Abstract
Recent studies have shown that inositol 1,4,5-trisphosphate 3-kinase isoform B (IP3KB) possesses important roles in the development of immune cells. IP3KB can be targeted to multiple cellular compartments, among them nuclear localization and binding in close proximity to the plasma membrane. The B isoform is the only IP3K that is almost ubiquitously expressed in mammalian cells. Detailed mechanisms of its targeting regulation will be important in understanding the role of Ins(1,4,5)P3 phosphorylation on subcellular calcium signaling and compartment-specific initiation of pathways leading to regulatory active higher phosphorylated inositol phosphates. Here, we identified an exportin 1-dependent nuclear export signal (134LQRELQNVQV) and characterized the amino acids responsible for nuclear localization of IP3KB (129RKLR). These two targeting domains regulate the amount of nuclear IP3KB in cells. We also demonstrated that the localization of IP3KB at the plasma membrane is due to its binding to cortical actin structures. Intriguingly, all three of these targeting activities reside in one small polypeptide segment (amino acids 104–165), which acts as a multitargeting domain (MTD). Finally, a hitherto unknown subnuclear localization of IP3KB could be demonstrated in rapidly growing H1299 cells. IP3KB is specifically enriched at nuclear invaginations extending perpendicular between the apical and basal surface of the nucleus of these flat cells. Such nuclear invaginations are known to be involved in Ins(1,4,5)P3-mediated Ca2+ signaling of the nucleus. Our findings indicate that IP3KB not only regulates cytoplasmic Ca2+ signals by phosphorylation of subplasmalemmal and cytoplasmic Ins(1,4,5)P3 but may also be involved in modulating nuclear Ca2+ signals generated from these nuclear envelope invaginations.
Keywords: Actin, Calcium Imaging, Inositol Phosphates, Nuclear Transport, Signal Transduction, Leptomycin B, Nuclear Export Signal, Nuclear Invagination, Nuclear Localization Signal
Introduction
It is well established that the highly inositol 1,4,5-trisphosphate (Ins(1,4,5)P3)2-specific IP3Ks remove the Ca2+-releasing second messenger Ins(1,4,5)P3 by phosphorylating it to Ins(1,3,4,5)P4 (1) supporting the formation of higher phosphorylated inositols in the cell (2). So far, three isoforms of IP3K, termed A (3), B (4), and C (5), have been identified in mammals. Full-length cDNAs encoding IP3K isoform B (IP3KB) were cloned from man (6) and rat (7). IP3KB was mapped to the telomeric end of human chromosome 1 (8, 9) and identified as a candidate gene in the pathogenesis of multiple sclerosis (10), Alzheimer disease (11), and malignant melanoma (12). But most importantly, IP3KB is a promising new target in immune disorders (13). Transgenic disruption of the IP3KB gene in mice causes a significant decrease in Ins(1,3,4,5)P4 levels in thymocytes leading to a block of positive selection and consequently to a severe T cell deficiency (14). Recent studies described additional functions of IP3KB in selection and survival of B cells (15, 16), signaling of neutrophils (17), and modulation of myelopoiesis (18). In mammalian cells, multiple intracellular localizations of IP3KB, namely to the plasma membrane, cytoskeleton, and endoplasmic reticulum, have been observed (19). Most likely, these intracellular targeting specificities of IP3KB are of regulatory importance for all processes mediated by IP3KB and its product Ins(1,3,4,5)P4. Therefore, the molecular basis of these targeting activities has been studied intensively, revealing an actin binding and nuclear targeting domain (7) as well as an endoplasmic reticulum membrane anchoring domain (20).
Our present study characterizes in detail the action of the multitargeting domain (amino acids 104–165) in the N-terminal region of the kinase that mediates both nucleo-cytoplasmic shuttling and actin targeting of IP3KB. In addition, we show here a hitherto unknown localization of IP3KB at nuclear invaginations extending deeply into and (in part) passing completely through the nucleus.
EXPERIMENTAL PROCEDURES
Materials
Cloning of the following EGFP fusion protein expression vectors is described elsewhere: Sp100 (21), coilin (22), Sam68 (23), Cbx4 (24, 25), Bmi-1 (26), Sc35 (27).
Cloning and Mutagenesis of EGFP Fusion Gene Expression Vectors
Cloning of full-length HsIP3KB cDNA and its subcloning into EGFP fusion protein expression vector pEGFP-N1 (Clontech) (generating HsIP3KB/EGFP) are described elsewhere (7). The cDNA of HsIP3KB was amplified using standard PCR techniques to create EGFP/HsIP3KB (primer pair: 5′-CAAGCTTCGATGGCTGTGTACTGCTATGCG-3′ and 5′-TCCGCGGTCAGGCGAGTGGGGCATCCTG-3′). The PCR primer introduced restriction sites (HindIII and SacII) were used for subcloning into pEGFP-C1. Deletion and substitution mutants (ΔNES, mNES, mNLS-1/2/3/4/5, mFEA-1/2, ΔMTD) were created by modified QuikChange site-directed mutagenesis (28) using appropriate primer pairs and HsIP3KB/EGFP as template. The isolated NES of HsIP3KB was cloned by oligonucleotide ligation. The primer pair 5′-TCGAGATGCGGATCTTGCAGCGCGAGTTGCAGAACGTGCAGGTGAACCAGCTGCA-3′ and 5′-GCTGGTTCACCTGCACGTTCTGCAACTCGCGCTGCAAGATCCGCATC-3′ was annealed and then ligated into pEGFP-N1 (digested with XhoI and PstI) to create NES/EGFP. For creation of EGFP/NES, the annealed primer pair 5′-CCGGATCTTGCAGCGCGAGTTGCAGAACGTGCAGGTGAACCAGTAAG-3′ and 5′-GATCCTTACTGGTTCACCTGCACGTTCTGCAACTCGCGCTGCAAGATCCGGG-TAC-3′ was ligated into pEGFP-C1 (digested with KpnI and BamHI). The same cloning technique was used to create NLS/EGFP (5′-TCGAGATGGAGGCCAAGAGGAAGCTGCGGATCTTGCCGC-3′ and 5′-GGCAAGATCCGCAGCTTCCTCTTGGCCTCCATC-3′; pEGFP-N1 digested with XhoI and SacII). Additionally, the cDNA of the multitargeting domain was amplified using standard PCR techniques to create MTD/EGFP (primer pair, 5′-CTCGAGATGGGTCGCCTGCGAGGGGACCGG-3′ and 5′-CCGCGGGCGGGGCGCTTGAATGGCGGAGCT-3′). The PCR primer introduced restriction sites (XhoI and SacII) were used for subcloning into pEGFP-N1. All constructs and further descriptions are summarized in Table 1.
TABLE 1.
Fusion genes and fusion gene derivates
Full-length cDNA and different fragments of HsIP3KB were cloned into the EGFP fusion gene expression vector pEGFP-N1. Subsequently, the full-length cDNA was altered by deletion and substitution mutagenesis.
| Construct | Description |
|---|---|
| HsIP3KB/EGFP | Full-length/WT |
| ΔNES | Δ 138–143 |
| mNES | 138LQNVQV→AQNAQA |
| NES/EGFP | 132–145 |
| EGFP/NES | 132–145 |
| mNLS-1 | 128KRKLR→QRKLR |
| mNLS-2 | 128KRKLR→KQKLR |
| mNLS-3 | 128KRKLR→KRQLR |
| mNLS-4 | 128KRKLR→KRKLQ |
| mNLS-5 | 128KRKLR→QQQLR |
| NLS/EGFP | 126–134 (126EAKRKLRIL) |
| mFEA-1 | 154IQAQS→PQAQP |
| mFEA-2 | 150FE→QA |
| ΔMTD | Δ103–165 |
| MTD/EGFP | 104–165 |
Cell Culture
All cell lines were cultured in a humidified atmosphere at 37 °C in the presence of 5% CO2. DMEM from Invitrogen was used for Mewo, H1299, HeLa, and L3.6pC cell lines. RPMI medium (Invitrogen) was used for MCF7, T47D, and IMR-32 cell line. Media were supplemented with 10% fetal bovine serum.
Immunofluorescence, Transient Gene Expression, and Fluorescence Microscopy
For immunofluorescence analysis, cells were washed with PBS, fixed with PBS containing 3% paraformaldehyde (for 10 min at 37 °C), permeabilized with methanol, prechilled to −20 °C (for 6 min at room temperature), blocked in PBS containing 5% BSA (for 30 min at room temperature), incubated with the primary antibody (for 2 h at room temperature), and then incubated with the secondary antibody (for 1 h at room temperature). Every step was followed by washing with PBS. For detection of IP3KB the primary antibody H00003707-M01 (Abnova) was used. The secondary α-mouse antibody was labeled with Alexa Fluor 568. For detection of Ins(1,4,5)P3 receptor type I, II, and III, the primary antibodies SC-6093, SC-7278, and SC-7277 (Santa Cruz Biotechnology) were used. The secondary α-goat antibody was labeled with Alexa Fluor 488. For detection of fibrillarin, the primary antibody ab5821 (Abcam) was used. The secondary α-rabbit antibody was labeled with Alexa Fluor 488. DAPI staining and examination of the intracellular localization by epifluorescence microscopy were previously described by Nalaskowski et al. (29). Transient transfection of eukaryotic cells was performed using the Metafectene reagent according to the manufacturer's instructions (Biontex Laboratories, Munich, Germany). Leptomycin B was purchased from Sigma and Alexis Biochemicals and added to the complete medium to a final concentration of 11 ng/ml (20 nm). TRITC-labeled phalloidin was purchased from Sigma, and staining was performed as described in Ref. 7.
Computational Optical Sectioning Microscopy and Three-dimensional Reconstruction
An Improvision imaging system (PerkinElmer Life Science) built around a fluorescence microscope (Leica DM IRE2) at 100-fold magnification (Leica objective type HCX APO ×100/1.3 oil U-V-I; numerical aperture, 1.3), was used. Illumination (DAPI, 355 nm; EGFP, 482 nm; FITC, 494 nm; TRITC, 546 nm) was carried out using a monochromator system (Polychromator IV, TILL Photonics). Images were taken at room temperature with a grayscale CCD camera (type C4742–95-12ER; Hamamatsu; operated in 12-bit mode; binning 1) on 100 consecutive horizontal planes (z), at a step width of 0.1 μm. Raw data images were stored on a hard disk. To obtain images of optical sections, mathematical deconvolution based on the point-spread function was carried out on up to 80 images of the z-stack using the volume deconvolution algorithm (10 neighbors) (Openlab software, version 3.0.9; PerkinElmer Life Science). The virtual pinhole for removal of stray light was set between 56 and 67%. Three-dimensional models from ∼65 consecutive images were constructed using NIH ImageJ 1.42q and ImageJ three-dimensional viewer 1.5.2 fcs software.
Ca2+ Imaging
For transfection, HeLa cells were seeded at low density on eight-well microslides (IBIDI, Martinsried, Germany). Transient transfection was performed as described above. Approximately 24 h after transfection, the Ca2+ indicator Fura-2/AM (Calbiochem) was added to the medium at a final concentration of 4 μm. After 30 min (37 °C), the cells were washed twice, and medium was replaced with buffer (140 mm NaCl, 5 mm KCl, 1 mm MgSO4, 1 mm CaCl2, 1 mm Na2HPO4, 5.5 mm glucose, and 20 mm Hepes, pH 7.4). Imaging of cells was performed using a Leica DM-IRBE fluorescence microscope with a 40× objective (1.3 numerical aperture). Alternating excitation at 340 and 380 nm was achieved using a monochromator system (Polychrome II; TILL Photonics, Graefelfing, Germany). Two images were acquired every 2–3 s with a grayscale CCD camera (type C4742-95-12NRB; Hamamatsu, Enfield, UK) in eight bit-mode. Raw data were stored to a hard drive, and ratio images (340:380 nm) were calculated from raw images using Openlab software (version 4.04; Perkin-Elmer Life Science). Cells positive for EGFP were selected as regions of interest, and the mean ratio of the regions of interest over time was used for further analysis.
Quantitation of Intracellular Distribution
At least three independent experiments were performed with each combination of construct, cell line, and leptomycin B (LMB) treatment. In each experiment, a minimum of 100 cells were randomly selected and examined. To determine the nuclear/cytoplasmic ratio, EGFP fluorescence intensities of six ROIs in both the nuclear and cytoplasmic areas in each cell were averaged to calculate the ratio of nuclear over cytoplasmic intensity using the ImageJ program (W. Rasband). To determine the plasma membrane/cytoplasmic ratio, EGFP fluorescence intensities of six ROIs both over the plasma membrane and in cytoplasmic areas (near the plasma membrane) in each cell were averaged to calculate the ratio of plasma membrane over cytoplasmic intensity. In selected experiments, background fluorescence was determined by selecting ROIs in areas without cells. Background correction was not generally performed because ratios were nearly unchanged after subtraction of background fluorescence. For evaluation of data, unpaired Student's t test and one-way analysis of variance were performed using GraphPad InStat (version 3.06; GraphPad Software). A value of p < 0.05 was considered statistically significant. Mean values and S.E. are given.
Western Blotting of IP3KB
Eucaryotic cells were harvested and lysed in M-PER mammalian protein extraction reagent from Pierce (1× Complete protease inhibitor mixture from Roche Applied Science, 100 mm PMSF and 200 kilo international units of trasylol were added before use). Equal amounts of cell lysates were separated by SDS-PAGE and transferred onto a nitrocellulose membrane using standard Western blotting techniques. For detection of IP3KB primary antibody, H00003707-M01 from Abnova was used. The secondary α-mouse antibody was conjugated with HRP.
RESULTS
Active Nuclear Export of IP3KB Is Mediated by a Leucine-rich Nuclear Export Signal
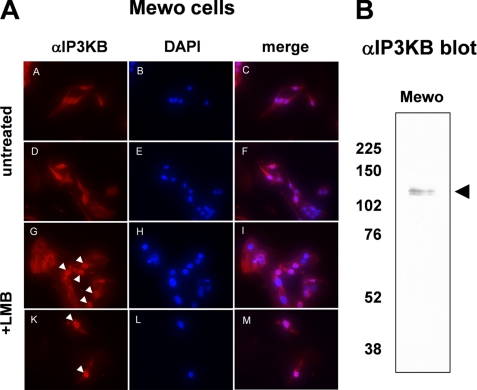
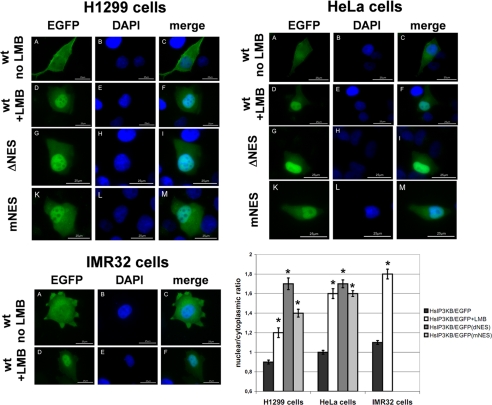
The C isoform of human and rat IP3K is actively imported into and actively exported out of the nucleus of mammalian cells (29, 30). Its nuclear export is mediated by leucine-rich nuclear export signals (NES). To investigate whether NES are also functional in nuclear export of IP3K isoform B, we investigated its nuclear export in Mewo cells. IP3KB was detected by immunofluorescence techniques using IP3KB-specific antibody H00003707-M01 (Abnova) in untreated control cells and in cells incubated with 20 nm LMB for 5 h. LMB is a widely used antifungal antibiotic that covalently modifies exportin 1 (31), thereby inhibiting nuclear export. In control cells, IP3KB was evenly distributed between nucleus and cytoplasm (Fig. 1A, untreated). In LMB-treated Mewo cells, a clear accumulation of IP3KB was observed in most of the nuclei (Fig. 1A, +LMB). Antibody specificity was confirmed by preincubation with a recombinantly expressed IP3KB fragment comprising the antibody binding domain. After this treatment, no IP3KB was detected in Mewo nuclei (data not shown). In additional Western blotting experiments using the same antibody, two protein bands at ∼125 kDa were detected (Fig. 1B). These data confirm the observations of Erneux et al. (6) who also detected IP3KB at 125 kDa, despite its calculated apparent molecular mass of 102 kDa. In subsequent studies, we used EGFP fusion proteins of IP3KB to identify the motif responsible for its LMB-sensitive nuclear export. In initial experiments, IP3KB fusion proteins with either C-terminal (HsIP3KB/EGFP) or N-terminal EGFP tag (EGFP/HsIP3KB) were transiently expressed in H1299 and HeLa cells. Both fusion proteins (Fig. 2 and data not shown) showed a very similar nucleocytoplasmic distribution of EGFP fluorescence (H1299: 0.9 ± 0.02 versus 1.0 ± 0.03; HeLa: 1.0 ± 0.02 versus 0.9 ± 0.03). Therefore, only fusion protein HsIP3KB/EGFP and its mutants were used in further experiments. Treatment of H1299 and HeLa cells with LMB for 2, 6, and 24 h always led to a significant (p < 0.01) nuclear accumulation of HsIP3KB/EGFP (data not shown). Because incubation with LMB for >6 h led to massive cell death, fluorescence was imaged after 6 h of LMB incubation in all subsequent experiments. A significant nuclear accumulation was detected in H1299 (0.9 ± 0.02 versus 1.2 ± 0.05), HeLa (1.0 ± 0.02 versus 1.6 ± 0.05), and IMR32 (1.1 ± 0.02 versus 1.8 ± 0.05) cells (Fig. 2). To identify the motif responsible for nuclear export, the amino acid sequence was searched using the relaxed consensus motif of leucine-rich NES ((LIVFM)-X2,3-(LIVFM)-X2,3-(LIVFM)-X-(LIVFM)) defined by La Cour and others (32, 33), and each predicted sequence motif obeying this consensus was deleted from the HsIP3KB/EGFP fusion protein. Only one deletion (ΔNES, Δ138–143) caused a significant accumulation in the nucleus in H1299 (1.7 ± 0.06) and HeLa (1.7 ± 0.04) cells. This putative NES was further investigated by functional inactivation using alanine substitution mutagenesis (mNES, 138LQNVQV→AQNAQA). Inactivation of the NES produced a significant nuclear accumulation of the fusion protein in H1299 (1.4 ± 0.04) and HeLa (1.6 ± 0.03) cells (Fig. 2). In conclusion, nuclear export of IP3KB is mediated by a single leucine-rich NES (134LQRELQNVQV) in the N-terminal region of the protein.
FIGURE 1.
Endogenous expression of IP3KB in mammalian cells. A, Mewo cells were either treated with 20 nm LMB for 6 h or mock treated. Then, intracellular localization of endogenous IP3KB was determined by immunofluorescence techniques using antibody H00003707-M01 (panels A, D, G, and K). Nuclei were stained with DAPI (panels B, E, H, and L), and overlays of the digitized images were created (panels C, F, I, and M). Nuclei showing a strong accumulation of IP3KB are marked by white arrowheads. B, cell lysates from Mewo cells were separated on SDS-polyacrylamide gels. Antibody H00003707-M01 was used to detect IP3KB using Western blot analysis. The black arrowhead indicates the expected protein band of endogenous IP3KB. The molecular mass of protein markers is indicated on the left in kDa.
FIGURE 2.
Exportin 1-dependent nuclear export of IP3KB. Different EGFP fusion genes were transiently expressed in H1299, HeLa, and IMR32 cells. After 24 h of expression, cells were fixed and nuclei stained with DAPI. EGFP fusion proteins (panels A, D, G, and K) and DAPI (panels B, E, H, and L) were visualized by fluorescence microscopy, and an overlay of the digitized images was created (panels C, F, I, and M). Cells expressing EGFP fusion proteins of full-length HsIP3KB (WT) were either untreated (no LMB) or treated for 6 h with 20 nm LMB (+LMB). In additional experiments, mutated forms of HsIP3KB lacking the NES (ΔNES) or possessing a functionally inactive NES (mNES) were expressed. In three independent experiments, at least 100 cells were analyzed for nuclear/cytoplasmic ratio of EGFP fluorescence (see “Experimental Procedures”). Results from one representative experiment are shown (mean ± S.E.) in the bar graph. For significance analysis, one-way analysis of variance and unpaired t test were used (*, p < 0.01; not significant, p > 0.05). Values were always compared with HsIP3KB/EGFP. For detailed information about the constructs, see Table 1.
Active Nuclear Import of IP3KB Is Mediated by a Noncanonical Monopartite Nuclear Localization Signal
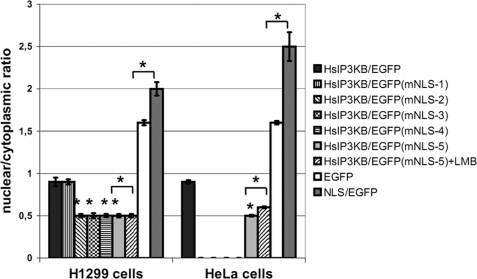
A nuclear localization signal mediating the nuclear import of IP3KB was first postulated by Brehm et al. (7) based on deletion studies. Remarkably, this sequence does not obey the canonical consensus motif of monopartite NLS (K-(KR)-X-(KR)) (34). For a more detailed characterization, single amino acids of the putative NLS (128KRKLR) were mutated in HsIP3KB/EGFP and the nuclear/cytoplasmic ratio of its intracellular distribution was investigated in H1299 and HeLa cells. Substitution of the basic residues Arg129 (mNLS-2), Lys130 (mNLS-3), or Arg132 (mNLS-4) in this motif by glutamine caused a significant (p < 0.01) decrease of nuclear accumulation of the fusion protein (Fig. 3). A triple mutant (mNLS-5, K128Q/R129Q/K130Q) showed an effect similar to that observed for the single mutants. No effect was seen by substitution of Lys128 with glutamine (mNLS-1). For example, the mNLS-5 mutation caused a significant decrease of the nuclear/cytoplasmic ratio from 0.9 ± 0.05 to 0.5 ± 0.02 in H1299 and from 0.9 ± 0.02 to 0.5 ± 0.01 in HeLa cells. Therefore, all three C-terminal basic amino acids in this motif but not Lys128 are necessary for active nuclear import of IP3KB. Cells that were expressing the mutated EGFP fusion protein mNLS-5 (inactive NLS) showed no significant (p > 0.05) change in its intracellular localization after 6 h of treatment with LMB (H1299, 0.5 ± 0.02; HeLa, 0.6 ± 0.01). Therefore, IP3KB does not possess any further active motifs mediating its nuclear import. In an additional experiment, the isolated NLS (126EAKRKLRIL) was fused to the N terminus of EGFP. This construct (NLS/EGFP) showed a significant increase of nuclear accumulation compared with EGFP alone (H1299, 2.0 ± 0.08 versus 1.6 ± 0.03; HeLa, 2.5 ± 0.17 versus 1.6 ± 0.02). Therefore, this sequence is sufficient for nuclear import of attached proteins. Furthermore, the import activity of this construct was compared with the activity of the isolated NLS of human inositol phosphate multikinase (35) revealing a similar strength (data not shown). In summary, the identified NLS (129RKLR) is necessary for the active nuclear import of human IP3KB and the isolated NLS is sufficient for active nuclear import of EGFP.
FIGURE 3.
NLS-mediated nuclear import of IP3KB. H1299 and HeLa cells were transfected with different EGFP fusion genes. After 24 h of expression, cells were fixed, and the nuclear/cytoplasmic ratio of EGFP fluorescence was determined. Constructs used encode EGFP fusion proteins of wild type HsIP3KB (WT) and forms with a mutated NLS (mNES-1/2/3/4/5). Additionally, EGFP alone (EGFP) and EGFP fused with the isolated NLS (NLS/EGFP) were expressed. In some experiments, cells were treated with 20 nm LMB for 6 h (+LMB). If not otherwise indicated, values were always compared with HsIP3KB/EGFP. For further details, see the legend to Fig. 2. For detailed information about the constructs, see Table 1.
Plasma Membrane Localization of IP3KB Is Due to Its Binding to Cortical Actin Structures
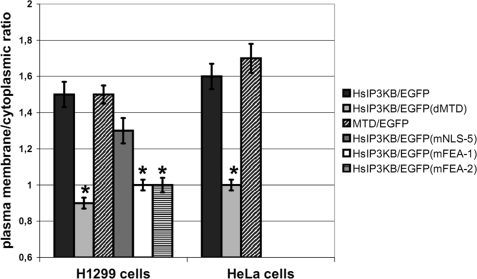
One additional aim of this study was to investigate the possibility that the previously observed plasma membrane localization of human IP3KB (19) is due to its binding to cortical actin structures via its actin binding motif (7). Determination of plasma membrane/cytoplasmic ratio of HsIP3KB/EGFP (WT) revealed a clear localization at the plasma membrane in both H1299 (1.5 ± 0.07) and HeLa (1.6 ± 0.07) cells (Fig. 4). Deletion of the actin binding motif led to complete abolishment of plasma membrane localization, translocating the mutant (ΔMTD, Δ103–165) to the cytoplasm (H1299, 0.9 ± 0.03; HeLa, 1.0 ± 0.03). The same effect was observed after substitution mutagenesis destroying a predicted α-helix (mFEA-1, 154IQAQS→PQAQP) or altering the phylogenetically highly conserved FEA sequence (mFEA-2, 150FE→QA) (prediction and phylogenetic data not shown) in the actin binding motif. Disruption of F-actin by treatment of H1299 cells with cytochalasin D (1 μm for 10 min) (36) also completely eliminated plasma membrane localization of IP3KB fused to EGFP (data not shown). An isolated fragment (amino acids 104–165) containing the actin binding motif and fused to EGFP (MTD/EGFP) showed a ratio (H1299, 1.5 ± 0.05; HeLa, 1.7 ± 0.08) similar to that of the full-length protein when fused to EGFP. In additional experiments, the actin fibers were directly identified by staining with TRITC-labeled phalloidin. HsIP3KB/EGFP and MTD/EGFP co-localize with cortical actin fibers in H1299 and HeLa cells (data not shown). In summary, we were able to show that the binding of human IP3KB to the plasma membrane is mediated by its binding to cortical actin structures.
FIGURE 4.
Plasma membrane localization of IP3KB mediated by its binding to cortical actin. Different EGFP fusion genes of HsIP3KB were transiently transfected into H1299 and HeLa cells. Cells were fixed 24 h post transfection, and EGFP was visualized by fluorescence microscopy. WT, wild type HsIP3KB; MTD, isolated multi-targeting domain; ΔMTD, multitargeting domain deletion mutant; mFEA-1/2, FEA motif substitution mutants. The plasma membrane/cytoplasmic ratio was determined as described under “Experimental Procedures.” For detailed information about the constructs, see Table 1; for further details, see the legend to Fig. 2. Values were compared with HsIP3KB/EGFP.
IP3KB Is Enriched at Nuclear Invaginations Traversing Deeply into Nucleoplasm
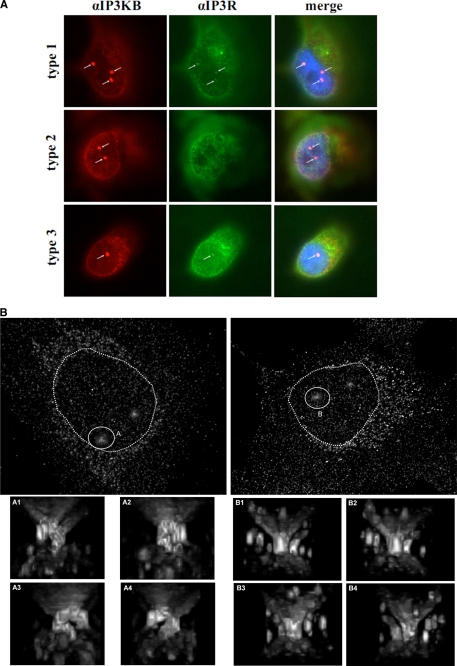
An intriguing, hitherto unknown localization of endogenous IP3KB at special nuclear substructures was detected in H1299, Mewo, MCF7, L3.6pC, and T47D cells by using immunofluorescence techniques (data not shown). In H1299 cells, at least one of these structures was observed in 78.8 ± 7.9% of all cells. More than one of these structures was observed in 26.3 ± 4.2% of all cells. In other cell lines examined, these structures were also detectable, but smaller than within H1299 cells. Overexpressed IP3KB/EGFP fusion proteins did not show this localization (data not shown). Using co-localization studies in H1299 cells, we showed that these nuclear structures are not identical with PML bodies (21), Cajal bodies (22), Sam68 nuclear bodies (23), polycomb bodies (24, 25, 26), or nuclear speckles (27) (data not shown). However, a co-localization with Ins(1,4,5)P3 receptor type I and III (Fig. 5A) was observed. Remarkably, in most of the H1299 cells (65 ± 5%) at least one of these nuclear structures is closely associated with a nucleolus visualized by fibrillarin immunofluorescence staining (data not shown). Data sets were obtained by fluorescence microscopy of αIP3KB immunofluorescence samples of H1299 cells and deconvolution. Subsequently sets consisting of 60–65 XY planes were used to construct a three-dimensional view of nuclear volumes using an ImageJ plug-in three-dimensional viewer (Fig. 5B and data not shown). These reconstructions show long, partly branching, intranuclear invaginations. These invaginations, which traverse deeply into the nucleoplasm, often are associated with nucleoli and in part reach through the whole nucleus as the nucleoplasm forms an annulus. Interestingly, all invaginations extend perpendicular to the plane of the cell substrate, between the basal and apical surface of the nucleus. No horizontally orientated invaginations are observed. Fig. 5B shows an H1299 cell with a branching invagination (termed structure A) ending in the nucleoplasm and another H1299 cell with an invagination (structure B) passing completely through the nucleus. Additional invaginations in these two cells are not marked.
FIGURE 5.
Enrichment of IP3KB at subnuclear structures (A) H1299 cells were fixed, and endogenous IP3KB (red) was detected by immunofluorescence techniques using antibody H00003707-M01. In addition, Ins(1,4,5)P3 receptor type I, II, and III (green) was detected using isoform-specific antibodies. Nuclei were stained with DAPI (blue), and an overlay of the digitized images was created. Nuclear structures showing an accumulation of IP3KB are marked by white arrows. B, endogenous IP3KB in H1299 cells was detected by immunofluorescence techniques. Then, microscopic data sets were obtained by fluorescence microscopy and subsequent deconvolution. Three-dimensional reconstructions were created using the ImageJ plug-in three-dimensional viewer. Reconstructions of two exemplary H1299 cells are shown. Exemplary nuclear invaginations are marked by white circles (A and B), and magnified images from different perspectives (A1–A4 and B1–B4) are shown below. Nuclei are marked by dotted circles.
Overexpression of IP3KB Fused to EGFP Represses Carbachol-induced Ca2+ Signals
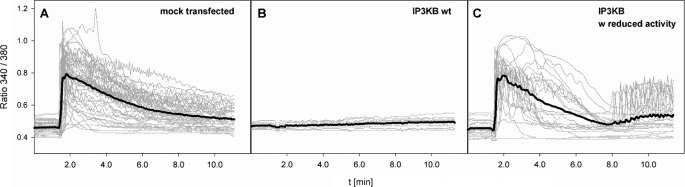
Effects of IP3K overexpression on Ca2+ signaling in eukaryotic cells have been discussed intensively (37, 38). To further investigate the impact of IP3KB, Ca2+ signals were recorded in carbachol-stimulated HeLa cells that were overexpressing IP3KB tagged with EGFP. Mock-transfected cells showed uniform transient Ca2+ signals after stimulation with 100 μm carbachol (Fig. 6A, 36 cells examined). Overexpression of wild type IP3KB completely prevented these Ca2+ signals in all examined cells (Fig. 6B, 10 cells). Interestingly, we observed this effect in all investigated cells independent of their IP3KB/EGFP expression level. Even cells with low expression of IP3KB/EGFP did not respond to carbachol stimulation. Overexpression of different mutants with altered intracellular targeting (mNES, mNLS, mFEA-1, mFEA-2; 10 or more cells; data not shown) also prevented Ca2+ signals induced by carbachol stimulation. Enzymatic activity is unaffected by these mutations (data not shown). Cells that were expressing an IP3KB mutant (D897N) with largely reduced enzymatic activity (1% or less of WT activity; data not shown) showed regular Ca2+ signals in response to carbachol stimulation (Fig. 6C, 20 cells). One-half of these cells (10/20) also showed an increase of cytoplasmic Ca2+ concentrations ∼360 s after stimulation. Oscillating signals were observed in five of these cells.
FIGURE 6.
IP3KB-mediated modulation of carbachol-induced Ca2+ signals in HeLa cells. HeLa cells were either mock transfected (A) or transfected with expression constructs for wild type (B) or mutant IP3KB with reduced enzymatic activity (C). Wild type and mutant IP3KB were fused with EGFP. Approximately 24 h post-transfection, cells were loaded with Fura-2/AM and subjected to single-cell Ca2+ imaging. 1.5 min after start of the recording, carbachol was added to the bath solution to a final concentration of 100 μm. Gray lines show single cell ratio tracings; black lines indicate the calculated means.
DISCUSSION
Nucleocytoplasmic Shuttling and Actin Binding
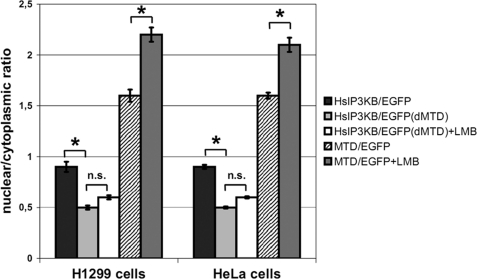
We show here that the previously described actin binding domain of IP3KB (7) not only comprises the critical amino acids for actin targeting but also contains signals for nuclear import and export of the kinase. The essential amino acids for all three targeting events are located in the sequence segment amino acids 104–165, for which a highly ordered secondary structure is predicted (7) and is now named the MTD of IP3KB (see Fig. 8). The deletion of this domain abrogated both nucleocytoplasmic shuttling (Fig. 7) and actin binding (see Fig. 4) of EGFP fusion proteins expressed in H1299 and HeLa cells (likewise shown earlier in NRK cells (7)). The fact that LMB treatment causes nuclear accumulation of the isolated MTD (H1299, 1.6 ± 0.06 versus 2.2 ± 0.07; HeLa, 1.6 ± 0.03 versus 2.1 ± 0.07) confirms the ability of this domain to shuttle actively between nucleus and cytoplasm. Point mutations that functionally inactivated the NES (39) led to nuclear accumulation of the EGFP tagged mutants showing that the identified motif (134LQRELQNVQV) is necessary for exportin 1-dependent nuclear export of human IP3KB. Therefore, the loss of nuclear uptake of deletion mutant ΔMTD is not due to changes of the gross secondary structure of IP3KB but to loss of its MTD. The observed nuclear accumulation of IP3KB requires an active nuclear import activity as its molecular size (102 kDa) rules out passive diffusion into the nucleus (40). The NLS motif identified by us earlier (7) was further characterized in this study, and a cluster of three basic amino acid residues (129RKLR) necessary for its functionality was identified. Because LMB treatment of cells that were transiently expressing a mutant with an inactivated NLS did not cause any accumulation in the nucleus of the cell, human IP3KB obviously possesses no further active motif mediating nuclear import. Fusion of the isolated NLS (amino acids 126–134) to EGFP clearly increased nuclear uptake of EGFP, further proving that this domain alone is sufficient for active nuclear import of proteins. In accordance with its actin binding activity, the isolated MTD of human IP3KB fused with EGFP (MTD/EGFP) still showed strong actin binding. Preservation of this activity in a 62-amino acid fragment indicates conservation of folding within the domain.
FIGURE 8.

Molecular structure of multitargeting domain. The amino acid sequence of the multitargeting domain (amino acids 104–165) in the N-terminal region of HsIP3KB is shown. The NLS is marked with boldface characters, the NES is marked with boldface/underlined characters, and the FEA motif is marked with boldface/gray characters. The one-letter amino acid code is used.
FIGURE 7.
Sequence motifs mediating nucleocytoplasmic shuttling and actin binding are located in the multitargeting domain. H1299 and HeLa cells transiently expressing EGFP fusion proteins were fixed, and the nuclear/cytoplasmic ratio was determined. Wild type IP3KB (WT), a mutant form lacking the multitargeting domain (ΔMTD), and the isolated multi-targeting domain fused with EGFP (MTD) were used. A final concentration of 20 nm LMB and an incubation time of 6 h were used (+LMB). Further details can be found in the legend to Fig. 2. For detailed information about the constructs, see Table 1.
In addition, substitution mutagenesis of residues shown to be necessary for actin binding in the rat homologue (7) and inactivation of the 150FEAH motif created IP3KB mutants without the ability to bind to the plasma membrane (see Fig. 4). Likewise, disruption of the cortical actin structures with cytocalasin D caused IP3KB to detach from the plasma membrane (data not shown). These data show that plasma membrane anchoring of IP3KB depends on binding to cortical actin via its multitargeting domain. Necessary for this targeting is the hitherto unknown sequence motif 150FEAH, which is highly conserved in vertebrate IP3KBs. This targeting immediately influences the fate of Ins(1,4,5)P3 released from the plasma membrane by agonist stimulated phospholipase C. IP3KB localized at cortical actin structures may metabolize part of the released Ins(1,4,5)P3 to Ins(1,3,4,5)P4 before it is able to reach its target receptors at the endoplasmic reticulum and thereby reduce the magnitude and duration of the Ca2+ release. However, it must be noted that Ins(1,3,4,5)P4 has a >10-fold higher affinity to inositol phosphate 5-phosphatase (41) than Ins(1,4,5)P3 and thus may protect Ins(1,4,5)P3 against dephosphorylation by 5-phosphatases. This would counteract a shortening of the Ins(1,4,5)P3 transient by the above mechanism. Recently, it was discussed that this inhibitory effect mediates IP3KA-regulated activation of store-operated Ca2+ entry (42) and therefore may prolong the Ca2+ release induced by Ins(1,4,5)P3.
In conclusion, NES, NLS, and actin binding motif of human IP3KB all reside in sequence segment amino acids 104–165 (Fig. 8) designated MTD of IP3KB. Obviously, this domain controls both nucleocytoplasmic shuttling and actin binding of the protein. Interplay between the MTD and the endoplasmic reticulum membrane anchoring domain of IP3KB (20), which is located in a more C-terminal part of the noncatalytic domain of IP3KB (36) may generate an even more complex and more dynamic intracellular targeting with sophisticated consequences on calcium signaling.
Localization at Invaginations of Nuclear Envelope
In this study, a hitherto unknown localization of IP3KB at invaginations of the nuclear envelope was detected using quasiconfocal immunofluorescence techniques (see “Results”). Interestingly, this localization cannot be observed when EGFP fusion proteins are highly overexpressed. It is known that nonphysiologically high levels of EGFP fusion protein can lead to a saturation of binding sites and to artificial changes of protein localization (43). These novel IP3KB binding sites at nuclear invaginations are probably saturated by endogenous protein or during overexpression of EGFP fusion protein. Therefore, EGFP fusion protein generally cannot bind to these invaginations, or a small amount of fusion protein bound to this localization is concealed by a huge amount of overexpressed protein accumulating in the cell.
Mammalian cell nuclei are known to contain long, dynamic tubular channels derived from the nuclear envelope that expand deeply into the nucleoplasm and may extend completely through the nucleus (44). These nuclear invaginations are not empty but contain cytoplasmic elements, in particular, F-actin (45), but mitochondria also have been shown to be contained within (46). Johnson et al. (47) have shown that actin-filled nuclear invaginations are indication for the degree of cell dedifferentiation. In their study, a highy dedifferentiated, invasive, mammary epithelial cancer cell line possessed the highest incidence of nuclear invaginations. Therefore, they speculated that the increased number of invaginations seen in highly dedifferentiated cells may provide an increased number of nuclear pore complexes necessary for rapid proliferation. However, Clubb and Locke (45) describe two kinds of nuclear invaginations. The first kind is horizontally orientated in the nuclei of adherent cells and contains F-actin, whereas the second kind is vertically orientated and lacks F-actin. Because all IP3KB containing invaginations run perpendicular to the substrate, they may be identical to the second form of invaginations devoid of F-actin.
Other studies have shown that nuclear invaginations are involved in nuclear Ca2+ signaling (46). Lui et al. (46) demonstrate that in histamine-stimulated HeLa cells, Ins(1,4,5)P3 diffuses very rapidly into these invaginations and evokes a Ca2+ release deep inside the nucleus of the cell by interaction with Ins(1,4,5)P3 receptors co-located on these invaginations. This mechanism enables the cell to regulate Ca2+ signals in localized subnuclear regions. Such localized release of Ca2+ has been shown to cause the translocation of nuclear protein kinase C to this region of the nucleus (48). Our finding of IP3KB and Ins(1,4,5)P3 receptor type I and III at nuclear invaginations of H1299 cells now provides a mechanism of terminating such nuclear Ca2+ signals by metabolizing Ins(1,4,5)P3 to Ins(1,3,4,5)P4. A similar mechanism is well known to terminate Ca2+ release by Ins(1,4,5)P3 receptors localized at the endoplasmic reticulum membrane (49). Malviya and Klein (50) have demonstrated that binding of Ins(1,3,4,5)P4 to a nuclear Ins(1,3,4,5)P4 receptor evokes an ATP-independent calcium uptake by isolated rat liver nuclei. Therefore, Ins(1,3,4,5)P4 formed by IP3KB in nuclear invaginations of H1299 cells might also regulate the reuptake of calcium by the nuclear envelope.
Effects of IP3KB Overexpression on Ca2+ Signaling
Effects of overexpression of IP3K and Ins(1,4,5)P3 5-phosphatase on Ins(1,4,5)P3-mediated Ca2+ signaling have been intensively investigated (Refs. 37 and 38 and references therein). We show here that Ca2+ signals in response to carbachol stimulation are effectively inhibited in HeLa cells by overexpression of IP3KB tagged with EGFP. Most likely, the enzymatic activity of overexpressed IP3KB leads to a fast and complete conversion of Ins(1,4,5)P3 to Ins(1,3,4,5)P4. Thus, Ins(1,4,5)P3 is being converted before it can activate Ins(1,4,5)P3 receptors and thereby stimulate Ca2+ release from the lumen of the endoplasmic reticulum. A similar effect was observed by Erneux and co-workers (19) in ATP-stimulated COS-7 cells overexpressing IP3KB. In contrast to the results reported by Erneux and co-workers (19), who observed Ca2+ signals in low expressing cells, in our experiments, Ca2+ signals were inhibited in all observed cells independent of the IP3KB/EGFP expression level. These differences might be due to the use of different transfection methods, cell lines, and agonists. A mutant form of IP3KB with strongly reduced enzymatic activity did not alter transient Ca2+ signals. In cells that were overexpressing this mutant, carbachol stimulation induced Ca2+ signals comparable to signals in untransfected or mock-transfected cells. Because of its reduced activity this mutant form most probably converts only a small fraction of Ins(1,4,5)P3 to Ins(1,3,4,5)P4, allowing for activation of Ins(1,4,5)P3 receptors and resulting in Ca2+ release. An additional increase in Ca2+ ∼360 s after carbachol stimulation was observed in one-half of the cells. Possibly, the moderate elevation of Ins(1,3,4,5)P4 caused by overexpression of the IP3KB mutant protects Ins(1,4,5)P3 by inhibiting Ins(1,4,5)P3 5-phosphatase (42). This protection mechanism may become increasingly important with time after stimulation because the amount of Ins(1,4,5)P3 newly released from the limited pool of phosphatidylinositol 4,5-bisphosphate, decreases.
Acknowledgments
Different plasmids encoding EGFP fusion proteins were kind gifts from H. Will (Sp100); G. Matera (coilin); J. J. He (Sam68); D. Allis, J. Winshell, S. de la Luna, and S. Aznar-Benitah (Cbx4); M. van Lohuizen and E. Verhoeven (Bmi-1); and V. K. Parnaik (Sc35). We thank S. Oetjen for help with quantification of intracellular localization.
This work was supported by Deutsche Forschungsgemeinschaft Grant MA 989/3-1 (to G. W. M.).
- Ins(1,4,5)P3
- inositol 1,4,5-trisphosphate
- Ins(1,3,4,5)P4
- inositol 1,3,4,5-tetrakisphosphate
- HsIP3KB
- human IP3KB
- IP3KB
- inositol 1,4,5-trisphosphate 3-kinase isoform B
- MTD
- multitargeting domain
- NLS
- nuclear localization signal
- NES
- nuclear export signal
- LMB
- leptomycin B
- EGFP
- enhanced GFP
- TRITC
- tetramethylrhodamine-5-isothiocyanate
- ROI
- region of interest.
REFERENCES
- 1. Berridge M. J., Irvine R. F. (1989) Nature 341, 197–205 [DOI] [PubMed] [Google Scholar]
- 2. Irvine R. F., Schell M. J. (2001) Nat. Rev. Mol. Cell Biol. 2, 327–338 [DOI] [PubMed] [Google Scholar]
- 3. Choi K. Y., Kim H. K., Lee S. Y., Moon K. H., Sim S. S., Kim J. W., Chung H. K., Rhee S. G. (1990) Science 248, 64–66 [DOI] [PubMed] [Google Scholar]
- 4. Takazawa K., Perret J., Dumont J. E., Erneux C. (1991) Biochem. J. 278, 883–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dewaste V., Pouillon V., Moreau C., Shears S., Takazawa K., Erneux C. (2000) Biochem. J. 352, 343–351 [PMC free article] [PubMed] [Google Scholar]
- 6. Dewaste V., Roymans D., Moreau C., Erneux C. (2002) Biochem. Biophys. Res. Commun. 291, 400–405 [DOI] [PubMed] [Google Scholar]
- 7. Brehm M. A., Schreiber I., Bertsch U., Wegner A., Mayr G. W. (2004) Biochem. J. 382, 353–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Erneux C., Roeckel N., Takazawa K., Mailleux P., Vassart G., Mattei M. G. (1992) Genomics 14, 546–547 [DOI] [PubMed] [Google Scholar]
- 9. Barrat F. J., Depetris D., Certain S., Mattei M. G., de Saint Basile G. (1997) Genomics 43, 111–113 [DOI] [PubMed] [Google Scholar]
- 10. Tajouri L., Mellick A. S., Tourtellotte A., Nagra R. M., Griffiths L. R. (2005) Brain Res. Brain Res. Protoc. 15, 79–91 [DOI] [PubMed] [Google Scholar]
- 11. Emilsson L., Saetre P., Jazin E. (2006) Neurobiol. Dis. 21, 618–625 [DOI] [PubMed] [Google Scholar]
- 12. Györffy B., Lage H. (2007) J. Invest. Dermatol. 127, 394–399 [DOI] [PubMed] [Google Scholar]
- 13. Huang Y. H., Hoebe K., Sauer K. (2008) Expert Opin. Ther. Targets 12, 391–413 [DOI] [PubMed] [Google Scholar]
- 14. Pouillon V., Hascakova-Bartova R., Pajak B., Adam E., Bex F., Dewaste V., Van Lint C., Leo O., Erneux C., Schurmans S. (2003) Nat. Immunol. 4, 1136–1143 [DOI] [PubMed] [Google Scholar]
- 15. Miller A. T., Sandberg M., Huang Y. H., Young M., Sutton S., Sauer K., Cooke M. P. (2007) Nat. Immunol. 8, 514–521 [DOI] [PubMed] [Google Scholar]
- 16. Maréchal Y., Pesesse X., Jia Y., Pouillon V., Pérez-Morga D., Daniel J., Izui S., Cullen P. J., Leo O., Luo H. R., Erneux C., Schurmans S. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 13978–13983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jia Y., Subramanian K. K., Erneux C., Pouillon V., Hattori H., Jo H., You J., Zhu D., Schurmans S., Luo H. R. (2007) Immunity 27, 453–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jia Y., Loison F., Hattori H., Li Y., Erneux C., Park S. Y., Gao C., Chai L., Silberstein L. E., Schurmans S., Luo H. R. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 4739–4744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dewaste V., Moreau C., De Smedt F., Bex F., De Smedt H., Wuytack F., Missiaen L., Erneux C. (2003) Biochem. J. 374, 41–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pattni K., Millard T. H., Banting G. (2003) Biochem. J. 375, 643–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sternsdorf T., Jensen K., Reich B., Will H. (1999) J. Biol. Chem. 274, 12555–12566 [DOI] [PubMed] [Google Scholar]
- 22. Hebert M. D., Matera A. G. (2000) Mol. Biol. Cell 11, 4159–4171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Henao-Mejia J., He J. J. (2009) Exp. Cell Res. 315, 3381–3395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Salichs E., Ledda A., Mularoni L., Albà M. M., de la Luna S. (2009) PLoS Genet 5, e1000397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bernstein E., Duncan E. M., Masui O., Gil J., Heard E., Allis C. D. (2006) Mol. Cell Biol. 26, 2560–2569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hernández-Muñoz I., Taghavi P., Kuijl C., Neefjes J., van Lohuizen M. (2005) Mol. Cell Biol. 25, 11047–11058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tripathi K., Parnaik V. K. (2008) J. Biosci. 33, 345–354 [DOI] [PubMed] [Google Scholar]
- 28. Wang W., Malcolm B. A. (1999) BioTechniques 26, 680–682 [DOI] [PubMed] [Google Scholar]
- 29. Nalaskowski M. M., Windhorst S., Stockebrand M. C., Mayr G. W. (2006) Biol. Chem. 387, 583–593 [DOI] [PubMed] [Google Scholar]
- 30. Nalaskowski M. M., Bertsch U., Fanick W., Stockebrand M. C., Schmale H., Mayr G. W. (2003) J. Biol. Chem. 278, 19765–19776 [DOI] [PubMed] [Google Scholar]
- 31. Wolff B., Sanglier J. J., Wang Y. (1997) Chem. Biol. 4, 139–147 [DOI] [PubMed] [Google Scholar]
- 32. la Cour T., Gupta R., Rapacki K., Skriver K., Poulsen F. M., Brunak S. (2003) Nucleic Acids Res. 31, 393–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. la Cour T., Kiemer L., Mølgaard A., Gupta R., Skriver K., Brunak S. (2004) Protein Eng. Des. Sel. 17, 527–536 [DOI] [PubMed] [Google Scholar]
- 34. Fontes M. R., Teh T., Kobe B. (2000) J. Mol. Biol. 297, 1183–1194 [DOI] [PubMed] [Google Scholar]
- 35. Nalaskowski M. M., Deschermeier C., Fanick W., Mayr G. W. (2002) Biochem. J. 366, 549–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yu J. C., Lloyd-Burton S. M., Irvine R. F., Schell M. J. (2005) Biochem. J. 392, 435–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dupont G., Erneux C. (1997) Cell Calcium 22, 321–331 [DOI] [PubMed] [Google Scholar]
- 38. Politi A., Gaspers L. D., Thomas A. P., Höfer T. (2006) Biophys. J. 90, 3120–3133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bogerd H. P., Fridell R. A., Benson R. E., Hua J., Cullen B. R. (1996) Mol. Cell Biol. 16, 4207–4214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Macara I. G. (2001) Microbiol. Mol. Biol. Rev. 65, 570–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hermosura M. C., Takeuchi H., Fleig A., Riley A. M., Potter B. V., Hirata M., Penner R. (2000) Nature 408, 735–740 [DOI] [PubMed] [Google Scholar]
- 42. Windhorst S., Fliegert R., Blechner C., Möllmann K., Hosseini Z., Günther T., Eiben M., Chang L., Lin H. Y., Fanick W., Schumacher U., Brandt B., Mayr G. W. (2010) J. Biol. Chem. 285, 5541–5554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zeidman R., Trollér U., Raghunath A., Påhlman S., Larsson C. (2002) Mol. Biol. Cell 13, 12–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fricker M., Hollinshead M., White N., Vaux D. (1997) J. Cell Biol. 136, 531–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Clubb B. H., Locke M. (1998) Tissue Cell 30, 684–691 [DOI] [PubMed] [Google Scholar]
- 46. Lui P. P., Chan F. L., Suen Y. K., Kwok T. T., Kong S. K. (2003) Biochem. Biophys. Res. Commun. 308, 826–833 [DOI] [PubMed] [Google Scholar]
- 47. Johnson N., Krebs M., Boudreau R., Giorgi G., LeGros M., Larabell C. (2003) Differentiation 71, 414–424 [DOI] [PubMed] [Google Scholar]
- 48. Echevarría W., Leite M. F., Guerra M. T., Zipfel W. R., Nathanson M. H. (2003) Nat. Cell Biol. 5, 440–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Millard T. H., Cullen P. J., Banting G. (2000) Biochem. J. 352, 709–715 [PMC free article] [PubMed] [Google Scholar]
- 50. Malviya A. N., Klein C. (2006) Can J. Physiol. Pharmacol. 84, 403–422 [DOI] [PubMed] [Google Scholar]