Abstract
The tumor suppressor p53 maintains genome stability and prevents malignant transformation by promoting cell cycle arrest and apoptosis. Both Mdm2 and Pirh2 have been shown to ubiquitylate p53 through their RING domains, thereby targeting p53 for proteasomal degradation. Using structural and functional analyses, here we show that the Pirh2 RING domain differs from the Mdm2 RING domain in its oligomeric state, surface charge distribution, and zinc coordination scheme. Pirh2 also possesses weaker E3 ligase activity toward p53 and directs ubiquitin to different residues on p53. NMR and mutagenesis studies suggest that whereas Pirh2 and Mdm2 share a conserved E2 binding site, the seven C-terminal residues of the Mdm2 RING directly contribute to Mdm2 E3 ligase activity, a feature unique to Mdm2 and absent in the Pirh2 RING domain. This comprehensive analysis of the Pirh2 and Mdm2 RING domains provides structural and mechanistic insight into p53 regulation by its E3 ligases.
Keywords: E3 Ubiquitin Ligase, p53, Protein-Protein Interactions, Ubiquitin-conjugating Enzyme (Ubc), Ubiquitylation, E2-E3 Interaction, Mutagenesis
Introduction
The tumor suppressor p53 acts as the “guardian of the genome” using transcription-dependent and -independent mechanisms. Through its sequence-specific DNA binding activity, p53 orchestrates cellular responses by regulating transcription of target genes that are involved in a number of cellular processes, including cell cycle arrest, apoptosis, senescence, DNA repair, and angiogenesis (1). p53 also triggers apoptosis by activating the intrinsic apoptotic pathway through direct interactions with BH3 proteins at the mitochondria (2, 3). Because p53 activation often leads to growth inhibition and cell death, p53 levels and activation are tightly regulated under normal cellular conditions in the absence of stress signals. Ubiquitylation of p53 mediated by several E3 ligases, including Mdm2, Pirh2, COP1, E6-AP, ARF-BP1, synoviolin, and the atypical E3 ligase E4F1, is the major mechanism that regulates p53 turnover, thereby modulating p53 protein levels in the cell (4–10).
MDM2, the murine double minute 2 gene (also denoted HDM2 for the human gene), encodes a p53-specific E3 ligase and is amplified in more than 7% of all cancers (11). The central role of Mdm2 in p53 regulation has been clearly demonstrated by the p53-dependent embryonic lethality in MDM2 knock-out mice, which can be rescued by deletion of the p53 gene (12, 13). p53 and Mdm2 form a negative feedback loop. Mdm2 down-regulates p53 function by directly inhibiting p53 transactivation, targeting p53 for proteasomal degradation and redistributing p53 from the nucleus to the cytoplasm, whereas the expression of Mdm2 is under the transcriptional control of p53. Mdm2-mediated p53 ubiquitylation requires coordination of multiple domains: 1) the N-terminal p53-binding domain, which binds with high affinity to the N-terminal p53 transactivation domain, 2) the central acidic domain, which recognizes the ubiquitylation signal presented in the p53 core domain, and 3) the C-terminal RING domain, which recruits a ubiquitin-conjugating enzyme E2. Deletion or mutation of any of these domains results in a loss or attenuation of p53 ubquitylation by Mdm2.
MdmX, a protein homologous to Mdm2, functions as another negative regulator of p53. Similar to Mdm2, it has been shown to act as a specific and essential inhibitor of p53 transactivation during embryonic development (14). Although MdmX contains all three domains of Mdm2 (i.e. the p53 binding domain, the acidic domain, and the RING domain), it lacks intrinsic E3 ligase activity and is incapable of ubiquitylating p53 or itself (15, 16). Mdm2 and MdmX are capable of forming homodimers as well as heterodimers through their C-terminal β-sheet extensions of their RING domain. Heterodimer formation of Mdm2 and MdmX has been demonstrated to stabilize Mdm2 and enhance its inhibitory effect on p53 (17, 18).
Among the newly emerging p53 E3 ligases, Pirh2 (p53-induced RING-H2 domain protein; also known as RCHY1) has been shown to regulate p53 functions in many ways similar to Mdm2. It represses p53-dependent transactivation and growth inhibition while being subject to p53 transcriptional control (6). Overexpression of Pirh2 has been reported in a number of human cancers, which correlates with an increase in p53 ubiquitylation and a decrease in p53 levels independent of Mdm2, suggesting that the Pirh2-mediated p53 inhibition could play a role in tumorigenesis (19, 20). Our recent structural study revealed that the interaction of p53 and Pirh2 employs a two-site binding mode, where the p53 DNA binding domain interacts with the Pirh2 N terminus, whereas the p53 tetramerization domain binds to the Pirh2 C terminus to enhance specificity in targeting the active, tetrameric form of p53 for ubiquitylation and proteasomal degradation (21).
Both Mdm2 and Pirh2 regulate p53 primarily through their E3 ubiquitin (Ub)3 ligase activity, which relies on the RING domains harbored in both proteins. RING domains are a special class of zinc finger, ∼60 amino acids in length. A typical RING domain binds two zinc ions and folds into a compact α/β topology. Unlike the HECT domain-containing E3 ligases that form a Ub-thioester intermediate prior to substrate ubiquitylation, the RING domain type E3 ligases serve to recruit the Ub-conjugating enzyme E2 as well as to stimulate the release and direct transfer of Ub from the E2 to a substrate lysine residue presented either on the E3 or the target substrate protein. Despite the fact that the RING domain structures of both Pirh2 and Mdm2 have been determined, the detailed molecular mechanisms by which these E3 ligases mediate p53 ubiquitylation are not well understood.
Resurrection of p53 activity in tumor cells that retain the wild type p53 with its function suppressed by oncogenic application of its negative regulators is an attractive strategy for potential anti-cancer therapy. Thus, p53 E3 ligases represent an attractive group of therapeutic targets. Structure and mechanistic understanding of these p53 E3 ligases would provide insight into the development of effective inhibitors of p53 ubiquitylation. One such example is that the small molecule Nutlin blocks Mdm2 interaction with p53, thereby inducing p53-mediated apoptosis in cancer cells with overexpressed Mdm2 (22, 23). In this study, we thoroughly examined the structural similarities and differences between the RING domains of Pirh2 and Mdm2 as well as characterizing the relative contributions of these structural features to p53 ubiquitylation and E2-E3 interactions. We investigated the activities of the respective RING domains in catalyzing E3 autoubiquitylation and ubiquitylation of p53, the residues essential for E2-E3 interactions, and the effects of RING domain dimerization on E3 ligase function. Our analysis aims to elucidate the mechanistic differences between Pirh2 and Mdm2 in ubiquitylation catalysis in order to provide a structural and biochemical basis for deciphering the different roles of Pirh2 and Mdm2 in Ub-mediated regulation of p53.
EXPERIMENTAL PROCEDURES
Protein Preparation
Pirh2 and Mdm2 proteins were prepared as described previously (21, 24–26). A panel of human E2 clones used in this study is listed in supplemental Table 1. The full-length E2s were amplified and cloned between NdeI and BamHI sites of the pET15 vector (Stratagene). The proteins were expressed in the Escherichia coli strain BL21 (Codon plusTM) (Stratagene) and induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside for 5 h at room temperature. All E2 proteins were purified with standard nickel affinity chromatography methods (Qiagen) and dialyzed against the E2 buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm PMSF, 1 mm DTT) overnight at 4 °C. The E2 proteins were frozen with 10% glycerol at −80 °C until use.
To prepare an Mdm2/MdmX heterodimer, the full-length GST-MdmX construct was cloned and expressed in the E. coli strain BL21 (Codon plusTM) (Stratagene). Purified full-length His-Mdm2 was incubated with the resin-bound GST-MdmX for 1 h. Following several washes, proteins were eluted from the GST resin using the elution buffer (50 mm Tris, 500 mm NaCl, 30 mm reduced glutathione, 10 mm β-mercaptoethanol, 10% glycerol) and immediately submitted to a second run of nickel affinity purification. The proteins were incubated with nickel metal affinity resin (Qiagen) for 1 h. After extensive washing with the wash buffer (50 mm Tris, 500 mm NaCl, 10 μm ZnCl2, 1 mm PMSF, 2 mm benzamidine, 5 mm β-mercaptoethanol, and 10 mm imidazole), the Mdm2/MdmX heterodimers were eluted from the resin with a buffer containing 50 mm Tris, 500 mm NaCl, 10 μm ZnCl2, 1 mm PMSF, 2 mm benzamidine, 5 mm β-mercaptoethanol, and 500 mm imidazole and stored at −80 °C until use.
NMR Spectroscopy and Chemical Shift Perturbation Experiments
NMR spectra were recorded at 25 °C on a Varian INOVA 500- or 600-MHz spectrometer equipped with triple resonance cold probes and a Bruker Avance 500-MHz spectrometer equipped with a cryoprobe. Spectra were processed using the NMRPipe software package (27) and analyzed with XEASY (28). The chemical shift mapping on the Pirh2 RING domain was performed by monitoring the 1H-15N HSQC spectra difference of the 15N-labeled Pirh2 RING domain alone and with an excess of unlabeled E2 proteins (molar ratio 1:3). The chemical shift perturbation experiments on UBE2D2 were carried out by monitoring the 1H-15N HSQC spectra difference of the 15N-labeled UBE2D2 alone and with an excess of unlabeled RING-H2 domain (molar ratio 1:2). The chemical shift differences were calculated using the formula, Δppm = ((δHN)2 + (δN/5)2)½.
In Vitro Ubiquitylation Assay
The ubiquitylation reaction was performed in a volume of 20 μl in a buffer of 50 mm Tris, pH 7.6, 5 mm MgCl2, 2 mm ATP, and 2 mm DTT. The reaction mixture typically contained E1 (50 ng) (Calbiochem), UBE2D2 (100 ng), ubiquitin (5 μg) (Sigma), and 0.5 μg of p53 for detection of p53 ubiquitylation. In these reactions, the concentration of E3 ligase was typically 0.5 μm, unless otherwise specified. The optimal conditions for E1, E2, Ub, and ATP were established through titration experiments to ensure that these reagents were in excess and would not be limiting factors in the reactions. After incubation at 30 °C for 90 min, the reactions were stopped by the addition of SDS-PAGE sample buffer and resolved on 7.5–10% SDS-polyacrylamide gels. Ubiquitylated proteins were visualized and evaluated by Western blot using a monoclonal antibody against GST (GE Healthcare) for Pirh2 autoubiquitylation, an antibody against p53 (PAb1801 or DO-1) (29) for p53 ubiquitylation, or an antibody against Mdm2 (SMP14, Santa Cruz Biotechnology, Inc. (Santa Cruz, CA)) for Mdm2 autoubiquitylation. The free ubiquitin in the reaction was detected with a monoclonal antibody against ubiquitin (Covance) and a Cy3-labeled secondary antibody. The free ubiquitin signal was quantified with a Typhoon imager (GE). All assays have been repeated independently more than three times using proteins from multiple preparations.
Mass Spectrometry
In-gel digests using l-1-tosylamido-2-phenylethyl chloromethyl ketone sequencing grade trypsin (Promega, Madison, WI) were performed described previously (30). Analytical columns (75-μm inner diameter) and precolumns (100 μm) for liquid chromatography-electrospray ionization-tandem mass spectrometry (LC-ESI-MS/MS) analysis were made in house from silica capillary tubing from InnovaQuartz (Phoenix, AZ) and packed in house with 5 μm 100-Å C18-coated silica particles (Magic, Michrom Bioresources, Auburn, CA).
Peptides were subjected to LC-ESI-MS/MS, using a 120-min RPLC (95% water, 95% acetonitrile, 0.1% formic acid) buffer gradient running at 400 nl/min on a Proxeon EASY-nLC pump in-line with a hybrid LTQ-Orbitrap mass spectrometer (Thermo Fisher Scientific, Waltham, MA). A parent ion scan was performed in the Orbitrap, using a resolving power of 60,000, and then the four most intense peaks were selected for MS/MS (minimum ion count of 1000 for activation), using standard collision-induced dissociation fragmentation. Fragment ions were detected in the LTQ. Dynamic exclusion was activated such that MS/MS of the same m/z (within a −0.1 and +2.1 Thomson (Th) window; exclusion list size = 500) detected three times within 45 s was excluded from analysis for 30 s.
For protein identification, Thermo. RAW files were converted to the. mzXML format using ReAdW software (31) and then searched using X!Tandem (32) against the human (Ensembl 44.36F) data base, supplemented with a curated custom data base containing the mature Ub sequence. X!Tandem search parameters were as follows: complete modifications, none; cysteine modifications, none; potential modifications, +114.04@K, +16@M and W, +32@M and W, +42@N terminus, +1@N and Q, −17@N-term Q, −18@N-term E; parent mass error, ±10 ppm; fragment error, 0.4 Da; maximum charge, 4+; missed cleavage sites, 3; semicleavage, no.
RESULTS
Structural Comparison of the Pirh2 and Mdm2 RING Domain
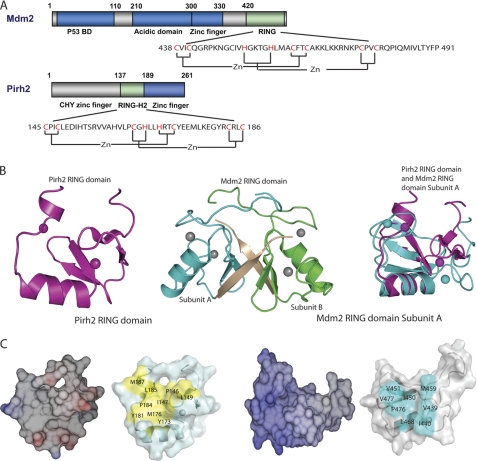
The recent completion of the three-dimensional structures of the Pirh2 and Mdm2 RING domains enabled structural comparison studies of these two p53 E3 ligases (21, 24, 25). Similar to the Pirh2 RING domain, the Mdm2 RING domain displays a compact α/β global fold. Both structures bear a shallow depression on the surface composed of multiple hydrophobic residues and bind two zinc ions with an interweaved zinc-binding motif (Fig. 1A). Pairwise structural comparison of the Pirh2 RING domain (Protein Data Bank code 2JRJ) and one subunit of the Mdm2 RING domain (Protein Data Bank code 2HDP) using DALI (33) showed a root mean square deviation value of 3.0 Å over 41 aligned Cα atoms, which is consistent with their membership in the RING domain superfamily.
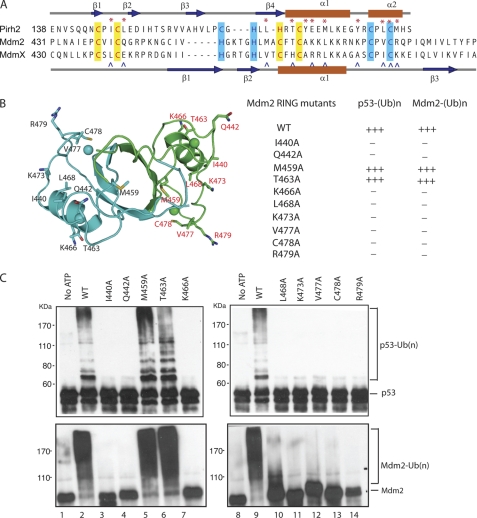
FIGURE 1.
Structural comparison of the Mdm2 and Pirh2 RING domains. A, schematic diagram showing the domain structure of Mdm2 and Pirh2 and the sequence of their RING domains. The cysteines and histidines involved in zinc coordination are labeled in red. B, ribbon representation of the Pirh2 RING, Mdm2 RING domain (Protein Data Bank code 2HDP), and overlay of the Pirh2 RING domain with the Mdm2 RING domain subunit A, the Pirh2 RING domain is shown in purple, and the Mdm2 RING domain is shown in cyan. The last seven residues of Mdm2 are shown in wheat. C, surface and electrostatic representation of the Pirh2 RING and Mdm2 RING domain subunit A. The orientation of the proteins in each case is the same as in B. The hydrophobic residues on the E2 binding surface of RING domains are indicated in yellow for Pirh2 and cyan for Mdm2. The figures were prepared with PyMOL (61) for the surface representation and Molmol (62) for electrostatic representation.
Despite the overall similarity in topology of the RING structures, the RING domains of Pirh2 and Mdm2 exhibit several important differences. Mdm2 forms a homodimer with its own RING domain or a heterodimer with the RING domain of MdmX. Dimerization is mediated by the C-terminal seven residues, which along with residues from the second β-strand of the opposite subunit form a very tight and small β-barrel (24). These C-terminal residues are not a part of the canonical RING domain but are necessary for maintaining the E3 ligase activity of Mdm2 (Fig. 1B) (26, 34). In contrast to Mdm2, analogous C-terminal residues are absent in Pirh2, which accounts for the fact that Pirh2 behaves as a monomer, as verified by size exclusion chromatography and small angle x-ray scattering (data not shown).
A second difference between the Mdm2 and Pirh2 RING domains is their zinc coordination schemes. The Mdm2 RING domain has C2H2C4 zinc coordination, with the residues Cys438, Cys441, Cys461, and Cys464 comprising zinc-binding site I and His452, His457, Cys475, and Cys478 for the second zinc (zinc-binding site II). Pirh2 is named after its RING-H2 domain, which has a C3H2C3 zinc-binding scheme. The Pirh2 RING domain binds the first zinc ion with the residues Cys145, Cys148, Cys164, and His166 and the second zinc with His169, Cys172, Cys183, and Cys186. The structure overlay of the two RING domains reveals substantial differences in the positions of the zinc ions, which is probably a result of permutation and spacing differences of the cysteine and histidine residues responsible for the zinc coordination in the two proteins (Fig. 1A).
Finally, the Mdm2 RING domain has a highly positively charged electrostatic surface with the majority of positive charge located at the α-helix and around the second zinc-binding site. The Pirh2 RING domain, on the other hand, contains several acidic residues (Glu174, Glu175, and Glu179) replacing the lysine residues in the Mdm2 RING domain (Fig. 1C). Thus, although the RING domain structures of Pirh2 and Mdm2 are similar in overall global folding and the interweaved zinc-binding motif, the disparities in their oligomeric states, surface electrostatic charge distribution, and zinc coordination may contribute to the differences in their ability to recruit E2 ligases and/or ubiquitylate their substrates.
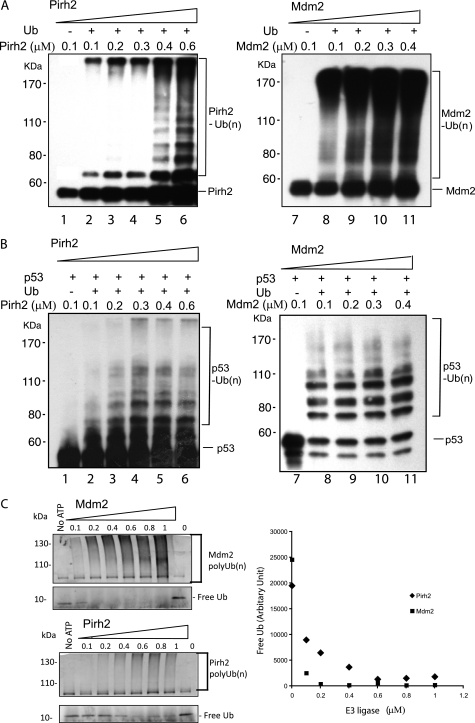
Pirh2 Appears to be Less Efficient than Mdm2 in Autoubiquitylation and Ubiquitylation of p53
Both Mdm2 and Pirh2 function as p53 E3 ligases. To better understand the relative contributions of the Mdm2 and Pirh2 RING domains toward p53 ubiquitylation, we carried out extensive biochemical assays and mutagenesis studies to characterize Pirh2- and Mdm2-mediated p53 ubiquitylation and their functional interactions with the Ub-conjugating enzyme E2s. To compare the E3 ligase activities of Pirh2 and Mdm2, we performed an in vitro ubiquitylation assays using recombinant GST-tagged Pirh2 and His6-tagged Mdm2 in a reaction containing E1, E2 (UBE2D2/UbcH5b), and His6-tagged Ub, with and without the recombinant p53 protein. Under all tested concentrations (0.1–0.6 μm), Mdm2 demonstrated greater E3 ligase activity than Pirh2 in both autoubiquitylation (Fig. 2A) and p53 ubiquitylation (Fig. 2B) assays, as judged by the amount of the Ub-conjugated products formed in these reactions (Fig. 2). Multiple Pirh2 protein preparations were utilized in these assays (all yielding similar activity), and in fact the same preparations were subjected to structure determination, indicating that this lower level of p53 ubiquitylation was not a result of protein misfolding. Slightly different p53 ubiquitylation patterns were also observed between Pirh2 and Mdm2 (Fig. 2B), suggesting that the two ligases may modify different lysine residues on p53. In order to unambiguously compare the E3 ligase activity of Pirh2 and Mdm2, a defined amount of free ubiquitin was used in the reactions with increasing amounts of Pirh2 or Mdm2. The free ubiquitin was observed to deplete at a much higher rate in Mdm2-containing reactions than those with Pirh2. Although we cannot distinguish between autoubiquitylation and p53 ubiquitylation in these experiments, these results indicate that Mdm2 conjugates ubiquitin more efficiently than Pirh2 (Fig. 2C). Although it remained possible that Pirh2 may be more active with other ubiquitin E2s (see below) or function with unknown co-factors in vivo, the results from our in vitro ubiquitylation assays suggest that Mdm2 is a more active E3 ligase.
FIGURE 2.
Comparison of E3 ligase activity of the Mdm2 and Pirh2 RING domains. A, autoubiquitylation of Pirh2 (lanes 1–6) and Mdm2 (lanes 7–11). GST-Pirh2 (E3) and His-Mdm2 (E3) were subjected to an in vitro ubiquitylation assay in the presence or absence of ATP, E1, E2 (UBE2D2), His-tagged ubiquitin, and p53, as indicated. After the reaction, the samples were resolved by SDS-PAGE and detected by Western blot. Pirh2 autoubiquitylation (left, lanes 1–6) was blotted with monoclonal antibody against GST. Autoubiquitylation of Mdm2 was evaluated using a specific antibody against Mdm2 (SMP14). B, p53 ubiquitylation by Pirh2 (lanes 1–6) and Mdm2 (lanes 7–11). Ubiquitylated p53 was detected using monoclonal antibody against p53 (PAB1801) in the reactions with increasing amounts of Pirh2 or Mdm2, as indicated. C, polyubiquitylation by Pirh2 and Mdm2. Left, total ubiquitylation catalyzed by Pirh2 and Mdm2 and the free ubiquitin in these reactions were subjected to fluorescence immunoblotting analysis and detected using a monoclonal antibody against ubiquitin and a Cy3-labeled mouse IgG. Right, the amount of free ubiquitin reported as an average fluorescence intensity of the free ubiquitin bands in the reactions of three repeated experiments quantified by a Typhoon imager.
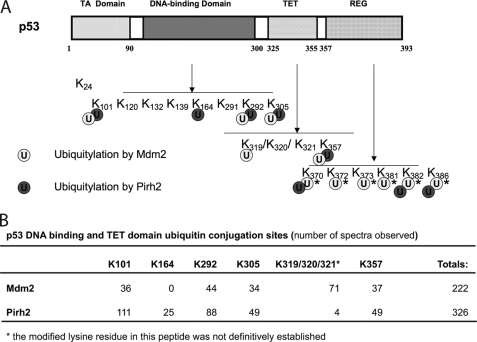
Pirh2 and Mdm2 Modify Different Lysine Residues in p53
To identify the p53 lysine residues targeted by these E3 ligases, the products of the in vitro ubiquitylation reactions containing p53, the ubiquitin E1, UBE2D2 and either Mdm2 or Pirh2 were subjected to mass spectrometric analysis, as described by Lallemand-Breitenbach et al. (35). Previous reports have indicated that p53 is modified by Mdm2 at lysine residues clustered in the C-terminal domain (36, 37). We observed the same p53 C-terminal residues modified in Pirh2-containing reactions (supplemental Fig. 1). We also found that p53 was ubiquitylated at a number of other lysine residues in these reactions, including Lys101, Lys164, Lys292, and Lys305 within the DNA-binding domain and Lys319* (we are unable to confidently assign the modified residue in this tryptic peptide to an individual lysine residue) and Lys357 in the tetramerization domain (Fig. 3A). Interestingly, whereas Lys101, Lys292, Lys305, and Lys357 appeared to be ubiquitylated by both Mdm2 and Pirh2 to a similar extent (calculated as a percentage of total spectral counts), p53 Lys164 was modified exclusively by Pirh2, and Lys319* was modified at levels 30-fold higher in our Mdm2 reactions (Fig. 3B). Thus, Pirh2 and Mdm2 appear to preferentially ubiquitylate different subsets of lysine residues in p53 in vitro. The biological significance of these observations remains to be determined.
FIGURE 3.
Pirh2 and Mdm2 favor different lysine linkage in p53 ubiquitylation and ubiquitylate different p53 lysine sites. A, schematic diagram showing the domain structure of p53 and the distribution of ubiquitylation target lysines of Pirh2 and Mdm2 within the p53 molecule. Lysine residues modified by Pirh2 and Mdm2 were identified by mass spectrometry of the p53 ubiquitin adducts produced by the in vitro ubiquitylation assay. The asterisks indicate the Ub sites reported in the literature (36, 37). B, the ubiquitin conjugation sites within the p53 DNA-binding and tetramerization (TET) domains detected by MS. In vitro reaction products were subjected to SDS-PAGE, and high molecular weight products were analyzed by LC-ESI-MS/MS. The numbers of spectra observed at the indicated sites are shown.
Pirh2 and Mdm2 Utilize Different Subsets of E2s for p53 Ubiquitylation
RING E3 ubiquitin ligases rely on the E2 ubiquitin-conjugating enzymes to transfer ubiquitin to substrate proteins. The E2s have been shown to play a major role in substrate modification in other E2/E3 systems (38, 39). We therefore asked whether utilization of different E2s would contribute to the functional differences observed between Pirh2 and Mdm2. A panel of E2s was tested for their ability to support Pirh2- and Mdm2-mediated autoubiquitylation and ubiquitylation of p53. As shown in supplemental Figs. 3 and 4, both Pirh2 and Mdm2 successfully catalyzed autoubiquitylation and p53 ubiquitylation with members of the UBE2D and UBE2E families. However, Mdm2 was also able to monoubiquitylate p53 in the presence of UBE2N (Ubc13), UBE2Q, the UBE2Q-like protein FLJ25076, UBE2A, and UBE2B (HRad6A and -B). It did not appear that monoubiquitylation of p53 was due to lower reactivity with these five E2s, because increased reaction times and reactions containing higher enzyme concentrations yielded similar results, which were not seen in the reactions in the absence of an E3 (supplemental Fig. 2). Thus, Mdm2 displayed a broader range of functional E2 partners for p53 ubiquitylation compared with Pirh2.
Mapping the E2 Binding Interface of the Pirh2 RING Domain Using NMR
Using NMR, we previously mapped the UBE2D2-binding site of Pirh2 to a shallow hydrophobic depression on the surface of the Pirh2 RING domain (21). To investigate whether all functionally interacting E2s identified in the above E2 screening assay bind to the same site of the Pirh2 RING, we titrated the 15N,1H-Pirh2 RING domain with UBE2D1,3,4 and UBE2E2. A consistent pattern of chemical shift perturbations was observed in the Pirh2 RING domain upon the addition of these E2s (supplemental Fig. 5). As with UBE2D2, the most perturbed sites were seen in two zinc coordination sites, including Ile147, Cys148, Leu185, and Met187, and residues from the central β-sheet and the helix α1 (residues Leu167, Leu168, His169, Thr171, Glu174, Met176, and Tyr181). The resonances of two residues (Leu149 and Glu179) were broadened beyond detection upon binding to E2s. When comparing the chemical shift perturbations of the Pirh2 RING domain observed for all titrations, we noticed that UBE2D1, -3, and -4 showed similar patterns but greater chemical shift changes than UBE2D2 and UBE2E2, suggesting that UBE2D1, -3, and -4 may bind the Pirh2 RING domain with higher affinity than UBE2D2 and UBE2E2. However, all tested E2s displayed binding saturation at ∼1 mm in the NMR titration experiments, suggesting that the binding affinities of these E2s with the Pirh2 RING domain are similar to UBE2D2 (Kd ∼0.2 mm (21)). In addition, chemical shift perturbations were also observed for more than half of the backbone resonances of the Pirh2 RING domain, suggesting that a conformational adjustment may have occurred upon binding to E2s.
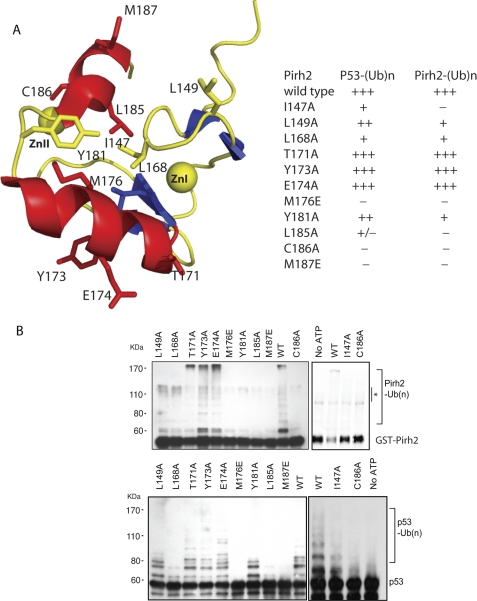
Identification of RING Domain-E2 Interaction Surfaces
To determine the contributions of key Pirh2 RING domain residues to ubiquitylation, we conducted site-directed mutagenesis on 10 residues (Ile147, Leu149, Leu168, Thr171, Tyr173, Glu174, Met176, Leu185, Cys186, and Met187) that showed the most significant chemical shift perturbations upon binding to E2s. All mutants were prepared as full-length GST-fusions in the same way as wild type Pirh2 to test their E3 ligase activity as well as in the context of the isolated RING domain to examine the effect on global protein folding using NMR HSQC. Of the 10 mutants, only C186A, a mutation of the zinc-coordinating residue, showed disruption of the RING structure, whereas the conformation of the remaining nine mutants was unaffected (data not shown). As shown in Fig. 4, T171A, Y173A, and E174A retained activity for autoubiquitylation and ubiquitylation of p53, suggesting that these residues are dispensable for sustaining the E3 ligase function. Notably, residues Thr171, Tyr173, and Glu174 are located in helix α1 with their side chains oriented away from the hydrophobic core (Fig. 4A). Thus, the 15N-1H chemical shift perturbations in these residues may reflect secondary effects caused by a conformational change in helix α1 upon binding an E2. Mutants I147A, L149A, and Y181A showed a significant attenuation of the autoubiquitylation activity while affecting p53 ubiquitylation to a lesser degree. These three residues are at the left and right borders of the hydrophobic core in the RING domain structure. The decreased activity may have resulted from reduced (but not disrupted) hydrophobicity in the RING domain structure of the mutants. The last group of mutants, L168A, M176E, L185A, and M187E, abolished both autoubiquitylation and p53 ubiquitylation activities. Residues Leu168, Met176, Leu185, and Met187 all reside at the central hydrophobic surface groove. The importance of these residues for E3 ligase activity, as demonstrated in the activity assays, suggests that they may be involved in direct interaction with E2s. Taken together, these results suggest that the residues that are part of the hydrophobic patch on the RING domain are essential for Pirh2 E3 ligase activity.
FIGURE 4.
Mutagenesis studies of the Pirh2 RING domain. A, schematic diagram of the Pirh2 RING domain showing the mutated residues in stick form. The table summarizes the activities of the Pirh2 RING domain mutants in mediating autoubiquitylation and ubiquitylation of p53. B, activity of the Pirh2 RING domain mutants in autoubiquitylation and p53 ubiquitylation was evaluated through an in vitro ubiquitylation assay in the presence of ATP, E1, E2 (UBE2D2), His-ubiquitin, Pirh2 mutants, and p53. Pirh2 autoubiquitylation was detected using an antibody against GST, and the ubiquitylated p53 was detected using monoclonal antibody against p53 (PAB1801). An asterisk indicates the GST-Pirh2 aggregates in the reaction, which are not ubiquitylated GST-Pirh2 products, as confirmed by the anti-ubiquitin immunoblot (not shown).
To investigate whether Mdm2 uses a similar E2 binding surface as Pirh2, the structurally aligned residues from the Mdm2 RING domain, namely Ile440, Gln442, Met459, Thr463, Lys466, Leu468, Lys473, Val477, and Arg479, were individually mutated to alanines in a set of full-length Mdm2 proteins (Fig. 5). The E3 ligase activity of these mutant Mdm2 proteins was then assessed. A known structural mutant (C478A (40)) was included as a negative control. As shown in Fig. 5C, wild type Mdm2 showed a high degree of autoubiquitylation and p53 ubiquitylation, whereas the control C478A revealed no activity. Mdm2 mutants M459A and T463A retained full ubiquitylation activity in as the wild type, whereas mutations at sites Ile440, Gln442, Lys466, Lys468, Lys473, Val477, and Arg492 abolished Mdm2 E3 ligase activity in both autoubiquitylation and p53 ubiquitylation. Due to poor solubility of the isolated Mdm2 RING domain under NMR conditions, the effect of the mutations on the overall structure of Mdm2 was evaluated by CD and compared with the spectrum of wild-type Mdm2. With the exception of the mutant C478A, all of the other nine mutants exhibited CD spectra very similar or identical to that of the wild type protein, suggesting a negligible effect of these mutations on Mdm2 structure (data not shown). Thus, the Mdm2 RING domain contains an interaction surface similar to that of the Pirh2 RING domain, which is essential to RING domain function in catalyzing ubiquitylation.
FIGURE 5.
Mutagenesis studies of the Mdm2 RING domain. A, Pirh2, Mdm2, and MdmX RING domain sequences aligned by conserved cysteine and histidine residues. Secondary structure topology of the Pirh2 RING is shown above, whereas that of Mdm2 and MdmX are shown below. α-Helices are colored red, and β-sheets are blue. The mutated residues are indicated by the asterisks (Pirh2) and carets (Mdm2). B, schematic diagram of the Mdm2 RING domain dimer showing the mutated residues in stick form. The table summarizes the activities of the Mdm2 RING domain mutants in autoubiquitylation and p53 ubiquitylation. C, activity of the Mdm2 RING domain mutants in autoubiquitylation and p53 ubiquitylation was evaluated through an in vitro ubiquitylation assay in the presence of ATP, E1, E2 (UBE2D2), His-ubiquitin, and p53. Mdm2 autoubiquitylation was detected using an antibody against Mdm2 (SMP14), and the ubiquitylated p53 was detected using monoclonal antibody against p53 (PAB1801).
Mdm2 Dimerization Interface Contributes to Ubiquitylation, a Feature Absent in Pirh2
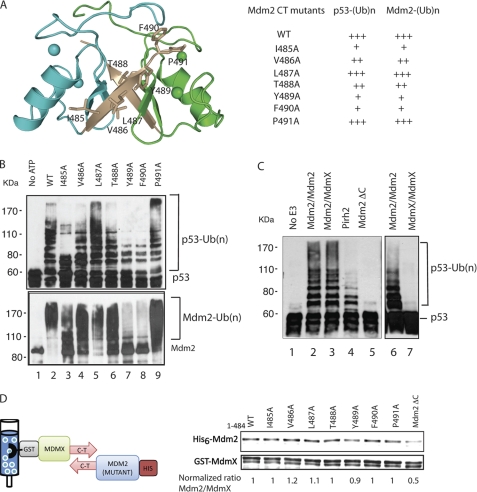
Mdm2 dimerization has been shown to be critical for its ubiquitylation function (26, 34), whereas Pirh2 does not require dimerization. The structure of Mdm2 suggests that dimerization could both stabilize the folded conformation of the RING domain and create an additional or larger surface for interactions with E2s or substrates (25, 26, 34). In order to dissect these potential roles of Mdm2 dimerization, we first investigated an Mdm2 deletion mutant missing all seven C-terminal residues (Mdm2 1–484, Mdm2 ΔC), which, as expected (25, 26), lacked the ability to ubiquitylate p53 (Fig. 6C). NMR analysis of the isolated Mdm2 RING (Mdm2 417–484, Mdm2RING ΔC) revealed that this mutant is conformationally disrupted and appears to be at least partially unfolded, suggesting that the last seven residues are required for maintenance of the structural integrity of the RING domain (supplemental Fig. 6).
FIGURE 6.
Mutagenesis studies of the Mdm2 RING last seven C-terminal residues. A, schematic diagram of the Mdm2 RING domain dimer showing the mutated residues in stick form. The table summarizes the activities of the Mdm2 RING C-terminal mutants in p53 ubiquitylation. B, activity of the His-tagged Mdm2 RING C-terminal mutants in autoubiquitylation and p53 ubiquitylation was evaluated through an in vitro ubiquitylation assay in the presence of ATP, E1, E2 (UBE2D2), His-ubiquitin, and p53. Mdm2 autoubiquitylation was detected using an antibody against Mdm2 (SMP14), and the ubiquitylated p53 was detected using monoclonal antibody against p53 (PAB1801). C, p53 ubiquitylation by Mdm2 homodimer, Mdm2/MdmX heterodimer, MdmX homodimer, Mdm2 ΔC, and Pirh2. The total concentration of Mdm2, MdmX, and Pirh2 was equivalent in each reaction mixture (0.5 μm). D, GST-pull-down analyses to assess the ability of Mdm2 C-terminal mutants to form a heterodimer with GST-MdmX. Mdm2 C-terminal mutants were detected with fluorescence immunoblotting using a monoclonal antibody against the His6 tag, and GST-MdmX was detected with an antibody against GST. The ability of each Mdm2 mutant to interact with GST-MdmX was quantified as the normalized ratio of Mdm2 mutants versus MdmX using Mdm2 WT/MdmX as 1.
Next, we assayed the Mdm2/MdmX heterodimer and compared its ubiquitylation activity to those of an equimolar subunit concentration of Mdm2 homodimer and Pirh2. To isolate the Mdm2/MdmX heterodimer, His6-tagged Mdm2 was first incubated with GST-tagged MdmX and then immobilized on glutathione-Sepharose resin. The final Mdm2/MdmX was prepared by successive glutathione-Sepharose and Ni2+ affinity chromatography to remove any MdmX or Mdm2 homodimers, a method described by Linke et al. (25) for the structural studies of the Mdm2/MdmX heterodimer. As shown in Fig. 6C, MdmX does not possess intrinsic E3 ligase activity, whereas the Mdm2 homodimer showed the highest p53 ubiquitylation activity. Both Mdm2 homo- and heterodimers demonstrated considerably higher activity than an equivalent amount of Pirh2. We also note that when comparing the activity of full-length proteins, one cannot attribute the differences in their ubiquitylation activities solely to the features of their respective RING domains. Mdm2 has much greater affinity for p53 (via the former's N-terminal domain), and this in itself might be the origin of greater activity (indeed, inhibition of this interaction can abolish Mdm2-mediated ubiquitylation of p53 (23)).
To further investigate the contribution of individual C-terminal residues to the structural integrity, dimerization, and catalysis of Mdm2 RING, we mutated each residue to alanine, namely I485A, V486A, L487A, T488A, Y489A, F490A, and P491A, in the full-length Mdm2. These mutants retained similar structural folding as the wild type Mdm2, as indicated by their identical circular dichroism spectra (data not shown). Next, we tested for the ability of these mutants to dimerize with GST-MdmX. His6-tagged Mdm2 mutants as well as the wild type and Mdm2 ΔC were incubated with glutathione resin-immobilized GST-MdmX and eluted with reduced glutathione. The amount of the His6-tagged Mdm2 mutants versus the wild type retained with GST-MdmX was quantified using fluorescence immunoblotting analysis. All C-terminal mutants were able to dimerize with GST-MdmX as well as the wild type Mdm2, whereas the ability of Mdm2 ΔC to interact with GST-MdmX was severely affected. The residual binding observed between Mdm2 ΔC and MdmX might be due to a weak interaction between the acidic domain and the RING domain (34, 41). Thus, although the Mdm2 C terminus is required for RING domain dimerization, a single alanine mutation is not sufficient to disrupt the RING dimer formation.
Because the Mdm2 C-terminal mutants maintain the ability to dimerize, they probably have a native-like conformation and are therefore useful for testing whether the C-terminal residues participate directly in ubiquitylation activity. Therefore, the activity of these mutants was assessed by an in vitro ubiquitylation assay (Fig. 6B). In contrast to what was observed for the Mdm2 RING domain mutants described above, none of the mutations in the C terminus of Mdm2 completely abolished Mdm2-mediated autoubiquitylation or ubiquitylation of p53. However, several mutants, including I485A, Y489A, and F490A, resulted in significant attenuation in ubiquitylation activity compared with the wild type Mdm2, whereas the activity of V486A and T488A was mildly affected. These results suggest that an intact C terminus not only mediates dimerization but also contributes directly to Mdm2 E3 ligase activity, a unique feature that is lacking in the Pirh2 RING domain.
Pirh2 and Mdm2 Bind a Conserved Interface of UBE2D2
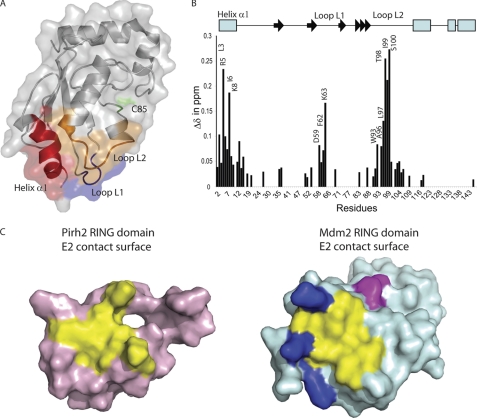
To better understand the molecular mechanism of the Mdm2 and Pirh2 E2-E3 interactions, we carried out NMR chemical shift perturbation experiments to define the residues of the E2s that are important for binding to the RING domain. Because the NMR resonance assignments for UBE2D2 were available (BMRB:6277), this protein was chosen as a model E2 to study the E2-E3 interaction with the Pirh2 RING domain (42). In agreement with what has been reported for UBE2D2 and CNOT4 interactions (43), the 15N amide resonances from three contiguous sites on the N15,H1-labeled UBE2D2 (helix α1 and Loops L1 and L2) were perturbed upon binding to the Pirh2 RING domain (Fig. 7, A and B), consistent with a conserved interaction surface for E3s of the RING domain class. The first interaction site (helix α1) exhibited chemical shift perturbations of residues Leu3, Arg5, Ile6, His7, Lys8, and Glu9 upon binding to the Pirh2 RING domain with the largest changes observed in residues Arg5 and Lys8. Analyses of other published E2-E3 complexes (UbcH7-c-Cbl and UBE2D2-cIAP2) (44, 45) revealed that helix α1 of UBE2D2 interacts with the first zinc-binding site of the RING domain. In addition to hydrophobic interactions, positively charged residues of the helix α1 (Arg5 and Lys8) of UBE2D2 may form salt bridges with Glu150 and Asp151 from the zinc-binding site I of the Pirh2 RING domain. Indeed, we observed chemical shift perturbations of Arg5 and Lys8 from UBE2D2 and Glu150 and Asp151 from the Pirh2 RING domain upon binding to each other.
FIGURE 7.
The chemical shift mapping of UBE2D2 upon binding to the Pirh2 RING domain. A, the regions with the greatest chemical shift changes induced upon binding to the Pirh2 RING domain are colored on the transparent surface representation of UBE2D2. The region of helix α1 is colored in red, Loop L1 is blue, and Loop L2 is orange. The catalytic Cys85 is colored in green. B, composite chemical shift changes versus residue number for UBE2D2 upon binding to the Pirh2 RING domain. The values shown were calculated by using the equation, Δcomp = (ΔδHN2 + (ΔδN/5)2)½. The secondary structure elements of UBE2D2 are shown at the top with an arrow for β-strands and a rectangle for α-helices. C, surface representation of Pirh2 (pink) and Mdm2 (pale blue) ring domain, indicating the conserved E2 binding site (yellow) and the extended E2 binding surface of Mdm2 contributed by the positively charged region (blue) and the C-terminal residues from the adjacent subunit (magenta).
The second cluster of residues affected was found to be the residues Asp59, Phe62, and Lys63 from Loop L1 of UBE2D2. In the structure of the c-Cbl-UbcH7 complex, the residues Pro and Phe from UbcH7 Loop L1 form hydrophobic interactions with the conserved Trp408 residue of the c-Cbl RING domain (45). Met176 of the Pirh2 RING domain and Leu476 of the Mdm2 RING are situated at the same position as Trp408 of c-Cbl. The mutagenesis data of Pirh2 Met176A and Mdm2 L476A support a critical role of this position in mediating interaction with UBE2D2. The last group of residues that showed significant chemical shift changes upon binding to the Pirh2 RING domain includes Trp93, Ser94, Ala96, Leu97, Thr98, Ile99, and Ser100 from Loop L2 of UBE2D2. This group of residues may form a third anchoring patch to assist docking of UBE2D2 onto the Pirh2 RING domain. As seen in the UbcH7-c-Cbl and UBE2D2-cIAP2 complexes, Loop L2 interacts with the second zinc-binding site (43–45). Our chemical shift perturbation data for Pirh2 RING and the mutational studies of Mdm2 and Pirh2 RINGs also support interactions of UBE2D2 with the second zinc-binding site of the RING domain. Due to limited solubility of the Mdm2 RING domain under NMR conditions and the weak interaction between E2 and E3, similar E2 NMR mapping with the Mdm2 RING could not be achieved.
Mdm2 Has an Extended E2 Interaction Surface
Integration of our structural, NMR, mutagenesis, and activity data for Pirh2 and Mdm2 allows a comparison of the apparent E2 interaction surfaces of their RING domains (Fig. 7C). Mutation of the residues involved in forming the hydrophobic depression on the surface of the RING domain and the zinc-binding sites resulted in attenuated or abolished activity in autoubiquitylation and ubiquitylation of p53 for both proteins. We also noticed that this conserved E2 binding site in Mdm2 is lined with four positively charged residues, whereas Pirh2 contains acidic and hydrophobic residues at these positions. Mutations in this region of the Mdm2 RING (Lys466 and Lys473; Fig. 5) abolished p53 ubiquitylation activity, whereas mutations in the same region of Pirh2 RING (Glu174 and Tyr181; Fig. 4) did not show much effect on its E3 ligase function, suggesting that Mdm2 may contain a more extended E2 binding surface than Pirh2 (blue residues in Fig. 7C). In addition, our mutational study of the Mdm2 RING domain C terminus suggests that the C terminus contributes to the optimal E3 ligase activity of Mdm2, apart from its role in dimerization (Fig. 7C, magenta). Specifically, the residues Tyr489 and Phe490 of Mdm2 RING have previously been shown to be critical for Mdm2-mediated ubiquitylation of p53 by serving as an extended contacting site for E2 binding (26, 34). The mutagenesis data on the C terminus of Mdm2 shown here further strengthen this notion. Taken together, our data support a three-point docking mode in which UBE2D2 binds to the Pirh2 RING domain via a hydrophobic, triangular, shallow groove on the surface of the RING domain. This interaction surface is conserved but extended in Mdm2, presumably contributing at least in part to the greater E3 ligase activity of Mdm2.
DISCUSSION
Ubiquitylation is thought to be the central cellular mechanism for p53 down-regulation that allows proliferation in unstressed cells. As the main regulators of p53 stability, p53 E3 ligases have attracted enormous interest. Overexpression of these negative regulators is found in a variety of human malignancies and therefore represent potential targets for the development of novel therapeutic compounds that induce growth-inhibitory and apoptotic effects of p53 in cancer cells. In this study, we have compared the in vitro ubiquitylation activities of Pirh2 and Mdm2 with a focus on the role of their RING domains in this process. We show that compared with Mdm2, Pirh2 is an intrinsically weaker E3 ligase in autoubiquitylation and ubiquitylation of p53 in vitro but is capable of ubiquitylating a distinct set of lysine residues of p53 as well as utilizing a smaller subset of E2 ligases. Using structure-guided mutagenesis and biophysical studies, we mapped the interaction surface between UBE2D2 and the RING domains of Pirh2 and Mdm2, showing that the mode of interaction is conserved with that of other known E2-E3 complexes but that Mdm2 has a more extended E2 interaction surface than Pirh2. The implications of these observations are discussed below.
The RING domains of Pirh2 and Mdm2 differ in their oligomeric states, the surface electrostatic charge distribution, and the zinc coordination schemes. Therefore, it is not surprising that there are differences in their interactions with the primary RING domain partners, the E2 enzymes. Like cIAPs (44), BRCA1-BARD (46), and Ring1b-Bmi1 (47), Mdm2 requires dimerization for ubiquitylation activity. However, Pirh2 and many other RING domain proteins, including c-Cbl (45), PML (48), and CNOT4 (43), facilitate E3 ligase function in a monomeric form. Thus, dimerization is not an essential feature of the RING domain type E3 ligases. Nevertheless, dimerization appears to be directly contributing to Mdm2 ubiquitylation activity by maintaining the proper folding of its RING domain. This is supported by the fact that the Mdm2 mutant lacking the dimerization sequence (Mdm2 ΔC) showed defective protein folding and inability to catalyze ubiquitylation. In addition, Mdm2 preferentially dimerizes with MdmX in the cell (17), and in vivo data suggest that dimerization of their RING domains inhibits Mdm2 autoubiquitylation, which exempts Mdm2 from proteasomal degradation and indirectly enhances Mdm2-mediated down-regulation of p53 (18, 49–51). Similar examples are found for the heterodimer of BRCA1-BARD and Ring1b-Bmi1, where BARD and Bmi1 RING domains have no detectable E3 ligase activity. The dimer formation of BRCA1 and Ring1b with their respective nonactive partner serves to stabilize the RING domain structure as well as inhibit E3 autoubiquitylation (52, 53). Another role for Mdm2 dimerization suggested by this study and others is that dimerization may create a more extended E2 interaction surface. Consistent with this model, the mutagenesis studies at the C terminus of Mdm2 attenuated Mdm2 E3 ligase activity without disrupting Mdm2 dimerization, suggesting that the C terminus of Mdm2 directly aids in catalysis (25, 26, 34).
All E2s contain a conserved core domain (UBC) and a catalytic Cys residue that accepts Ub from a ubiquitin-activating enzyme E1. There are over 30 E2s in the human genome, suggesting potential differences in their regulation of cellular processes and functional diversity in ubiquitylation. In this study, we tested a set of E2s that may interact with Pirh2 and Mdm2 to catalyze ubiquitylation. Surprisingly, in addition to the two closely related subfamilies UBE2D and UBE2E, which were utilized by both Mdm2 and Pirh2 for p53 polyubiquitylation, Mdm2 also catalyzed p53 monoubiquitylation in the presence of UBE2A/B, UBE2N, and UBE2Q/Q-like. The effects of UBE2A/B in p53 monoubiquitylation have been reported and are implicated in regulating DNA damage response, p53 activity, and localization (54). UBE2N also has been shown to promote Lys63-linked p53 ubiquitylation, which attenuates Mdm2-mediated p53 polyubiquitylation when UBE2N is overexpressed in the cell (55, 56). The function of UBE2Q proteins is not clear, and their role in Mdm2-mediated p53 ubiquitylation therefore warrants further investigation.
In addition to recruitment of the E2 enzyme by the RING domains of Pirh2 and Mdm2, our data provide insight into the differential in vitro activity of the full-length proteins. The p53 interaction domains of these E3s are very different, in terms of their structure arrangements and p53 binding affinities. Mdm2 binds to the N terminus of p53 in the nanomolar range (57), whereas the Pirh2-p53 binding constant is at least an order of magnitude less in vitro (21). This is likely to contribute to the lower p53 E3 ligase activity of Pirh2. Perhaps more interesting, however, are the differences in ubiquitylated p53 products generated by Pirh2 and Mdm2. Mdm2 preferentially ubiquitylates the lysine residues at the C terminus of p53 (36, 37). However, in addition to C-terminal residues, Pirh2 and Mdm2 can also ubiquitylate lysine residues located in the p53 core DNA binding domain and tetramerization domain. The preferential ubiquitylation of Lys164 by Pirh2 is of particular interest because this residue has recently been identified to be a specific acetylation site for p300/CBP and essential for p53-mediated cell growth arrest and apoptosis (58). It is consistent with the role of Pirh2 in regulation of p53 transcription (6). Another site, Lys320, was exclusively ubiquitylated by Mdm2. This may be explained by the distinctly different interaction modes of Mdm2 and Pirh2 with p53. Although both E3 ligases interact with p53 in a bivalent manner, Mdm2 primarily binds to the p53 N terminus with its N-terminal domain that subsequently induces a secondary interaction between the p53 core domain and its central acidic region (59), whereas Pirh2 uses its N-terminal domain for binding to the p53 core domain and its C-terminal domain to bind to the p53 tetramerization domain. Presumably, the different geometry of the Pirh2 and Mdm2 interactions causes differential presentation of lysine residues in the core and linker regions of p53 for ubiquitylation.
The apparent weaker intrinsic activity of Pirh2 is in line with a secondary role of Pirh2 in modulating p53 protein levels in relation to that of Mdm2 (6). Substantial heterogeneity has been observed in Pirh2-mediated p53 responses in different cell lines, depending on tissue type and Pirh2 levels, whereas Mdm2 has been shown to be the primary E3 ligase for p53 in all systems (6, 60). In conclusion, our studies of the biochemical properties of the Mdm2 and Pirh2 RING domains provide insight into the molecular mechanisms of p53 ubiquitylation by its E3 ligases and provide a structural basis for future biochemical and cellular investigation of this system.
Supplementary Material
Acknowledgments
We thank the staff in the laboratory of Dr. Sam Benchimol, especially Weili Ma, Dr. Keith Wheaton, and Dr. Yinping Lin, for technical help and insightful discussions. We thank Irene Wang and Dr. Masoud Vedadi for help with biophysical characterization.
This work was supported by funding from the Canadian Cancer Society, the National Cancer Institute of Canada (to C. H. A.), the Canada Research Chairs program (to C. H. A.), the Ontario Ministry of Health and Long Term Care (to B. R.), the Leukemia and Lymphoma Society of Canada (a postdoctoral fellowship) (to Y. S.), and the Banting Research Foundation (to Y. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Figs. 1–6.
- Ub
- ubiquitin
- ESI
- electrospray ionization.
REFERENCES
- 1. Levine A. J. (1997) Cell 88, 323–331 [DOI] [PubMed] [Google Scholar]
- 2. Chipuk J. E., Green D. R. (2003) J. Clin. Immunol. 23, 355–361 [DOI] [PubMed] [Google Scholar]
- 3. Moll U. M., Wolff S., Speidel D., Deppert W. (2005) Curr. Opin. Cell Biol. 17, 631–636 [DOI] [PubMed] [Google Scholar]
- 4. Le Cam L., Linares L. K., Paul C., Julien E., Lacroix M., Hatchi E., Triboulet R., Bossis G., Shmueli A., Rodriguez M. S., Coux O., Sardet C. (2006) Cell 127, 775–788 [DOI] [PubMed] [Google Scholar]
- 5. Honda R., Tanaka H., Yasuda H. (1997) FEBS Lett. 420, 25–27 [DOI] [PubMed] [Google Scholar]
- 6. Leng R. P., Lin Y., Ma W., Wu H., Lemmers B., Chung S., Parant J. M., Lozano G., Hakem R., Benchimol S. (2003) Cell 112, 779–791 [DOI] [PubMed] [Google Scholar]
- 7. Dornan D., Wertz I., Shimizu H., Arnott D., Frantz G. D., Dowd P., O'Rourke K., Koeppen H., Dixit V. M. (2004) Nature 429, 86–92 [DOI] [PubMed] [Google Scholar]
- 8. Chen D., Kon N., Li M., Zhang W., Qin J., Gu W. (2005) Cell 121, 1071–1083 [DOI] [PubMed] [Google Scholar]
- 9. Scheffner M., Huibregtse J. M., Vierstra R. D., Howley P. M. (1993) Cell 75, 495–505 [DOI] [PubMed] [Google Scholar]
- 10. Yamasaki S., Yagishita N., Sasaki T., Nakazawa M., Kato Y., Yamadera T., Bae E., Toriyama S., Ikeda R., Zhang L., Fujitani K., Yoo E., Tsuchimochi K., Ohta T., Araya N., Fujita H., Aratani S., Eguchi K., Komiya S., Maruyama I., Higashi N., Sato M., Senoo H., Ochi T., Yokoyama S., Amano T., Kim J., Gay S., Fukamizu A., Nishioka K., Tanaka K., Nakajima T. (2007) EMBO J. 26, 113–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Toledo F., Wahl G. M. (2006) Nat. Rev. Cancer 6, 909–923 [DOI] [PubMed] [Google Scholar]
- 12. Montes de Oca Luna R., Wagner D. S., Lozano G. (1995) Nature 378, 203–206 [DOI] [PubMed] [Google Scholar]
- 13. Jones S. N., Roe A. E., Donehower L. A., Bradley A. (1995) Nature 378, 206–208 [DOI] [PubMed] [Google Scholar]
- 14. Parant J., Chavez-Reyes A., Little N. A., Yan W., Reinke V., Jochemsen A. G., Lozano G. (2001) Nat. Genet. 29, 92–95 [DOI] [PubMed] [Google Scholar]
- 15. Marine J. C., Francoz S., Maetens M., Wahl G., Toledo F., Lozano G. (2006) Cell Death Differ. 13, 927–934 [DOI] [PubMed] [Google Scholar]
- 16. Francoz S., Froment P., Bogaerts S., De Clercq S., Maetens M., Doumont G., Bellefroid E., Marine J. C. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 3232–3237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gu J., Kawai H., Nie L., Kitao H., Wiederschain D., Jochemsen A. G., Parant J., Lozano G., Yuan Z. M. (2002) J. Biol. Chem. 277, 19251–19254 [DOI] [PubMed] [Google Scholar]
- 18. Stad R., Little N. A., Xirodimas D. P., Frenk R., van der Eb A. J., Lane D. P., Saville M. K., Jochemsen A. G. (2001) EMBO Rep. 2, 1029–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Duan W., Gao L., Druhan L. J., Zhu W. G., Morrison C., Otterson G. A., Villalona-Calero M. A. (2004) J. Natl. Cancer Inst. 96, 1718–1721 [DOI] [PubMed] [Google Scholar]
- 20. Logan I. R., Gaughan L., McCracken S. R., Sapountzi V., Leung H. Y., Robson C. N. (2006) Mol. Cell. Biol. 26, 6502–6510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sheng Y., Laister R. C., Lemak A., Wu B., Tai E., Duan S., Lukin J., Sunnerhagen M., Srisailam S., Karra M., Benchimol S., Arrowsmith C. H. (2008) Nat. Struct. Mol. Biol. 15, 1334–1342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tovar C., Rosinski J., Filipovic Z., Higgins B., Kolinsky K., Hilton H., Zhao X., Vu B. T., Qing W., Packman K., Myklebost O., Heimbrook D. C., Vassilev L. T. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 1888–1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Vassilev L. T., Vu B. T., Graves B., Carvajal D., Podlaski F., Filipovic Z., Kong N., Kammlott U., Lukacs C., Klein C., Fotouhi N., Liu E. A. (2004) Science 303, 844–848 [DOI] [PubMed] [Google Scholar]
- 24. Kostic M., Matt T., Martinez-Yamout M. A., Dyson H. J., Wright P. E. (2006) J. Mol. Biol. 363, 433–450 [DOI] [PubMed] [Google Scholar]
- 25. Linke K., Mace P. D., Smith C. A., Vaux D. L., Silke J., Day C. L. (2008) Cell Death Differ. 15, 841–848 [DOI] [PubMed] [Google Scholar]
- 26. Poyurovsky M. V., Priest C., Kentsis A., Borden K. L., Pan Z. Q., Pavletich N., Prives C. (2007) EMBO J. 26, 90–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 28. Bartels C. X., Billeter M., Cuntert P., Wuthrich K. (1995) J. Biomol. NMR 5, 1–10 [DOI] [PubMed] [Google Scholar]
- 29. Banks L., Matlashewski G., Crawford L. (1986) Eur. J. Biochem. 159, 529–534 [DOI] [PubMed] [Google Scholar]
- 30. Makhnevych T., Sydorskyy Y., Xin X., Srikumar T., Vizeacoumar F. J., Jeram S. M., Li Z., Bahr S., Andrews B. J., Boone C., Raught B. (2009) Mol. Cell 33, 124–135 [DOI] [PubMed] [Google Scholar]
- 31. Pedrioli P. G., Eng J. K., Hubley R., Vogelzang M., Deutsch E. W., Raught B., Pratt B., Nilsson E., Angeletti R. H., Apweiler R., Cheung K., Costello C. E., Hermjakob H., Huang S., Julian R. K., Kapp E., McComb M. E., Oliver S. G., Omenn G., Paton N. W., Simpson R., Smith R., Taylor C. F., Zhu W., Aebersold R. (2004) Nat. Biotechnol. 22, 1459–1466 [DOI] [PubMed] [Google Scholar]
- 32. Craig R., Beavis R. C. (2004) Bioinformatics 20, 1466–1467 [DOI] [PubMed] [Google Scholar]
- 33. Holm L., Sander C. (1995) Trends Biochem. Sci. 20, 478–480 [DOI] [PubMed] [Google Scholar]
- 34. Uldrijan S., Pannekoek W. J., Vousden K. H. (2007) EMBO J. 26, 102–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lallemand-Breitenbach V., Jeanne M., Benhenda S., Nasr R., Lei M., Peres L., Zhou J., Zhu J., Raught B., de Thé H. (2008) Nat. Cell Biol. 10, 547–555 [DOI] [PubMed] [Google Scholar]
- 36. Lohrum M. A., Woods D. B., Ludwig R. L., Bálint E., Vousden K. H. (2001) Mol. Cell. Biol. 21, 8521–8532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rodriguez M. S., Desterro J. M., Lain S., Lane D. P., Hay R. T. (2000) Mol. Cell. Biol. 20, 8458–8467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Christensen D. E., Brzovic P. S., Klevit R. E. (2007) Nat. Struct. Mol. Biol. 14, 941–948 [DOI] [PubMed] [Google Scholar]
- 39. Weissman A. M. (2001) Nat. Rev. Mol. Cell Biol. 2, 169–178 [DOI] [PubMed] [Google Scholar]
- 40. Fang S., Jensen J. P., Ludwig R. L., Vousden K. H., Weissman A. M. (2000) J. Biol. Chem. 275, 8945–8951 [DOI] [PubMed] [Google Scholar]
- 41. Dang J., Kuo M. L., Eischen C. M., Stepanova L., Sherr C. J., Roussel M. F. (2002) Cancer Res. 62, 1222–1230 [PubMed] [Google Scholar]
- 42. Houben K., Dominguez C., van Schaik F. M., Timmers H. T., Bonvin A. M., Boelens R. (2004) J. Mol. Biol. 344, 513–526 [DOI] [PubMed] [Google Scholar]
- 43. Dominguez C., Bonvin A. M., Winkler G. S., van Schaik F. M., Timmers H. T., Boelens R. (2004) Structure 12, 633–644 [DOI] [PubMed] [Google Scholar]
- 44. Mace P. D., Linke K., Feltham R., Schumacher F. R., Smith C. A., Vaux D. L., Silke J., Day C. L. (2008) J. Biol. Chem. 283, 31633–31640 [DOI] [PubMed] [Google Scholar]
- 45. Zheng N., Wang P., Jeffrey P. D., Pavletich N. P. (2000) Cell 102, 533–539 [DOI] [PubMed] [Google Scholar]
- 46. Brzovic P. S., Meza J. E., King M. C., Klevit R. E. (2001) J. Biol. Chem. 276, 41399–41406 [DOI] [PubMed] [Google Scholar]
- 47. Buchwald G., van der Stoop P., Weichenrieder O., Perrakis A., van Lohuizen M., Sixma T. K. (2006) EMBO J. 25, 2465–2474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Borden K. L., Boddy M. N., Lally J., O'Reilly N. J., Martin S., Howe K., Solomon E., Freemont P. S. (1995) EMBO J. 14, 1532–1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Marine J. C., Dyer M. A., Jochemsen A. G. (2007) J. Cell Sci. 120, 371–378 [DOI] [PubMed] [Google Scholar]
- 50. Jackson M. W., Berberich S. J. (2000) Mol. Cell. Biol. 20, 1001–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Marine J. C., Jochemsen A. G. (2004) Cell Cycle 3, 900–904 [PubMed] [Google Scholar]
- 52. Li Z., Cao R., Wang M., Myers M. P., Zhang Y., Xu R. M. (2006) J. Biol. Chem. 281, 20643–20649 [DOI] [PubMed] [Google Scholar]
- 53. Brzovic P. S., Rajagopal P., Hoyt D. W., King M. C., Klevit R. E. (2001) Nat. Struct. Biol. 8, 833–837 [DOI] [PubMed] [Google Scholar]
- 54. Lyakhovich A., Shekhar M. P. (2003) Mol. Cell. Biol. 23, 2463–2475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Laine A., Topisirovic I., Zhai D., Reed J. C., Borden K. L., Ronai Z. (2006) Mol. Cell. Biol. 26, 8901–8913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Topisirovic I., Gutierrez G. J., Chen M., Appella E., Borden K. L., Ronai Z. A. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 12676–12681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kussie P. H., Gorina S., Marechal V., Elenbaas B., Moreau J., Levine A. J., Pavletich N. P. (1996) Science 274, 948–953 [DOI] [PubMed] [Google Scholar]
- 58. Tang Y., Zhao W., Chen Y., Zhao Y., Gu W. (2008) Cell 133, 612–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ma J., Martin J. D., Zhang H., Auger K. R., Ho T. F., Kirkpatrick R. B., Grooms M. H., Johanson K. O., Tummino P. J., Copeland R. A., Lai Z. (2006) Biochemistry 45, 9238–9245 [DOI] [PubMed] [Google Scholar]
- 60. Duan W., Gao L., Wu X., Zhang Y., Otterson G. A., Villalona-Calero M. A. (2006) Exp. Cell Res. 312, 3370–3378 [DOI] [PubMed] [Google Scholar]
- 61. DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific LLC, San Carlos, CA [Google Scholar]
- 62. Koradi R., Billeter M., Wuthrich K. (1996) J. Mol. Graph. 14, 51–55, 29–32 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.