnRNPH, and hnRNPF.
To take this finding further, we have asked whether the ISE enhances the DM20 5′ss independently from G1 by in vivo splicing assays. We have measured the PLP/DM20 ratio derived from minigene constructs in which the ISE replaces M2 and flanking sequences with the natural G1 (ISE-ISE) or G1 mutated to polyT (G1MT/ISE-ISE) (Figs. 1 and 7B). The PLP/DM20 ratio derived from ISE-ISE is 0.013 ± 0.003 and that from G1MT/ISE-ISE is 0.04 ± 0.01 compared with 0.34 ± 0.02 from the WT, suggesting that the ISE strongly enhances DM20 splicing even in the absence of G1 (Fig. 7C). Next, we have assessed whether knockdown of hnRNPH/F reduce the ISE-mediated enhancement of DM20 splicing. hnRNPH/F are the major transacting factors that mediate G1M2 regulation of DM20 splicing (27). The PLP/DM20 ratio derived from ISE-ISE was increased 15-fold (0.2 ± 0.06 versus 0.013 ± 0.003) and that from G1MT/ISE-ISE was increased 60-fold (2.53 ± 0.05 versus 0.04 ± 0.01) in siF/H-treated Oli-neu cells versus untreated cells (Fig. 7C). The data indicate that hnRNPH/F regulate the ISE-dependent splicing of DM20 5′ss and have a greater impact when G1 is mutated.
To determine whether differences in spliceosomal complex proteins may account for the extension and splicing data, we have examined proteins assembled onto the G1ISE, G1MTISE, and WT by RNA affinity precipitations (Fig. 7D). More SRp40 and ASF/SF2 were detected in G1ISE precipitates compared with G1MTISE (Fig. 7D, compare lane 2 with lane 3). The amount of SRp40 pulled down with G1ISE was also greater than with WT (Fig. 7D, compare lane 2 with lane 1). By contrast, hnRNPH/F and hnRNPL were detected in similar amounts with all templates (Fig. 7D). The stronger presence of SR proteins in complexes associated with G1ISE may account for the greater DM20 splicing induced by the ISE compared with M2 and the lower impact that the hnRNPH/F knockdown has on the PLP/DM20 ratio derived from the ISE-ISE compared with G1M2.
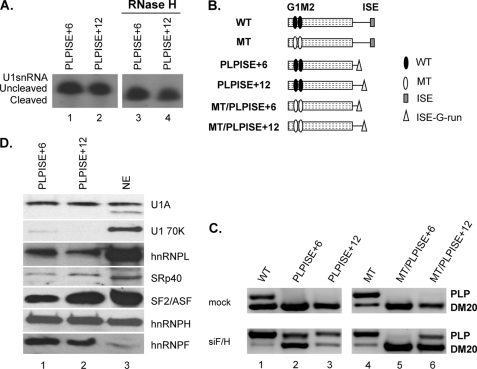
Next, we have asked whether the ISE close to the PLP 5′ss also recruits U1snRNA independently from the 5′ end. The ISE G runs, normally located 32 nucleotides downstream of the PLP 5′ss, were positioned 6 nucleotides (PLPISE+6) and 12 nucleotides (PLPISE+12) downstream of the PLP 5′ss (Table 1), the distances of G1 and M2 from the DM20 5′ss, respectively. The amount of uncleaved and cleaved U1snRNA bound to the PLPISE+6 and PLPISE+12 was similar, indicating that recruitment of U1snRNA is independent from the 5′ end of the U1snRNA (Fig. 8A, compare lanes 3 and 4 with lanes 1 and 2). The data support the interpretation that the distance from the 5′ss plays a critical role in the G run-mediated recruitment of the U1snRNA.
FIGURE 8.
The function of the ISE is dependent on context and distance from the PLP 5′ splice site. A, representative primer extension analysis of U1snRNA bound to the PLP RNAs. Untreated (lanes 1 and 2) and RNase H-treated (lanes 3 and 4) HeLa nuclear extracts were incubated with the biotinylated RNA templates indicated (sequences in Table 1). The RNA was extracted from the streptavidin bead precipitates and assayed by reverse transcription with a primer complementary to loop 2 of the U1sn RNA (n = 2). The extension products from cleaved and uncleaved U1snRNA are shown. B, schematic representation of all constructs (see also Fig. 1). C, representative RT-PCR analysis of PLP and DM20 products amplified in RNA isolated from Oli-neu cells transfected with WT, PLPISE+6, PLPISE+12, MT, MT/PLPISE+6, and MT/PLPISE+12 and treated with siF/H (n = 3) (30 PCR cycles). Mock are cells transfected with the same plasmids and treated with scrambled siRNA (30 PCR cycles). D, representative Western blot analysis of RNA affinity precipitates of PLPISE+6 and PLPISE+12 (lanes 1 and 2, respectively) (sequences in Table 1) incubated with untreated HeLa nuclear extract (n = 2). Lane 3 is the HeLa nuclear extract. The precipitates were separated by SDS-PAGE, blotted, and probed with antibodies to U1A, U170K, SF2/ASF, SRp40, hnRNPL, hnRNPH, and hnRNPF.
Next, we examined the function of the ISE by in vivo splicing assays with PLP-neo constructs carrying the ISE G runs at +6 (PLPISE+6) and +12 (PLPISE+12) relative to the PLP 5′ss (Figs. 1 and 8B). The PLP/DM20 ratio derived from PLPISE+6 was 0.01 ± 0 and that from PLPISE+12 was 0.04 ± 0.01 versus 0.34 ± 0.02 from the WT, suggesting that PLP splicing is severely reduced (Fig. 8C, compare lanes 2 and 3 with lane 1). To determine whether the dramatic decrease in PLP splicing is due to the loss of ISE function versus a gain of silencing function, we mutated G1M2 (MT) in the PLPISE+6 and +12 constructs (MT/PLPISE+6 and MT/PLPISE+12). The PLP/DM20 ratio derived from MT/PLPISE+6 was 0.02 ± 0.005 and that from MT/PLPISE+12 was 0.17 ± 0.03 compared with 4 ± 1 from the MT (Fig. 8C, lanes 5 and 6 versus lane 4). These data suggest that the ISE G runs positioned close to the PLP 5′ss function as silencers.
We have shown that hnRNPH/F bind to the ISE, and their knockdown affects the ISE enhancer function in its natural position, albeit modestly (27). Here, we asked whether hnRNPH/F knockdown affects the PLP/DM20 ratio derived from PLPISE+6 and PLPISE+12 (Fig. 8C). The PLP/DM20 ratio derived from PLPISE+6 was increased by 70-fold (0.74 ± 0.15 versus 0.01 ± 0) and that from PLPISE+12 was increased 45-fold (1.8 ± 0.12 versus 0.04 ± 0.01) in siF/H-treated versus untreated Oli-neu cells (Fig. 8C, lanes 2 and 3) compared with 10-fold increase with WT (3.59 ± 0.23 versus 0.34 ± 0.02) (Fig. 6C, lane 1) (27). The PLP/DM20 ratio derived from MT/PLPISE+6 was increased by 10-fold (0.21 ± 0.1 versus 0.02 ± 0.005) and MT/PLPISE+12 was increased by ∼3-fold (0.48 ± 0.06 versus 0.17 ± 0.03) in siF/H-treated compared with untreated Oli-neu cells (Fig. 8C, lanes 5 and 6). The PLP/DM20 ratio derived from MT decreased after knockdown of hnRNPH/F (3.33 ± 0.01 versus 4 ± 1), reflecting the weak enhancer function of the ISE in its natural position (Fig. 8C, lane 4) (27). The fold changes in the PLP/DM20 ratio are higher with the PLPISE+6 and +12 than with the MT/ISE+6 and +12 and reflect the G1M2-dependent regulation of DM20 5′ss by hnRNPH/F (27). Together, these data support the interpretation that, in close proximity to the PLP 5′ss, the ISE G runs act as silencers, and hnRNPH/F mediate the silencing effect. Interestingly, the G runs are stronger silencers when are placed within the first 10 nucleotides from the 5′ss (see “Discussion”).
Collectively, the data show that the ISE, close to both DM20 and PLP 5′ss, recruits U1snRNA independently of the 5′ end of the U1snRNA. However, it has the opposite functional effect on splicing outcome, enhancing the DM20 5′ss but repressing the PLP 5′ss.
To determine whether differences in spliceosomal complex proteins may account for the silencing function, we have examined proteins assembled onto the PLPISE+6 and +12 and compared with WT by RNA affinity precipitations (Fig. 8D). U1A, U170K, hnRNPH, hnRNPF, hnRNPL, SF2/ASF, and SRp40 are detected in similar amounts with the PLPISE+6 and +12 (Fig. 8D, lanes 1 and 2) and do not appear to be significantly different from those assembled onto WT, except for U1 70K, which associated in greater amount with the WT (compare Fig. 8D with Fig. 6B, lane 2). Thus, differences in the composition of the U1snRNP complexes assembled onto the PLP substrates do not seem to account for the ISE-mediated silencing of PLP 5′ss.
DISCUSSION
The G-rich enhancers, G1M2 and ISE, play a critical role in the selection of the DM20 and PLP 5′ splice sites, respectively, and regulate the developmental increase in the PLP/DM20 ratio and the abundance of these major central nervous system myelin proteins (25, 27, 30). Tight regulation of the PLP/DM20 ratio is critical for brain development and function (26, 28, 29). Deletion of the ISE impairs the developmental increase in the PLP/DM20 ratio in vivo, resulting in reduction of the PLP product, which causes a neurological disorder in humans, known as Pelizaeus-Merzbacher disease, and in mice (26, 30). To date, mutations of G1 and/or M2 have not been identified in Pelizaeus-Merzbacher disease patients, suggesting that loss of these enhancers may have deleterious consequences incompatible with brain development. Understanding the mechanisms by which G1M2 and ISE regulate the PLP/DM20 ratio has important implications for human disease.
In the current study, we have identified significant differences in the roles that G1M2 and ISE play in the initial steps of DM20 and PLP 5′ splice site recognition. G1M2 binds directly to the U1snRNA; this interaction is largely independent of the U1snRNA 5′ end and accounts almost entirely for recruitment of the U1snRNA to the DM20 5′ss. G1M2 promotes the ATP-independent formation of E complex, and its absence significantly impairs E complex formation, delays the ATP-dependent formation of subsequent spliceosomal complexes, and dramatically reduces overall splicing efficiency. Interestingly, the G1M2-mediated mechanism is distinct from that of G runs of the HIV tat gene, which regulate ATP-dependent spliceosomal complex formation but have no effect on the formation of E complex (39). The data indicate that multiple mechanisms underlie G run-mediated splicing regulation and spliceosomal assembly.
Direct base pairing between the 5′ end of U1snRNA and the DM20 5′ss plays a limited role during the initial recognition of the DM20 5′ splice site, supporting the notion that G1M2 is critical for U1snRNA recruitment and subsequent DM20 splicing. In addition, we directly demonstrate that G1M2 is sufficient to support DM20 splicing even when the U1snRNA is cleaved. Collectively, these data support a model in which G1M2 directly recruits U1snRNA, regulates the efficiency of E complex formation, and promotes DM20 splicing. The mechanism by which G1M2 recruits U1snRNA directly remains to be elucidated. Mutations of either G1 or M2 abolish recruitment of U1snRNA, supporting the notion that both G runs are required. It is possible that G1 and M2 interact with each other, forming G stacks and a secondary structure that permit interactions with U1snRNA. Furthermore, hnRNPH/F are similarly reduced in the complexes assembled on either G1MT or M2MT, but the impact on U1snRNA recruitment is greater with M2MT, which also reduces ASF/SF2 binding. The latter suggests that the association of U1snRNP with G1M2 may involve a complex interaction, possibly through direct U1snRNA binding to G stacks, and splicing factor-mediated recruitment. Collectively, the data suggest that multimeric complexes formed by hnRNPH/F and SR proteins on G1 and M2 may recruit U1snRNP and that the two G runs interact cooperatively. Another important finding is that mutations that increase the strength of the DM20 5′ss do not affect the initial recruitment of U1snRNA, whose association with G1M2 remains independent of the U1snRNA 5′ end (Fig. 4A). However, the strength of the 5′ splice site appears to be an important determinant of DM20 splicing efficiency (Fig. 4C), possibly through mechanisms that involve later stages of spliceosomal assembly rather than the initial recognition of the 5′ splice site. G1M2 regulates the efficiency of E complex formation; thus, it is conceivable that the strength of the 5′ splice site plays a role in subsequent ATP-dependent stages of spliceosomal assembly.
The ISE, on the other hand, does not appear to contribute significantly to the initial recognition of the PLP 5′ss by the U1snRNA nor to the assembly of the spliceosomal proteins. U1snRNA associates with the PLP 5′ss in the presence or absence of the ISE mostly through the 5′ end of U1snRNA consistent with direct base pairing. These data show that recruitment of the U1snRNA to the PLP 5′ss depends on base pairing interactions and that the ISE is dispensable for this initial step in 5′ splice site recognition. Interestingly, the U1snRNA associates with the ISE largely through base pairing of its 5′ end with the ISE, and when this interaction is abolished by deletion of the ISE, more U1snRNA is recruited to the PLPISEdel template (Fig. 6A). Although the greater amount of U1snRNA bound to this RNA could reflect differences in RNA sequences and secondary structure, it is tempting to speculate that a competition for U1snRNA binding takes place between the ISE and the PLP 5′ss. The ISE contains a 5′ splice site-like sequence, which may participate in the interaction of the U1snRNA and may serve regulatory functions on PLP splicing regulation during brain development. In other genes, pseudo 5′ splice sites embedded within regulatory sequences play a critical role in alternative splicing regulation (24, 34, 40). The 5′ splice site-like sequence in the ISE does not lead to a productive splicing reaction either in the normal gene context in vivo3 or in the case of mutations that abolish or severely reduce the PLP 5′ss strength (28). Interestingly, accumulation of an intermediate PLP transcript in which intron 3 is retained was detected in the developing brain, suggesting that intron 3 splicing may be temporally regulated (41). It is tempting to speculate that the ISE may serve a function in the temporal regulation of intron 3 splicing of the PLP transcript, possibly by stalling the U1snRNA. Binding of splicing factors to the ISE during brain development may help displace the U1snRNA from the ISE and shift it to the PLP 5′ss, leading to increased PLP splicing in the mature brain. Alternatively, the interaction of the U1snRNA with the ISE may create a “balance system” that regulates later stages of spliceosomal assembly when U5/U6 replaces U1snRNP.
In close proximity to either the PLP or the DM20 5′ss, the ISE recruits the U1snRNA independently of the U1snRNA 5′ end, suggesting that distance is of critical importance for this interaction. Functionally, however, the ISE acts as an enhancer for the DM20 5′ splice site, whereas it acts as a silencer of PLP splicing. This functional difference could be explained by the intronic position of the G runs. It was previously shown that intronic GGGG motifs positioned between 6 and 10 nucleotides downstream of the 5′ss act as silencer by forming a complex with exonic sequences (23). In keeping with these data, we observe a stronger silencing effect from the G runs positioned at +6 compared with +12 (Fig. 8C). The mechanism by which the G runs act as silencers of PLP remains to be elucidated. We did not detect significant differences in proteins and splicing factors in the U1snRNP complex assembled by the ISE in close proximity to DM20 and PLP 5′ splice sites compared with its natural position. Although it is possible that changes may occur in proteins not examined in this study, the current data suggest that differences in the complex proteins are not likely to explain the dramatic change in functional outcome. An intriguing possibility is that the 5′ end-independent recruitment of U1snRNA by the ISE creates a more stable association of U1snRNA, which may block splicing of the 5′ splice site. A mechanism similar to that caused by hyperstabilization of U1snRNA shown to inhibit splicing in yeast could explain the inhibition of PLP splicing (42). Finally, hnRNPH/F mediate the silencing effect of the ISE positioned close to the PLP 5′ss. Interestingly, although knockdown of hnRNPH/F has only a modest effect on the enhancer function of the ISE (27), it has a tremendous impact on the silencing effect of the ISE. These data suggest that in close proximity to the PLP 5′ss, hnRNPH/F directly recruit U1snRNP and may help to stabilize its interaction with the 5′ss.
In close proximity to the DM20 5′ss, the ISE directly recruits the U1snRNA to the DM20 splice site, indicating that the position and distance of the G runs from the splice site are major determinants of U1snRNA recruitment by G runs. In this context, the ISE acts as an enhancer, similar to but stronger than M2, and its enhancer effect correlates with binding of SRp40. These data suggest that binding of SRp40, in addition to SF2/ASF, which is normally present in the complex assembled on G1M2, might enhance the SR protein-mediated regulation of DM20 splicing, resulting in a lower PLP/DM20 ratio both in basal conditions and after knockdown of hnRNPH/F. Interestingly, G1 and M2 are both necessary for DM20 splicing, and a mutation in either sequence has a strong effect on the PLP/DM20 ratio. By contrast, G1 and ISE appear to be independent from each other. Mutation of G1 has minimal impact on the U1snRNA recruitment and on the PLP/DM20 ratio when the ISE is in place of M2. To identify features of G1M2 and ISE that would help in predicting differences in function, we have analyzed the sequences using the G-rich sequences database, which predicts the unimolecular formation of G quadruplex (43). The G runs of ISE are predicted to form a quadruplex without any interaction with an additional downstream G triplet, whereas the G1 and M2 engage a downstream G group. As part of the same paired structure, it is not surprising that both G1 and M2 are necessary for the recognition of the splice site.
Collectively, the data support the conclusion that the distance from the 5′ splice site is an important determinant of the mechanism of U1snRNA recruitment by G runs and point to a complex relationship of G runs with context sequences in determining the functional outcome on splicing. If our findings can be extended to other G runs, our data may also have a general relevance for G run-mediated recognition of 5′ splice sites.
Supplementary Material
Acknowledgments
We thank Dr. Martha Peterson for critical reading of the manuscript, Dr. Black for the hnRNPF antibody, Dr. McCullough for the HeLa nuclear extract used for primer extension and RNA affinity precipitations, and Dr. E. Buratti for the PY7NS minigene construct.
This work was supported, in whole or in part, by National Institutes of Health Grants RO1NS053905 (to F. C.) and RO1GM62287 (to K. J. H.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
E. Wang, unpublished observations.
- ISE
- intronic splicing enhancer(s)
- PLP
- proteolipid protein
- ss
- splice site(s)
- snRNP
- heterogeneous nuclear ribonuclear protein
- hnRNP
- heterogeneous nuclear ribonuclear protein
- SR
- serine arginine proteins.
REFERENCES
- 1. Black D. L. (2003) Annu. Rev. Biochem. 72, 291–336 [DOI] [PubMed] [Google Scholar]
- 2. Ladd A. N., Cooper T. A. (2002) Genome Biol. 3, 0008.1–0008.16 [Google Scholar]
- 3. Brow D. A. (2002) Annu. Rev. Genet 36, 333–360 [DOI] [PubMed] [Google Scholar]
- 4. Nilsen T. W. (2003) Bioessays 25, 1147–1149 [DOI] [PubMed] [Google Scholar]
- 5. Staley J. P., Guthrie C. (1998) Cell 92, 315–326 [DOI] [PubMed] [Google Scholar]
- 6. Hwang D. Y., Cohen J. B. (1996) Genes Dev. 10, 338–350 [DOI] [PubMed] [Google Scholar]
- 7. Madhani H. D., Guthrie C. (1994) Annu Rev Genet 28, 1–26 [DOI] [PubMed] [Google Scholar]
- 8. Green M. R. (1991) Annu. Rev. Cell Biol. 7, 559–599 [DOI] [PubMed] [Google Scholar]
- 9. Du H., Rosbash M. (2001) RNA 7, 133–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Du H., Rosbash M. (2002) Nature 419, 86–90 [DOI] [PubMed] [Google Scholar]
- 11. McCullough A. J., Berget S. M. (2000) Mol. Cell Biol. 20, 9225–9235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Carlo T., Sierra R., Berget S. M. (2000) Mol. Cell Biol. 20, 3988–3995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Carlo T., Sterner D. A., Berget S. M. (1996) RNA 2, 342–353 [PMC free article] [PubMed] [Google Scholar]
- 14. McCullough A. J., Berget S. M. (1997) Mol. Cell Biol. 17, 4562–4571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nussinov R. (1987) Biochim. Biophys. Acta 910, 261–270 [DOI] [PubMed] [Google Scholar]
- 16. Nussinov R. (1988) J. Theor. Biol. 133, 73–84 [DOI] [PubMed] [Google Scholar]
- 17. Nussinov R. (1989) J. Biomol. Struct. Dyn. 6, 985–1000 [DOI] [PubMed] [Google Scholar]
- 18. Sirand-Pugnet P., Durosay P., Brody E., Marie J. (1995) Nucleic Acids Res. 23, 3501–3507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xiao X., Wang Z., Jang M., Nutiu R., Wang E. T., Burge C. B. (2009) Nat. Struct. Mol. Biol. 16, 1094–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yeo G., Hoon S., Venkatesh B., Burge C. B. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 15700–15705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Caputi M., Zahler A. M. (2002) EMBO J. 21, 845–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Galarneau A., Richard S. (2005) Nat. Struct. Mol. Biol. 12, 691–698 [DOI] [PubMed] [Google Scholar]
- 23. Han K., Yeo G., An P., Burge C. B., Grabowski P. J. (2005) PLoS Biol. 3, e158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hastings M. L., Wilson C. M., Munroe S. H. (2001) RNA 7, 859–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang E., Dimova N., Cambi F. (2007) Nucleic Acids Res. 35, 4164–4178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hobson G. M., Huang Z., Sperle K., Stabley D. L., Marks H. G., Cambi F. (2002) Ann. Neurol. 52, 477–488 [DOI] [PubMed] [Google Scholar]
- 27. Wang E., Cambi F. (2009) J. Biol. Chem. 284, 11194–11204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hobson G. M., Huang Z., Sperle K., Sistermans E., Rogan P. K., Garbern J. Y., Kolodny E., Naidu S., Cambi F. (2006) Hum. Mutat. 27, 69–77 [DOI] [PubMed] [Google Scholar]
- 29. Wang E., Huang Z., Hobson G. M., Dimova N., Sperle K., McCullough A., Cambi F. (2006) J. Cell Biochem. 97, 999–1016 [DOI] [PubMed] [Google Scholar]
- 30. Wang E., Dimova N., Sperle K., Huang Z., Lock L., McCulloch M. C., Edgar J. M., Hobson G. M., Cambi F. (2008) Exp. Neurol. 214, 322–330 [DOI] [PubMed] [Google Scholar]
- 31. Buratti E., Baralle M., De Conti L., Baralle D., Romano M., Ayala Y. M., Baralle F. E. (2004) Nucleic Acids Res. 32, 4224–4236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pagani F., Buratti E., Stuani C., Bendix R., Dörk T., Baralle F. E. (2002) Nat. Genet. 30, 426–429 [DOI] [PubMed] [Google Scholar]
- 33. Buratti E., Brindisi A., Pagani F., Baralle F. E. (2004) Am. J. Hum. Genet. 74, 1322–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dhir A., Buratti E., van Santen M. A., Luhrmann R., Baralle F. E. (2010) EMBO J. 29, 749–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lim S. R., Hertel K. J. (2004) Mol. Cell 15, 477–483 [DOI] [PubMed] [Google Scholar]
- 36. Das R., Reed R. (1999) RNA 5, 1504–1508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Buratti E., Muro A. F., Giombi M., Gherbassi D., Iaconcig A., Baralle F. E. (2004) Mol. Cell Biol. 24, 1387–1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lynch K. W., Weiss A. (2001) J. Biol. Chem. 276, 24341–24347 [DOI] [PubMed] [Google Scholar]
- 39. Schaub M. C., Lopez S. R., Caputi M. (2007) J. Biol. Chem. 282, 13617–13626 [DOI] [PubMed] [Google Scholar]
- 40. Cloutier P., Toutant J., Shkreta L., Goekjian S., Revil T., Chabot B. (2008) J. Biol. Chem. 283, 21315–21324 [DOI] [PubMed] [Google Scholar]
- 41. Vouyiouklis D. A., Barrie J. A., Griffiths I. R., Thomson C. E. (2000) J. Neurochem. 74, 940–948 [DOI] [PubMed] [Google Scholar]
- 42. Staley J. P., Guthrie C. (1999) Mol. Cell 3, 55–64 [DOI] [PubMed] [Google Scholar]
- 43. Kostadinov R., Malhotra N., Viotti M., Shine R., D'Antonio L., Bagga P. (2006) Nucleic Acids Res. 34, D119–D124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nagai K., Muto Y., Pomeranz Krummel D. A., Kambach C., Ignjatovic T., Walke S., Kuglstatter A. (2001) Biochem. Soc. Trans. 29, 15–26 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.