Abstract
Heightened DJ-1 (Park7) expression is associated with a reduction in chemotherapeutic-induced cell death and poor prognosis in several cancers, whereas the loss of DJ-1 function is found in a subgroup of Parkinson disease associated with neuronal death. This study describes a novel pathway by which DJ-1 modulates cell survival. Mass spectrometry shows that DJ-1 interacts with BBS1, CLCF1, MTREF, and Cezanne/OTUD7B/Za20d1. Among these, Cezanne is a known deubiquitination enzyme that inhibits NF-κB activity. DJ-1/Cezanne interaction is confirmed by co-immunoprecipitation of overexpressed and endogenous proteins, maps to the amino-terminal 70 residues of DJ-1, and leads to the inhibition of the deubiquitinating activity of Cezanne. Microarray profiling of shRNA-transduced cells shows that DJ-1 and Cezanne regulate IL-8 and ICAM-1 expression in opposing directions. Similarly, DJ-1 enhances NF-κB nuclear translocation and cell survival, whereas Cezanne reduces these outcomes. Analysis of mouse Park7−/− primary cells confirms the regulation of ICAM-1 by DJ-1 and Cezanne. As NF-κB is important in cellular survival and transformation, IL-8 functions as an angiogenic factor and pro-survival signal, and ICAM-1 has been implicated in tumor progression, invasion, and metastasis; these data provide an additional modality by which DJ-1 controls cell survival and possibly tumor progression via interaction with Cezanne.
Keywords: Cancer Tumor Promoter, Deubiquitination, Interleukin, NF-kappa B, Parkinson's Disease, Cezanne, DJ-1, ICAM-1
Introduction
Lung cancer is the leading cause of cancer-related mortality in the world. It is estimated that over 215,000 new cases were diagnosed in the United States in 2008 alone (1). Of the two major types of lung cancer, 85% are non-small cell lung carcinomas (NSCLC)2 (2). With NSCLC, the best chance for a long term cure is surgery. Unfortunately, 70% of NSCLC patients are initially contraindicated for surgery due to locally invasive or metastatic disease (3). For these patients, the current treatment options are a combination of chemotherapy and radiation. With treatment, NSCLC patients have a 5-year survival rate of 15%. It is therefore imperative to investigate new pathways to target in NSCLC. Our work in the H157 NSCLC squamous cell line has focused on the role of DJ-1.
DJ-1 is associated with a spectrum of human disease. Loss of DJ-1 activity, due to inactivation by mutation or genetic deletion, causes early onset Parkinson disease with high penetrance (4). Due to the protective effects of DJ-1 against toxic insults, this association is hypothesized to be due to a protective role of DJ-1 in dopaminergic neurons of the substantia nigra (5–7). As loss of DJ-1 activity leads to neuronal cell death, increased DJ-1 activity can promote cell survival, leading to its association with human malignancies. Initial research on the role of DJ-1 in cancer showed that although DJ-1 can weakly transform cells alone, it can act synergistically in combination with Ras (8). DJ-1 is highly expressed in a number of cancer types including acute leukemia, breast, prostate, ovarian, and thyroid cancer, and NSCLC and is associated with tumor progression in esophageal and cervical cancer (9–15). Our laboratory and others have shown that increased DJ-1 expression confers protection against toxic insults including oxidative and endoplasmic reticulum stress, proteasome inhibition, and chemotherapeutic agents (16–18). This protection is partially mediated by the ability of DJ-1 to inhibit p53 and Phosphatase and Tensin homolog (PTEN), an inhibitor of the PI3K cell survival pathway, and to positively regulate the Nrf2 detoxification pathway (15, 19, 20). Here we examine a novel pathway in which DJ-1 binds to and inhibits Cezanne, a negative regulator of the cell survival-associated transcription factor NF-κB.
Cezanne is a member of the A20 family of deubiquitination enzymes (21). Similar to A20, Cezanne has been shown to inhibit NF-κB through modulation of the ubiquitination state of two of its positive regulators, RIP-1 and TRAF6 (21, 22). Cezanne is implicated in cancer biology as a 93-kb sequence containing Cezanne was shown to be duplicated in acute lymphoblastic leukemia and Burkitt lymphoma (23). Although the precise role of Cezanne in cancer is unknown, its inhibition of NF-κB would suggest that Cezanne may act as a tumor suppressor.
Although typically associated with regulating the inflammatory response to infection or injury, NF-κB subunits are constitutively activated in a number of human carcinomas (24–26). This family of transcription factors plays a role in cancer cell survival, angiogenesis, invasion, progression, and metastasis. Two important targets of NF-κB-mediated transcription in relation to cancer are interleukin 8 (IL-8) and intercellular adhesion molecule 1 (ICAM-1). These two proteins are involved in the homing and extravasation of neutrophils from the blood into inflamed tissues but have additional functions in cancer biology. IL-8 expression has been associated with cancer progression, invasion, angiogenesis, and metastasis (27–33). It is also secreted by certain NSCLC cell lines, including H157 cells, in response to Paclitaxel treatment, activating a pro-survival-positive feedback loop via signaling through the CXCR1 receptor (34, 35). Blockade of IL-8 binding to its receptor causes a decrease in tumor size, angiogenesis, tumor growth, and metastasis while promoting cancer cell apoptosis (36). ICAM-1 is highly expressed in multiple human cancers as well (37–39). Although it has not been associated with effects on cancer cell survival, ICAM-1 has been linked to both tumor progression and metastasis through its ability to mediate cell adhesion and extravagation (33, 40, 41).
In this study, we identify Cezanne as a binding partner of DJ-1. This interaction can be mediated by the amino terminus of DJ-1 and leads to the inhibition of the deubiquitinating activity of Cezanne in vitro. Microarray analysis on H157 cells shows that DJ-1 and Cezanne shRNAs regulate IL-8 and ICAM-1 gene expression, albeit in opposing directions. DJ-1 and Cezanne shRNA treatments also result in reciprocal phenotypes in chemotherapeutic-induced cell death and NF-κB nuclear localization in vitro. Finally, we analyzed ICAM-1 expression in Park7−/− mouse embryonic fibroblasts (MEF) to verify that this pathway is intact in a primary cell line. These findings identify a novel pathway through which DJ-1 modulates a number of critical aspects of cancer cell biology as well as uncovering a potential role of DJ-1 in leukocyte migration and inflammation. This work emphasizes the importance of future research on pharmacologic inhibitors of the DJ-1/Cezanne pathway and the role of DJ-1 in cancer biology in vivo.
EXPERIMENTAL PROCEDURES
Cell Culture, Treatments, and Plasmid Constructs
H157 cells were grown in RPMI 1640 (Invitrogen) with 10% FCS (HyClone). HEK293T cells and MEFs were grown in high glucose DMEM (Invitrogen) with 10% FCS. MEFs were isolated from 13.5-day embryos and used within three passages from initial isolation. All mammalian cell cultures were incubated at 37 °C with 5% CO2 in the presence of penicillin and streptomycin. H2O2 (Sigma) was added to cells at 100 μm for 1 h. TNFα (eBioscience) was used at 5–50 ng/ml for 6 h before sample harvest. For cell death assays, Paclitaxel and U0126 (Sigma) reconstituted in dimethyl sulfoxide (DMSO) were used at 500 nm and 10 μm, respectively.
Human Cezanne ORF was subcloned into the V5/His-tagged pcDNA3.1D-Topo plasmid (Invitrogen) using the primers 5′-CAC CAT GAC CCT GGA CAT GGA TGC TG-3′ and 5′-GAA CCT GTG CAC CAG GAG CT-3′. Directionality and expression were verified by sequencing and immunoblot analysis, respectively. The FLAGTM-DJ-1 construct was generously given to us by Y. Hod (12). The point and truncation mutants of FLAG-DJ-1 were produced using QuikChange mutagenesis (Stratagene) and verified by sequencing. MEF cultures were transfected using Lipofectamine 2000 (Invitrogen) at a 4:1 reagent to DNA ratio, and HEK293T cultures were transfected using FuGENE 6 (Roche Applied Science) at a 3:1 ratio as per the manufacturer's instructions.
Q-TOF Mass Spectrometry
Park7−/− MEFs were transfected with either FLAG-DJ-1 or C016A-FLAG. After H2O2 or vehicle control treatment, samples were lysed in 0.5% Triton X-100 lysis buffer and immunoprecipitated with anti-FLAG M2-agarose (Sigma). Samples were eluted with FLAG peptide and desalted using a C4 reverse-phase media-loaded micropipette tip (Millipore). The desalted proteins were eluted from the tip using a solution of 80% methanol with 0.1% formic acid and then lyophilized. After reconstitution in 25 mm ammonium bicarbonate, the protein samples were enzymatically digested with sequencing grade trypsin (Promega). The peptides were analyzed by nano-LC-MS/MS using a Waters API-US Q-TOF, equipped with a Waters capLC system and a 75-μm inner diameter × 15-cm PepMap C18 column (Dionex). The Q-TOF data were processed using the Waters MassLynx and ProteinLynx software and searched against the NCBInr mouse database for possible protein matches using the Mascot (Boston, MA) database search engine. The search parameters include a peptide mass tolerance of ± 200 ppm, fragment mass tolerance of ± 0.2 Da, and trypsin as the enzyme with one possible missed cleavage.
Lentiviral Vector Construction and Production
Oligonucleotides containing 19-mers targeting DJ-1 (5′-GAC CCA GUA CAG UGU AGC C-3′), human Cezanne (5′-GAT CAT GAA TGG AGG AAT A-3′ and 5′-GCA GCA AGC TCA AGA AGA A-3′), mouse Cezanne (5′-GGC GGA AGG AGA AGT CAA A-3′), or non-silencing controls (5′-GAC GCT GAA GAC TCT TGG C-3′, 5′-GAT GAA GTA AGC ACG TAA A-3′, and 5′-GCA CCT ACC ACT ACA TGT A-3′) with appropriate start (5′-TCC GCT CGA GAA AAA-3′), loop (5′-TCT CTT GAA-3′), antisense, and termination (5′-GGT GTT TCG TCC TTT CCA CAA GAT ATA TAA AGC C-3′) sequences were synthesized (Integrated DNA Technologies) and cloned into FG12 vectors (42). Lentiviral particles were then produced by co-expressing the FG12 constructs with lentiviral VSVg, Rev, and GAGPOL vectors in HEK293T cells (42, 43). Lentiviral transduction and gene expression were assessed by GFP fluorescence and semiquantitative RT-PCR for DJ-1, Cezanne, and β-actin, respectively.
Affymetrix Analysis
RNA was harvested from H157 cells transduced with DJ-1, Cezanne, or mutant control shRNA lentivirus using an RNeasy kit (Qiagen). RNA quality was assessed by an Agilent bioanalyzer (Agilent). CDNA production, biotinylated cRNA generation and fragmentation, array hybridization, washing, staining, and scanning were done as shown previously (19). Final filtering and analysis of the GeneChip Human Gene 1.0 ST array (Affymetrix) raw data were performed using GeneSpring GX (Agilent). We eliminated dots with <20 percentile intensity, averaged the dot intensities of the three mutant control samples, and calculated -fold changes on hits shared between the DJ-1 and Cezanne shRNA-treated experimental groups. The -fold change cut off was set at >1.7.
In Vitro Deubiquitination Assay
Vectors for production of recombinant DJ-1 and Cezanne were produced by cloning DJ-1 and Cezanne ORFs into the His6-tagged pQE-82L (Qiagen) and pET101D (Invitrogen) vectors, respectively. Recombinant protein was produced in BL21 star (Invitrogen) Escherichia coli stimulated with 1 mm isopropyl β-d-1-thiogalactopyranoside (Sigma). The bacteria were lysed by sonication in the presence of hen egg lysozyme (Sigma) and Benzonase (Novagen), and the target recombinant proteins were purified using a nickel-nitrilotriacetic acid agarose (Invitrogen) column. Recombinant protein expression and purity were assessed by Coomassie Blue staining and immunoblot. The K48-linked ubiquitin chains (Ub2–16) and recombinant IsoT were purchased (BIOMOL). Each sample containing 1 μg of ubiquitin chains and indicated proteins was incubated at 37 °C for 6 h in deubiquitination buffer (50 mm Hepes, pH 7.8, 0.5 mm EDTA, 0.01% Brij 35, 3 mm DTT), and ubiquitin chain degradation was assessed by immunoblot.
Immunoblot Analysis and Immunoprecipitation
Immunoblot analysis was performed as reported previously (19). Primary antibodies used for blotting were anti-FLAG M2-HRP (Sigma), anti-V5-HRP (Invitrogen), anti-β-actin-HRP C11 (Santa Cruz Biotechnology), Cezanne rabbit polyclonal, DJ-1 rabbit polyclonal, HDAC2 (Santa Cruz Biotechnology), anti-ubiquitin P4D1 (Santa Cruz Biotechnology), and anti-ICAM-1 EP1442Y (Abcam). Goat anti-mouse-HRP and goat anti-rabbit HRP antibodies were used as secondary antibodies (Santa Cruz Biotechnology). Nuclear and cytoplasmic fractionation was performed using the NE-PER fractionation kit (Pierce) as per the manufacturer's instructions. Samples for immunoprecipitation were lysed in 0.5% Triton X-100 lysis buffer, cleared by centrifugation, and precipitated overnight with appropriate beads at 4 °C. V5-tagged and FLAG-tagged proteins were precipitated using V5- (Invitrogen) or FLAG-(Sigma) agarose, respectively, whereas endogenous DJ-1 was precipitated using DJ-1 4D1.3 mouse monoclonal antibody and protein A/G beads (Pierce). After overnight incubation, the samples were washed in lysis buffer and eluted in 2× LDS loading buffer with DTT at 90 °C. Input and eluate samples were then analyzed by immunoblot for expression and protein association.
Real-time and Reverse Transcriptase Semiquantitative PCR
Real-Time PCR was performed using an ABI 7900HT PCR system (Applied Biosystems) in a 384-well, 15-μl sample format with TaqMan universal PCR master mix (Applied Biosystems). Prevalidated TaqMan primer and probe sets against human/mouse ICAM-1, mouse GAPDH, human IL-8, and human β-actin were purchased from Applied Biosystems. Real-time runs and data analysis were performed using the SDS software (Applied Biosystems). Total RNA was isolated from cells using RNeasy kits (Qiagen) and quantified using a Nanodrop ND-1000 (Nanodrop). cDNA was produced with Moloney murine leukemia virus reverse transcriptase from total RNA primed with oligo(dT) (Integrated DNA Technology) in the presence of RNasin (Promega). Semiquantitative PCR was carried out using primers against human DJ-1 (5′-ATG TCA TGA GGC GAG CTG-3′, 5′-ATT TTG TCT TTA GCA AGA GGG-3′), human Cezanne (5′-TGG CAG ACA CCA TGC TGA GGG-3′, 5′-CGC TTT GAC TTC TCC TTC CGC-3′), human β-actin (5′-ATC TGG CAC CAC ACC TTC TAC AAT GAG CTG CG-3′, 5′ CAT ACT CCT GCT TGC TGA TCC ACA TC-3′), mouse ICAM-1 (5′-CAG TCC GCT GTG CTT TGA GAA CTG T-3′, 5′-GGT ATA TCC GAG CTT CAG AGG CAG G-3′), mouse DJ-1 (5′-GGA GCA GAG GAG ATG GAG ACA GTG A-3′, 5′-CGC GGC TCT CTG AGT AGC TGT AGT GA-3′), and mouse GAPDH (5′-CCA CTC ACG GCA AAT TCA ACG GCA CAG-3′, 5′-GTG GCA GTG ATG GCA TGG ACT GTG GTC-3′). The number of cycles run on the Mastercycler (Eppendorf) was dependent on the target (GAPDH/β-actin, 17; DJ-1, 21; Cezanne/ICAM-1, 25).
Luciferase Reporter Assay
HEK293T cells transduced with Cezanne, DJ-1, or control shRNA-containing lentivirus were plated in 6-well dishes at 4 × 105/well. The following day, the cells were transfected with either 100 ng of an SV40-positive control or 100 ng of an NF-κB-responsive luciferase reporter construct using FuGENE 6. After 18 h, the samples were washed with PBS, lysed with reporter lysis buffer with one freeze/thaw cycle, and cleared using centrifugation. Luciferase activity was assessed in each sample using an LMax microplate luminometer (Molecular Devices) (19). Data are shown as luciferase relative light units in NF-κB reporter construct containing sample normalized to a matched SV40 control. Experiments were performed in triplicate with error bars representing S.D. in Figs. 4 and 5.
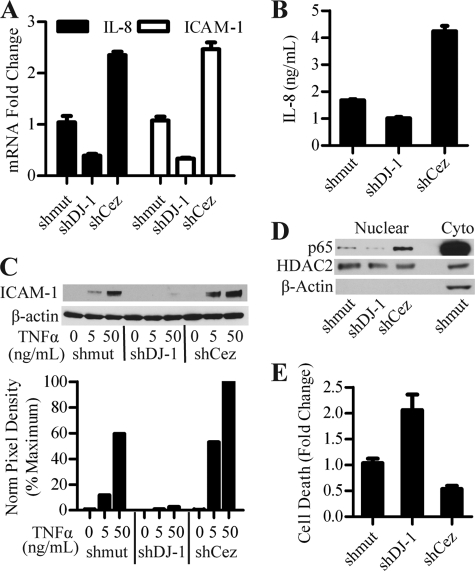
FIGURE 4.
DJ-1 and Cezanne regulate IL-8 and ICAM-1 through modulation of NF-κB nuclear localization. A, real-time PCR for IL-8 and ICAM-1 in H157 cells treated with DJ-1 and Cezanne shRNA. The ΔΔCt method was used to calculate -fold changes based on differences in cycle number as compared with matched β-actin controls and mutant shRNA (shmut) samples. Error bars denote S.D. of three separate samples within an experiment repeated three times. B, transduced H157 cultures were incubated with fresh media for 6 h, supernatants were collected, and secreted IL-8 levels were assessed by ELISA. Error bars denote S.D. of three separate samples within an experiment repeated three times. C, H157 cultures transduced with lentivirus were treated with increasing doses of TNFα for 6 h and assessed for ICAM-1 protein levels by immunoblot. Norm, normal. D, nuclear and cytoplasmic (Cyto) fractions collected from transduced H157 cultures were assessed for nuclear p65 by immunoblot. HDAC2 and β-actin were used as a loading control and fractionation control, respectively. E, transduced H157 cultures were treated with 500 nm Paclitaxel and 10 μm U0126 for 24 h. After treatment, the samples were collected and tested for cell death using a histone-based cell death ELISA. Untreated samples were used to control for cell turnover. Data were expressed as -fold change as compared with mutant shRNA control samples. Error bars denote S.D. of data from three normalized experiments.
FIGURE 5.
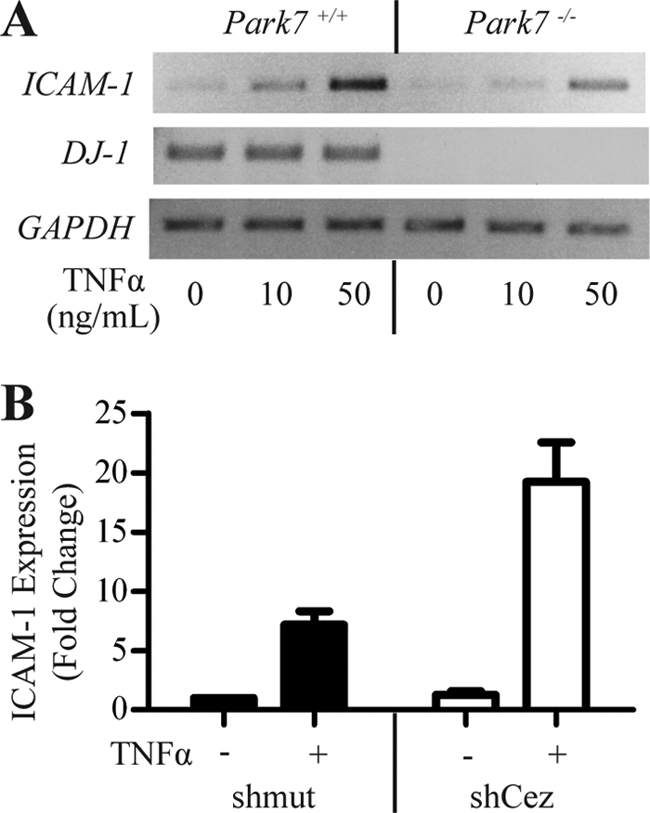
ICAM-1 is regulated by DJ-1 and Cezanne in primary MEFs. A, ICAM-1 expression is decreased in Park7−/− MEFs. Park7+/+ and Park7−/− MEFs were plated and treated with 0, 10, or 50 ng/ml TNFα for 6 h. After treatment, RNA was harvested, and ICAM-1 levels were assessed by RT-PCR using DJ-1 to verify the genotype and GAPDH to show equal loading. B, real-time PCR assessing the effects of Cezanne shRNA on ICAM-1 expression in primary MEFs. Park7+/+ MEFs were transduced with Cezanne or mutant control shRNA (shmut). The cultures were treated with 50 ng/ml TNFα for 6 h, RNA was harvested, and real-time PCR was performed to assess ICAM-1 expression and induction. -Fold changes were calculated by the ΔΔCt method using GAPDH as a reference control in matched samples. Error bars denote S.D. of three normalized experiments.
RESULTS
Q-TOF Mass Spectrometric Identification of DJ-1-interacting Proteins
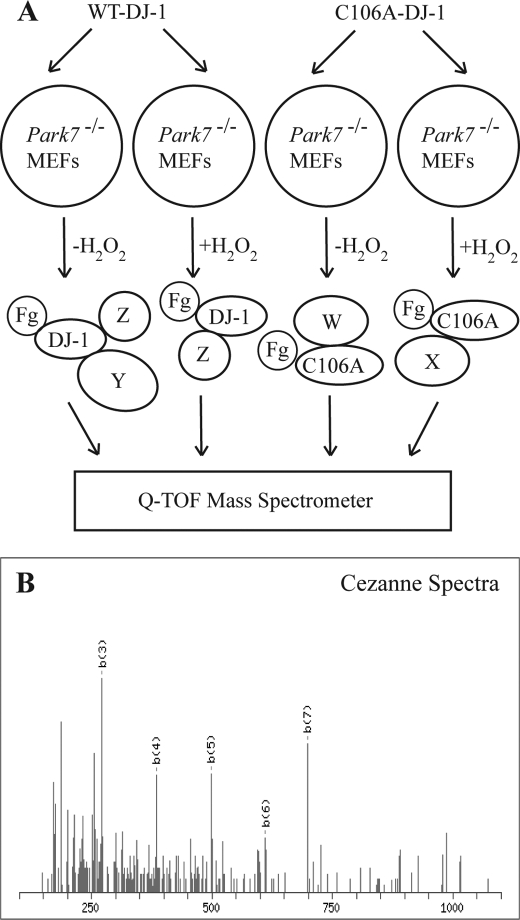
To understand the mechanism of action of DJ-1, we used Q-TOF mass spectrometry to identify binding partners of DJ-1. Several controls were built into the experimental design to minimize artifacts (Fig. 1A). First, we designed our screen to identify proteins that bound DJ-1 under oxidative and non-oxidative conditions as previously published data highlight a role of oxidation in DJ-1 function (44). Second, we used Park7−/− MEFs to prevent endogenous DJ-1 from binding proteins of interest and decreasing assay sensitivity. To start, we transfected MEFs with either FLAG-tagged DJ-1 or FLAG-tagged C106A mutant DJ-1. Because the Cys-106 residue is known to be oxidized and affect DJ-1 function, the C106A mutant was used to assess the oxidative dependence of binding as alanine is non-oxidizable (45). To mimic physiologic oxidative stress, the transfected samples were then treated with H2O2 or left untreated. After treatment, the samples were immunoprecipitated with FLAG-agarose and eluted with FLAG peptide. These eluates were trypsinized and run on a Q-TOF mass spectrometer. Using a mock-transfected control to remove any proteins that bound nonspecifically to the FLAG beads, we identified four proteins that bound specifically to DJ-1 (Table 1). We selected Cezanne for further study due to its known ability to regulate NF-κB and therefore potential to affect cell survival, a topic relevant to cancer biology and Parkinson disease (Fig. 1B).
FIGURE 1.
Overview of Q-TOF mass spectrometry results. A, schematic of Q-TOF MS experimental design. Park7−/− MEFs were transfected with FLAG-tagged DJ-1 (Fg-DJ-1) or C106A mutant DJ-1 (Fg-C106A) and either left untreated or treated with 100 μm H2O2 for 1 h. After treatment, the samples were immunoprecipitated with FLAG-agarose, eluted with FLAG peptide, and analyzed by mass spectrometry. X, Y, and Z are putative interacting proteins. B, peptide spectra for Cezanne, one of the four proteins found to interact with DJ-1 (shown in Table 1).
TABLE 1.
Identified DJ-1-binding proteins
Proteins found to bind DJ-1 by Q-TOF mass spectrometry after removal of nonspecific protein interactions using a control without DJ-1 overexpression are listed.
| Gene | Name | Known functions |
|---|---|---|
| BBS1 | Bardet-Biedl syndrome 1 | Associated with syndrome characterized by retinitis pigmentosa, polydactyly, and obesity |
| CLCF1 | Cardiotrophin-like cytokine factor 1 | Cytokine in interleukin 6 family |
| MTREF | MTREF domain containing 1 | Mitochondrial transcription termination factor |
| OTUD7B | Cezanne/Za20d1 | Deubiquitination enzyme, inhibits NF-κB |
DJ-1 Binds to Cezanne and Inhibits Its Deubiquitinating Activity
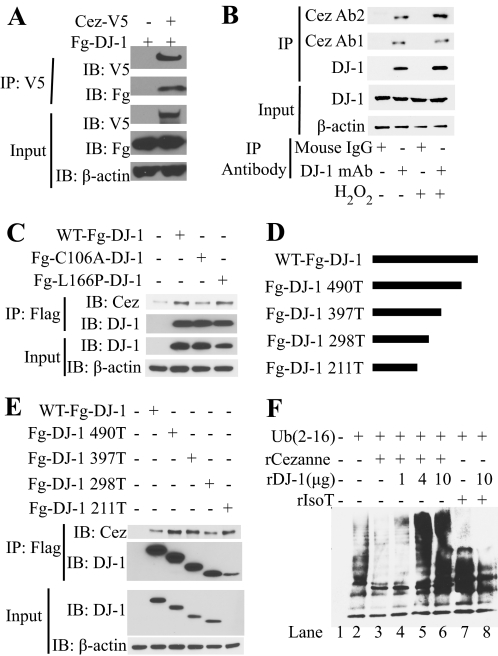
To confirm the interaction between DJ-1 and Cezanne, we performed a series of co-immunoprecipitation experiments. The interaction was first confirmed using an overexpression system (Fig. 2A). FLAG-DJ-1 was overexpressed in HEK293T cells in the presence or absence of V5-epitope tagged Cezanne. After immunoprecipitation of Cezanne, DJ-1 was co-immunoprecipitated in the presence of Cezanne but not in its absence as detected by immunoblot. To confirm an endogenous interaction between DJ-1 and Cezanne (Fig. 2B), we first treated HEK293T cells with H2O2 or left them untreated, and then immunoprecipitated endogenous DJ-1. Western blot of the immunoprecipitation samples showed the presence of endogenous Cezanne only in the samples where DJ-1 was immunoprecipitated. A modest increase in this interaction was detected with H2O2 treatment with one anti-Cezanne antibody (Cez Ab1) suggesting that there might be a form of Cezanne that is recognized by antibody 2 (Cez Ab2) that more efficiently interacts with oxidized DJ-1.
FIGURE 2.
DJ-1 binds to Cezanne and inhibits the deubiquitinating activity of Cezanne. A, overexpression co-immunoprecipitation (IP) to verify interaction between DJ-1 and Cezanne. Cells were transfected with FLAG-DJ-1 (Fg-DJ-1) in the presence or absence of Cezanne-V5 (Cez-V5). After immunoprecipitation with anti-V5-agarose, input controls and the immunoprecipitated samples were analyzed by immunoblot (IB) with the antibodies indicated. B, endogenous co-immunoprecipitation to verify an endogenous interaction of DJ-1 and Cezanne. Cells were either left untreated or treated with H2O2 and immunoprecipitated with either DJ-1 mouse monoclonal antibody (DJ-1 mAb) or mouse control IgG. After immunoprecipitation, input controls and immunoprecipitated samples were analyzed by immunoblot with the indicated antibodies. Cez Ab1, Cezanne antibody 1; Cez Ab2, Cezanne antibody 2; Ab1, antibody 1; Ab2, antibody 2. C, semi-endogenous co-immunoprecipitation to assess the effect of the C106A or L166P mutation on the interaction of DJ-1 with endogenous Cezanne. Wild type and the DJ-1 point mutants were overexpressed in HEK293T cells. Following H2O2 treatment, the wild type and DJ-1 mutants were immunoprecipitated with FLAG-agarose, and input controls and immunoprecipitated samples were assessed by immunoblot. D, schematic of DJ-1 carboxyl-terminal truncation mutants. E, semi-endogenous co-immunoprecipitation to identify which portion of DJ-1 binds endogenous Cezanne. DJ-1 truncation mutants were overexpressed in HEK293T cells. After H2O2 treatment, the samples were immunoprecipitated with FLAG-agarose, and immunoblot was used to assess the input controls and immunoprecipitated samples. F, in vitro deubiquitination assay. Synthetic ubiquitin chains were incubated with recombinant Cezanne (rCezanne) or IsoT (rIsoT) in the presence or absence of DJ-1 at 37 °C in assay buffer. After incubation, the samples were analyzed for ubiquitin chain degradation by immunoblot.
To ascertain the effect of DJ-1 Cys-106 residue oxidation on DJ-1/Cezanne binding, we tested the ability of overexpressed wild type DJ-1, non-oxidizable C106A mutant DJ-1, or the Parkinson disease-associated L166P mutant DJ-1 to bind endogenous Cezanne in the presence of H2O2 (Fig. 2C). Each of the three constructs was overexpressed in HEK293T cells treated with H2O2. As expected, lower expression of L166P DJ-1 was seen due to its known decreased stability (46). After immunoprecipitation for the FLAG-tagged DJ-1 proteins, immunoblot analysis was performed to detect endogenous Cezanne. Cezanne was co-immunoprecipitated with wild type DJ-1 and the L166P mutant. The C106A mutant DJ-1 co-precipitated Cezanne at a reduced level as compared with wild type DJ-1 but more than the untransfected negative control. This suggests that oxidation of the Cys-106 residue enhances the interaction of DJ-1 and Cezanne in this cell type. To identify the structural element of DJ-1 that binds Cezanne, we produced a series of truncation mutant constructs by sequentially removing ∼100 bp off of the 3′ end of the 570-bp open reading frame of DJ-1 in a FLAG-tagged construct (Fig. 2D). This method was used due to the lack of multiple domain structures within DJ-1. These constructs were transfected into HEK293T cells treated with H2O2, and the samples were immunoprecipitated with FLAG-agarose. Expression of the smaller amino-terminal fragments of DJ-1 showed a progressively reduced expression, which is consistent with the smaller sizes of the truncation constructs DJ-1 490T to 298T, except for DJ-1 211T, which displayed a reduction in expression that cannot be accounted for by its reduced size, which may be due to reduced expression or decreased stability (47). All of the truncation constructs immunoprecipitated Cezanne as well as the wild type control. The efficient interaction of 211T DJ-1 and endogenous Cezanne shows that the amino-terminal 70 residues of DJ-1 are sufficient for interaction with Cezanne. Residues encoded by nucleotides 298–211 might be inhibitory of this binding as their removal results in more efficient binding, considering that the expression of DJ-1 211T is much less than DJ-1 298T (Fig. 2E).
Due to the known deubiquitinating activity of Cezanne in vitro and the interaction between DJ-1 and Cezanne, we produced recombinant DJ-1 and analyzed its effect on the deubiquitination activity of Cezanne in a cell-free system (Fig. 2F). To assess the deubiquitinating activity of Cezanne, we incubated synthetic ubiquitin chains with recombinant Cezanne in the presence or absence of DJ-1 (21). After incubation, we analyzed the reduction in ubiquitin chain size by immunoblot. Although Cezanne caused a decrease in both long and medium length ubiquitin chains (lanes 2 and 3), DJ-1 significantly inhibited the activity of Cezanne in a dose-dependent manner (lanes 4–6). The inhibition of the deubiquitination activity of Cezanne by DJ-1 was found to be specific as DJ-1 did not inhibit the deubiquitinating activity of IsoT. Although a very modest activation of IsoT activity on medium length ubiquitin chains was seen with the addition of DJ-1, DJ-1 did not inhibit the deubiquitination activity of IsoT on long length ubiquitin chains (lanes 7 and 8).
Identification of IL-8 and ICAM-1 as Downstream Targets by Affymetrix Microarray
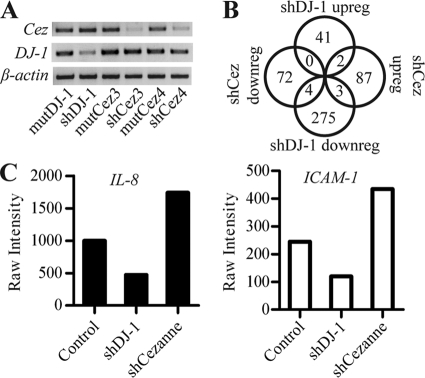
Due to the known effects of DJ-1 and Cezanne on the transcriptome, we assessed the effect of their interaction on global transcription using microarray. To identify possible downstream transcripts of DJ-1 and Cezanne, we generated a recombinant lentivirus encoding shRNA oligonucleotides specific for DJ-1 or Cezanne and mutant negative controls. After transduction of H157 cells with recombinant lentivirus, we extracted RNA and assessed targeted gene expression using semiquantitative RT-PCR (Fig. 3A). These samples showed significant reduction of Cezanne and DJ-1 gene expression as compared with matched negative controls. The RNA was then validated for microarray, used to produce cRNA, and hybridized on a GeneChip Human Gene 1.0 ST array. The normalized intensities for the three mutant controls were averaged. We then filtered out any readings below 20th percentile intensity and calculated -fold changes comparing the DJ-1 and Cezanne knockdown samples against the control using a 1.7-fold change cutoff. Due to our interest in genes regulated by both DJ-1 and Cezanne, we cross-referenced the lists of genes regulated by DJ-1 or Cezanne (Fig. 3B). This search showed that IL-8 and ICAM-1 were reciprocally controlled by DJ-1 and Cezanne (Fig. 3C). Ingenuity pathway analysis (Ingenuity Systems) was used to identify any role of Cezanne in regulation of Nrf2 downstream targets. Out of the 43 Nrf2 targets assessed, none of the targets were changed significantly enough to meet our -fold change cut off, suggesting no or little role for Cezanne in the regulation of Nrf2 (supplemental Fig. S1).
FIGURE 3.
Affymetrix microarray analysis of cells with DJ-1 or Cezanne shRNA. A, semiquantitative RT-PCR confirming reduced expression with DJ-1 (shDJ-1) or Cezanne (shCez3 and shCez4) shRNA and control shRNAs (mutDJ-1, MutCez3, and MutCez4) in appropriate samples transduced with lentivirus. B, Venn diagram showing the number of genes up-regulated (upreg) or down-regulated (downreg) with DJ-1 or Cezanne shRNA. The raw intensities of replicate samples were averaged, and filters were set for >20 percentile intensity using a cutoff of >1.7-fold as compared with the averaged negative control sample. C, graph showing the raw intensity results from the microarray analysis for IL-8 and ICAM-1, the only two characterized targets differentially regulated by DJ-1 and Cezanne.
DJ-1 and Cezanne Regulate IL-8 and ICAM-1 through Modulation of NF-κB Nuclear Localization
To verify the microarray results, we used real-time PCR to verify the regulation of IL-8 and ICAM-1 by DJ-1 and Cezanne. After transducing H157 cells with control, DJ-1, or Cezanne shRNA, real-time PCR was performed for human β-actin, IL-8, and ICAM-1 (Fig. 4A). This analysis confirmed a significant decrease in IL-8 and ICAM-1 expression with DJ-1 shRNA and an increased expression of these genes with Cezanne shRNA, consistent with the Affymetrix results. To assess the effect of DJ-1 and Cezanne on IL-8 secretion, supernatants from lentiviral transduced H157 cell lines were analyzed using IL-8 ELISA. The datum showed that DJ-1 shRNA decreased IL-8 production, whereas Cezanne shRNA increased IL-8, matching the observations seen at the transcript level (Fig. 4B). Immunoblot analysis of transduced H157 cells treated with TNFα showed a decrease in ICAM-1 protein levels with DJ-1 shRNA and an increase in ICAM-1 levels with Cezanne shRNA consistent with the microarray, real-time PCR, and IL-8 ELISA data (Fig. 4C). As NF-κB is a known target of Cezanne as well as an inducer of IL-8 and ICAM-1 transcription, we made several attempts at analyzing the effects of DJ-1 and Cezanne on NF-κB activity using luciferase reporter constructs. Using overexpression of DJ-1 in this system yielded no significant difference in NF-κB reporter activity, most likely due to the high endogenous expression of DJ-1 (not shown). We then switched to an endogenous system using shRNA-mediated knockdown of DJ-1, which resulted in a modest but reproducible decrease in NF-κB reporter activity with reduced DJ-1 levels (supplemental Fig. S2). We next used a more physiologic assay and evaluated the nuclear localization of NF-κB in DJ-1 and Cezanne shRNA-treated H157 cells. After transduction with DJ-1 or Cezanne shRNA, nuclear and cytoplasmic fractions were collected and analyzed by immunoblot. Immunoblot for the p65 NF-κB subunit showed that DJ-1 shRNA caused a decrease in nuclear p65 as compared with the shRNA control, whereas Cezanne shRNA-treated samples caused increased nuclear p65 (Fig. 4D). This effect of DJ-1 on NF-κB nuclear localization also affects the transcriptional activity of NF-κB. Immunoblots for histone deacetylase 2 (HDAC2) and β-actin were used to show equal loading and nuclear fraction purity, respectively. Due to the known pro-survival effect of increased DJ-1 expression in cancer cells undergoing chemotherapeutic treatment (16), we assessed the effect of DJ-1 and Cezanne shRNA on chemotherapeutic-induced H157 cell death. We treated lentiviral transduced H157 cells with Paclitaxel and MEK inhibitor for 24 h, a combination treatment shown to be effective in H157 cells (16). After treatment, cell death was analyzed using a histone-based cell death ELISA (Roche Applied Science). Untreated samples were used to account for homeostatic cell turnover, and -fold changes in cell death were calculated as compared with the mutant shRNA control. A statistically significant increase in cell death was seen with DJ-1 shRNA, whereas a decrease was seen in the Cezanne shRNA samples (Fig. 4E).
ICAM-1 Is Regulated by DJ-1 and Cezanne in Primary MEFs
The above results were obtained with a transformed cell line; thus it was important to analyze the expression of ICAM-1 in a primary cell line such as MEFs. Only ICAM-1 was assayed because mouse cells do not express IL-8. Park7+/+ and Park7−/− MEFs were treated with increasing levels of TNFα to induce ICAM-1 expression. After treatment, the levels of ICAM-1, DJ-1, and GAPDH were assessed by semiquantitative RT-PCR (Fig. 5A). DJ-1 was expressed in Park7+/+ but not Park7−/− cells, and the expression of ICAM-1 was reduced in the Park7−/− cells relative to GAPDH control. This finding confirms the positive regulatory role of DJ-1 in ICAM-1 transcript expression in primary MEFs. To assess the effect of Cezanne on ICAM-1 expression in MEFs, ICAM-1 expression was measured by real-time PCR in TNFα-treated or untreated Park7+/+ MEFs transduced with Cezanne shRNA or a matched mutant control (Fig. 5B). Cezanne shRNA caused an increase in ICAM-1 expression in TNFα-treated MEFs. These data confirmed an activating role of DJ-1 and an inhibitory role of Cezanne on ICAM-1 expression in MEFs.
DISCUSSION
Due to the prominent link between DJ-1 genetic inactivation and early onset Parkinson disease in humans, a large portion of the DJ-1 literature has focused on the role of DJ-1 in neuronal cell death (48, 49). That work uncovered a broader role of DJ-1 in cell survival outside of the nervous system. DJ-1 has been shown to be a central molecule in cell survival by positively regulating Nrf2-dependent detoxification pathways (19), inhibiting p53-mediated cell cycle checkpoints (20) possibly via caspase-6 (51), and inducing Akt activity through the inhibition of Phosphatase and Tensin homolog (PTEN) (15). Likely due to its effects on cell survival, DJ-1 has been found to be dysregulated in several human cancers (8, 11, 12, 16). Increased expression of DJ-1 has been shown in a number of human carcinomas and is associated with negative survival outcomes and resistance to chemotherapeutics.
In this study, we identify a novel pathway by which DJ-1 modulates IL-8 and ICAM-1 levels through the regulation of Cezanne activity and NF-κB nuclear localization. Using a proteomic approach, we identify four binding partners of DJ-1. One of these DJ-1 binding partners, Cezanne, is a member of the A20 family of immunoregulatory deubiquitinating enzymes. Similar to A20, Cezanne is known to inhibit NF-κB through modulation of the ubiquitination status of RIP-1 and TRAF6 (21, 22). NF-κB, a cell survival-associated transcription factor, is known to be dysregulated in a variety of cancer types including NSCLC (24–26, 50). After confirming endogenous interaction of DJ-1 and Cezanne, we showed that the DJ-1/Cezanne interaction is enhanced by oxidation of the Cys-106 residue of DJ-1 and that the amino terminus of DJ-1 is sufficient to mediate this interaction. The interaction also leads to the inhibition of the deubiquitinating activity of Cezanne in vitro. We used microarray analysis to identify the transcriptional targets and thus the pathways regulated by the DJ-1/Cezanne interaction. DJ-1 and Cezanne shRNA were found to reciprocally regulate ICAM-1 and IL-8. As ICAM-1 and IL-8 are targets of NF-κB, this datum suggests a role of DJ-1 and Cezanne in NF-κB-mediated transcription, whereas no role of Cezanne was seen in Nrf2-mediated transcription as downstream targets of Nrf2 were unchanged in the presence of Cezanne shRNA. This link between DJ-1 and NF-κB was confirmed as nuclear p65 was found to be decreased in the presence of DJ-1 shRNA, making this work the first to identify DJ-1 as a positive regulator of NF-κB.
IL-8 is released by a subset of NSCLC cell lines in response to chemotherapy treatment (34). As IL-8 is a pro-survival signal, this IL-8 release can inhibit chemotherapeutic efficacy (32). Acting in an autocrine or paracrine fashion to activate NF-κB through stimulation of the CXCR1 receptor, IL-8 causes its continued release, completing a pro-survival-positive feedback loop (34, 35). Through inhibition of DJ-1 or activation of Cezanne, we can disrupt this IL-8-controlled pro-survival loop, thus increasing the sensitivity of IL-8 secreting tumors to chemotherapeutics (36). Aside from its effects on cell survival, IL-8 has been shown to promote angiogenesis, cancer progression, invasion, and metastasis (27, 29–33). Concordant with IL-8, ICAM-1 dysregulation has been associated with tumor progress, invasion, and metastasis (33, 40, 41). Through regulation of IL-8 and ICAM-1, DJ-1 and Cezanne may impact angiogenesis and tumor progression, leading to changes in tumor growth and survival in in vivo cancer models separate from their effects on cell survival and drug efficacy seen in vitro.
As a whole, we identify DJ-1 as a positive regulator of cancer cell survival via a cell type-specific pathway that controls IL-8 and ICAM-1 expression. The pro-cancer activities of these molecules are well documented. However, IL-8 is also a potent chemotactic molecule for infiltrating inflammatory cells such as neutrophils. Therefore IL-8 has manifold roles during carcinogenesis. In an autocrine role, it contributes to cell, survival but in a cancer cell-autonomous role, it facilitates the recruitment of infiltrating inflammatory cells such as neutrophils. Future studies will need to be conducted to confirm that this pathway is mediated through the deubiquitination of RIP-1 and TRAF6 by Cezanne as well as identify other NF-κB targets regulated by DJ-1 under cell stimulation. Finally, mouse tumor models will be necessary to examine the effects of DJ-1 and Cezanne expression on tumor growth and inflammation in vivo.
Due to the central role of DJ-1 in transcriptional regulation, identification of DJ-1 inhibitors may lead to novel treatment regimens that modulate cancer hallmarks including cancer cell growth, survival, angiogenesis, progression, invasion, and metastasis as well as boosting the efficacy of current chemotherapeutics.
Supplementary Material
Acknowledgments
We thank Dr. Willie June Brickey for assistance with microarray data analysis and the University of North Carolina (UNC) Functional Genomics and UNC Proteomics Centers for assistance on the microarray and mass spectrometry experiments, respectively.
This work was supported by the Lineberger Comprehensive Cancer Center, the University Cancer Research Fund, and UNC Cancer Cell Biology Training Grant 5T32CA071341-15. This work was also supported by a grant from the Crohn's and Colitis Foundation of America (to B. K. D.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- NSCLC
- non-small cell lung carcinomas
- ICAM-1
- intercellular adhesion molecule 1
- MEF
- mouse embryonic fibroblast
- Nrf2
- nuclear factor (erythroid-derived 2)-like 2
- Q-TOF
- quadrupole-time-of-flight
- IsoT
- Isopeptidase T.
REFERENCES
- 1. Jemal A., Siegel R., Ward E., Hao Y., Xu J., Murray T., Thun M. J. (2008) CA Cancer J. Clin. 58, 71–96 [DOI] [PubMed] [Google Scholar]
- 2. Sher T., Dy G. K., Adjei A. A. (2008) Mayo Clin. Proc. 83, 355–367 [DOI] [PubMed] [Google Scholar]
- 3. Pfister D. G., Johnson D. H., Azzoli C. G., Sause W., Smith T. J., Baker S., Jr., Olak J., Stover D., Strawn J. R., Turrisi A. T., Somerfield M. R. (2004) J. Clin. Oncol. 22, 330–353 [DOI] [PubMed] [Google Scholar]
- 4. Bonifati V., Rizzu P., van Baren M. J., Schaap O., Breedveld G. J., Krieger E., Dekker M. C., Squitieri F., Ibanez P., Joosse M., van Dongen J. W., Vanacore N., van Swieten J. C., Brice A., Meco G., van Duijn C. M., Oostra B. A., Heutink P. (2003) Science 299, 256–259 [DOI] [PubMed] [Google Scholar]
- 5. Inden M., Taira T., Kitamura Y., Yanagida T., Tsuchiya D., Takata K., Yanagisawa D., Nishimura K., Taniguchi T., Kiso Y., Yoshimoto K., Agatsuma T., Koide-Yoshida S., Iguchi-Ariga S. M., Shimohama S., Ariga H. (2006) Neurobiol. Dis. 24, 144–158 [DOI] [PubMed] [Google Scholar]
- 6. Lev N., Ickowicz D., Barhum Y., Lev S., Melamed E., Offen D. (2009) J. Neural Transm. 116, 151–160 [DOI] [PubMed] [Google Scholar]
- 7. Aleyasin H., Rousseaux M. W., Marcogliese P. C., Hewitt S. J., Irrcher I., Joselin A. P., Parsanejad M., Kim R. H., Rizzu P., Callaghan S. M., Slack R. S., Mak T. W., Park D. S. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 3186–3191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nagakubo D., Taira T., Kitaura H., Ikeda M., Tamai K., Iguchi-Ariga S. M., Ariga H. (1997) Biochem. Biophys. Res. Commun. 231, 509–513 [DOI] [PubMed] [Google Scholar]
- 9. Yuen H. F., Chan Y. P., Law S., Srivastava G., El-Tanani M., Mak T. W., Chan K. W. (2008) Cancer Epidemiol. Biomarkers Prev. 17, 3593–3602 [DOI] [PubMed] [Google Scholar]
- 10. Liu H., Wang M., Li M., Wang D., Rao Q., Wang Y., Xu Z., Wang J. (2008) Biochem. Biophys. Res. Commun. 375, 477–483 [DOI] [PubMed] [Google Scholar]
- 11. Le Naour F., Misek D. E., Krause M. C., Deneux L., Giordano T. J., Scholl S., Hanash S. M. (2001) Clin. Cancer Res. 7, 3328–3335 [PubMed] [Google Scholar]
- 12. Hod Y. (2004) J. Cell. Biochem. 92, 1221–1233 [DOI] [PubMed] [Google Scholar]
- 13. Davidson B., Hadar R., Schlossberg A., Sternlicht T., Slipicevic A., Skrede M., Risberg B., Flørenes V. A., Kopolovic J., Reich R. (2008) Hum. Pathol. 39, 87–95 [DOI] [PubMed] [Google Scholar]
- 14. Zhang H. Y., Wang H. Q., Liu H. M., Guan Y., Du Z. X. (2008) Endocr. Relat. Cancer 15, 535–544 [DOI] [PubMed] [Google Scholar]
- 15. Kim R. H., Peters M., Jang Y., Shi W., Pintilie M., Fletcher G. C., DeLuca C., Liepa J., Zhou L., Snow B., Binari R. C., Manoukian A. S., Bray M. R., Liu F. F., Tsao M. S., Mak T. W. (2005) Cancer Cell 7, 263–273 [DOI] [PubMed] [Google Scholar]
- 16. MacKeigan J. P., Clements C. M., Lich J. D., Pope R. M., Hod Y., Ting J. P. (2003) Cancer Res. 63, 6928–6934 [PubMed] [Google Scholar]
- 17. Taira T., Saito Y., Niki T., Iguchi-Ariga S. M., Takahashi K., Ariga H. (2004) EMBO Rep. 5, 213–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yokota T., Sugawara K., Ito K., Takahashi R., Ariga H., Mizusawa H. (2003) Biochem. Biophys. Res. Commun. 312, 1342–1348 [DOI] [PubMed] [Google Scholar]
- 19. Clements C. M., McNally R. S., Conti B. J., Mak T. W., Ting J. P. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 15091–15096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fan J., Ren H., Jia N., Fei E., Zhou T., Jiang P., Wu M., Wang G. (2008) J. Biol. Chem. 283, 4022–4030 [DOI] [PubMed] [Google Scholar]
- 21. Evans P. C., Smith T. S., Lai M. J., Williams M. G., Burke D. F., Heyninck K., Kreike M. M., Beyaert R., Blundell T. L., Kilshaw P. J. (2003) J. Biol. Chem. 278, 23180–23186 [DOI] [PubMed] [Google Scholar]
- 22. Enesa K., Zakkar M., Chaudhury H., Luong le A., Rawlinson L., Mason J. C., Haskard D. O., Dean J. L., Evans P. C. (2008) J. Biol. Chem. 283, 7036–7045 [DOI] [PubMed] [Google Scholar]
- 23. La Starza R., Crescenzi B., Pierini V., Romoli S., Gorello P., Brandimarte L., Matteucci C., Kropp M. G., Barba G., Martelli M. F., Mecucci C. (2007) Cancer Genet. Cytogenet. 175, 73–76 [DOI] [PubMed] [Google Scholar]
- 24. Bargou R. C., Emmerich F., Krappmann D., Bommert K., Mapara M. Y., Arnold W., Royer H. D., Grinstein E., Greiner A., Scheidereit C., Dörken B. (1997) J. Clin. Invest. 100, 2961–2969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sovak M. A., Bellas R. E., Kim D. W., Zanieski G. J., Rogers A. E., Traish A. M., Sonenshein G. E. (1997) J. Clin. Invest. 100, 2952–2960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Brown M., Cohen J., Arun P., Chen Z., Van Waes C. (2008) Expert Opin. Ther. Targets 12, 1109–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rubie C., Frick V. O., Pfeil S., Wagner M., Kollmar O., Kopp B., Graber S., Rau B. M., Schilling M. K. (2007) World J. Gastroenterol. 13, 4996–5002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chavey C., Mühlbauer M., Bossard C., Freund A., Durand S., Jorgensen C., Jobin C., Lazennec G. (2008) Mol. Pharmacol. 74, 1359–1366 [DOI] [PubMed] [Google Scholar]
- 29. Masuya D., Huang C., Liu D., Kameyama K., Hayashi E., Yamauchi A., Kobayashi S., Haba R., Yokomise H. (2001) Cancer 92, 2628–2638 [DOI] [PubMed] [Google Scholar]
- 30. Karashima T., Sweeney P., Kamat A., Huang S., Kim S. J., Bar-Eli M., McConkey D. J., Dinney C. P. (2003) Clin. Cancer Res. 9, 2786–2797 [PubMed] [Google Scholar]
- 31. Millar H. J., Nemeth J. A., McCabe F. L., Pikounis B., Wickstrom E. (2008) Cancer Epidemiol. Biomarkers Prev. 17, 2180–2187 [DOI] [PubMed] [Google Scholar]
- 32. Singh R. K., Lokeshwar B. L. (2009) Mol. Cancer 8, 57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dong C., Slattery M. J., Liang S., Peng H. H. (2005) Mol. Cell. Biomech. 2, 145–159 [PMC free article] [PubMed] [Google Scholar]
- 34. Collins T. S., Lee L. F., Ting J. P. (2000) Cancer Immunol. Immunother 49, 78–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ginestier C., Liu S., Diebel M. E., Korkaya H., Luo M., Brown M., Wicinski J., Cabaud O., Charafe-Jauffret E., Birnbaum D., Guan J. L., Dontu G., Wicha M. S. (2010) J. Clin. Invest. 120, 485–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Huang S., Mills L., Mian B., Tellez C., McCarty M., Yang X. D., Gudas J. M., Bar-Eli M. (2002) Am. J. Pathol. 161, 125–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ke J. J., Shao Q. S., Ling Z. Q. (2006) World J. Gastroenterol. (2006) 12, 3609–3611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kammerer S., Roth R. B., Reneland R., Marnellos G., Hoyal C. R., Markward N. J., Ebner F., Kiechle M., Schwarz-Boeger U., Griffiths L. R., Ulbrich C., Chrobok K., Forster G., Praetorius G. M., Meyer P., Rehbock J., Cantor C. R., Nelson M. R., Braun A. (2004) Cancer Res. 64, 8906–8910 [DOI] [PubMed] [Google Scholar]
- 39. Torii A., Harada A., Nakao A., Nonami T., Ito M., Takagi H. (1993) J. Surg. Oncol. 53, 239–242 [DOI] [PubMed] [Google Scholar]
- 40. Lin Y. C., Shun C. T., Wu M. S., Chen C. C. (2006) Clin. Cancer Res. 12, 7165–7173 [DOI] [PubMed] [Google Scholar]
- 41. Rosette C., Roth R. B., Oeth P., Braun A., Kammerer S., Ekblom J., Denissenko M. F. (2005) Carcinogenesis 26, 943–950 [DOI] [PubMed] [Google Scholar]
- 42. Qin X. F., An D. S., Chen I. S., Baltimore D. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 183–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Taxman D. J., Livingstone L. R., Zhang J., Conti B. J., Iocca H. A., Williams K. L., Lich J. D., Ting J. P., Reed W. (2006) BMC Biotechnol. 6, 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Blackinton J., Lakshminarasimhan M., Thomas K. J., Ahmad R., Greggio E., Raza A. S., Cookson M. R., Wilson M. A. (2009) J. Biol. Chem. 284, 6476–6485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Waak J., Weber S. S., Görner K., Schall C., Ichijo H., Stehle T., Kahle P. J. (2009) J. Biol. Chem. 284, 14245–14257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Anderson P. C., Daggett V. (2008) Biochemistry 47, 9380–9393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ooe H., Maita C., Maita H., Iguchi-Ariga S. M., Ariga H. (2006) Neurosci. Lett. 406, 165–168 [DOI] [PubMed] [Google Scholar]
- 48. Martinat C., Shendelman S., Jonason A., Leete T., Beal M. F., Yang L., Floss T., Abeliovich A. (2004) PLoS Biol. 2, e327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kim R. H., Smith P. D., Aleyasin H., Hayley S., Mount M. P., Pownall S., Wakeham A., You-Ten A. J., Kalia S. K., Horne P., Westaway D., Lozano A. M., Anisman H., Park D. S., Mak T. W. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 5215–5220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mukhopadhyay T., Roth J. A., Maxwell S. A. (1995) Oncogene 11, 999–1003 [PubMed] [Google Scholar]
- 51. Giaime E., Sunyach C., Druon C., Scarzello S., Robert G., Grosso S., Auberger P., Goldberg M. S., Shen J., Heutink P., Pouysségur J., Pagès G., Checler F., Alves da Costa C. (2010) Cell Death Differ. 17, 158–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.