Abstract
RNA 3′-phosphate cyclase (Rtc) enzymes are a widely distributed family that catalyze the synthesis of RNA 2′,3′-cyclic phosphate ends via an ATP-dependent pathway comprising three nucleotidyl transfer steps: reaction of Rtc with ATP to form a covalent Rtc-(histidinyl-N)-AMP intermediate and release PPi; transfer of AMP from Rtc to an RNA 3′-phosphate to form an RNA(3′)pp(5′)A intermediate; and attack by the terminal nucleoside O2′ on the 3′-phosphate to form an RNA 2′,3′-cyclic phosphate product and release AMP. The chemical transformations of the cyclase pathway resemble those of RNA and DNA ligases, with the key distinction being that ligases covalently adenylylate 5′-phosphate ends en route to phosphodiester synthesis. Here we show that the catalytic repertoire of RNA cyclase overlaps that of ligases. We report that Escherichia coli RtcA catalyzes adenylylation of 5′-phosphate ends of DNA or RNA strands to form AppDNA and AppRNA products. The polynucleotide 5′ modification reaction requires the His309 nucleophile, signifying that it proceeds through a covalent RtcA-AMP intermediate. We established this point directly by demonstrating transfer of [32P]AMP from RtcA to a pDNA strand. RtcA readily adenylylated the 5′-phosphate at a 5′-PO4/3′-OH nick in duplex DNA but was unable to covert the nicked DNA-adenylate to a sealed phosphodiester. Our findings raise the prospect that cyclization of RNA 3′-ends might not be the only biochemical pathway in which Rtc enzymes participate; we discuss scenarios in which the 5′-adenylyltransferase of RtcA might play a role.
Keywords: ATP, Enzyme Catalysis, Histidine, Nucleic Acid Enzymology, RNA Modification
Introduction
RNA 2′,3′-cyclic phosphate ends play important roles in RNA metabolism, especially as substrates for RNA ligases during tRNA splicing (1–4). There are two enzymatic routes to generate RNA 2′,3′-cyclic phosphate termini: (i) via transesterification, whereby an internal ribose 2′-OH attacks the adjacent 3′-5′ phosphodiester and expels a 5′-OH terminated RNA-leaving strand; or (ii) de novo cyclization of an RNA 3′-phosphomonoester in a three-step reaction that consumes ATP (5).
Escherichia coli RtcA2 exemplifies the large family of RNA 3′-terminal phosphate cyclase enzymes that catalyze de novo cyclization, with family members distributed widely among bacterial, archaeal, and eukaryal taxa (6). Cyclization occurs via a series of three nucleotidyl transfer reactions (6–12). In the first step, RtcA reacts with ATP and a divalent cation to form a covalent Rtc-AMP intermediate and liberate PPi. Adenylate is linked via a phosphoamide bond to the His309 Nϵ atom (13). In the second step, the adenylate is transferred from RtcA-AMP to the RNA 3′-phosphate terminus to form an activated phosphoanhydride intermediate, RNA(3′)pp(5′)A. In the third step, the terminal RNA ribose 2′-OH attacks the 3′-phosphate of RNA(3′)pp(5′)A to generate an RNA 2′,3′-cyclic phosphate product and release AMP.
The RNA cyclase pathway is reminiscent of the three nucleotidyl transfer steps catalyzed by RNA and DNA ligases, also via enzyme-AMP and polynucleotide-adenylate intermediates (14–18). The cyclase and ligase pathways differ in that: (i) ligases rely on lysine rather than histidine as the nucleophile for enzyme adenylylation during step 1; (ii) ligases transfer the adenylate to a polynucleotide 5′-phosphate end during step 2 to form an A(5′)pp(5′)RNA/DNA intermediate; (iii) attack by a nucleoside 3′-OH on the adenylylated end during ligation step 3 generates a 3′,5′-phosphodiester linkage between two polynucleotide ends rather than a cyclic phosphodiester terminus on one polynucleotide. The lack of structural similarity between RNA cyclases and RNA/DNA ligases, either globally or with respect to their active site architectures (13, 19), attests that Nature has independently evolved diverse solutions to perform related chemical transformations at polynucleotide ends.
Despite the depth of understanding of RtcA structure and mechanism (12, 13, 20, 21), the physiological role of RNA cyclase remains mysterious. RtcA is inessential in E. coli (11). Budding yeast have no active RNA cyclase in their proteomes (22). Because RtcA-type enzymes were purified based on their cyclization activity in vitro (9, 11), it is naturally assumed that cyclization is their true function, indeed their only function.
In the present study, we test this view and query whether the catalytic repertoire of RNA cyclase overlaps that of RNA/DNA ligases. We report that E. coli RtcA catalyzes adenylylation of 5′-phosphate ends of DNA or RNA strands to form AppDNA and AppRNA products. We delineate the reaction requirements and demonstrate the capacity of RtcA to adenylylate the 5′-phosphate at a 3′-OH/5-PO4 nick in duplex DNA. Whereas RtcA executes the first two steps of the polynucleotide ligase pathway, it is unable to convert the nicked DNA-adenylate into a sealed phosphodiester.
EXPERIMENTAL PROCEDURES
Enzyme Purifications
Wild-type E. coli RtcA and mutant H309A were produced as N-terminal His10-tagged fusion proteins and purified from soluble bacterial extracts by sequential Ni-agarose, anion-exchange, and gel-filtration chromatography steps as described (13). Wild-type Chlorella virus DNA ligase and mutant K27A were produced as N-terminal His10-tagged fusions and purified from soluble bacterial extracts by Ni-agarose and phosphocellulose chromatography (23). Protein concentrations were determined by using the Bio-Rad dye reagent with bovine serum albumin as the standard.
Assay of Polynucleotide 5′-Adenylylation
The polynucleotide substrates used in the adenylylation assays were 5′-32P end-labeled and then gel-purified as described (24). Adenylylation reaction mixtures (10 μl) containing 50 mm Tris acetate (pH 6.0), 1 mm DTT, 2 mm MgCl2, 1 mm ATP, 3 pmol (300 nm) 5′-32P-labeled 12-mer DNA oligonucleotide 5′-pCAATTGCGACCC (or other polynucleotides as specified), and RtcA as specified were incubated at 37 °C for 30 min. The reactions were quenched by adding 5 μl of 90% formamide, 50 mm EDTA, 0.1% bromphenol blue. The samples were heated at 95 °C for 1 min and then analyzed by electrophoresis (at 7 W for ∼100 min) through a 15-cm 24% polyacrylamide gel containing 8 m urea in 0.45 mm Tris borate, 1.2 mm EDTA. The radiolabeled products were visualized by autoradiography. The extents of DNA adenylylation were quantified by scanning the gel with a Fujix BAS2500 imager.
Analysis of the Adenylylated Product
Reaction mixtures (50 μl) containing 50 mm Tris acetate (pH 6.0), 1 mm DTT, 2 mm MgCl2, 1 mm ATP, 300 nm 12-mer DNA substrate, and either 80 nm RtcA (RtcA+) or no added enzyme (RtcA−) were incubated at 37 °C for 30 min. Aliquots (2 μl) of the reaction mixtures were then subjected to digestion with various analytical enzymes, either calf intestine alkaline phosphatase (CIP), nuclease P1, or nucleotidyl pyrophosphatase (NPPase). The CIP reaction mixtures (10 μl) containing 50 mm Tris-HCl (pH 7.9), 10 mm MgCl2, 100 mm NaCl, 1 mm DTT, and 0.1 unit CIP (New England Biolabs) were incubated for 1 h at 37 °C. Nuclease P1 reaction mixtures (10 μl) containing 50 mm sodium acetate (pH 5.2), 1 mm ZnCl2, and 0.1 unit nuclease P1 (Sigma) were incubated for 20 min at 50 °C. For double digestions with nuclease P1 and CIP, the samples were initially digested with nuclease P1 as described above, then the reactions were adjusted to CIP buffer and subsequently treated with CIP as above. For double digestions with nuclease P1 and NPPase, the P1-treated samples were adjusted to 50 mm Tris-HCl (pH 8.0), 10 mm MgCl2, 100 mm NaCl and then incubated with 0.001 unit NPPase (Sigma) for 20 min at 37 °C. For triple digestions, the P1/NPPase double-digested samples were adjusted to CIP buffer and then incubated with 0.1 unit CIP for 1 h at 37 °C. The digestion reactions were quenched by adding 2 μl of 5 m formic acid. Aliquots (6 μl) of the quenched samples were then spotted on a polyethyleneimine (PEI)-cellulose TLC plate (Merck) alongside unlabeled marker nucleotides CTP, CDP, and CMP. Ascending TLC was performed with 0.75 m LiCl as the mobile phase. The radiolabeled species were visualized by autoradiography of the TLC plate. Unlabeled marker nucleotides were located by UV illumination.
Preparation of RtcA-[32P]AMP and Transfer of [32P]AMP to 5′-Phosphate DNA
An in vitro adenylylation reaction mixture (70 μl) containing 50 mm Tris-HCl (pH 7.4), 10 mm MgCl2, 1 mm DTT, 50 μm [α-32P]ATP, 250 μm pyrophosphate, and 16 μm RtcA was incubated at 37 °C for 20 min. The resulting RtcA-[32P]AMP complex was recovered by centrifugal gel filtration through a 0.5-ml spin desalting column (7-kDa cutoff; Pierce) equilibrated with 50 mm Tris acetate (pH 6.0), 1 mm DTT. The gel filtrate was stored at −80 °C. An aliquot of the gel filtrate was analyzed by electrophoresis through a 12% polyacrylamide gel containing 0.1% SDS. The RtcA-[32P]AMP adduct was quantified by scanning the gel with a Fujix BAS-2500 imager in parallel with serial dilution of the [α-32P]ATP stock solution.
[32P]AMP transfer reaction mixtures (10 μl) containing 50 mm Tris acetate (pH 6.0), 1 mm DTT, 200 fmol (20 nm) RtcA-[32P]AMP, and either 1 pmol (100 nm) cold 5′-phosphate-terminated 12-mer DNA (5′-pCAATTGCGACCCOH), 1 pmol (100 nm) cold 5′-hydroxyl-terminated 12-mer DNA (5′-HOCAATTGCGACCCOH), or no added polynucleotide were incubated at 37 °C for 30 min. The reactions were quenched with formamide-EDTA, and the reaction products were resolved by urea-PAGE.
RESULTS
RtcA Adenylylates a Polynucleotide 5′-Phosphate End
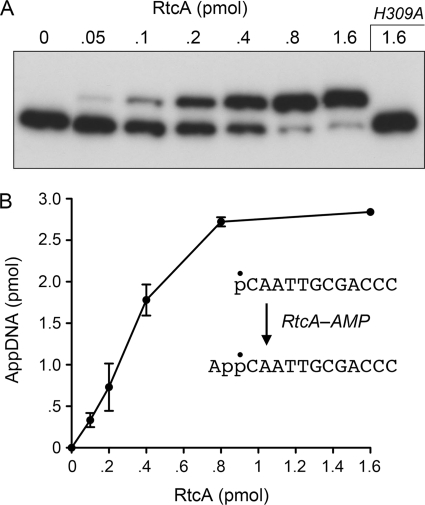
Reaction of purified recombinant E. coli RtcA with a 5′-32P-labeled 12-mer DNA oligonucleotide in the presence of ATP and magnesium resulted in conversion of the radiolabeled substrate to a novel product that migrated ∼1.5 nucleotide steps slower during electrophoresis through a denaturing polyacrylamide gel (Fig. 1A). The extent of product formation was proportional to the level of input RtcA; virtually all of the substrate was converted to the larger product at saturating enzyme (Fig. 1, A and B). From the slope of the titration curve, we estimated that ∼4 pmol of product was generated per pmol of RtcA (Fig. 1B). By contrast, RtcA mutant H309A, which lacks the histidine to which AMP becomes covalently attached during the RNA 3′-cyclization pathway (12, 13), was unreactive with the 5′-32P-labeled 12-mer DNA at a level of input protein that was saturating for product formation by wild-type RtcA (Fig. 1A). We surmise that the novel reaction evinced by this experiment entails transfer of adenylate from the RtcA-AMP covalent intermediate to the DNA 5′-monophosphate to form a DNA-adenylate, A(5′)pp(5′)DNA (Fig. 1B).
FIGURE 1.
RtcA adenylylates a 5′-phosphate DNA end. A, reaction mixtures containing 50 mm Tris acetate (pH 6.0), 1 mm DTT, 2 mm MgCl2, 1 mm ATP, 3 pmol (300 nm) 5′-32P-labeled 12-mer DNA (5′-pCAATTGCGACCCOH), and RtcA as specified were incubated for 30 min at 37 °C. The products were analyzed by urea-PAGE and visualized by autoradiography. B, the extents of conversion of the pDNA strand to AppDNA product were quantified and plotted as a function of input RtcA. Each datum is the average of three independent enzyme titration experiments ± S.E. (error bars).
Similar experiments performed with a 5′-32P-labeled 12-mer RNA oligonucleotide revealed an RtcA concentration-dependent conversion of the pRNA substrate to a single, slower migrating product (supplemental Fig. S1). About 2.5 pmol of this RNA product was formed per pmol of RtcA (supplemental Fig. S1). RtcA-H309A was unable to perform the putative RNA 5′-adenylylation reaction. These results reveal the capacity of RtcA to catalyze a ligase-like 5′-end activation reaction at either pDNA or pRNA ends.
5′-Adenylylation Reaction Requirements
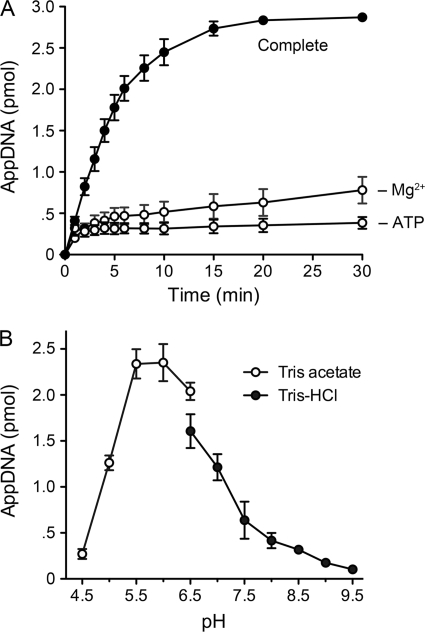
In the experiments here and below, we used the 12-mer pDNA substrate to characterize the 5′-adenylylation activity of RtcA. The kinetic profile of the reaction of 80 nm RtcA with 300 nm pDNA in the presence of ATP and magnesium revealed steady accumulation of product between 1 and 10 min; the reaction reached an end point at 20–30 min with 96% conversion to AppDNA (Fig. 2A), signifying that the input RtcA had catalyzed multiple rounds of product formation. The rate of product formation (i.e. the slope of the curve), declined progressively as the substrate was consumed (Fig. 2A). Omission of ATP resulted in a different kinetic pattern, whereby an end point was achieved within minutes with only 11–12% conversion of pDNA to AppDNA (Fig. 2A). ATP-independent transadenylylation reflects a single-turnover reaction catalyzed by the substantial fraction of preadenylylated RtcA in the enzyme preparation (13). From the extent of AppDNA formed by 80 nm total RtcA protein in the −ATP reaction, we estimated that active RtcA-AMP comprised at least 45% of the RtcA preparation. Omission of magnesium also curtailed the reaction at a low end point, but there appeared to be a biphasic kinetic pattern, whereby an initial burst of adenylylation (to ∼12% conversion at 3 min) was followed by a slower accumulation of product, to an extent of 26% at 30 min (Fig. 2).
FIGURE 2.
Requirements for DNA 5′-adenylylation by RtcA. A, kinetic profile. The “complete” reaction mixture (130 μl) containing 50 mm Tris acetate (pH 6.0), 1 mm DTT, 2 mm MgCl2, 1 mm ATP, 300 nm 5′-32P-labeled 12-mer DNA, and 80 nm RtcA was incubated at 37 °C. Divalent cation and ATP were omitted from reaction mixtures, −Mg2+ and −ATP, respectively. Aliquots (10 μl, containing 3 pmol DNA) were withdrawn at the times specified and quenched immediately with formamide-EDTA. The time 0 sample was withdrawn prior to adding the RtcA. The extents of conversion of the pDNA strand to AppDNA product are plotted as a function of reaction time. Each datum is the average of three independent experiments ± S.E. (error bars). B, pH profile. Reaction mixtures (10 μl) containing either 50 mm Tris acetate (pH 4.5, 5.0, 5.5 6.0, or 6.5) or Tris-HCl (pH 6.5, 7.0, 7.5, 8.0, 8.5, 9.0, or 9.5), 1 mm DTT, 2 mm MgCl2, 1 mm ATP, 3 pmol 5′-32P-labeled 12-mer DNA, and 0.6 pmol RtcA were incubated at 37 °C for 30 min. The extents of conversion of the pDNA strand to AppDNA product are plotted as function of pH. Each datum is an average of three independent experiments ± S.E.
The pH profile of the multiturnover 5′-adenylation reaction of E. coli RtcA was noteworthy for its optimum at mildly acidic pH (5.5–6.0 in Tris acetate buffer) and a sharp decline in product formation at alkaline pH (≥8.0 in Tris-HCl) (Fig. 2B). The multiturnover RNA 3′-cyclization reaction of RtcA displays a very different pH profile, with a sharp optimum at pH 8.0–8.5 and virtually no activity at pH ≤6.0 (11). In the case of the single-turnover 5′-adenylylation reaction of RtcA (without ATP or magnesium), the yields of AppDNA were similar across the entire range tested, from pH 4.5 to 9.5 (data not shown). In subsequent experiments, we assayed multiturnover 5′-adenylylation at pH 6.0. Single-turnover 5′-adenylylation was performed at pH 6.0 or 7.4.
The results in Fig. 2A highlight the importance of ATP and magnesium in promoting a multiturnover process of 5′-adenylylation under conditions of pDNA excess, presumably via their contributions to the reaction of RtcA apoenzyme with ATP to regenerate the RtcA-AMP intermediate. They also suggest that ATP and magnesium are not strictly essential for the subsequent step of adenylate transfer from RtcA-AMP to the pDNA end. We verified this inference by examining the adenylylation reaction in the absence of magnesium and ATP under conditions in which the concentration of RtcA-AMP was in 12-fold excess over that of the pDNA substrate (supplemental Fig. S2). AppDNA accumulated with pseudo-first-order kinetics (kobs 0.18 min−1), and the reaction reached an end point at 20–40 min with 95% conversion of pDNA to AppDNA (supplemental Fig. S2). We considered the possibility that the RtcA preparation might contain residual divalent cations, but found that the extent of AppDNA formation under single-turnover conditions in RtcA-AMP excess was unaffected by inclusion of 10 mm EDTA in the reaction mixture (data not shown).
Product Analysis
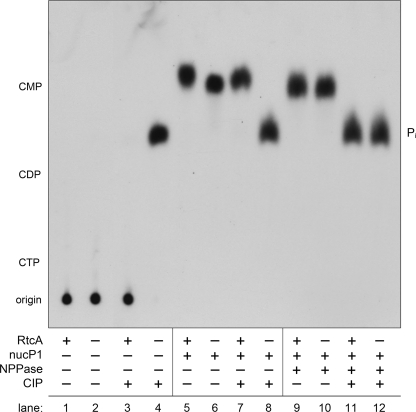
The identity of the putative AppDNA product was verified by digestion of the product with various hydrolytic enzymes, CIP, NPPase, or nuclease P1, singly or sequentially. The 32P-labeled digestion products were analyzed by ascending PEI-cellulose TLC in parallel with untreated input pDNA substrate and untreated RtcA reaction product (Fig. 3). The pDNA substrate (lane 2) and the RtcA product (lane 1) remained at the chromatographic origin, as expected for polyanionic oligonucleotides. Whereas the 5′-[32P]phosphate of the pDNA substrate was converted quantitatively by CIP to 32Pi (lane 4), the RtcA reaction product was resistant to CIP (lane 3), signifying that the product 5′-end was blocked. Digestion of the substrate with nuclease P1 (which cleaves the 3′-O-P bond) liberated a single species, predicted to be 5′-dCMP, that co-migrated with the marker mononucleotide 5′-CMP (lane 6) and was quantitatively converted by CIP to 32Pi (lane 8). By contrast, nuclease P1 digestion of the RtcA reaction product liberated a single species (lane 5; migrating slightly faster than 5′-dCMP), that was resistant to CIP (lane 7). However, when the nuclease P1-digested RtcA product, predicted to be a dinucleotide App(dC), was treated with NPPase, it was converted to a CIP-sensitive species (lane 9; co-migrating with the 5′-dCMP mononucleotide) that was hydrolyzed quantitatively to 32Pi during subsequent CIP treatment (lane 11). These results are fully consistent with our designation of the RtcA reaction product as A(5′)pp(5′)DNA.
FIGURE 3.
Analysis of the adenylylated product. Reaction mixtures (50 μl) containing 50 mm Tris acetate (pH 6.0), 1 mm DTT, 2 mm MgCl2, 1 mm ATP, 300 nm 12-mer DNA substrate, and either 80 nm RtcA (RtcA+) or no added enzyme (RtcA−) were incubated at 37 °C for 30 min. Aliquots of the reaction mixtures were then subjected to digestion with various analytical enzymes, either CIP, nuclease P1 (nucP1), or NPPase, where indicated by +. The digested and undigested samples were analyzed by PEI-cellulose TLC. An autoradiograph of the chromatogram is shown. The origin and positions of unlabeled nucleotide markers (CTP, CDP, CMP) are indicated on the left. The species corresponding to 32Pi is indicated on the right.
Influence of Polynucleotide Length on 5′-Adenylylation by RtcA
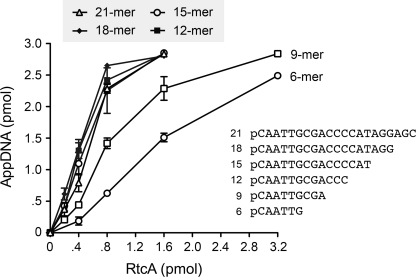
A series of 5′-32P-labeled DNAs of varying length, but common 5′-nucleotide sequence, were tested as substrates for RtcA at equivalent oligonucleotide concentrations. Enzyme titration experiments revealed similar specific activities for 21-mer, 18-mer, 15-mer, and 12-mer pDNAs (Fig. 4). By contrast, 9-mer and 6-mer pDNAs were 2-fold and 4-fold less reactive, respectively, than the 12-mer substrate (Fig. 4).
FIGURE 4.
Influence of DNA length on 5′-adenylylation. Reaction mixtures containing 50 mm Tris acetate (pH 6.0), 1 mm DTT, 2 mm MgCl2, 1 mm ATP, 3 pmol (300 nm) 5′-32P-labeled 21-mer, 18-mer, 15-mer, 12-mer, 9-mer, or 6-mer DNAs, and RtcA as specified were incubated for 30 min at 37 °C. The products were analyzed by urea-PAGE. The extents of conversion of the pDNA strands to AppDNA products were quantified and plotted as a function of input RtcA. Each datum is the average of three independent enzyme titration experiments ± S.E. (error bars).
Demonstration of 32P-Adenylate Transfer from RtcA to pDNA
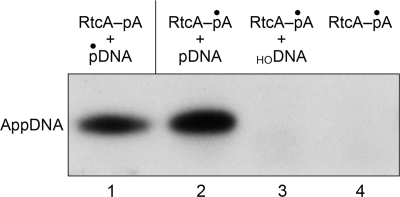
The proposed reaction scheme for 5′-adenylylation by RtcA, based on the stimulation of the multiturnover reaction by ATP and the requirement for the His309 nucleophile, is that an AMP covalently attached to RtcA is the source of the blocking nucleotide of the AppDNA product. We tested this prediction by adenylylating RtcA in vitro by reaction with [α-32P]ATP in the presence of magnesium and 250 μm PPi. (PPi enhances the yield of AMP labeling by virtue of converting preformed cold RtcA-AMP to RtcA apoenzyme (13).) The RtcA-[32P]AMP intermediate was recovered by gel filtration under native conditions, and the level of radiolabeled enzyme was determined by SDS-PAGE analysis and quantification of the covalent RtcA-[32P]AMP adduct. The isolated RtcA-[32P]AMP intermediate was then incubated with either cold 5′-monophosphate-terminated 12-mer DNA (5′-pCAATTGCGACCCOH), cold 5′-OH-terminated 12-mer DNA (5′-HOCAATTGCGACCCOH), or no added polynucleotide. The products of these reactions were analyzed by urea-PAGE (Fig. 5). The salient finding was that RtcA-[32P]AMP transferred the labeled adenylate to the pDNA strand to yield a [32P]AMP-labeled oligonucleotide product (lane 2) that co-migrated with the AppDNA species formed by reaction of cold RtcA with 5′-32P-labeled 12-mer DNA (lane 1). The formation of the [32P]AMP-labeled oligonucleotide depended on inclusion of the 12-mer pDNA strand (lane 4). Moreover, RtcA-[32P]AMP was unreactive with an otherwise identical 12-mer DNA with a 5′-OH terminus (lane 3). These results support the following two-step reaction pathway.
 |
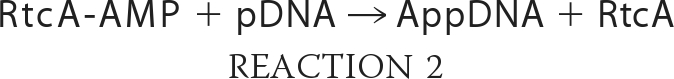 |
The results also establish the specificity for a terminal phosphomonoester as the nucleophile in the second step of the 5′-adenylylation reaction, the same requirement that pertains to the second adenylate transfer step of the 3′-phosphate cyclization pathway.
FIGURE 5.
Transfer of [α-32P]AMP from RtcA-AMP to a DNA 5′-phosphate end. Reaction mixtures containing 200 fmol RtcA-[32P]AMP and either 1 pmol cold 5′-phosphate-terminated 12-mer DNA (5′-pCAATTGCGACCCOH) (lane 2), 1 pmol cold 5′-hydroxyl-terminated 12-mer DNA (5′-HOCAATTGCGACCCOH) (lane 3), or no added polynucleotide (lane 4) were incubated at 37 °C for 30 min. The reaction products were analyzed by urea-PAGE in parallel with the product formed by reaction of cold RtcA with 5′-32P-labeled 12-mer DNA (lane 1). The radiolabeled oligonucleotides were visualized by autoradiography.
RtcA Can Adenylylate (but Not Seal) a 5′-PO4/3′-OH Nick
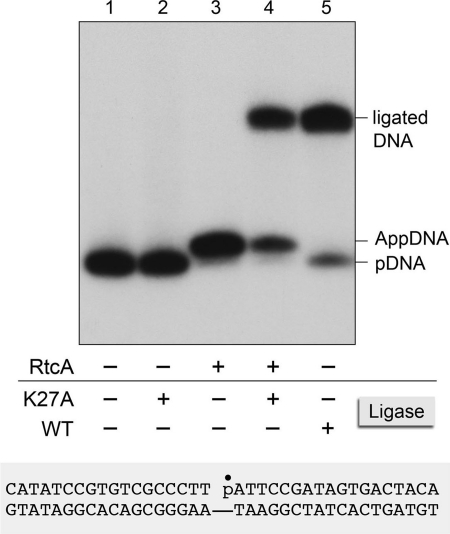
The capacity of RtcA to catalyze ligase-like 5′-end-activation reactions raises the question of whether RtcA might also be capable of phosphodiester synthesis, akin to the third step of the ligase pathway. A corollary issue is whether the 5′-adenylyltransferase activity of RtcA is restricted to single-stranded pDNA and pRNA acceptors or more broadly accepting of duplex structures such as those repaired by canonical DNA ligases. We addressed these questions by presenting RtcA with a singly nicked 36-bp duplex formed by annealing a 5′-32P-labeled 18-mer strand and an unlabeled 18-mer 3′-OH strand to a complementary 36-mer (Fig. 6). Control experiments verified that reaction of this nicked duplex with Chlorella virus DNA ligase resulted in efficient conversion of the radiolabeled 18-mer pDNA strand into a sealed 36-mer product (Fig. 6, compare lanes 1 and 5). By contrast, reaction of RtcA with the nicked duplex resulted in near quantitative conversion of the labeled pDNA strand to a novel species that migrated ∼1.5 nucleotide steps slower during urea-PAGE, said species corresponding to a 5′-adenylylated 18-mer AppDNA (Fig. 6, lane 3). There was no detectable 36-mer ligation product in the RtcA reaction. To verify the identity and integrity of the putative nicked DNA-adenylate formed by RtcA, we reacted the RtcA product with a mutated version of Chlorella virus DNA ligase (K27A) that lacks the lysine to which AMP is covalently attached during the ligase adenylylation reaction (23). The K27A protein is inert in sealing a 5′-PO4/3′-OH nick (Fig. 6, lane 2) but is active in catalyzing phosphodiester synthesis at a preadenylylated nick (23). The instructive finding was that the K27A ligase sealed the adenylylated nick formed by RtcA to yield a 36-mer ligation product (Fig. 6, lane 4). Thus, we conclude that RtcA can generate the same covalently activated nick as DNA ligase but cannot proceed with phosphodiester synthesis (at least not under the conditions surveyed here).
FIGURE 6.
RtcA adenylylates the 5′-phosphate strand at a DNA nick. Reaction mixtures (10 μl) containing 50 mm Tris HCl (pH 7.4), 5 mm DTT, 50 nm singly nicked 36-bp DNA substrate (illustrated at the bottom of the figure, with the 32P-labeled nick 5′-phosphate denoted by ●) and either 13.9 μm RtcA (RtcA+) or no added enzyme (RtcA−) were incubated at 37 °C for 30 min. The mixtures were then supplemented as specified with 10 mm MgCl2 and either 0.27 μm wild-type Chlorella virus DNA ligase (WT+), 0.27 μm K27A mutant ligase (K27A+), or no ligase (Ligase−) and then incubated further at 22 °C for 30 min. The radiolabeled products were analyzed by urea-PAGE. An autoradiograph of the gel is shown, with the positions of the 18-mer pDNA strand, the AppDNA strand, and the ligated 36-mer product indicated on the right.
DISCUSSION
We report here a novel polynucleotide 5′-adenylylation activity of E. coli RtcA that highlights its similarities to RNA and DNA ligases, notwithstanding the lack of structural homology between the cyclase and ligase protein families. It is now evident that RtcA does not discriminate strictly between polynucleotide 3′-monophosphate and polynucleotide 5′-monophosphate ends as nucleophiles capable of attacking the AMP phosphate of the RtcA-AMP intermediate. Whereas an activated RNA(3′)pp(5′)A is thought be a transient intermediate that is instantly converted via intramolecular attack to an RNA 2′,3′-cyclic phosphate end product (7, 11), the activated A(5′)pp(5′)DNA/RNA species reported here is itself a stable end product, reflecting the absence of a vicinal hydroxyl nucleophile in cis. The versatility of RtcA in generating AppDNA and AppRNA strands in excellent yield recommends its use as a reagent for polynucleotide synthesis and RNA cloning, especially in strategies that employ preadenylylated linkers to selectively tag one RNA end (25).
The liberal use of either DNA or RNA strands for 5′-adenylylation has interesting implications for the mode of polynucleotide acceptor recognition. The crystal structures of the RtcA-AMP intermediate and the RtcA apoenzyme revealed an oxyanion-binding pocket adjacent to the adenylate site that was proposed to mediate the binding of the 3′-monophosphate and penultimate phosphodiester of the RNAp substrate for the cyclization reaction (13, 20). A parsimonious view of the present findings with respect to 5′-adenylylation is that pDNA/RNA strands bind to this same site on the enzyme, albeit with opposite polarity, such that the sites occupied by two sulfates in the RtcA-AMP structure (13) would demarcate the positions of the 5′-monophosphate and adjacent phosphodiester in the 5′-adenylyltransferase substrate. A definitive evaluation of this model must await co-crystallization of RtcA with bound polynucleotide substrates or adenylylated polynucleotide products/intermediates.
The biological functions and physiological substrates of RNA cyclases are uncharted. The present study raises the interesting prospect that cyclization of RNA 3′-ends might not be the only biochemical pathway in which they participate. We envision a number of biological scenarios in which the 5′-adenylyltransferase of RtcA might play a role. These include: (i) adenylylation of DNA or RNA 5′-ends as a means of protecting them against exonucleolytic decay, analogous to the function of the eukaryal mRNA cap; (ii) installing an inverted 5′-adenylate as a “mark” that targets RNA or DNA molecules for downstream events, e.g. localization, translation, recombination, etc; or (iii) the generation of AppDNA or AppRNA ends that serve as substrates for subsequent DNA/RNA sealing reactions catalyzed by separate repair enzymes. Finally, it is now apparent that Nature has evolved at least two distinct structural solutions to the chemical steps of 5′-nucleotidylation, exemplified by the ligase/capping enzyme superfamily and the Rtc superfamily, respectively, both of which rely on covalent catalysis via a phosphoramidate enzyme-NMP intermediate.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant GM46330.

The on-line version of this article (available at http://www.jbc.org) contains Figs. S1 and S2.
- RtcA
- RNA 3′-phosphate cyclase
- CIP
- calf intestine alkaline phosphatase
- NPPase
- nucleotidyl pyrophosphatase.
REFERENCES
- 1. Konarska M., Filipowicz W., Gross H. J. (1982) Proc. Natl. Acad. Sci. U.S.A. 79, 1474–1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Greer C. L., Peebles C. L., Gegenheimer P., Abelsonm J. (1983) Cell 32, 537–546 [DOI] [PubMed] [Google Scholar]
- 3. Filipowicz W., Shatkin A. J. (1983) Cell 32, 547–557 [DOI] [PubMed] [Google Scholar]
- 4. Laski F. A., Fire A. Z., RajBhandary U. L., Sharp P. A. (1983) J. Biol. Chem. 258, 11974–11980 [PubMed] [Google Scholar]
- 5. Filipowicz W., Konarska M., Gross H. J., Shatkin A. J. (1983) Nucleic Acids Res. 11, 1405–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Genschik P., Billy E., Swianiewicz M., Filipowicz W. (1997) EMBO J. 16, 2955–2967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Filipowicz W., Strugala K., Konarska M., Shatkin A. J. (1985) Proc. Natl. Acad. Sci. U.S.A. 82, 1316–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Reinberg D., Arenas J., Hurwitz J. (1985) J. Biol. Chem. 260, 6088–6097 [PubMed] [Google Scholar]
- 9. Vicente O., Filipowicz W. (1988) Eur. J. Biochem. 176, 431–439 [DOI] [PubMed] [Google Scholar]
- 10. Filipowicz W., Vicente O. (1990) Methods Enzymol. 181, 499–510 [DOI] [PubMed] [Google Scholar]
- 11. Genschik P., Drabikowski K., Filipowicz W. (1998) J. Biol. Chem. 273, 25516–25526 [DOI] [PubMed] [Google Scholar]
- 12. Billy E., Hess D., Hofsteenge J., Filipowicz W. (1999) J. Biol. Chem. 274, 34955–34960 [DOI] [PubMed] [Google Scholar]
- 13. Tanaka N., Smith P., Shuman S. (2010) Structure 18, 449–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shuman S., Lima C. D. (2004) Curr. Opin. Struct. Biol. 14, 757–764 [DOI] [PubMed] [Google Scholar]
- 15. Pascal J. M., O'Brien P. J., Tomkinson A. E., Ellenberger T. (2004) Nature 432, 473–478 [DOI] [PubMed] [Google Scholar]
- 16. Nandakumar J., Shuman S., Lima C. D. (2006) Cell 127, 71–84 [DOI] [PubMed] [Google Scholar]
- 17. Nandakumar J., Nair P. A., Shuman S. (2007) Mol. Cell 26, 257–271 [DOI] [PubMed] [Google Scholar]
- 18. Nair P. A., Nandakumar J., Smith P., Odell M., Lima C. D., Shuman S. (2007) Nat. Struct. Mol. Biol. 14, 770–778 [DOI] [PubMed] [Google Scholar]
- 19. Shuman S. (2009) J. Biol. Chem. 284, 17365–17369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Palm G. J., Billy E., Filipowicz W., Wlodawer A. (2000) Structure 8, 13–23 [DOI] [PubMed] [Google Scholar]
- 21. Tanaka N., Shuman S. (2009) RNA 15, 1865–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Billy E., Wegierski T., Nasr F., Filipowicz W. (2000) EMBO J. 19, 2115–2126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sriskanda V., Shuman S. (1998) Nucleic Acids Res. 26, 525–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shuman S., Ru X. M. (1995) Virology 211, 73–83 [DOI] [PubMed] [Google Scholar]
- 25. Pfeffer S., Sewer A., Lagos-Quintana M., Sheridan R., Sander C., Grässer F. A., van Dyk L. F., Ho C. K., Shuman S., Chien M., Russo J. J., Ju J., Randall G., Lindenbach B. D., Rice C. M., Simon V., Ho D. D., Zavolan M., Tuschl T. (2005) Nat. Methods 2, 269–276 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.