Abstract
The S-adenosylmethionine (AdoMet) salvage enzyme 5′-methylthioadenosine phosphorylase (MTAP) has been implicated as both a cancer target and a tumor suppressor. We tested these hypotheses in mouse xenografts of human lung cancers. AdoMet recycling from 5′-methylthioadenosine (MTA) was blocked by inhibition of MTAP with methylthio-DADMe-Immucillin-A (MTDIA), an orally available, nontoxic, picomolar transition state analogue. Blood, urine, and tumor levels of MTA increased in response to MTDIA treatment. MTDIA treatment inhibited A549 (human non-small cell lung carcinoma) and H358 (human bronchioloalveolar non-small cell lung carcinoma cells) xenograft tumor growth in immunodeficient Rag2−/−γC−/− and NCr-nu mice. Systemic MTA accumulation is implicated as the tumor-suppressive metabolite because MTDIA is effective for in vivo treatment of A549 MTAP−/− and H358 MTAP+/+ tumors. Tumors from treated mice showed increased MTA and decreased polyamines but little alteration in AdoMet, methionine, or adenine levels. Gene expression profiles of A549 tumors from treated and untreated mice revealed only modest alterations with 62 up-regulated and 63 down-regulated mRNAs (≥3-fold). MTDIA antitumor activity in xenografts supports MTAP as a target for lung cancer therapy.
Keywords: Cancer Therapy, DNA Methylation, Gene Expression, Polyamines, S-Adenosylmethionine, MTAP, Lung Cancer, Transition State Analogue
Introduction
Disruption of pathways linked to polyamine synthesis and S-adenosylmethionine (AdoMet)2 salvage provides metabolic targets in anticancer therapy based on the essential roles of these metabolites in cell growth. AdoMet is the major methyl donor for biosynthetic methylation reactions, a precursor for polyamine synthesis, and the source of methyl groups for DNA methylation. Targeting polyamine metabolism directly at l-ornithine decarboxylase by α,α-difluoromethylornithine has had limited anticancer success (1). Two AdoMet molecules are converted to 5′-methylthioadenosine (MTA) in spermine synthesis, and 5′-methylthioadenosine phosphorylase (MTAP) recycles MTA by phosphorolysis to permit subsequent resynthesis of AdoMet (Fig. 1). Our working hypothesis was that inhibition of MTAP would affect cellular MTA and AdoMet metabolism with downstream effects on protein, DNA methylation, polyamine synthesis, and polyamine-dependent enzyme reactions. We targeted MTAP by transition state analysis and developed methylthio-DADMe-Immucillin-A (MTDIA), an orally available transition state analogue inhibitor of MTAP (2). Others have proposed that MTAP is a tumor suppressor gene (3), and experiments here explore the effects of MTAP inhibition in human lung cancer xenografts.
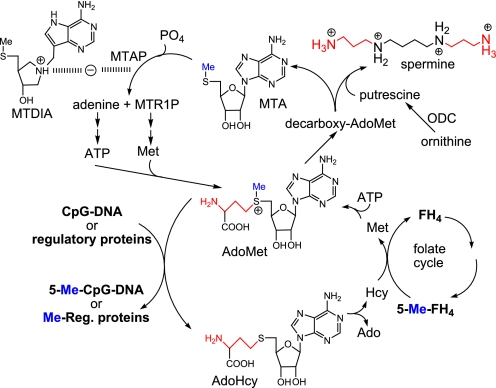
FIGURE 1.
MTA metabolism and action of MTDIA. Spermine biosynthesis forms 2 mol of MTA as a dead-end metabolite. Its sole fate in humans is recycling to AdoMet via MTAP and downstream enzymes. Two propylamino groups (red) are used for spermine synthesis, and the methylthio groups (blue) are conserved. AdoMet methyltransferases produce AdoHcy that is recycled using 5-methyltetrahydrofolate (5-Me-FH4). MTDIA blocks MTAP action causing MTA accumulation.
We previously demonstrated that treatment of human FaDu head and neck tumors in mouse xenografts with MTDIA prevented tumor growth with no apparent toxicity to the mice (4). Here, we report that both MTAP-positive (H358) and MTAP-deleted (A549) human lung cancer cell lines are also sensitive to MTAP inhibition in mouse xenografts. The mechanism is probed by the metabolic and genetic consequences of MTDIA administration. In culture, MTDIA in combination with MTA slows A549 cell growth but induces apoptosis in H358 cells.
Lung cancer is the leading cause of cancer-related deaths worldwide (5). Patients diagnosed at an advanced stage have a median survival of less than 12 months (6–8). Thus, development of novel therapeutics for lung cancer is a research priority. MTDIA demonstrated significant suppression of tumor growth with human lung cancer A549 and H358 cells in mouse xenografts. Low toxicity, oral availability, and significant tumor suppression by MTDIA all support additional evaluation as an agent for the treatment of lung cancers.
EXPERIMENTAL PROCEDURES
Cell Lines
Human non-small cell lung adenocarcinoma (NSCLC) cell line A549 and prostate carcinoma cell line PC3 were obtained from the American Type Culture Collection (Manassas, VA). Human bronchioloalveolar NSCLC cell line H358 was provided by Dr. J. Locker (Albert Einstein College of Medicine, Bronx, NY). Human breast carcinoma cell lines, MCF7, and a stable MTAP transfectant were provided by Dr. J. Martignetti (Mount Sinai School of Medicine, New York). MTDIA was synthesized by published methods (2).
Cell Culture
A549 and PC3 cells were maintained in Dulbecco's modified Eagle's medium/F-12 medium (50:50) with 10% fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin. H358 cells were maintained in RPMI 1640 medium with 10% fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin. MCF7 (wild type) cells were maintained in Eagle's medium supplemented with 10% fetal bovine serum, 100 units/ml penicillin, 100 μg/ml streptomycin, 0.1 mm nonessential amino acids, and 1 mm sodium pyruvate.
Cytotoxicity and Apoptotic Assay
Cell viability was evaluated using the Alamar Blue assay. Cells were seeded onto 96-well plates at a density of 2000 cells/well and incubated with increasing concentrations of MTDIA (10 pm to 1 mm) for 5 days at fixed MTA concentrations (0 and 20 μm). IC50 was determined following the manufacturer's instructions (Biotium, Inc., Hayward, CA). Apoptosis was measured by fluorescence-activated cell sorter analysis. Cells (both free and attached) were harvested, and cell cycle analyses were done with a FACScan flow cytometer (BD Biosciences). Cell cycle distributions and sub-G1 fractions were calculated using FlowJo software (Tree Star, Inc., Ashland, OR).
MTAP Activity Assay
Two groups of C57BL/6 mice (n = 3) were treated with either a single oral dose of water or 50 μg of MTDIA (2 mg/kg). A separate group of C57BL/6 mice received an intraperitoneal injection of 50 μg of MTDIA. At specific time points, blood samples were collected from the tail vein, mixed 1:1 with 0.6% Triton X-100 in PBS, and stored at −80 °C until analysis for MTAP activity. Labeled MTA was synthesized from AdoMet using [8-14C]ATP (2). Whole blood lysed with Triton X-100 was added to a 300-μl reaction mixture containing 50 mm HEPES, pH 7.4, 50 μm MTA, and 2.5 × 106 cpm [14C]MTA. Products of the MTAP reaction were resolved on TLC silica plates with 1 m ammonium acetate, pH 7.55, containing 5% isopropyl alcohol. Adenine spots were excised and analyzed by scintillation counting.
Determination of Polyamines
Polyamines from 0.5 m perchloric acid extracts were analyzed as the dansyl derivatives (4). The pH of the sodium carbonate used for derivatization was adjusted to 9.3, and the concentration of dansyl chloride added to samples was adjusted to 100 mm. Dansyl-polyamines were quantitated by HPLC/fluorescence on a Waters Millennium system. Elution from a Phenomenex Luna 5μ C18 (2) column used a mobile phase of 30% acetonitrile in a 50 mm ammonium acetate buffer, pH 6.8 (eluent A), and 100% acetonitrile (eluent B) with a gradient of 80% eluent A to 95% eluent B from 2 to 20 min. Fluorescence detection was by excitation at 338 nm and emission at 500 nm.
Quantitation of MTA, Adenine, AdoMet, and Methionine Levels
Solid tumor samples were mixed with 7 pmol of [2,8-2H]adenosine (9), 7 pmol of [S-methyl-2H3]AdoMet (CDN Isotopes), 7 pmol of [2-2H]methionine (CDN Isotopes), 7 pmol of [U-13C]adenine (Moravek Biochemicals and Radiochemicals) and ground in water. An aliquot was removed for protein quantification. Immediately, samples were treated with 0.5 m HClO4 at 1:6 (v/v), vigorously mixed, and incubated for 20 min at 4 °C. Samples were neutralized with 5 m KOH for 20 min at 4 °C and filtered through a MultiScreen® filter plate with Ultracel®-10 Membrane (Millipore). Metabolite levels were quantified by ultra performance liquid chromatography-MS/MS analyses using an Acquity Ultra Performance LC system coupled to a Xevo TQ mass spectrometer (Waters). The separation of MTA, adenine, and methionine was achieved with an Acquity HSS T3 column (2.1 × 100 mm, 1.8 μm, Waters). The gradient system was composed of 5 mm ammonium formate in water (eluent A) and 5 mm ammonium formate in methanol (eluent B) at a flow rate of 0.6 ml/min. The column temperature was 60 °C, and the auto-sampler was maintained at 4 °C. The injection volume for both calibrators and biological samples was 5 μl. The gradient started at 2% B and increased to 30% B over 0.9 min, changing to 80% B over 0.5 min, and decreased to 2% B over 0.5 min. The separation of AdoMet was achieved with an Acquity BEH C18 column (2.1 × 50 mm, 1.7 μm, Waters) with slight modification of the method described by Kirsch et al. (10). The gradient system was composed of 0.1% formic acid in water (A) and 100% acetonitrile (B) at a flow rate of 0.35 ml/min. The column temperature was 30 °C, and the auto-sampler was maintained at 4 °C. Quantitative determination was performed in electrospray ionization positive-ion mode using multiple-reaction monitoring mode. For electrospray ionization-MS/MS analysis, the following ion transitions, cone voltage (CV), and collision energy (CE) were used: MTA m/z 298.2 >136.1 (CV, 24 V; CE, 16 eV); [2,8-2H]adenosine m/z 270.1 >138.1 (CV, 24 V; CE, 18 eV); adenine m/z 136.0 >119.0 (CV, 34 V; CE, 20 eV); [U-13C]adenine m/z 141.1 >124.1 (CV, 38 V; CE, 12 eV); methionine m/z 150.0 >104.0 (CV, 16 V; CE, 10 eV); [2-2H]methionine m/z 151.0 >105.0 (CV, 18 V; CE, 10 eV); AdoMet m/z 399.4 >250.1 (CV, 20 V; CE, 16 eV); and [S-methyl-2H3]AdoMet m/z 402.4 >250.1 (CV, 20 V; CE, 16 eV). The electrospray ionization capillary voltage was 0.3 kV; source temperature was set at 150 °C, and desolvation temperature was 450 °C. Data acquisition was carried out by MassLynx version 4.1 and QuanLynx software. The concentration of the metabolites was calculated by interpolation of the observed analyte/internal standard peak area ratio with the corresponding calibration curve. Protein concentration was determined with a bicinchoninic acid (BCA) protein assay kit according to the manufacturer's instruction (Pierce) using BSA as a standard. Data from biological samples were normalized to the protein content, and all data are presented as means ± S.D.
Quantitation of Mouse Plasma, Blood Cells, and Urine MTA Levels
C57BL/6 mice in sterile metabolic cages were given water, water containing 250 μm MTDIA, pH 6.4 (at 0 h), and 250 μl of 4 mm MTDIA or 250 μl of 4 mm MTA + 4 mm MTDIA double intraperitoneal dose (at 0 and 24 h). Fluid consumption was recorded for both treatment groups. Mice were sacrificed 36 h after treatment was initiated, and whole blood was collected via cardiac puncture into heparinized tubes and immediately centrifuged to separate plasma and blood cells. Plasma, blood cell pellets, and urine from 36-h samples were frozen at −80 °C until analysis. Samples were processed and analyzed by LC-MS/MS as described above using [2,8-2H]adenosine as the internal standard.
Immunoblot Analysis
Cells were harvested, washed with phosphate-buffered saline (PBS), and lysed using M-PER buffer (Pierce) with complete protease inhibitor mixture (Sigma). Tumor tissues were homogenized and washed in PBS, pelleted, and lysed using T-PER buffer (Pierce) with inhibitor mixture. Anti-MTAP antibody was produced by Harlan Bioproducts for Science (Indianapolis, IN) and used as described previously (4). Lysates were subjected to SDS-PAGE, transferred to polyvinylidene difluoride membrane, and immunoblotted with primary antibody and horseradish peroxidase-conjugated secondary antibody. The blot was developed using the ECL kit (Amersham Biosciences). Secondary anti-rabbit antibody and antibody for actin were purchased from Cell Signaling, Inc. (Beverly, MA) and Sigma, respectively.
Xenograft Study
Male Rag2−/−γC−/− and Ncr-Nu mice (6–8 weeks old) were obtained from Taconic Farms and NCI, National Institutes of Health. Animal experiments were conducted in accordance with approved protocol guidelines of the Animal Committee of the Albert Einstein College of Medicine. For Rag2−/−γC−/− mice, A549 xenografts were established for 6 or 18 days, and for H358 a xenograft was established 9 days followed by treatment with MTDIA at 9 mg/kg (for A549) and 16 mg/kg (for H358) body weight in drinking water or by daily intraperitoneal injections of 5 mg/kg (for A549) and 10.2 mg/kg body weight, and for Ncr-Nu mice, oral treatment was started after 9 and 23 days after A549 tumor inoculation. After tumors were established, mice were randomly assigned to treatment or control groups of five animals each. Tumor volume (V) was determined as follows: V = (4/3)·(22/7)·1/8·(length·width·height). Differences between treatment cohorts were determined using the Student's t test. Mice were weighed every 4–5 days, monitored for hair loss, loss of appetite, vomiting, and diarrhea. Mice were sacrificed with age-matched controls, and tissues (tumors, liver, and lung) were collected.
Histology and Immunohistochemistry
Primary tumors and lungs were removed from the sacrificed animals, fixed in formalin, embedded in paraffin, sectioned, and subjected to histological or immunohistochemical analysis. A549 tumors were also examined as frozen sections. Lung sections were examined for metastasis. Proliferation was detected using antibody to Ki-67 (Dako North America, Inc.). The TUNEL assay was performed using ApopTag® plus peroxidase in situ apoptosis kit (Chemicon) according the manufacturer's instructions. Tumor blood vessels were detected in frozen sections with an antibody to CD31 or in paraffin sections with an antibody to CD34 (both from Pharmingen).
Identification of Differentially Expressed Genes Using cDNA Microarray
Total RNA from fresh-frozen tissue was extracted using TRIzol (Invitrogen) as per the supplier's instructions. Linear amplification of total RNA and subsequent fluorescent labeling of corresponding cDNA with Cy5 and Cy3 dyes were carried out using the MessageAmp T7 linear amplification kit (Ambion), and cDNA labeling protocols were developed at the AECOM microarray facility (11). Red (Cy5) and green (Cy3) signal intensities for each element on the array were calculated using GenePix Pro 3.0 software (Axon Instruments) package as described previously (11). cDNA arrays consisted of 27,323 sequence-verified human cDNA clones, representing both known genes and expressed sequence tags. Hybridization was overnight at 50 °C in hybridization buffer (30% formamide, 3× SSC, 0.75% SDS, and 100 ng of human Cot-1 DNA). Following hybridization, slides were dipped into a solution of 2× SSC, 0.1% SDS, followed by 20 min at room temperature in 0.2× SSC, 0.1% SDS and then for 20 min in 0.1× SSC. Arrays were spun dry and immediately scanned using the GenePix 4000B microarray scanner. Data designated to be of poor quality or that did not achieve a signal to noise ratio of at least 2-fold were discarded from subsequent analysis. RNA from treated cells was compared with untreated controls as the ratio of calculated Cy5/Cy3 fluorescence intensities, which represents a measure of differential gene expression between the two samples. In this study, genes with normalized Cy5/Cy3 signal ratios >1.7 are defined as up-regulated, and those with Cy5/Cy3 signal ratios <0.58 were defined as down-regulated.
Analysis using BLAST and the web server Clone/Gene ID converter mapped clone identifiers to annotated genes. Regulated mRNAs were assigned to KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways and to functional descriptors from Gene Ontology (GO) using GATHER (Gene Annotation Tool to Help Explain Relationships).
Quantitative Real Time RT-PCR
cDNA expression array data were quantitated for selected mRNA species by real time quantitative RT-PCR amplification. The primers used for qRT-PCR are shown in supplemental Table S1. SDS 2.0 software (Applied Biosystems) was used to analyze the results, and the 2− ΔΔCT method was used to calculate the relative gene expression. RNA was first isolated from solid tumors from untreated and MTDIA-treated mice with TRIzol (Invitrogen). Complementary DNA was then synthesized from 2 μg of RNA starting template using Superscript III RT-PCR kit (Invitrogen). This cDNA was used for quantitative real time PCR using Power SYBR Green master mix (Applied Biosystems) and ABI 7900HT sequence detection system (Applied Biosystems, Foster City, CA) for product quantitation. β-Actin was used to normalize individual gene expression. The cycling parameters were initial denaturation at 95 °C for 10 min followed by 40 cycles at 95 °C for 10 s, 60 °C for 20 s, and 72 °C for 30 s.
Microarray Profiling of CpG Island DNA Methylation
DNA from fresh-frozen tissue was isolated with DNeasy tissue kit (Qiagen). Global profiling of aberrant DNA methylation of CpG islands in A549 solid tumor xenograft genomic DNA was carried out using Human Methylation 27 DNA Analysis BeadChip (Illumina Inc., San Diego). Illumina BeadStudio software was used for data set assembly. We interrogated 27,578 CpG loci covering more than 14,000 genes at a single nucleotide resolution. The methylation level at each CpG locus on the BeadChip was determined by measuring ∼20 independent measures of the methylation fraction (beta), defined as the fraction of methylated signal over the total signal (unmethylated + methylated fractions) in each genomic DNA sample. This value ranged continuously from 0 (unmethylated) to 1 (fully methylated) for each CpG locus.
DNA Methyltransferase Assay
The activity of DNA methyltransferases was assayed using intact nuclei isolated from tumor and tissue culture samples using a nuclear extraction kit from New England Biolabs (Ipswich, MA). After suspending the nuclei in 100 μl of reaction buffer (25 mm HEPES, pH 7.6, 50 mm KCl, 5% glycerol, 0.1% DTT), 10-μl aliquots were removed in duplicate and then 20 μl of nuclear lysis buffer was added, and the nuclei were lysed according to the instructions of the nuclear extraction kit (New England Biolabs). The nuclear extract was analyzed for protein content by the Bradford assay (Bio-Rad) in comparison with bovine serum albumin standards. DNA methyltransferase activity was assayed on 10-μl aliquots of the suspension of intact nuclei in triplicate. Reactions contained 5 μg of poly(dIdC) (Sigma) and 1.65 μCi of [methyl-3H]AdoMet (70.8 Ci/mmol, PerkinElmer Life Sciences) and were performed in a 40-μl total volume in reaction buffer. Reactions were incubated at 37 °C on a shaker rotating at 200 rpm for 2 h. Reactions were then diluted with 500 μl of water and quenched with 500 μl of 25:24:1 phenol/chloroform/isoamyl alcohol following the addition of 75 μg of salmon testes carrier DNA (Sigma). Following inversion, extractions were spun at 15,000 ×g for 5 min. 450 μl of the aqueous phase was carefully removed, which was extracted a second time and spun for 10 min. 350 μl of aqueous phase was removed, and the DNA was precipitated by the addition of 35 μl of 3 m NaOAc, pH 5.5, and 700 μl of ethanol. Tubes were inverted and incubated at room temperature for 10 min and then spun for 10 min. The supernatant was decanted, and the DNA pellet washed with ice-cold 70% ethanol, which was spun for 5 min and decanted. After drying overnight, the pellet was resuspended in 50 μl of TE buffer, pH 8.0. 1 ml of Liquiscint scintillation fluid (Fisher) was added, and the counts/min were measured on a TriCarb 2910TR liquid scintillation counter (PerkinElmer Life Sciences). Signals were background-corrected using a control in which no poly(dIdC) was added and were normalized to the total protein content of each sample according to the Bradford assays.
Statistical Method
Statistical analysis was conducted using the Student's t test.
RESULTS
MTDIA Induces Apoptosis in H358 but Not A549 in Cell Culture
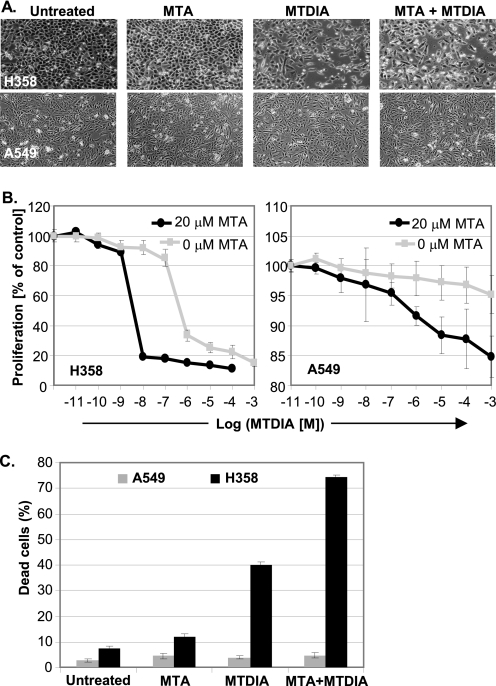
Cultured H358 cell growth is insensitive to MTA, but MTDIA reduced viability (Fig. 2). MTDIA in combination with MTA reduced viability to give IC50 values from 850 nm (no MTA) to 6 nm with 20 μm MTA (Fig. 2B (left panel)). Treated H358 cells arrested in sub-G1 and led to over 70% apoptosis in the presence of MTDIA and MTA (Fig. 2C and supplemental Fig. S1). The MTAP−/− A549 cell line showed modest cell growth inhibition in response to the same treatment with no significant apoptotic cell death (Fig. 2, A, lower panel, and B, right panel, and supplemental Fig. S1).
FIGURE 2.
Cell culture response to MTDIA and MTA. Cultured H358 cells show no response to MTA (20 μm), but MTDIA (1 μm) and MTA + MTDIA in combination kills cells (upper panel of A, left panel of B, and C). Cultured A549 cells show a weak response to MTA (20 μm), MTDIA (1 μm) (lower panel of A), but their combination causes moderate growth inhibition with no significant cell killing (right panel of B and C). H358 cell killing is dose-responsive to both MTDIA and MTA (left panel of B and C).
MTDIA Oral Availability
A single dose of 50 μg of MTDIA (∼2.5 mg/kg, oral or intraperitoneal) to mice inhibits blood MTAP activity with rapid onset of inhibition (supplemental Fig. S2). MTAP activity was regained with t½ of 98–124 h depending on route of administration. Minimum doses of 9 mg/kg daily oral and 5 mg/kg daily intraperitoneally were selected to ensure sustained MTAP inhibition.
MTA Levels in Blood and Urine
MTDIA treatment increased MTA in the blood and urine of C57BL/6 mice. MTA was below 0.05 μm (the level of detection) in normal mouse plasma and present at 3.6 ± 2.5 μm (n = 3) in urine. Plasma levels increased to 1.1 ± 0.3 μm (n = 3) with urinary levels reaching 88.7 ± 21.3 μm (n = 3) in response to oral treatment with MTDIA (p < 0.001 to 0.01). By 36 h after double intraperitoneal dose of MTDIA and MTDIA + MTA, plasma and blood cell levels of MTA returned to control levels, but urinary MTA was 164.6 ± 45.2 and 109.6 ± 53.9 μm, respectively (n = 3, p < 0.003 to 0.03).
MTDIA Suppression of A549 and H358 Growth in Mouse Xenografts
A549 Rag2−/−γC−/− Mouse Model
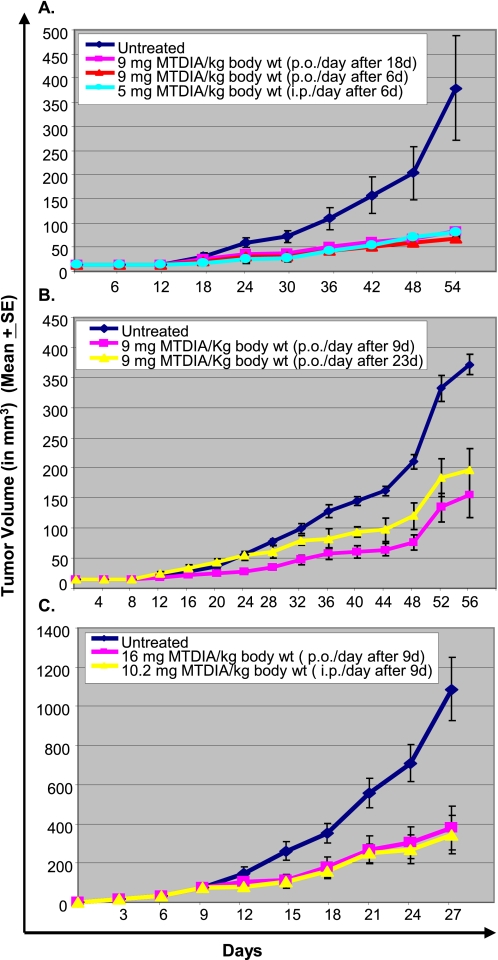
Growth of A549 cells in the dorsum hind foot of Rag2−/−γC−/− mice responded significantly to both oral and intraperitoneal once-a-day dosing of MTDIA (Fig. 3A). Tumor growth in animals treated with MTDIA was significantly slower than in controls (p < 0.005). Treatments starting at 6 or 18 days post tumor implant gave comparable inhibition.
FIGURE 3.
A549 and H358 tumors respond to MTDIA in mouse xenografts. A549 tumor growth in mouse hind foot dorsum is suppressed by oral or intraperitoneal MTDIA in Rag2−/−γC−/− mice when treatment was started at 6 or 18 days after tumor implant (A). A549 tumor growth is also suppressed by oral MTDIA in NCr-nu mice with treatment starting 9 or 23 days after tumor implant (B). Subcutaneous H358 tumor growth in mouse flank is suppressed by oral or intraperitoneal MTDIA in Rag2−/−γC−/− mice with treatment starting 9 days after tumor implant (C).
A549 Nude (NCr-nu) Mouse Model
Growth of dorsum hind foot A549 tumors in NCr-nu mice was suppressed by MTDIA (Fig. 3B). Suppression of tumor growth by oral treatment was significant for treatments initiated at 9 or 23 days post tumor implants (p < 0.003 and 0.005, respectively).
H358 Rag2−/−γC−/− Mouse Model
Subcutaneous growth of H358 cells in the flank of Rag2−/−γC−/− mice showed a significant response to both oral and intraperitoneal MTDIA treatments initiated 9 days post tumor implant (p < 0.005 and 0.006, respectively) (Fig. 3C). In the three xenograft models, MTDIA administration gave no apparent toxicity to mice because there was no weight loss, and gross appearance and organ histology were normal.
Effect of MTDIA on Polyamine Levels in Vivo
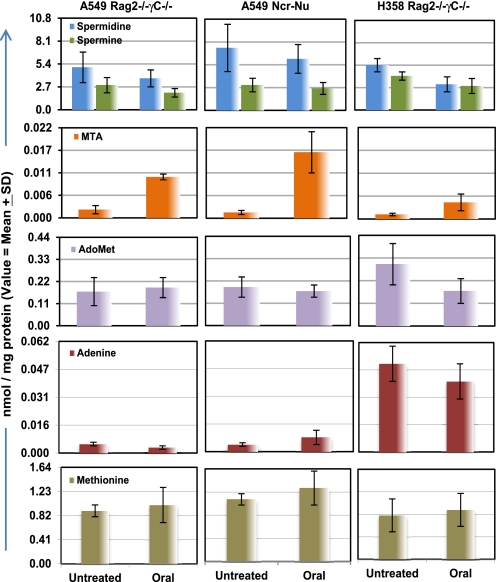
H358 and A549 xenograft tumors treated with MTDIA showed modest decreases in spermidine and spermine levels (Fig. 4). This finding is consistent with the known product inhibition of polyamine biosynthesis by MTA (12, 13).
FIGURE 4.
Tumor metabolite levels from treated versus untreated mice. Oral MTDIA treatment decreased polyamine levels in isolated tumors from mice. Spermidine levels were reduced in A549 tumors from Rag2−/−γC−/− mice (p < 0.07) and in H358 Rag2−/−γC−/− mice (p < 0.04). Spermine levels were reduced in A549 tumors from Rag2−/−γC−/− mice (p < 0.03). MTA levels increased significantly in all treatment groups (p < 0.002). AdoMet decreased significantly only in treated H358 tumors (p < 0.07). There are no significant changes seen in adenine and methionine levels after treatment.
MTAP Expression in A549 and H358
A549 and H358 are reported to be MTAP−/− and MTAP+/+, respectively. Cultured cell extracts and solid tumors from A549 and H358 xenografts were analyzed for MTAP protein expression (supplemental Fig. S3). A549 cultured cells and tumor lysates were MTAP− but extracts of H358 cells or tumors were MTAP+ (supplemental Fig. S3).
MTA Accumulation in MTDIA-treated Solid Tumors
Solid tumor extracts were subjected to LC-MS/MS analysis to reveal a 4–12-fold increase of MTA in treated tumors (p < 0.008 and 0.002) (Fig. 4). MTA did not accumulate in untreated A549 tumors, despite their MTAP−/− status. Thus, the normal MTAP of bystander tissue, blood, and stroma efficiently removes MTA from these MTAP−/− tumors.
MTDIA Suppresses Metastatic Lung Tumor Growth
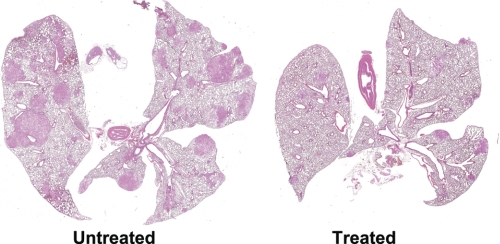
Rag2−/−γC−/− animals bearing subcutaneous A549 xenografts gave primary tumors of 1.78 ± 0.35 compared with 0.52 ± 0.09 g (p < 0.0005) for treated mice (42 days of oral MTDIA). Lungs from untreated mice were heavy due to metastases (1.35 ± 0.04 g) relative to mice treated with MTDIA (0.49 ± 0.03 g) (p < 0.00005), and the number of lung metastases was lower in treated mice (71 ± 3.9 versus 10 ± 2.2, p < 0.00001) (Fig. 5). Rag2−/−γC−/− animals bearing subcutaneous H358 xenografts (18 days oral or intraperitoneal MTDIA) were analyzed for metastases (large primary tumors in untreated mice necessitated termination after 18 days therapy). Micrometastases of 3–10 cells were apparent in lungs of all untreated mice (3.5 ± 1.7 per full lung cross-section/mouse). Oral or intraperitoneal treatment significantly reduced the incidence of metastasis (oral = 1.0 ± 0.8, p < 0.04 and intraperitoneal = 0.8 ± 1.0, p < 0.03). Thus MTDIA administration reduced lung metastasis and primary tumor growth.
FIGURE 5.
MTDIA inhibits A549 lung metastatic growth in mouse xenografts. Representative cross-sections of inflated formalin-fixed lungs from untreated (left) and orally treated (right) Rag2−/−γC−/− mice bearing hind foot dorsum primary tumors. Lungs from untreated animals are highly metastatic at 54 days. Treatment reduced the average number and size of metastases (p < 0.00001). The photograph scanned at ×1.
Histopathological Analysis of Tumors
Late stage tumors showed changes consistent with the reduced growth (supplemental Fig. S4). Both A549 and H358 grew as poorly differentiated carcinomas with a minimal fibrovascular stroma. A549 tumors had highly proliferative peripheral zones and large central zones of necrosis. In untreated tumors, the demarcation of these zones was gradual, with numerous proliferating cells in a transitional region. In contrast, the treated tumors had an abrupt transition to necrosis. Thus, the untreated tumors had a much higher rate of proliferation in the interior tumor regions (supplemental Fig. S4B). H358 tumors also had central zones of necrosis and the highest rates of proliferation in peripheral zones. The width of the proliferating zone was reduced by treatment, which also increased the relative proportion of necrotic areas (supplemental Fig. S4F). The effects were not explained by differences in angiogenesis or apoptosis. Both A549 and H358 had limited tumor microvasculature, confined to fibrovascular septae (supplemental Fig. S4, C and G). No differences were observed after treatment. Similarly, both tumors had scattered apoptotic cells, but the numbers were not increased by treatment (supplemental Fig. S4, D and H).
Gene Expression in A549 Tumors
Gene expression profiles focused on alterations caused by MTDIA treatment in A549 xenograft tumors, detected with cDNA microarrays of 27,323 clones (11). 125 mRNAs with the greatest changes, defined by a >3.0 or <0.33 expression ratios relative to untreated tumors (supplemental Table S3A), represented many different functional categories. A treeview structure of these 125 experiments demonstrated their consistency among tumors (supplemental Fig. S5A).
Two regulators of one-carbon metabolism, dihydrofolate reductase and AHCYL1, were strongly down-regulated by MTDIA, apparently a direct response to its metabolic effects. Expression of TGFβ, a growth factor that frequently inhibits growth, was also prominent. The most striking inductions, however, were of two IGF-like peptides (IGFL2 and IGFL3), growth factors of unknown function. Pro-platelet basic protein (chemokine (CXC motif) ligand 7), a proinflammatory chemokine, was also strongly induced. Among the 3-fold regulated genes, the heterogeneous changes indicate altered signal transduction, induced and suppressed expression of a few transcription factors, and various responses to cell injury. However, no responses were obvious causes rather than effects of altered tumor growth.
A larger set of 1502 cDNA clones showed altered expression of >2.0 or <0.50 at the end of treatment. 698 clones mapped to annotated genes, 426 and 272 down- and up-regulated, respectively (supplemental Table S3B). The mRNAs were assigned to KEGG pathways and to functional descriptors from Gene Ontology (GO). This analysis related altered gene expressions to the following five GO biological processes correlated with cancer (supplemental Fig. S5B): (a) cell proliferation (GO: 0008283), 55 down- and 27 up-regulated; (b) cell cycle (GO: 0007049), 44 down- and 15 up-regulated; (c) regulation of cell cycle (GO: 0000074), 23 down- and 11 up-regulated; (d) mitotic cell cycle (GO: 0000278), with 19 down- and 3 up-regulated; and (e) G-protein-coupled receptor protein signaling pathways (GO: 0007186) with 3 down- and 7 up-regulated.
The GO correlations indicated changes in many cell cycle regulators. We therefore studied effects on genes that show cell cycle-controlled patterns of expression (14) to look for an inhibitory mechanism that targeted a specific part of the cell cycle (supplemental Table S3C). MTDIA treatment altered expression of 152 cell cycle-regulated genes either >1.5- or <0.66-fold. The effects were uniform throughout the cell cycle. The expression of MYC, a classical G1/S regulator reduced by 0.48, typifies these changes. Polo-like kinase-1 (PLK1), a G2/M phase regulator was reduced slightly more, to 0.38.
Validation of A549 Candidate Genes by qRT-PCR
Selected candidate genes from the expression microarray data and other mRNAs related to AdoMet methyltransferase reactions were validated by qRT-PCR using intron-spanning primers and β-actin controls (Table 1). Genes with ≥2.5-fold decreased expression include DNMT1, DNMT3A, DNMT3B, AMD1 (AdoMetDC), ODC1, SMS, DHFR, MTHFD2, AHCYL1, MYC, HDAC1, H2AFY, PLK1, and HMGA1. As on the microarrays, the MTDIA-induced changes in MYC and PLK1 mRNA were similar, but qPCR indicated a more substantial reduction. In addition, qRT-PCR confirmed significant increases in the expression of TGFB2, TGFBR1, and IGFL3.
TABLE 1.
MTDIA-dependent changes in gene expression of A549 xenografts by quantitative real time PCR
Total RNA isolation, RT, and real time PCR were performed as described under “Experimental Procedures.” The list of primers used for real time RT-PCR is shown in supplemental Table 1.
| Unigene ID | Symbol | Mean fold change |
|---|---|---|
| Hs.202672 | DNMT1 | 0.18 ± 0.02 |
| Hs.515840 | DNMT3A | 0.16 ± 0.01 |
| Hs.713611 | DNMT3B | 0.29 ± 0.27 |
| Hs.159118 | AMD1 | 0.27 ± 0.05 |
| Hs.467701 | ODC1 | 0.20 ± 0.02 |
| Hs.288487 | SMS | 0.40 ± 0.05 |
| Hs.648635 | DHFR | 0.13 ± 0.01 |
| Hs.469030 | MTHFD2 | 0.20 ± 0.01 |
| Hs.705418 | AHCYL1 | 0.22 ± 0.06 |
| Hs.202453 | MYC (c-myc) | 0.08 ± 0.01 |
| Hs.88556 | HDAC1 | 0.32 ± 0.03 |
| Hs.696013 | H2AFY | 0.35 ± 0.01 |
| Hs.592049 | PLK1 | 0.07 ± 0.01 |
| Hs.518805 | HMGA1 | 0.10 ± 0.01 |
| Hs.133379 | TGFB2 | 2.58 ± 0.21 |
| Hs.494622 | TGFBR1 | 1.93 ± 0.28 |
| Hs.365496 | IGFL3 | 26.76 ± 6.46 |
MTDIA and DNA Methyltransferase Activity
Treatment of cultured A549 cells with MTDIA and MTA caused a small (1.3-fold) increase in apparent DNMT activity (supplemental Fig. S6A). Addition of 2 μm MTDIA caused no DNMT inhibition when assayed directly from nuclear extracts isolated from treated and control tumors (supplemental Fig. S6B). Despite the reduced mRNA levels for DNA methyltransferases from tumors of treated mice (Table 1), DNA methylation activity showed a 1.7-fold increase in methyl group incorporation into DNA of treated and control tumors (supplemental Fig. S6B). Increased methyl transfer in this assay could be caused by increased DNMT catalytic capacity or the result from globally under-methylated DNA causing an increase in sites for methyl transfer. Thus, microarray profiling was initiated.
Microarray Profiling of CpG DNA Methylation in A549 Tumors
DNA methylation at CpG islands was evaluated in A549 tumors using Illumina Human Methylation27 DNA Analysis BeadChips. Genomic DNA samples were analyzed for changes in methylated CpG loci (average beta value of 0.7 or greater). Control and treated A549 tumors indicated 5716 and 5816 methylated CpG loci, respectively. Replicate data sets with differences in beta values set at >0.2 and ≤0.2 identified 38 candidate genes in which DNA methylation was altered in response to MTDIA treatment. Of these, 18 gained and 20 lost methylation (supplemental Table S2). Using GATHER, several hypermethylated genes were assigned to GO biological processes correlated with cancer as follows: (a) cell proliferation, five genes (GO: 0008283), and (b) apoptosis, four genes (GO: 0006915). Among the hypomethylated genes, three were assigned to cell proliferation and three were assigned to apoptosis (supplemental Table S2). No corresponding changes in the gene expression array data were observed. MTDIA treatment does not cause global demethylation in A549 xenografts, and the reduction of tumor growth is more likely related to changes in metabolite-linked mRNA expression patterns.
DISCUSSION
AdoMet Pathways as Targets
We selected MTAP as a cancer target because its function is linked to polyamine synthesis, AdoMet recycling from MTA, resynthesis of methionine, salvage of adenine, and removal of MTA as a product inhibitor of the polyamine pathway (Fig. 1). MTDIA administration caused whole-animal inhibition of MTAP as evidenced by the metabolic accumulation of MTA and its surprisingly robust renal clearance. MTA accumulation is the most apparent metabolic change as a consequence of MTDIA administration in tumors with a smaller change in polyamines and no significant changes in AdoMet, methionine, or adenine. Metabolite changes are independent of MTAP+/+ and MTAP−/− tumor status, and thus the MTA is degraded by bystander tissue when the tumor is unable to metabolize it. Deletion of the MTAP gene often occurs together with p16 (CDKN2A, a tumor suppressor gene) and is common in human tumor-derived cell lines and in primary tumors (15). This co-deletion has led to speculation that MTAP is also a tumor suppressor (16, 17). Although MTAP genetic knock-out in mice is embryonic lethal (day 8), heterozygotes (MTAP+/−) are prone to age-related leukemia, also supporting the postulate that MTAP is a tumor suppressor gene (3). Here, we use MTDIA to test the effect of inhibitor-based elimination of MTAP function and demonstrate inhibition of human NSCLC xenograft growth by systemic MTAP inhibition.
MTAP−/− cells are unable to form adenine from MTA and have therefore been predicted to be more dependent on de novo purine synthesis. This metabolic aberration has been exploited using inhibitors of purine synthesis specifically in MTAP−/− tumors. Despite the logic, two multicenter phase I/II clinical trials of l-alanosine were suspended early because of inadequate efficacy of the drug treatment (18). The results here show adenine levels in MTAP−/− tumors to be significantly decreased compared with MTAP+/+ tumors. However, levels in MTAP−/− tumors are readily measured, indicating that transport or diffusion from neighboring tissues contributes to tumor purine metabolism and contributes to the limited antitumor action of l-alanosine in MTAP−/− cancers.
The polyamine pathway is thought to be subject to regulation by MTA feedback-product inhibition, but the changes in polyamine levels are significant but relatively small between treated, untreated, and MTAP−/− tumor tissues. The blood and tissue levels of MTA increase only to the low micromolar levels in mice because of the robust renal clearance of MTA. Blocking renal clearance of MTA would be expected to increase blood and tumor MTA levels and possibly result in enhanced antitumor effects. Although experimental inhibitors of the polyamine pathway have shown limited value, altered MTA may provide a more effective metabolic regulatory mechanism (19–22).
MTA Metabolism in MTAP−/− Cells and in Vivo
MTAP−/− cancer cells in xenografts do not accumulate MTA; thus, it is removed by the neighboring MTAP+/+ blood, tissue, and stroma (Fig. 4). Accumulation of MTA does occur in MTAP−/− tumors when MTAP is systemically inhibited in tumor-bearing mice, but at levels comparable with those in MTAP+/+ tumors. MTA is nontoxic to human, mice, and rats at high doses over extended periods (23). The metabolite alone is not toxic to either A549 or H358 cells in culture, suggesting host-influenced metabolism or genetic factors to be involved with MTA action at the target.
Treatment of mice with MTDIA inhibits MTAP activity causing increased MTA in the blood and excretion in the urine. No drug toxicity was observed. The lack of lung toxicity is apparent in Fig. 5. MTA levels in plasma and tumors increased an order of magnitude upon MTDIA treatment with even greater increase in urine. The MTA level in urine indicates a robust metabolic flux through MTAP in normal metabolism, resulting in MTA accumulation when MTAP is blocked. The MTA level in human urine is reported to be 5.8 μm and increases severalfold in leukemia or other malignancies (24). MTA in normal human serum is ∼3 nm (25). Thus, mice and humans have similar MTA in plasma and urine. MTDIA alone had a slight inhibitory effect on A549 cells in culture, apparent as a 5% reduction in growth at 1 mm. Because MTAP is not a component of fetal bovine serum, and A549 is MTAP−/−, the inhibition is presumably an off-target effect that occurs at concentrations far above therapeutic levels.
MTDIA and Tumor Growth in Mice
A549 and H358 human lung tumor growth in Rag2−/−γC−/− and Ncr-Nu immunodeficient mouse models was suppressed by MTDIA without apparent toxicity in other tissues. A549 cells responded moderately to MTA or MTDIA treatment in culture, but the response was more substantial in mice; thus, the presence of host factors in vivo distinguishes these conditions. Cancer cell lines and growth environments elicit specific metabolic and gene expression signatures. We postulate that different cell line responses in culture and mice may result from distinct gene expression patterns with distinct responses to MTA. Altered MTA/AdoMet ratios resulting from MTA accumulation may disrupt metabolic and/or gene expression changes needed for tumor progression. Indirect evidence for these effects comes from tumor histology where the proliferating zone of tumors is reduced by treatment but without apparent changes in microvasculature.
Gene Regulation and DNA Methylation
qRT-PCR analysis of mRNA from tumor samples demonstrate significant changes in expression of the DNA methyltransferases, enzymes in the polyamine biosynthetic pathway, enzymes in the folate/AdoMet cycle, and seven mRNAs linked to cell cycle regulation (Fig. 1 and Table 1) (14). MYC and Polo-like kinase-1 (PLK1) mRNA levels are down-regulated to 7.8 and 7.2% of their value in untreated tumors. Because these proteins are both transcriptionally regulated at different phases of the cell cycle, the similar down-regulation suggests a global decrease in cell proliferation. The oncogenic transcription factor MYC is frequently overexpressed in human cancers. Activation of MYC occurs through various mitogenic signals such as Wnt, Shh, and EGF (via the MAPK/ERK pathway), which results in numerous biological effects like driving cell proliferation (by up-regulating cyclins and down-regulating p21), regulating cell growth (by up-regulating ribosomal RNA and proteins), apoptosis (by down-regulating Bcl-2), differentiation, and stem cell self-renewal. MYC transactivates many target genes, including genes involved in polyamine metabolism, ODC, AMD1, and SRM, all of which work together to produce spermidine (26). The inhibition of all three of these genes therefore reflects a signature of MYC down-regulation. Moreover, inhibition of MYC triggers rapid regression of lung tumors (27).
The PLK1 protein is a serine/threonine kinase that serves to trigger the G2/M transition, is overexpressed in cancers, and regulates the proteins that function to accomplish mitosis. Its deletion causes decreased cell viability and apoptosis in cancer cells and is a target for anticancer drug design (28). Decreased PLK1 expression may contribute to in vivo tumor suppression.
Induction of apoptosis by DNA damage signals through p53. A549 cells have wild type p53, whereas H358 cells are p53 null (29). The antitumor effect of MTDIA therefore appears independent of p53, consistent with our previous study of head and neck cancer (4). Both H358 and A549 cells have mutated KRAS, a change found in about 30% of lung cancer. There are no effective targeted therapies for the KRAS mutant at present, although dual inhibition of EGF receptor and MEK (MAPK/ERK kinase) has been proposed to treat KRAS-mutant lung cancers (30). Our earlier study showed that MTDIA inhibited cancer cell lines FaDu and CAL27, which both lack mutations of KRAS, HRAS, or NRAS (4). Inhibition by MTDIA therefore does not depend on KRAS mutation, suggesting that use of this agent would be complementary to treatment with kinase inhibitors.
Decreased DNA methyltransferase mRNAs in response to treatment suggested global DNA methylation changes. DNA promoter hypomethylation may cause increased expression of tumor suppression genes (31). 5-Azacytidine causes similar effects, and MTA may also be a DNA methyltransferase inhibitor (31–34). Direct assay of DNMT catalytic activity indicated no change in an in vitro assay, consistent with the minimal change in global CpG methylation with MTDIA treatment. The small subsets of differentially methylated CpG sites indicate small, local DNA methylation changes. Of sites with increased methylation (silencing), four are associated with apoptosis and four are associated with cell proliferation. Thus, no distinct pattern of growth inhibition from CpG methylation is apparent.
cDNA and Microarray Analysis
The significant decrease in mRNA levels of c-MYC, ODC, AdoMetDC, and SMS upon MTDIA treatment indicates a metabolic-genetic link to the pathways involving AdoMet and MTA (Table 1). Increased MTA and inhibition of AdoMet recycling and decreased expression of these enzymes are expected to decrease polyamine biosynthesis. AdoMet is the primary methyl group donor in humans and is essential as a substrate for DNA (cytosine-5)-methyltransferases (DNMT1, -3A, and -3B) at CpG islands (Fig. 1). S-Adenosylhomocysteine hydrolase-like 1 (AHCYL1), dihydrofolate reductase, MTHFD2, and AHCYL1 (Fig. 1) are all related to the one-carbon methionine metabolic cycle linked to AdoMet formation. Folate depletion alone or combined with methionine deficiency in human cells is known to impact the AdoMet and nucleotide pools to cause global DNA hypomethylation (35). Treatment with MTDIA acts differently by causing only small numbers of specific CpG alterations. Thus, direct metabolic effects of MTA or yet unknown mechanisms of altered mRNA expression are implicated in suppression of NSCLC cell growth in vivo.
cDNA microarray analysis from treated tumors revealed down-regulation of cell cycle regulators, including CDC34, CKS1B, and CKS2 (cell cycle progression); CDC25C, CDCA5, and PLK1 (entry into mitosis, centrosome maturation and spindle assembly); and MCM3, MCM4, and MCM7 (minichromosome maintenance complex components). Many of these are responsible for the initiation of DNA replication and cell cycle control of genome duplication and may reflect a response to cell cycle changes induced by increased MTA levels.
Quantitation by qRT-PCR demonstrated low mRNA of PLK1, a key regulatory protein in mitosis. Proteins targeted by PLK1 include p53, Cdc25C, and cyclin B1 (36). In turn, PLK1 is activated by the actions of Bora and Aurora kinase (37). H2AFY shows reduced expression and is involved in regulation of inducible gene transcription and the metastatic progression of cancer cells. MTDIA treatment caused up-regulation of several genes in the TGFB signaling pathway, namely TGFB2, TGFBR1, BMP2, THBS3, LTBP2, and RSPO3. These changes may also contribute to the repression of tumor growth and metastases in A549 xenografts.
Alteration of AdoMet Recycling
MTA recycling to AdoMet is a significant step in purine salvage because the human genetic deficiency of adenine phosphoribosyltransferase results in ∼20% of the normal uric acid pool being diverted to 2,8-dihydroxyadenine (38, 39). MTAP activity is the only known metabolic source of adenine in humans. Inhibition of MTAP blocks recycling of MTA to AdoMet. Because AdoMet levels change little in treated and untreated tumor tissue, this result suggests that synthesis of AdoMet through the folate cycle and/or AdoMet synthetase must be increased. Although DNA methylation is reported to be linked in tumorigenesis (40), the MTA metabolite effect is more likely to act via MTA or polyamine changes directly linked to metabolism and metabolite-linked gene expression changes.
MTA as an Active Agent
MTAP is expressed in normal cells but many malignant cells lack MTAP activity and excrete MTA (15, 16). The chromosomal location of MTAP on human chromosome 9p21 follows the gene order p15–p16–MTAP–IFN-α–IFN-β, and all or part of this region can be deleted in tumors (41). The therapeutic effect of MTA accumulation is independent of MTAP expression in A549 cells in vivo because systemic host inhibition of MTAP caused equivalent MTA accumulation in MTAP−/− or MTAP+/+ tumors from mouse xenografts.
The value of MTAP as a therapeutic target has been questioned, and MTA in blood has been suggested as a biomarker for cancer (17). Here, we demonstrate a robust suppression of lung cancer growth and metastases in mouse xenografts by systemic inhibition of MTAP leading to increased MTA in blood, tumors, and urine. An important difference between whole-organism inhibition of MTAP and gene deletion in a specific cancer cell line is that MTA does not accumulate in vivo in p16-MTAP-negative cancer cells because of the bystander effect of MTAP in neighboring tissues. Treatment with MTDIA alone causes dramatic alteration of MTA metabolism such that urinary excretion increases by over 100-fold, and the blood and tissue levels increase by approximately an order of magnitude. Because the sole source of MTA is AdoMet metabolism, MTAP inhibition perturbs MTA, polyamine, and AdoMet metabolism. These are linked to the growth suppression of lung tumors. The low toxicity, oral availability, and robust tumor suppression by MTDIA provides promise as a new agent in lung cancer therapy.
Supplementary Material
Acknowledgments
We thank Drs. Lorenzo Agoni for FACS study assistance, David S. Neufeld for microscopy, and Lei Li and Andrew Murkin for labeled nucleoside synthesis.
This work was supported, in whole or in part, by National Institutes of Health Grants CA135405 and CA85953 and Pilot Project Award P30 CA013330. Conflict of interest: C. G., G. B. E., and V. L. S. are advisors to Pico Pharmaceuticals, Inc.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S3 and Figs. S1–S6.
- AdoMet
- S-adenosylmethionine
- NSCLC
- non-small cell lung carcinoma
- MTA
- 5′-methylthioadenosine
- MTAP
- 5′-methylthioadenosine phosphorylase
- MTDIA
- (3R,4S)-1-[(9-deazaadenin-9-yl)methyl]-3-hydroxy-4-(methylthiomethyl) pyrrolidine
- DFMO
- difluoromethylornithine
- DNMT
- DNA methyltransferase
- dansyl
- 5-dimethylaminonaphthalene-1-sulfonyl
- qRT
- quantitative RT
- CV
- cone voltage
- CE
- collision energy.
REFERENCES
- 1. Meyskens F. L., Jr., Gerner E. W. (1999) Clin. Cancer Res. 5, 945–951 [PubMed] [Google Scholar]
- 2. Evans G. B., Furneaux R. H., Lenz D. H., Painter G. F., Schramm V. L., Singh V., Tyler P. C. (2005) J. Med. Chem. 48, 4679–4689 [DOI] [PubMed] [Google Scholar]
- 3. Kadariya Y., Yin B., Tang B., Shinton S. A., Quinlivan E. P., Hua X., Klein-Szanto A., Al-Saleem T. I., Bassing C. H., Hardy R. R., Kruger W. D. (2009) Cancer Res. 69, 5961–5969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Basu I., Cordovano G., Das I., Belbin T. J., Guha C., Schramm V. L. (2007) J. Biol. Chem. 282, 21477–21486 [DOI] [PubMed] [Google Scholar]
- 5. Jemal A., Siegel R., Ward E., Hao Y., Xu J., Thun M. J. (2009) CA Cancer J. Clin. 59, 225–249 [DOI] [PubMed] [Google Scholar]
- 6. Klastersky J., Paesmans M. (2001) Lung Cancer 34, S95–S101 [DOI] [PubMed] [Google Scholar]
- 7. Crino L., Cappuzzo F. (2002) Semin. Oncol. 29, 9–16 [DOI] [PubMed] [Google Scholar]
- 8. Tanoue L. T., Detterbeck F. C. (2009) Expert Rev. Anticancer Ther. 9, 413–423 [DOI] [PubMed] [Google Scholar]
- 9. Sajiki H., Esaki H., Aoki F., Maegawa T., Hirota K. (2005) Synlett. 9, 1385–1388 [Google Scholar]
- 10. Kirsch S. H., Knapp J. P., Geisel J., Herrmann W., Obeid R. (2009) J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 877, 3865–3870 [DOI] [PubMed] [Google Scholar]
- 11. Belbin T. J., Singh B., Smith R. V., Socci N. D., Wreesmann V. B., Sanchez-Carbayo M., Masterson J., Patel S., Cordon-Cardo C., Prystowsky M. B., Childs G. (2005) Arch. Otolaryngol. Head Neck Surg. 131, 10–18 [DOI] [PubMed] [Google Scholar]
- 12. Pajula R. L., Raina A. (1979) FEBS Lett. 99, 343–345 [DOI] [PubMed] [Google Scholar]
- 13. Pegg A. E. (1983) Methods Enzymol. 94, 294–297 [DOI] [PubMed] [Google Scholar]
- 14. Whitfield M. L., Sherlock G., Saldanha A. J., Murray J. I., Ball C. A., Alexander K. E., Matese J. C., Perou C. M., Hurt M. M., Brown P. O., Botstein D. (2002) Mol. Biol. Cell 13, 1977–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. García-Castellano J. M., Villanueva A., Healey J. H., Sowers R., Cordon-Cardo C., Huvos A., Bertino J. R., Meyers P., Gorlick R. (2002) Clin. Cancer Res. 8, 782–787 [PubMed] [Google Scholar]
- 16. Christopher S. A., Diegelman P., Porter C. W., Kruger W. D. (2002) Cancer Res. 62, 6639–6644 [PubMed] [Google Scholar]
- 17. Stevens A. P., Spangler B., Wallner S., Kreutz M., Dettmer K., Oefner P. J., Bosserhoff A. K. (2009) J. Cell. Biochem. 106, 210–219 [DOI] [PubMed] [Google Scholar]
- 18. Kindler H. L., Burris H. A., 3rd, Sandler A. B., Oliff I. A. (2009) Invest. New Drugs 27, 75–81 [DOI] [PubMed] [Google Scholar]
- 19. Abeloff M. D., Slavik M., Luk G. D., Griffin C. A., Hermann J., Blanc O., Sjoerdsma A., Baylin S. B. (1984) J. Clin. Oncol. 2, 124–130 [DOI] [PubMed] [Google Scholar]
- 20. Abeloff M. D., Rosen S. T., Luk G. D., Baylin S. B., Zeltzman M., Sjoerdsma A. (1986) Cancer Treat. Rep. 70, 843–845 [PubMed] [Google Scholar]
- 21. Gerner E. W., Meyskens F. L., Jr. (2004) Nat. Rev. Cancer 4, 781–792 [DOI] [PubMed] [Google Scholar]
- 22. Casero R. A., Pegg A. E. (2009) Biochem. J. 421, 323–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Avila M. A., García-Trevijano E. R., Lu S. C., Corrales F. J., Mato J. M. (2004) Int. J. Biochem. Cell Biol. 36, 2125–2130 [DOI] [PubMed] [Google Scholar]
- 24. Keppner S., Proschak E., Schneider G., Spänkuch B. (2009) Chem. Med. Chem. 4, 1806–1809 [DOI] [PubMed] [Google Scholar]
- 25. Lee S.-H., Cho Y.-D. (1997) J. Biochem. Mol. Biol. 30, 403–409 [Google Scholar]
- 26. Gerner E. W. (2010) Cancer Prev. Res. 3, 125–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Soucek L., Whitfield J., Martins C. P., Finch A. J., Murphy D. J., Sodir N. M., Karnezis A. N., Swigart L. B., Nasi S., Evan G. I. (2008) Nature 455, 679–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Strebhardt K. (2010) Nat. Rev. Drug Discov. 9, 643–660 [DOI] [PubMed] [Google Scholar]
- 29. Gu J., Zhang L., Swisher S. G., Liu J., Roth J. A., Fang B. (2004) Oncogene 23, 1300–1307 [DOI] [PubMed] [Google Scholar]
- 30. Yoon Y. K., Kim H. P., Han S. W., Oh do Y., Im S. A., Bang Y. J., Kim T. Y. (2010) Mol. Carcinog. 49, 353–362 [DOI] [PubMed] [Google Scholar]
- 31. Kurdistani S. K. (2007) Br. J. Cancer 97, 1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Enouf J., Lawrence F., Tempete C., Robert-Gero M., Lederer E. (1979) Cancer Res. 39, 4497–4502 [PubMed] [Google Scholar]
- 33. Woodcock D. M., Adams J. K., Allan R. G., Cooper I. A. (1983) Nucleic Acids Res. 11, 489–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Miyaji K., Tani E., Nakano A., Ikemoto H., Kaba K. (1995) J. Neurosurg. 83, 690–697 [DOI] [PubMed] [Google Scholar]
- 35. Bistulfi G., Diegelman P., Foster B. A., Kramer D. L., Porter C. W., Smiraglia D. J. (2009) FASEB J. 23, 2888–2897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Toyoshima-Morimoto F., Taniguchi E., Shinya N., Iwamatsu A., Nishida E. (2001) Nature 410, 215–220 [DOI] [PubMed] [Google Scholar]
- 37. Seki A., Coppinger J. A., Jang C. Y., Yates J. R., Fang G. (2008) Science 320, 1655–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lu S. C. (2000) Int. J. Biochem. Cell Biol. 32, 391–395 [DOI] [PubMed] [Google Scholar]
- 39. Sahota A., Tischfield J. A., Kamatani N., Simmonds H. A. (2001) in Adenine Phosphoribosyltransferase Deficiency and 2,8-Dihydroxyadenine Lithiasis (Scriver C. R., Beaudet A. L., Sly W. S., Valle D. eds) 8th Ed., pp. 2571–2584, McGraw-Hill, Inc., New York [Google Scholar]
- 40. Szyf M., Pakneshan P., Rabbani S. A. (2004) Cancer Lett. 211, 133–143 [DOI] [PubMed] [Google Scholar]
- 41. Olopade O. I., Pomykala H. M., Hagos F., Sveen L. W., Espinosa R., 3rd, Dreyling M. H., Gursky S., Stadler W. M., Le Beau M. M., Bohlander S. K. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 6489–6493 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.