Abstract
The unfolded protein response (UPR) is a homeostatic signaling mechanism that balances the protein folding capacity of the endoplasmic reticulum (ER) with the secretory protein load of the cell. ER protein folding capacity is dependent on the abundance of chaperones, which is increased in response to UPR signaling, and on a sufficient ATP supply for their activity. An essential branch of the UPR entails the splicing of XBP1 mRNA to form the XBP1 transcription factor. XBP1 has been shown to be required during adipocyte differentiation, enabling mature adipocytes to secrete adiponectin, and during differentiation of B cells into antibody-secreting plasma cells. Here we find that adenylate kinase 2 (AK2), a mitochondrial enzyme that regulates adenine nucleotide interconversion within the intermembrane space, is markedly induced during adipocyte and B cell differentiation. Depletion of AK2 by RNAi impairs adiponectin secretion in 3T3-L1 adipocytes, IgM secretion in BCL1 cells, and the induction of the UPR during differentiation of both cell types. These results reveal a new mechanism by which mitochondria support ER function and suggest that specific mitochondrial defects may give rise to impaired UPR signaling. The requirement for AK2 for UPR induction may explain the pathogenesis of the profound hematopoietic defects of reticular dysgenesis, a disease associated with mutations of the AK2 gene in humans.
Keywords: Adipocyte, ATP, Endoplasmic Reticulum (ER), Lymphocyte, Mitochondria, Protein Secretion, ER Stress, Unfolded Protein Response
Introduction
Cellular homeostasis depends on the continuous synthesis of transmembrane and secretory proteins to maintain cellular integrity and an appropriate extracellular environment. These proteins are translocated into the endoplasmic reticulum (ER)2 for post-translational processing, involving the function of numerous chaperones that catalyze ATP-dependent protein folding. When the need for endoplasmic reticulum folding capacity rises (ER stress), a transient inhibition of protein synthesis is triggered followed by an increase in chaperone levels. This coordinated response is known as the unfolded protein response (UPR). When the UPR fails to resolve ER stress, a global response is triggered that can lead to apoptosis and is linked to multiple human pathologies (1–5).
The UPR is initiated through three known signaling pathways, the PERK, ATF6, and IRE1 pathways (2, 6, 7). PERK activation transiently decreases protein synthesis, and ATF6 is proteolytically cleaved to form a transcription factor that induces chaperone transcription. IRE1 is a bifunctional kinase/site-specific endoribonuclease that catalyzes XBP1 mRNA splicing, generating a stable mRNA product (sXBP1) from which the transcription factor XBP1 is translated. Like ATF6, XBP1 induces chaperone transcription. An important feature of IRE1 is that, unlike other autophosphorylating kinases, IRE1 is not activated by self-phosphorylation but rather by the occupancy of its ATP binding site (8).
Given the requirement for ATP in chaperone function and IRE1 activation, the initiation and maintenance of the UPR are likely to be influenced by cellular energy levels. Several observations are consistent with the need for adequate energy metabolism to support the induction of the UPR and ER function. For example, ischemia, which is associated with reduced nutrient delivery and decreased ATP production, affects the ER stress response both in brain and in myocardium (9, 10). Also, adipose tissue from obese animals displays evidence of enhanced PERK activity (11), possibly resulting from obesity-related hypoxia (12). A specific, major function of adipose tissue is to secrete adiponectin, a 247-amino acid polypeptide that undergoes extensive folding to form a trimer stabilized through disulfide bonds (13, 14). Multimers of the trimer are secreted into the circulation, reaching concentrations of 10–20 mg/ml, suggesting that adiponectin secretion comprises a substantial load on the adipocyte ER (15). Adiponectin secretion by 3T3-L1 adipocytes is compromised by mitochondrial poisons and enhanced by increased mitochondrial capacity (16), suggesting a high energy requirement for processing.
We have previously reported that differentiation of 3T3-L1 preadipocytes is accompanied by a robust increase in mitochondrial biogenesis (17), which coincides with the initiation of adiponectin secretion. Additionally, it has been shown that the UPR is up-regulated during 3T3-L1 adipocyte differentiation (18). These findings suggest the possibility that these two events are functionally linked, with mitochondrial function being required for UPR induction and to sustain adiponectin secretion in differentiated adipocytes.
Mitochondria play an essential role in fulfilling the energetic needs of the cell, as well as numerous other roles required for cell function. This multiplicity of roles is reflected in the variation in the mitochondrial proteome seen among different tissues (19, 20) or during differentiation (17). Here we report that in addition to a general increase in mitochondrial mass and oxidative phosphorylation (17), adipocyte differentiation is associated with a striking increase in the levels of adenylate kinase 2 (AK2), an enzyme localized in the mitochondrial intermembrane space that controls adenine nucleotide levels. AK2 is a member of an ancient family of proteins (EC. 2.7.4.3), present from bacteria to humans, that catalyze the reversible reaction ATP + AMP = 2ADP. The function of AK is classically described to be the maintenance of a constant concentration and fixed ratio of adenine nucleotides and the monitoring of cellular energy state through nucleotide sensing and signaling (21). The single isoform of adenylate kinase present in bacteria and lower eukaryotes is essential for life (22–24), and AK2 is required for Drosophila development (25). Moreover, mutations in AK2 are the basis for a profound failure of lymphopoiesis in humans (26, 27). Based on these precedents, the induction of AK2 with adipocyte differentiation may signify an important role for this enzyme in energy homeostasis in the mature adipocyte.
To investigate the relationship between mitochondrial function and the induction of the UPR, we focused on two aspects of mitochondrial energy metabolism: the generation of ATP through oxidative phosphorylation and AK2-catalyzed nucleotide conversion. Previous work from our laboratory has shown that Tfam, a factor involved in mitochondrial DNA transcription and replication (28), can be silenced during 3T3-L1 adipocyte differentiation, mitigating the increase in oxidative phosphorylation that accompanies mitochondrial biogenesis (29). Thus, we investigated the effects of both Tfam and AK2 depletion on the induction of the UPR. We find that although impairment in oxidative phosphorylation has little effect, AK2 depletion strongly compromises the induction of the UPR in adipocytes, and more so in B cells, which represent professional secretory cells. These results reveal a novel mechanism by which mitochondrial energy metabolism and ER homeostasis are linked, which may explain the profound hematopoietic defects associated both with UPR deficiency (30) and with genetic deficiency of AK2 (26, 27).
EXPERIMENTAL PROCEDURES
Reagents
Primary antibodies used were: anti-ACRP30/adiponectin (Affinity Bioreagents, Inc.); anti-BiP (BD Transduction Laboratories); anti-cytochrome c (BD Pharmingen); anti-CHOP (Santa Cruz Biotechnology); anti-calreticulin (Calbiochem); anti-mouse IgM (μ-chain-specific) and anti-β-actin (Sigma); anti-phospho-AMPKα (product number 40H9) and anti-phospho-acetyl-CoA carboxylase (Cell Signaling Technology). Chicken IgY against AK2 was generated by immunization with full-length purified mouse AK2. Anti-ERO1-La and XBP1 was kindly provided by Dr. Fumihiko Urano.
Cell Culture
All cells were obtained from American Type Culture Collection and grown under 10% CO2. 3T3-L1 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 units of penicillin/ml, 100 μg of streptomycin/ml, and 50 μg of Normocin/ml (InvivoGen), which was replaced every 48 h unless otherwise stated. 3T3-L1 cells were grown on 150-mm dishes. 3 days after reaching confluence (day 0), medium was replaced with culture medium containing 0.5 mm 3-isobutyl-1-methylxanthine (Sigma), 0.25 μm dexamethasone (Sigma), and 1 μm insulin (Sigma). 72 h later, medium was replaced with culture medium. For chemical treatments, cells were treated with 6 μm tunicamycin, 10 μg/ml oligomycin, or 5 μm FCCP for times indicated. BCL1 cells were cultured in RPMI 1640 medium supplemented with 10 mm Hepes, 1 mm sodium pyruvate, 2.5 g/liters d-glucose, 10% fetal bovine serum (FBS), 100 units of penicillin/ml, 100 μg of streptomycin/ml, and 55 μm B-mercaptoethanol. Cells were induced to differentiate by stimulating with 20 μg/ml lipopolysaccharide (LPS).
Immunoblot Analysis
Cell monolayers were washed twice with ice-cold phosphate-buffered saline (PBS) and scraped into boiling 1% sodium dodecyl sulfate (SDS) in PBS. Protein concentration was determined by BCA (Pierce), and equal amounts of protein were separated on SDS-PAGE and transferred to nitrocellulose. Samples of the cell culture medium were taken every 24 h just prior to replacement, spun for 5 min at 500 × g, and equal volumes were separated on SDS-PAGE and transferred to nitrocellulose. The membranes were blocked with 5% nonfat-milk in Tris-buffered saline with 0.1% Tween and incubated with primary antibodies overnight. They were then incubated with anti-rabbit or anti-mouse IgG horseradish peroxidase-conjugated antibodies (Promega) or anti-chicken IgY horseradish peroxidase-conjugated antibodies (Sigma) and detected using enhanced chemiluminescence (PerkinElmer Life Sciences). Films were scanned, and band intensities were quantified by using Photoshop software as follows. Images were inverted to render the bands white; bands were next selected by using the rectangular marquee tool and then delineated by using the color range tool until the whole band was selected. The histogram function was used to determine the number of pixels in the selection, which was multiplied by the average pixel intensity.
siRNA Transfection of 3T3-L1 Cells
siRNA oligonucleotides to mouse AK2 were obtained and used as a SMARTpool from Dharmacon. siRNA against mouse Tfam mRNA was purchased from Dharmacon in a duplex form with 3′-dTdT protective overhangs (sense sequence is 5′-GAAUGUGGAUCGUGCUAAAdTdT-3′). Experiments were performed 48 h after transfection or as indicated in the legends for Figs. 1–6. For day 2 transfections, medium was collected, and cells were lifted with 1× trypsin supplemented with 1% collagenase and transfected with either 5 nmol of Tfam-directed or scrambled siRNA or with 20 nmol of AK2 or scrambled siRNA into ∼107 cells by electroporation. Cells were resuspended in the collected day 2 medium and replated; 24 h later, the differentiation medium was replaced by complete medium, and the medium was refreshed every 2 days afterward. For day 5 transfections, cells were lifted with 1× trypsin with 1% collagenase, and 20 nmol of siRNA was introduced into ∼107 cells in suspension by electroporation. Cells were replated, and 48 h later, experiments were performed.
FIGURE 1.

3T3-L1 differentiation-induced UPR accompanies increased secretion. A–C, 3T3-L1 cells were harvested at full confluence (day −2), just prior to the addition of hormonal mixture (day 0), and at the indicated days thereafter. A, cell extracts were analyzed by Western blotting with antibodies to the proteins indicated. B, culture medium was analyzed by Western blotting with anti-adiponectin (Acrp30). C, cell extracts were analyzed by Western blotting with antibodies to the proteins indicated. CytC, cytochrome c. D, Western blotting of mouse tissues with an antibody to mouse AK2. W, white adipose tissue; M, muscle; L, liver; B, brain; H, heart; P, pancreas; D, diaphragm; Lu, lung; K, kidney; Sp, spleen; St, stomach.
FIGURE 2.
Tfam depletion does not inhibit induction of UPR. Cells were transfected with scrambled (Scr) or Tfam targeting (Tfam) siRNA at day 2 of differentiation. Experiments were performed on day 5 of differentiation. A, level of Tfam mRNA determined by qRT-PCR. B, level of mtDNA copy number. C, -fold change of mRNAs for specific UPR proteins determined by qRT-PCR upon depletion of Tfam relative to scrambled. D, time course of IRE1 activation by tunicamycin, measured by qRT-PCR of spliced XBP1 (sXBP1). E, time course of BiP accumulation upon tunicamycin treatment determined by qRT-PCR of BiP mRNA. * indicates p < 0.05 by paired two-tailed Student's t test. All values shown are means of a minimum of three independent experiments. Error bars indicate S.E.
FIGURE 3.
AK2 depletion leads to inhibition of UPR activation and adiponectin secretion. A, B, and E–F, cells were transfected with scrambled (Scr) or AK2-targeting (AK2) siRNA at day 2 of differentiation, and experiments were performed at day 5. A, the degree of knockdown was determined for the protein level by Western blot (top panel) and for mRNA expression by qRT-PCR (lower panel). B, -fold change of mRNAs for adipocyte proteins determined by qRT-PCR upon depletion of AK2 relative to scrambled. C and D, cells were transfected with scrambled or AK2-targeting siRNA at day 5 of differentiation and experiments were performed at day 7. C, -fold change of basal glucose transport assessed by 2-deoxyglucose (2-DOG) uptake. D, -fold change of oxygen consumption relative to untreated (Con) scrambled cells. Olig, oligomycin; Gluc, glucose; Palm, palmitate. E, -fold change of mRNAs for mitochondrial proteins determined by qRT-PCR upon depletion of AK2 relative to scrambled. F, -fold change of mRNAs for specific UPR proteins determined by qRT-PCR upon depletion of AK2 relative to scrambled. G, level of protein expression in cell lysates was determined by Western blot analysis with antibodies against the proteins indicated. Calretic., calreticulin; Acrp30, anti-adiponectin. H, culture medium was removed at days 4, 5, and 6 of differentiation as indicated, and secretion of adiponectin (Acrp30) over the previous 24 h was determined by Western blot. A representative blot is shown (top panel) as well as the average of all experiments (bottom panel). * indicates p < 0.05 by paired two-tailed Student's t test. All values shown are means of a minimum of three independent experiments. Error bars indicate S.E.
FIGURE 4.
AK2 depletion mitigates ER stress signaling. Cells were transfected at day 5 of differentiation with scrambled (Scr) or AK2-directed (AK2) siRNA and analyzed 48 h later. A, the degree of knockdown was determined for the mRNA expression by qRT-PCR (left panel), and the protein level was determined by Western blot (right panel). * indicates p < 0.05 by paired two-tailed Student's t test. B, time course of IRE1 activation by tunicamycin, measured by qRT-PCR of spliced XBP1 (sXBP1). Paired two-tailed Student's t tests yielded a p = 0.00058, with means of 35.5 and 23.1 times basal values (t = 0) for scrambled and AK2 groups, respectively. C, time course of BiP accumulation upon tunicamycin (Tuni) treatment measured by Western blotting of BiP with β-actin shown as a loading control (upper panel) and qRT-PCR of BiP mRNA (bottom panel); p = 0.0029, with means of 5.8 and 3.6 times basal values for scrambled and AK2 groups. S, scrambled siRNA-transfected cells; A, AK2-directed siRNA-transfected cells. D, cell lysates and culture medium from multiple experiments were analyzed by Western blot with antibodies to AK2 and adiponectin (Acrp30). Band intensities were expressed as the ratio between AK2 and scrambled and plotted against each other. Regression analysis yielded an adjusted R2 of 0.326 and a significance of 0.012. E, time course of CHOP accumulation upon tunicamycin treatment measured by Western blotting (upper panel) and qRT-PCR of CHOP mRNA (bottom panel). p = 0.00028, with means of 3.4 and 2.2 times basal values for scrambled and AK2 groups. All values shown are means of a minimum of three independent experiments. Error bars indicate S.E.
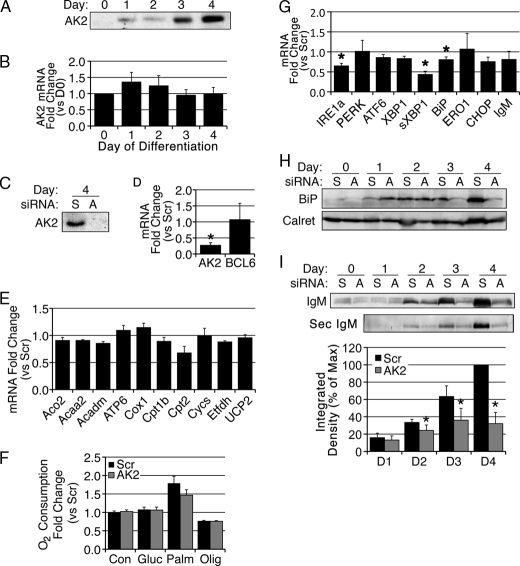
FIGURE 5.
AK2 depletion leads to inhibition of UPR activation and IgM secretion. A, Western blot analysis of AK2 protein expression in BCL1 cells during LPS induced differentiation at days indicated. B, AK2 mRNA expression by qRT-PCR in BCL1 cells during LPS-induced differentiation at days indicated. D0, day 0. C–I, BCL1 cells were nucleofected with either scrambled (Scr) or AK2-directed (AK2) siRNA and then induced to differentiate 24 h later by incubation with LPS. C, Western blot analysis for AK2 protein expression in scrambled (S) or AK2-directed (A) siRNA-transfected cells. D, -fold change of mRNAs for AK2 and BCL6 determined by qRT-PCR upon depletion of AK2 relative to scrambled at day 4 of differentiation. E, -fold change of mRNAs for mitochondrial proteins determined by qRT-PCR upon depletion of AK2 relative to scrambled at day 4 of differentiation. F, -fold change in oxygen consumption relative to untreated (Con) scrambled cells. Olig, oligomycin; Gluc, glucose; Palm, palmitate. G, -fold change of mRNAs for UPR proteins and IgM determined by qRT-PCR upon depletion of AK2 relative to scrambled at day 4 of differentiation. H, Western blot analysis with antibodies against BiP and calreticulin (Calret) in scrambled or AK2-directed siRNA-transfected cells during BCL1 differentiation. I, Western blot analysis for IgM protein expression in cell lysate (top panel) or culture medium (middle panel) in scrambled or AK2-directed or siRNA-transfected cells during BCL1 differentiation. The bottom panel shows the quantification of IgM secretion through analysis by densitometry. % of Max, percentage of maximum; D1–D4, days 1–4. * indicates p < 0.05 by paired two-tailed Student's t test. All values shown are means of a minimum of three independent experiments. Error bars indicate S.E.
FIGURE 6.
IRE1 activation is highly sensitive to ATP levels. A–C, level of adenine nucleotides isolated from cell lysates by TCA extraction expressed as relative luminescent units (RLU). A, 3T3-L1 cells were transfected with scrambled (Scr) or AK2-targeting (AK2) siRNA at day 2 of differentiation, and nucleotides were extracted at day 5. B, 3T3-L1 cells were transfected with scrambled or AK2-targeting siRNA at day 5 of differentiation, and nucleotides were extracted at day 7. C, BCL1 cells were transfected with scrambled or AK2-targeting siRNA prior to differentiation, and nucleotides were extracted at day 4. D, 3T3-L1 adipocytes at day 7 of differentiation were exposed to oligomycin (Olig) or FCCP for 2–4 h, and ATP levels in whole cell extracts were measured. Values are expressed as a function of the value in untreated cells (Con). E, -fold change of spliced XBP1 (sXBP1) mRNA levels was measured by qRT-PCR for 3T3-L1 adipocytes at day 7 of differentiation treated with oligomycin or FCCP for 2–4 h as compared with untreated cells. F, cells were treated with tunicamycin (Tuni), with oligomycin present from the start (t = 0 h) or added after 5.5 h of incubation. Spliced XBP1 was then measured by qRT-PCR at the times indicated. Values are expressed as a function of untreated levels. * indicates p < 0.05 by paired two-tailed Student's t test. All values shown are means of a minimum of three independent experiments. Error bars indicate S.E.
siRNA Nucleofection of BCL1 Cells
Scrambled siRNA or siRNA oligonucleotides to mouse AK2 were obtained and used as a SMARTpool from Dharmacon. Cells were suspended, and 0.5 nmol of siRNA was introduced into ∼2.5 × 106 cells using the Amaxa Nucleofector® kit V and program G-015. Cells were replated, and 24 h later, medium was replaced and cells were induced to differentiate.
Quantitative RT-PCR
Total RNA was extracted by using TRIzol reagent (Invitrogen). For quantitative mRNA analysis, 2 μg of the total RNA was reverse-transcribed by using an iScript cDNA synthesis kit (Bio-Rad). 10% of each RT reaction was subjected to quantitative real-time PCR analysis using an iQ SYBR Green supermix kit and real-time PCR detection system following the manufacturer's instructions (MyiQ, Bio-Rad). Ferritin heavy chain or β-actin were used as an internal housekeeping gene. Relative gene expression was calculated by the 2−ΔΔCT method (31).
Mitochondrial DNA Measurement
Total DNA was extracted by the DNeasy® tissue kit. Primer pairs corresponding to Nd1 (mitochondrial) and Actb (nuclear) were used to amplify a mitochondrial and nuclear DNA fragment, respectively. The 2−ΔΔCT method was used to analyze the relative mtDNA level.
Adenine Nucleotide Assay
Levels of adenine nucleotides were measured by a coupled enzymatic assay (32). Cells were washed twice with 1× PBS and then scraped into 20 mm Hepes with 3 mm MgCl2, pH 7.75. An equal volume of 10% TCA was added, and lysate was incubated for 3 min and then spun at 5000 × g for 5 min. Supernatant was transferred to a new tube and neutralized by diluting 1:5 with 1 m Tris, pH 7.5. 10 μl of sample was added to 12 wells each of an opaque 96-well plate. 90 μl of 20 mm Hepes with 3 mm MgCl2, pH 7.75, was added to each well. To eight wells of each sample, 1.5 mm phosphoenolpyruvate (PEP) and 2.3 units/ml pyruvate kinase (PK) were added to convert ADP to ATP. To four of those wells, 36 units/ml adenylate kinase were added to determine AMP levels. The plate was incubated for 30 min, and then 100 μl of the CellTiter-Glo® luminescent assay reagent (Promega) was added and incubated for an additional 15 min. Luminescence was measured using a Safire2 multimode microplate spectrophotometer (Tecan). Relative ADP levels were determined by subtracting the wells with PEP/PK from those without. Relative AMP levels were determined by subtracting the wells with PEP/PK/AK from those with just PEP/PK.
2-Deoxyglucose Uptake Assay
Cells were plated in a 24-well dish. Cells were starved for 2 h in Krebs-HEPES buffer (with 2 mm sodium pyruvate and 1% BSA), and uptake was initiated by the addition of [3H]2-deoxy-d-glucose to a final assay concentration of 100 μm for 5 min at 37 °C. Assays were terminated by three washes with ice-cold phosphate-buffered saline, the cells were solubilized with 0.4 ml of 1% Triton X-100, and 3H content was determined by scintillation counting. Nonspecific deoxyglucose uptake was measured in the presence of 20 μm cytochalasin B and subtracted from each determination to obtain specific uptake.
Oxygen Consumption
Cells were trypsinized and resuspended in Krebs-HEPES buffer. Approximately 7.5 × 104 cells in 150 μl of Krebs-HEPES buffer (with 2 mm sodium pyruvate and 1% BSA) were added to wells in a BD oxygen biosensor system plate (BD Biosciences). 10 μg/ml oligomycin, 5 μm FCCP, 2.5 mm glucose, or 10 μm palmitate was added to wells in triplicate. Plates were sealed, and fluorescence intensity was recorded for 120 min (2-min intervals) at an excitation wavelength of 485 nm and emission wavelength of 630 nm on a SAFIRE multimode microplate spectrophotometer (Tecan) maintained at 37 °C.
RESULTS
UPR Induction Correlates with Mitochondrial Biogenesis and Remodeling in 3T3-L1 Cells
Consistent with previous findings (18), we find that the levels of XBP1 and several proteins involved in protein folding, such as BiP and ERO1a, are up-regulated early during adipocyte differentiation (Fig. 1A). This induction precedes secretion of adiponectin/Acrp30 (Fig. 1B) and parallels mitochondrial biogenesis as assessed by cytochrome c protein expression (Fig. 1C, upper panel). In addition to increased mitochondrial mass, differentiation is accompanied by mitochondrial remodeling, where mitochondrial proteins specific for adipocyte function are induced (17, 19, 20). We find that one of these proteins is adenylate kinase 2, (Fig. 1C, lower panel), which is also highly expressed in white adipose tissue, liver, pancreas, and kidney (Fig. 1D) (27, 33).
Effect of Tfam Depletion on UPR Induction
We have previously shown that siRNA-mediated depletion of Tfam, a nuclear-encoded transcription factor for mitochondrial DNA, leads to impaired respiratory chain function (29) without changes in overall mitochondrial mass. This alteration of respiratory chain function did not impair secretory function as adiponectin secretion was unaffected (29). Consistent with these previous findings, siRNA-targeted depletion led to a decrease in Tfam mRNA expression and mtDNA content by 60 and 40%, respectively (Fig. 2, A and B). This depletion in Tfam expression did not impair basal sXBP1 levels, nor the basal levels of key elements of the UPR, including PERK, ATF6, and IRE1α (Fig. 2C). Analysis of proteins involved in the control of post-translational processing in the ER revealed a decrease in mRNA levels for ERO1a, which reoxidizes protein disulfide isomerase to support disulfide bond formation (34). ERO1a levels are sensitive to oxygen tension (35), suggesting that its decrease may reflect alterations in redox state caused by impairment in respiratory chain function in Tfam-depleted cells. Taken together these data indicate that decreased respiratory chain function through Tfam depletion does not significantly impair the induction of the UPR induced by differentiation in 3T3-L1 cells.
To determine whether impaired respiratory chain function would affect the acute induction of the UPR in fully differentiated cells, we examined the effects of tunicamycin, which blocks N-linked glycosylation, causing the rapid accumulation of unfolded proteins. The integrated magnitude of tunicamycin-induced XBP1 splicing (Fig. 2D) and induction of BiP mRNA (Fig. 2E) over a 24-h period were not diminished by Tfam depletion. These experiments suggest that respiratory chain activity and oxidative phosphorylation are not rate-limiting for acute UPR induction in fully differentiated 3T3-L1 adipocytes. These results are also consistent with previous reports of a lack of requirement for electron transport for BiP induction in response to thapsigargin-induced ER stress (36).
Effect of AK2 Depletion during 3T3-L1 Adipocyte Differentiation
To analyze the role of AK2, RNAi oligonucleotides directed against AK2 were introduced at day 2 of differentiation to match the experimental conditions used in the analysis of Tfam shown above. At day 5 of differentiation, AK2 mRNA levels were decreased by 90%, and protein levels were decreased by an average of 60% (Fig. 3A). Depletion of AK2 did not measurably affect differentiation as estimated by lipid accumulation (data not shown), nor by the mRNA levels of genes involved in adipocyte differentiation such as peroxisome proliferator-activated receptor-γ or reflective of differentiation such as Glut4 (Fig. 3B). To determine whether AK2 depletion affected adipocyte energy homeostasis, we measured glucose uptake, lactate production, and oxygen consumption as indicators of glycolytic rate and oxidative capacity. A small increase in basal glucose uptake (Fig. 3C) was seen, together with a decrease in oxygen consumption (Fig. 3D), observed in the presence of either glucose or fatty acids as substrates and also when maximal consumption was elicited by uncoupling the respiratory chain with FCCP. AK2 depletion was also accompanied by decreased mRNA levels of several mitochondrial proteins involved in oxidative phosphorylation and fatty acid transport (Fig. 3E) but not by an overall decrease in mitochondrial gene expression. These results indicate that depletion of AK2 results in changes in adipocyte energy homeostasis, but these effects do not detectably impair key adipocyte properties such as mitochondrial biogenesis and triglyceride storage. The functional effects of these alterations on adenine nucleotide levels are described later in Fig. 6.
We then examined the effects of AK2 depletion on the expression of genes associated with the UPR. IRE1a, PERK, and ATF6 mRNA levels were decreased by 40–50%, and total and spliced XBP1 were decreased by 80% in AK2-depleted cells (Fig. 3F). The expression level of the transcription factor CHOP, which is responsive to sustained UPR signaling, was also significantly decreased (Fig. 3F). We were not able to detect changes in protein levels of IRE1a, PERK, or ATF6, possibly due to antibody sensitivity, but significant decreases in protein levels of XBP1 and of BiP and ERO1a, which are downstream of UPR signaling, were consistently seen (Fig. 3G). In contrast, the calcium-binding protein calreticulin, which is not regulated by UPR signaling, was unaffected. To determine whether the perturbations in differentiation-induced UPR signaling translated into alterations in ER function, we measured adiponectin secretion in AK2-depleted cells. Despite unaltered levels of total cellular adiponectin seen in Western blotting of cell extracts (Fig. 3G, bottom panel), adiponectin secretion into the medium was diminished (Fig. 3H). Thus, the decreased expression level of ER components induced by AK2 depletion appears sufficient to result in impaired secretion.
Effect of AK2 Depletion on the Acute Induction of the UPR
To determine whether AK2 might play a role in the acute induction of the UPR in mature adipocytes, we silenced AK2 at day 5 of differentiation and analyzed the effects of tunicamycin 48 h later, when AK2 mRNA and protein levels were depleted by >80% (Fig. 4A). XBP1 splicing in response to tunicamycin was delayed and reached a lower maximal in AK2-depleted cells (Fig. 4B). This decrease was physiologically relevant as BiP mRNA and protein accumulation was also significantly impaired (Fig. 4C), as was adiponectin secretion (Fig. 4D). A later, smaller response to tunicamycin treatment, CHOP induction, was also diminished in AK2-depleted cells (Fig. 4E). These experiments indicate that AK2 activity is necessary for the full induction of the UPR both during differentiation and in response to an acute increase in protein misfolding.
Effect of AK2 Depletion during BCL1 Cell Differentiation
If indeed AK2 is important for the induction of the UPR, its role should also be evident in professional secretory cells. The UPR, and in particular splicing of XBP1, are required for terminal differentiation of B cells into antibody-producing plasma cells (30, 37). Thus, we examined the role of AK2 in BCL1 cells, a mouse B cell lymphoma line that can differentiate in vitro in response to LPS. Western blotting of extracts from BCL1 cells revealed a marked induction of AK2 protein upon LPS-induced differentiation (Fig. 5A), similar to that seen in 3T3-L1 adipocytes (Fig. 1C). Interestingly, AK2 mRNA levels did not change during this period, indicating that AK2 protein expression is controlled by post-transcriptional mechanisms (Fig. 5B).
To examine the role of AK2 in B cell differentiation, siRNA to AK2 was introduced prior to LPS stimulation. After 4 days, AK2 protein was undetectable (Fig. 5C), and mRNA levels were decreased by 75% (Fig. 5D). AK2 depletion did not impair LPS-induced differentiation as BCL6, a canonical marker of B cell differentiation, was unchanged, as was the level of IgM mRNA. To determine the effects of AK2 depletion on energy homeostasis in this cell line, we measured mRNA levels of mitochondrial proteins as well as mitochondrial oxidative function. No differences were seen in mitochondrial protein mRNA levels, on oxygen consumption (Fig. 5F), or on lactate production (data not shown) in response to AK2 depletion. Thus, AK2 depletion does not globally affect mitochondrial levels or oxidative function in BCL1 cells.
To determine the effect of AK2 depletion on the induction of the UPR, we analyzed the mRNA levels of relevant ER proteins. A significant decrease in mRNA levels of IRE1a, sXBP1, and BiP was seen (Fig. 5G), and protein levels of BiP, but not calreticulin, were markedly reduced (Fig. 5H). To determine whether these changes are functionally relevant, we measured the levels of IgM in whole cell extracts, as well as in the amount secreted into the medium. A profound decrease in both antibody levels and secretion was seen in AK2-depleted cells (Fig. 5I). These experiments indicate that AK2 is required for the induction of the UPR during B cell differentiation and for the establishment of secretory function.
Relationship between Adenine Nucleotide Levels and UPR Induction
These results shown above raise the question of the mechanisms by which AK2 can exert these effects on endoplasmic reticulum function. This enzyme catalyzes the conversion of adenine nucleotides in the mitochondrial intermembrane space, through the reversible reaction 2ADP = ATP + AMP. Adenylate kinase function has been hypothesized to comprise a phospho-relay mechanism that enables efficient coupling of mitochondrial ATP production with cytoplasmic ATP-consuming processes (38, 39). Thus, it is possible that the lack of AK2 results in an inadequate supply of ATP to the endoplasmic reticulum to sustain protein folding. Alternatively, an imbalance in adenine nucleotide levels caused by insufficient AK2 activity may lead to activation of compensatory mechanisms, such as activation of nucleotide-sensitive protein kinases (21), which in turn could affect endoplasmic reticulum function.
To better define the alterations in adenine nucleotide levels caused by AK2 depletion, we measured ATP, ADP, and AMP levels in 3T3-L1 adipocytes in which AK2 was silenced before (Fig. 6A) or after (Fig. 6B) differentiation and in BCL1 cells after LPS induction (Fig. 6C). A small decrease in total ATP levels was seen in all cases, and an increase in AMP levels was seen in mature adipocytes in response to AK2 depletion. We analyzed the effects of these changes in adenine nucleotide levels on AMP-activated protein kinase (AMPK) and its substrate acetyl-CoA carboxylase by Western blotting with phospho-specific antibodies. We were unable to detect any evidence for increased phosphorylation (data not shown), suggesting that AMPK activation does not underlie the impairment of secretion seen under these conditions.
To better understand the relationship between ATP levels and UPR function, we manipulated ATP levels using FCCP or oligomycin, which impairs oxidative phosphorylation by dissipating the mitochondrial proton gradient or by directly inhibiting the mitochondrial ATPase, respectively. At concentrations at which oligomycin caused a decrease in ATP levels comparable with that seen in AK2 knockdown cells, a small increase in basal levels of sXBP1 was observed (Fig. 6E) (16), but the >20-fold induction of XBP1 splicing in response to tunicamycin was completely blocked (Fig. 6F). Similar results were seen in response to FCCP (not illustrated). Moreover, the addition of oligomycin to cells following tunicamycin treatment rapidly stopped ongoing XBP1 splicing activity (Fig. 6F). These results indicate that mitochondrial ATP production is required for the induction and maintenance of the UPR in response to an acute increase in misfolded protein load. AK2 may be important for coupling ATP production in mitochondria with ATP utilization by the ER.
DISCUSSION
The UPR is an important cellular homeostatic mechanism that is elicited in response to increased folding needs in the ER. The UPR has also been proposed to function as a sensor for whole cell energy status (40), being responsive to both glucose and oxygen deprivation. In fact, the UPR was first identified as a response to glucose limitation, which leads to an accumulation of misfolded proteins by impairing protein glycosylation. Interestingly, activation of PERK, one of the key pathways of the UPR, is stimulated in some cases in response to glucose deprivation by pathways that are independent of misfolded protein accumulation (41). These precedents underscore the important functional link between energy homeostasis and the UPR, mediated through various yet unknown molecular mechanisms.
In this study, we find that adenylate kinase, a core component of adenine nucleotide metabolic homeostasis from prokaryotes to eukaryotes, is linked to the induction of the UPR in mammalian cells. Specifically we find that depletion of AK2, the isoform of adenylate kinase localized to the mitochondrial intermembrane space, impairs the induction of the UPR seen upon the differentiation of 3T3-L1 adipocytes and B cells. In addition, AK2 is required for the full induction of the UPR in response to acute accumulation of misfolded proteins in mature adipocytes.
AK2 is a member of a conserved, ancient protein family involved in the interconversion of adenine nucleotides through the reversible reaction ATP + AMP = 2ADP (33, 42, 43). Several isoforms of AK exist in humans, and they differ in subcellular localization and tissue distribution (44). The most studied is AK1, a cytoplasmic protein expressed specifically in muscle and heart. Studies in knock-out mice have suggested that AK1 functions to maximize ATP utilization efficiency during muscle contraction by distributing ATP from sites of production to sites of consumption via phosphotransfer reactions (39, 45, 46). Although the specific role of AK2 in the regulation of adenine nucleotide metabolism and energy homeostasis in mammalian cells is to our knowledge unexplored, our results suggest that AK2 may mediate energetic coupling between mitochondria and ER.
The specific location in the mitochondrial intermembrane space positions AK2 to locally supply ATP to the ER under a range of cellular energetic conditions. Under conditions of diminished oxidative phosphorylation, accumulating ADP levels can drive the AK2 reaction toward ATP and AMP production (Fig. 7). ATP thus formed can sustain ATP-dependent ER functions. This model may explain why Tfam depletion fails to impair the UPR, whereas AK2 depletion has a pronounced effect. AK2 can also maximize the pool of high energy adenine nucleotides. For example, under conditions when oxidative phosphorylation is functioning at high capacity, ADP can become rate-limiting for ATP synthesis. Under these conditions, high ATP levels can drive the AK2 reaction toward ADP production by recruiting low energy AMP. In this way, AK2 has a unique capacity to supply ATP under low fuel conditions and to maximize energy production under high fuel conditions. Through these mechanisms, AK2 can ensure a constant adequate supply of ATP to the ER.
FIGURE 7.
Model for the interactions between AK2 and IRE1. AK2, localized to the mitochondrial intermembrane space, catalyzes the reversible interconversion of adenine nucleotides. In this location, AK2 can draw AMP into the high energy nucleotide pool under conditions of high ATP production and ADP limitation. Conversely, under conditions of ADP accumulation, such as after FCCP-mediated uncoupling or oligomycin-mediated ATPase inhibition, AK2 can catalyze the synthesis of ATP from ADP to support IRE1 activity and other essential functions. In addition, AK2 may be necessary for channeling ATP from its site of synthesis to sites of contact with the ER, facilitating energy flux to IRE1. MOM, mitochondrial outer membrane; MIM, mitochondrial inner membrane.
The possibility that the regulation of adenine nucleotide balance by AK2 may be compartmentalized may explain the finding that AMPK activity was not detectably altered in response to AK2 depletion. This result could be explained if the pool of adenine nucleotides that can be sensed by AMPK is functionally separate from that sensed by AK2. Alternatively, the degree to which the AMP/ATP ratio is altered by AK2 depletion, which may be less than that seen in response to stimuli that activate AMPK in 3T3-L1 cells (47, 48), may be insufficient to produce a detectable change.
AK2 gene mutations have been recently identified as the cause of reticular dysgenesis, a rare form of human severe combined immunodeficiency in humans (26, 27). Individuals with reticular dysgenesis are almost completely lacking in mature lymphoid cells as well as granulocytes. The unfolded protein response is required for the differentiation of mature lymphoid cells, such as B cells and T cells (37, 49). Moreover, pro-B cells of mice lacking IRE1 are deficient in VDJ recombination of Ig genes and unable to express B cell receptor, indicating a role for components of the UPR earlier in B lymphopoiesis (30). Therefore, the mechanism for which AK2 mutations may impair lymphopoiesis may be through a failure in the induction of the UPR and subsequent cell differentiation.
Acknowledgments
We thank Drs. Michael Czech and Fumihiko Urano for valuable discussions during the progress of this work and Catherine Bue, Deanna Navaroli, Kunal Gurav, Deborah Leonard, and Olga Gealikman for helpful comments on the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant DK60837 (to S. C.) and core services supported by the National Institutes of Health Diabetes Endocrinology Research Center Grant DK32520.
- ER
- endoplasmic reticulum
- UPR
- unfolded protein response
- AMPK
- AMP-activated protein kinase
- FCCP
- carbonyl cyanide p-trifluoromethoxyphenylhydrazone
- AK
- adenylate kinase
- PEP
- phosphoenolpyruvate
- PK
- pyruvate kinase
- qRT
- quantitative RT.
REFERENCES
- 1. Lin J. H., Walter P., Yen T. S. (2008) Annu. Rev. Pathol. 3, 399–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhao L., Ackerman S. L. (2006) Curr. Opin. Cell Biol. 18, 444–452 [DOI] [PubMed] [Google Scholar]
- 3. Lin J. H., Li H., Yasumura D., Cohen H. R., Zhang C., Panning B., Shokat K. M., Lavail M. M., Walter P. (2007) Science 318, 944–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ron D., Walter P. (2007) Nat. Rev. Mol. Cell Biol. 8, 519–529 [DOI] [PubMed] [Google Scholar]
- 5. Rutkowski D. T., Arnold S. M., Miller C. N., Wu J., Li J., Gunnison K. M., Mori K., Sadighi Akha A. A., Raden D., Kaufman R. J. (2006) PLoS Biol. 4, e374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Harding H. P., Calfon M., Urano F., Novoa I., Ron D. (2002) Annu. Rev. Cell Dev. Biol. 18, 575–599 [DOI] [PubMed] [Google Scholar]
- 7. Schröder M., Kaufman R. J. (2005) Annu. Rev. Biochem. 74, 739–789 [DOI] [PubMed] [Google Scholar]
- 8. Papa F. R., Zhang C., Shokat K., Walter P. (2003) Science 302, 1533–1537 [DOI] [PubMed] [Google Scholar]
- 9. Morimoto N., Oida Y., Shimazawa M., Miura M., Kudo T., Imaizumi K., Hara H. (2007) Neuroscience 147, 957–967 [DOI] [PubMed] [Google Scholar]
- 10. Szegezdi E., Duffy A., O'Mahoney M. E., Logue S. E., Mylotte L. A., O'brien T., Samali A. (2006) Biochem. Biophys. Res. Commun. 349, 1406–1411 [DOI] [PubMed] [Google Scholar]
- 11. Ozcan U., Cao Q., Yilmaz E., Lee A. H., Iwakoshi N. N., Ozdelen E., Tuncman G., Görgün C., Glimcher L. H., Hotamisligil G. S. (2004) Science 306, 457–461 [DOI] [PubMed] [Google Scholar]
- 12. Hosogai N., Fukuhara A., Oshima K., Miyata Y., Tanaka S., Segawa K., Furukawa S., Tochino Y., Komuro R., Matsuda M., Shimomura I. (2007) Diabetes 56, 901–911 [DOI] [PubMed] [Google Scholar]
- 13. Berg A. H., Combs T. P., Scherer P. E. (2002) Trends Endocrinol. Metab. 13, 84–89 [DOI] [PubMed] [Google Scholar]
- 14. Scherer P. E., Williams S., Fogliano M., Baldini G., Lodish H. F. (1995) J. Biol. Chem. 270, 26746–26749 [DOI] [PubMed] [Google Scholar]
- 15. Hoffstedt J., Arvidsson E., Sjölin E., Wåhlén K., Arner P. (2004) J. Clin. Endocrinol. Metab. 89, 1391–1396 [DOI] [PubMed] [Google Scholar]
- 16. Koh E. H., Park J. Y., Park H. S., Jeon M. J., Ryu J. W., Kim M., Kim S. Y., Kim M. S., Kim S. W., Park I. S., Youn J. H., Lee K. U. (2007) Diabetes 56, 2973–2981 [DOI] [PubMed] [Google Scholar]
- 17. Wilson-Fritch L., Burkart A., Bell G., Mendelson K., Leszyk J., Nicoloro S., Czech M., Corvera S. (2003) Mol. Cell. Biol. 23, 1085–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Basseri S., Lhoták S., Sharma A. M., Austin R. C. (2009) J. Lipid Res. 50, 2486–2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Forner F., Kumar C., Luber C. A., Fromme T., Klingenspor M., Mann M. (2009) Cell Metab. 10, 324–335 [DOI] [PubMed] [Google Scholar]
- 20. Pagliarini D. J., Calvo S. E., Chang B., Sheth S. A., Vafai S. B., Ong S. E., Walford G. A., Sugiana C., Boneh A., Chen W. K., Hill D. E., Vidal M., Evans J. G., Thorburn D. R., Carr S. A., Mootha V. K. (2008) Cell 134, 112–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dzeja P., Terzic A. (2009) Int. J. Mol. Sci. 10, 1729–1772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Couñago R., Shamoo Y. (2005) Extremophiles 9, 135–144 [DOI] [PubMed] [Google Scholar]
- 23. Konrad M. (1993) J. Biol. Chem. 268, 11326–11334 [PubMed] [Google Scholar]
- 24. Miron S., Munier-Lehmann H., Craescu C. T. (2004) Biochemistry 43, 67–77 [DOI] [PubMed] [Google Scholar]
- 25. Fujisawa K., Murakami R., Horiguchi T., Noma T. (2009) Comp. Biochem. Physiol. B Biochem. Mol. Biol. 153, 29–38 [DOI] [PubMed] [Google Scholar]
- 26. Lagresle-Peyrou C., Six E. M., Picard C., Rieux-Laucat F., Michel V., Ditadi A., Demerens-de Chappedelaine C., Morillon E., Valensi F., Simon-Stoos K. L., Mullikin J. C., Noroski L. M., Besse C., Wulffraat N. M., Ferster A., Abecasis M. M., Calvo F., Petit C., Candotti F., Abel L., Fischer A., Cavazzana-Calvo M. (2009) Nat. Genet. 41, 106–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pannicke U., Hönig M., Hess I., Friesen C., Holzmann K., Rump E. M., Barth T. F., Rojewski M. T., Schulz A., Boehm T., Friedrich W., Schwarz K. (2009) Nat. Genet. 41, 101–105 [DOI] [PubMed] [Google Scholar]
- 28. Scarpulla R. C. (2008) Physiol. Rev. 88, 611–638 [DOI] [PubMed] [Google Scholar]
- 29. Shi X., Burkart A., Nicoloro S. M., Czech M. P., Straubhaar J., Corvera S. (2008) J. Biol. Chem. 283, 30658–30667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang K., Wong H. N., Song B., Miller C. N., Scheuner D., Kaufman R. J. (2005) J. Clin. Invest. 115, 268–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Livak K. J., Schmittgen T. D. (2001) Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 32. Detimary P., Jonas J. C., Henquin J. C. (1995) J. Clin. Invest. 96, 1738–1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Noma T., Song S., Yoon Y. S., Tanaka S., Nakazawa A. (1998) Biochim. Biophys. Acta 1395, 34–39 [DOI] [PubMed] [Google Scholar]
- 34. Sevier C. S., Kaiser C. A. (2008) Biochim. Biophys. Acta 1783, 549–556 [DOI] [PubMed] [Google Scholar]
- 35. Gess B., Hofbauer K. H., Wenger R. H., Lohaus C., Meyer H. E., Kurtz A. (2003) Eur. J. Biochem. 270, 2228–2235 [DOI] [PubMed] [Google Scholar]
- 36. Kwong J. Q., Henning M. S., Starkov A. A., Manfredi G. (2007) J. Cell Biol. 179, 1163–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Reimold A. M., Iwakoshi N. N., Manis J., Vallabhajosyula P., Szomolanyi-Tsuda E., Gravallese E. M., Friend D., Grusby M. J., Alt F., Glimcher L. H. (2001) Nature 412, 300–307 [DOI] [PubMed] [Google Scholar]
- 38. Carrasco A. J., Dzeja P. P., Alekseev A. E., Pucar D., Zingman L. V., Abraham M. R., Hodgson D., Bienengraeber M., Puceat M., Janssen E., Wieringa B., Terzic A. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 7623–7628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dzeja P. P., Terzic A. (2003) J. Exp. Biol. 206, 2039–2047 [DOI] [PubMed] [Google Scholar]
- 40. Kaufman R. J., Scheuner D., Schröder M., Shen X., Lee K., Liu C. Y., Arnold S. M. (2002) Nat. Rev. Mol. Cell Biol. 3, 411–421 [DOI] [PubMed] [Google Scholar]
- 41. Gomez E., Powell M. L., Bevington A., Herbert T. P. (2008) Biochem. J. 410, 485–493 [DOI] [PubMed] [Google Scholar]
- 42. Fukami-Kobayashi K., Nosaka M., Nakazawa A., Go M. (1996) FEBS Lett. 385, 214–220 [DOI] [PubMed] [Google Scholar]
- 43. Noma T. (2005) J. Med. Invest. 52, 127–136 [DOI] [PubMed] [Google Scholar]
- 44. Van Rompay A. R., Johansson M., Karlsson A. (1999) Eur. J. Biochem. 261, 509–517 [DOI] [PubMed] [Google Scholar]
- 45. Janssen E., Dzeja P. P., Oerlemans F., Simonetti A. W., Heerschap A., de Haan A., Rush P. S., Terjung R. R., Wieringa B., Terzic A. (2000) EMBO J. 19, 6371–6381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Janssen E., de Groof A., Wijers M., Fransen J., Dzeja P. P., Terzic A., Wieringa B. (2003) J. Biol. Chem. 278, 12937–12945 [DOI] [PubMed] [Google Scholar]
- 47. Gauthier M. S., Miyoshi H., Souza S. C., Cacicedo J. M., Saha A. K., Greenberg A. S., Ruderman N. B. (2008) J. Biol. Chem. 283, 16514–16524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Liu Q., Gauthier M. S., Sun L., Ruderman N., Lodish H. (2010) FASEB J. 24, 4229–4239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kamimura D., Bevan M. J. (2008) J. Immunol. 181, 5433–5441 [DOI] [PMC free article] [PubMed] [Google Scholar]