Abstract
Staphylococcus aureus causes life-threatening pneumonia in hospitals and deadly superinfection during viral influenza. The current study investigated the role of surfactant protein A (SP-A) in opsonization and clearance of S. aureus. Previous studies showed that SP-A mediates phagocytosis via the SP-A receptor 210 (SP-R210). Here, we show that SP-R210 mediates binding and control of SP-A-opsonized S. aureus by macrophages. We determined that SP-A binds S. aureus through the extracellular adhesin Eap. Consequently, SP-A enhanced macrophage uptake of Eap-expressing (Eap+) but not Eap-deficient (Eap−) S. aureus. In a reciprocal fashion, SP-A failed to enhance uptake of Eap+ S. aureus in peritoneal Raw264.7 macrophages with a dominant negative mutation (SP-R210(DN)) blocking surface expression of SP-R210. Accordingly, WT mice cleared infection with Eap+ but succumbed to sublethal infection with Eap- S. aureus. However, SP-R210(DN) cells compensated by increasing non-opsonic phagocytosis of Eap+ S. aureus via the scavenger receptor scavenger receptor class A (SR-A), while non-opsonic uptake of Eap− S. aureus was impaired. Macrophages express two isoforms: SP-R210L and SP-R210S. The results show that WT alveolar macrophages are distinguished by expression of SP-R210L, whereas SR-A−/− alveolar macrophages are deficient in SP-R210L expressing only SP-R210S. Accordingly, SR-A−/− mice were highly susceptible to both Eap+ and Eap− S. aureus. The lungs of susceptible mice generated abnormal inflammatory responses that were associated with impaired killing and persistence of S. aureus infection in the lung. In conclusion, alveolar macrophage SP-R210L mediates recognition and killing of SP-A-opsonized S. aureus in vivo, coordinating inflammatory responses and resolution of S. aureus pneumonia through interaction with SR-A.
Keywords: Inflammation, Innate Immunity, Lung, Macrophage, Pulmonary Surfactant
Introduction
There is limited knowledge about host factors that facilitate eradication of Staphylococcus aureus infection in the lung. Methicillin-resistant S. aureus has remained a major cause of hospital- and health care-associated pneumonia since its appearance over 40 years ago and has recently become a more prominent etiology in community acquired pneumonia. Colonization of nasal epithelium with S. aureus, a normal occurrence in over 20% of the population, increases the risk for the development of staphylococcal pneumonia (1). Furthermore, S. aureus co-infections are a major complication contributing to high morbidity and mortality during both pandemic and seasonal influenza virus pneumonia (2). S. aureus deploys a combination of virulence factors, including adhesins, toxins, and immunomodulatory molecules, that facilitate infection of different host tissues (3, 4).
Surfactant protein A (SP-A)3 is a crucial component of the pulmonary innate immune system in the alveolar spaces (5, 6). SP-A is the major protein constituent of pulmonary surfactant; it is involved in organization of large aggregate surfactant phospholipids lining the alveolar surface and acts as an opsonin for pathogens (7). SP-A is incorporated in the tubular myelin fraction of pulmonary surfactant that covers the alveolar lining fluid of the distal airway epithelium. The presence of pathogen-derived molecules may trigger reorganization of surfactant lipids (8–11) and exposure of SP-A to bind pathogens at points of entry on the surfactant interface. Alveolar macrophages in the aqueous hypophase may then patrol areas of disturbance on the surfactant layer binding SP-A-opsonized bacteria. SP-A binds pathogens via a carboxyl-terminal carbohydrate recognition domain in a calcium-dependent manner. Amino-terminal collagen-like and coiled-coil domains form trimers, whereas intermolecular disulfide bonds contribute to oligomerization of trimers into decaoctamers. The presence of calcium results in SP-A aggregation that enables carbohydrate recognition domains to bind multiple carbohydrate ligands on the surface of microorganisms. SP-A is a member of the collectin family of proteins, which include surfactant protein D (SP-D) in lung and mannose-binding lectin (MBL) in blood circulation. SP-D and MBL are specific for carbohydrate ligands (6). However, the carbohydrate recognition domain of SP-A is more generic, having a wider spectrum of microbial ligands that include lipid and protein moieties (12–14). Previous studies determined that SP-A is an opsonin for the Gram-positive S. aureus-enhancing phagocytosis of this pathogen by macrophages (15–17). On the other hand, binding of SP-A to S. aureus does not appear to involve lipoteichoic acid (LTA) or peptidoglycan, the major cell wall glycoconjugates of Gram-positive bacteria (18).
Previous studies established that SP-A modulates macrophage phagocytosis and a host of pro- and anti-inflammatory responses that help in eradication of infection first and then resolution of inflammation in vivo (7, 16, 19–24). Several macrophage receptors have been implicated in the ability of SP-A to coordinate clearance of pathogens and apoptotic cells and temporal control of inflammation in the lungs (6). The SP-A receptor SP-R210 was identified as cell surface isoforms of unconventional Myo18A (25). The Myo18A gene encodes two alternatively spliced SP-R210 isoforms, SP-R210L and SP-R210S. The longer 230–240-kDa SP-R210L isoform contains an amino-terminal PDZ protein interaction module that is absent from the shorter 210-kDa SP-R210S (25). SP-R210S is highly expressed in both mature macrophages and in immature monocytic cells. However, SP-R210L is only expressed in mature macrophages (25). Earlier studies showed that SP-R210 mediates phagocytosis and killing of SP-A-opsonized Mycobacterium bovis BCG (SP-A-BCG) by bone marrow-derived macrophages (23). These studies showed that ligation of SP-R210 with SP-A-BCG complexes enhanced expression of TNFα and nitric oxide that enabled macrophages to control mycobacterial growth (23, 26). On the other hand, SP-R210 can control the level of inflammatory cells and mediators in the presence of mycobacterial extracts, suggesting a secondary role of SP-R210 in immune homeostasis (27).
The present studies establish that SP-R210 is an opsonic phagocytic receptor of SP-A-opsonized S. aureus. Phagocytosis of SP-A-opsonized S. aureus via SP-R210 was coordinated with secretion of TNFα and suppression of bacterial growth in macrophages. Furthermore, the present work determined the mechanism of SP-A binding to S. aureus; expression of the staphylococcal adhesin Eap is necessary for both SP-A binding and enhanced phagocytosis of SP-A-opsonized bacteria by SP-R210. Correspondingly, infection of mice with Eap expressing (Eap+) and Eap-deficient (Eap−) strains indicate that mice are highly susceptible to infection with the SP-A-resistant Eap− S. aureus. Finally, additional experiments revealed a previously unknown interaction between expression of SP-R210 isoforms and the scavenger receptor SR-A. In this model, SP-R210 mediates opsonic bacterial clearance, whereas the interaction of SP-R210 with SR-A is necessary for temporal control of inflammation.
EXPERIMENTAL PROCEDURES
Reagents
The following monoclonal antibodies or isotype-matched IgGs were purchased: for M5/114.15.2 (PE-conjugated anti-mouse MHC-II, rat IgG2b), 2.4G2 (anti-mouse CD16/CD32 Fc block, rat IgG2b), and N418 (PE/Cy5-conjugated anti-mouse CD11c, Armenian hamster IgG) (all from eBiosciences (San Diego, CA)); for 2F8 (FITC-conjugated anti-mouse SR-A) (AbD Serotec (Raleigh, NC)). The generation and purification of rabbit polyclonal antibodies against either the carboxyl-terminal (anti-SP-R210ct) or SP-A binding neck domains of SP-R210/Myo18A (anti-SP-R210n) and preimmune IgG were obtained as described previously (25, 28). A commercial affinity-purified anti-SP-R210/Myo18A antibody raised against a carboxyl-terminal peptide was from ProteinTech (Chicago, IL). Secondary Alexa647-conjugated goat anti-rabbit antibody was from Molecular Probes, Inc. (Eugene, OR). Chemicals, antibiotics, and buffers were from Sigma-Aldrich or Fisher unless noted otherwise. Fetal bovine serum (FBS) was from Atlanta Biologicals (Atlanta, GA). The Improm-II RT-PCR kit was from Promega (Milwaukee, WI). Taq polymerase was obtained from New England Biolabs (Beverly, MA). Native human serum lipoprotein (LDL) labeled with 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine (DiI) was purchased from Biomedical Technologies (Stoughton, MA). Human SP-A was isolated from therapeutic lung lavage from alveolar proteinosis patients (provided by Dr. Bruce Trapnell, Rare Disease Consortium (Cincinnati, OH)), as described previously (25). LPS contamination was below 0.02 pg/μg SP-A. LPS contamination was tested using a limulus amebocyte lysate LPS detection kit (BioWhitaker). SP-A used for ligand blot analyses was first amino-terminally biotinylated as described previously (29). The staphylococcal adhesin Eap was purified by LiCl extraction and column chromatography from S. aureus Newman (30), and recombinant Eap proteins from S. aureus strain Mu50 were obtained as described in detail previously (31, 32). SP-A used in binding assays was radiolabeled with 125I using a Chloramine-T-based procedure (25, 29). Cell culture media and reagents were obtained from CellGro (Manassas, VA). Phosphate-buffered saline (PBS) without or with CaCl2 and MgCl2 (CM-PBS) was purchased from Sigma-Aldrich. Sulfo-NHS-LC-LC-biotin and streptavidin-agarose were from Thermo Scientific Pierce. A rabbit polyclonal anti-SP-A antibody was obtained from Seven Hills Bioreagents (Cincinnati, OH). Fluorescent FITC-conjugated S. aureus and Escherichia coli bioparticles were obtained from Molecular Probes.
Cells
Human THP-1 premonocytic cells, mouse Raw264.7 peritoneal macrophages, L-929 fibroblasts, and monkey COS-1 fibroblasts were obtained from ATCC (Manassas, VA). Cells were cultured in DMEM or RPMI medium supplemented with 10% FBS. The generation of stable COS-1 cells transfected with control or SP-R210S expression plasmid was described previously (25). Mouse bone marrow-derived macrophages (MBMM) were obtained as described previously (25). Briefly, mouse femurs were flashed in DMEM containing 10% FBS, and macrophages were then differentiated in medium supplemented with 20% L-cell conditioned medium as a source of macrophage colony-stimulating factor. Media were changed on day 4, and cells were used on day 8 after culture in L-cell conditioned media. One day prior to experiments, MBMM monolayers were lifted using a trypsin/EDTA solution and subcultured overnight in DMEM, 5% FBS at a density of 300,000 cells/well in 24-well plates.
Bacteria
S. aureus strain Newman was grown for 16–18 h in 50 ml of tryptic soy broth (TSB). Bacteria were then washed in PBS, and 40-μl aliquots containing 200 or 400 × 106 bacteria were stored frozen at −80 °C until use. Bacterial cultures were quantified by spectrophotometry at 600 nm. Bacterial viability was determined by counting colony-forming units after culture of serially diluted stocks on TSB-agar plates. The generation and culture of Eap-deficient S. aureus Newman, the Eap-deficient strain mAH12, and the Eap-complemented strain mAH12(pCXEap) were described in detail previously (33).
Identification of SP-A-binding Proteins on S. aureus Cell Wall
S. aureus bacteria were grown in TSB for 18 h, washed, and then treated with either lysostaphin or 0.4% SDS to extract cell wall-anchored and cell wall-associated adhesins, respectively. Proteins were resolved on 10% SDS-PAGE gels and either stained with Colloidal Blue (Bio-Rad) or electroblotted to nitrocellulose. Blots were blocked in TBS-T containing 5% milk for 1 h and then incubated overnight with 30 μg/ml biotinylated SP-A in TBS-T. Blots were then washed and blotted for an additional 30 min with HRP-conjugated streptavidin (Calbiochem). Bound SP-A was then visualized by chemiluminescence using the Western Lightning ECL plus kit (PerkinElmer Life Sciences). Corresponding gel bands containing SP-A-binding proteins were then excised and subjected to in-gel trypsin digestion. Proteins were identified by MALDI fingerprint analysis of peptide digests (25). MALDI analysis resulted in identification of Eap as the main SP-A-binding protein. SP-A binding to Eap was then verified in solid phase assays using purified or recombinant Eap. Ligand blot analysis, carried out as described above, determined SP-A binding to LiCl-extracted cell wall protein from parental (Eap+), Eap-deficient (Eap−), or Eap-complemented (cEap, mAH12) S. aureus Newman.
For solid phase assays, 1 μg of purified native or recombinant Eap was coated overnight at 4 °C in 0.1 m sodium carbonate buffer on 96-well microtiter plates. Control wells were coated with BSA. Next, nonspecific binding was blocked in buffer composed of 5 mm Hepes, pH 7.4, 150 mm NaCl, and 5 mg/ml BSA for 1 h at room temperature with gentle agitation. Human SP-A (0–20 μg/ml) was added in blocking buffer, and plates were incubated for 1 h at 37 °C. After washing, plates were incubated with 1:5000 dilutions of rabbit anti-SP-A polyclonal antibodies followed by washing and incubation with 1:10,000 dilution of an HRP-conjugated anti-rabbit antibody. Bound protein was visualized colorimetrically at 450 nm using tetraethyl benzidine as the HRP substrate.
Subsequently, binding assays using 125I-labeled SP-A determined the specificity of Eap-mediated opsonization of S. aureus by SP-A. Binding assays were carried out in 0.2 ml of PBS containing 5% BSA and either 50 × 106 cfu/ml Eap+ or Eap− S. aureus Newman. Bacteria were incubated with increasing concentrations of SP-A (0–18 nm) for 1 h at 37 °C. Complexes of bacteria and 125I-SP-A were separated over a mixture of mineral and silicone oil, and bound SP-A was determined using a γ-counter.
RT-PCR
Total RNA from alveolar macrophages was isolated using TRIzol (Invitrogen) according to the manufacturer's directions. The mRNA in 1 μg of total RNA was primed using oligo(dT), and single strand cDNA synthesis was carried out using the Improm II kit (Promega). Amplifications of SP-R210 cDNA representing the carboxyl terminus (Myo18Act) domain of SP-R210 were carried out using Taq polymerase (New England Biolabs) with these site-specific primers: sense Myo18Act primer, 5′-GAGGATGAGATGGAAAGTGAC-3′; antisense Myo18Act primer, 5′-CACTGGTCTCTGTCAGCTTG-3′. The PCRs were performed in a 15-μl total volume containing 1.5 μl of 10× Taq polymerase buffer (New England Biolabs), 2 mm MgCl2, 10 μm each dNTPs, 200 nm Myo18A primers, 1 μl of cDNA, and 1 unit of Taq polymerase. The cycling conditions were as follows: denaturation at 94 °C for 2 min, 30–40 cycles of 94 °C for 30 s, 60 °C for 30 s, 72 °C for 30 s, and extension for 7 min at 72 °C. PCR products were separated on 2% agarose gels made in TAE buffer and visualized by EtBr staining. The sequence of PCR products was verified commercially at SeqWright (Houston, TX).
Dominant Negative Disruption of SP-R210 in Raw264.7 Macrophages
The 300-bp region of SP-R210 expressing the unique carboxyl-terminal domain of SP-R210 (28) was amplified from alveolar macrophage total RNA by RT-PCR as described above using primers containing 5′ and 3′ NcoI and NotI restriction sites. The 5′ sense primer sequence was 5′-gaattcccatgGAGGATGAGATGGAAAG-3′, and the 3′ antisense primer sequence was 5′-GACAGAGACCAGTGCAgcggccgcataaactat-3′. Amplified cDNA was digested with NcoI and NotI, gel-purified, and subcloned into the pTRIEX-2-neo expression vector (Novagen) between NcoI and NotI sites as described previously (28). Vector insert was sequenced, propagated in JM109-competent E. coli, and purified using the EndoFree plasmid purification kit (Qiagen). To obtain stably transfected cells, the Raw264.7 (ATCC) macrophage cell line was transfected with empty vector or vector containing SP-R210ct cDNA (SP-R210(DN)) using GeneJuice (Novagen), and stable cells were selected and propagated in RPMI medium supplemented with 10% FBS and 600 μg/ml neomycin sulfate, as described previously (25). Expression of truncated SP-R210 was verified by RT-PCR and Western blot analysis.
Cell Surface Biotinylation
The Raw264.7 control and SP-R210(DN) cell lines were seeded in 75-mm2 tissue culture plates at 3–4 × 106 cells/plate for 48–60 h. Macrophage monolayers, cultured as described above, were washed three times in cold CM-PBS adjusted to pH 8.0 and subsequently incubated for 30 min at 4 °C in CM-PBS, pH 8.0, containing 1 mg/ml sulfo-NHS-LC-LC-biotin. Cells were then washed once in 50 mm Tris, pH 8.0, and once in PBS. Labeled cells were detached in a cell stripper and washed, and cell extracts were generated in lysis buffer (containing 1% Nonidet P-40, 50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1 mm PMSF, 2 μg/ml leupeptin, 2 μg/ml aprotinin, 10 μg/ml chymostatin, and 10 μg/ml trypsin-chymotrypsin) by repeated freeze and thaw cycles as described previously (27, 34). Cell lysates were then incubated for 1 h on a rotator at 4 °C with 100 μl of a 50% suspension of streptavidin-agarose. Bound protein was then washed three times in lysis buffer at 15,000 × g. Bound protein was then identified by Western blot analysis using anti-SP-R210 antibodies.
Preparation of Lung Homogenates
Lungs form WT and SR-A−/− mice were perfused in 10 ml of cold PBS to remove blood cells. The lungs were then excised and homogenized in 2.0 ml of lysis buffer. Lysates were processed through three freeze and thaw cycles as described previously (34), and insoluble material was removed by centrifugation at 15,000 × g. Protein was quantified using the BCA assay. Expression of SP-R210 was assessed by Western blot analysis using affinity-purified anti-Myo18A (Protein Tech) antibodies.
SP-A Binding Assays
Binding assays were carried out using radiolabeled SP-A as described previously (29). Control and SP-R210(DN) macrophages were cultured for 2 days until 90% confluent and lifted in a non-enzymatic cell dissociation buffer (Invitrogen). Binding assays were carried out in 0.1 ml of binding buffer (29), containing 1.6 million cells/ml, with increasing concentration of SP-A. Cell-associated SP-A was separated by centrifugation over oil and quantified using a γ-counter. Nonspecific binding was determined in the presence of 5 mm EDTA.
Flow Cytometry
Alveolar macrophages were incubated in blocking buffer containing 1× phosphate-buffered saline, pH 7.4, supplemented with 1% BSA, 2% heat-inactivated FBS or normal goat serum, and 10 μg/ml Fc block for 1 h on ice. Then 100 μl cell aliquots containing 100,000–200,000 cells were dispensed into 1.5-ml Eppendorf tubes and incubated with 0.5 μg of PE-conjugated MHC-II, 0.5 μg PE/Cy5-conjugated CD11c, and 0.2 μg of affinity-purified anti-SP-R210 antibody for 30 min on ice. The cells were then washed twice in 1.0 ml of blocking buffer and incubated with 1:1000 dilutions of Alexa647-conjugated anti-rabbit IgG for 15 min on ice. Control and SP-R210(DN) Raw264.7 macrophages were stained with FITC-conjugated anti-SR-A antibody. The stained cells were washed twice in 1.0 ml of blocking buffer, resuspended in PBS, and analyzed by flow cytometry using a BD FACSCalibur flow analyzer (BD Pharmingen). Cells were separated according to forward and side scatter properties and gated to eliminate events from cellular debris and dead cells. A voltage adjustment was applied on unstained cells to set autofluorescent cells as negative events. Quadratic or linear gating was used to determine the percentage of single or double positive cells expressing SP-R210 or CD11c compared with background staining with isotype control antibodies. Linear gating over fluorescent Gaussian histograms was used to obtain mean fluorescence.
Uptake of DiI-labeled Native Serum LDL (DiI-LDL) or Acetylated LDL (DiI-AcLDL)
Control Raw/Triex or SP-R210(DN) Raw264.7 cells were cultured as described above in 24-well plates and incubated for 30 min at 37 °C with 0.8 μg/ml of DiI-LDL or DiI-AcLDL. The cells were then washed and analyzed by flow cytometry.
Binding and Phagocytosis of S. aureus
Live S. aureus Newman or fluorescent S. aureus Wood bioparticles, 50 × 106 bacteria/ml, were preincubated with 5, 10, or 20 μg/ml SP-A in opsonization buffer, respectively. Mixtures were rotated for 1 h at 37 °C in a humidified 5% CO2 chamber. The opsonization buffer was composed of Hanks' balanced salt solution buffered with 10 mm Hepes, pH 7.4, and 1% BSA as blocking agent.
Control and SP-R210S-COS-1 cells were cultured overnight in DMEM, 10% FBS at a density of 200,000 cells/well in 24-well plates. The media were then replaced with opsonization buffer containing a 50:1 ratio of unopsonized or SP-A-opsonized fluorescent S. aureus bioparticles and incubated for 1 h at 37 °C. Cells were then washed and harvested, and the percentage of cells containing bound bioparticles was determined by flow cytometry. Cells were viewed using A Nikon TE-100 fluorescent microscope under a ×40 phase-contrast lens adjusting bright field illumination to visualize both cells and attached fluorescent S. aureus bioparticles. Images were captured using a Sensicam Monochrome camera (3I Imaging).
Attachment of live SP-A-opsonized S. aureus was assessed in undifferentiated THP-1 monocytic cells. THP-1 cells were suspended in opsonization buffer at a concentration of 500,000 cells/ml and rotated with 1.5 × 106 cfu/ml unopsonized or SP-A-opsonized S. aureus Newman for 1 h at 37 °C. The role of SP-R210 was evaluated after treatment of THP-1 with 50 μg/ml control IgG or neutralizing anti-SP-R210n antibodies for 2 h prior to infection. After incubation with bacteria, cells were washed and lysed in 0.1% SDS containing 1% BSA. Bacterial cfu were enumerated following culture of serial dilutions of cell extracts on TSB-agar plates.
The role of SP-R210 in growth of S. aureus and secretion of TNFα was then assessed in matured MBMM. Eight-day-old MBMM were trypsinized and subcultured overnight in 24-well dishes in DMEM, 5% FBS at a density of 300,000 cells/well. Cells were maintained in serum-free medium for 1–4 h before infection. Cells were then incubated with 1.5 × 106 cfu of unopsonized or SP-A (10 μg/ml)-opsonized S. aureus. The role of SP-R210 was determined following a 2–3-h preincubation with 20 μg/ml control IgG or anti-SP-R210n antibody. Infected cells were washed and lysed 3 and 8 h after infection to enumerate cfu by serial dilution of cell lysates on TSB-agar plates. The concentration of TNFα was measured in media by ELISA at 3 h after infection. Control and SP-R210(DN) Raw264.7 macrophages were infected with a 3:1 ratio of unopsonized or SP-A (5 μg/ml)-opsonized Eap+ and Eap− S. aureus as above to assess the effect of deficiencies in either SP-A opsonization or SP-R210.
The phagocytic activity of control and SP-R210(DN) macrophages was assessed using FITC-labeled S. aureus, E. coli, or yeast zymosan bioparticles. Control and SP-R210(DN) macrophages (350,000 cells/well) were cultured overnight in RPMI, 10% FCS medium. The cells were then incubated with a 10:1 ratio of indicated bioparticles for 30 min at 37 °C in a humidified 5% CO2 chamber. Cells were washed in cold PBS and incubated with a trypsin/EDTA solution for 1–2 min. After the addition of 1% FBS in PBS, the cells were placed on ice for 1 h and detached from the plates by triturating using a 1-ml pipette. Cell suspensions were analyzed by flow cytometry before or after the addition of a 0.05% trypan blue solution to distinguish between surface and internalized bioparticles. The phagocytic index was then calculated as a percentage of internalized bacteria.
Mice
Specific pathogen-free C57BL/6 mice 6–8 weeks of age were purchased from Jackson Laboratories (Bar Harbor, ME) or NCI-Frederick. Transgenic SR-A-deficient (SR-A−/−) mice (35, 36) in the C57BL/6 background were maintained in the University of Texas Health Science Center animal facility under pathogen-free conditions. All mice were housed in microisolator cages and were provided autoclaved water and standard mouse chow ad libitum. The Institutional Animal Care and Use Committee of the University of Texas Health Science Center at Tyler approved all animal procedures.
S. aureus Pneumonia
Anesthetized WT or SR-A−/− mice 6–8 weeks of age were infected intranasally with either acute 200 × 106 or sublethal doses of 300 × 106 cfu of Eap+ and Eap− S. aureus. Bacteria were delivered using a pipette in 40 μl of PBS as described previously (37). Uninfected controls received PBS only. Anesthesia was induced via intraperitoneal injection of a ketamine (100 mg/kg)/xylazine (10 mg/kg) mixture. Infected mice were observed at 12-h intervals to monitor survival from intranasal pneumonia. Moribund mice were euthanized immediately using an overdose of Beuthanasia-D followed by cervical dislocation and counted as dead.
Kinetics of pulmonary bacterial clearance and inflammatory responses were determined in at 4 h and then at daily intervals after subacute infection with S. aureus strains. Infected lungs from 8–10 mice/group were lavaged with 6 ml of PBS. Lavaged lungs were then homogenized in 2.5 ml of PBS and stored frozen at −80 °C until further analysis. The lavage was centrifuged at 250 × g on a table top centrifuge for 10 min. Supernatants were then aliquoted in 1-ml portions and stored frozen at −80 °C until further use. Cell pellets were resuspended in 1 ml of PBS and counted using a hemacytometer. Inflammatory cells were identified morphologically as macrophages or neutrophils by differential staining of cytospins using a HEMA-3 staining kit (Fisher). Serial dilution of lavage and postlavage homogenates on TSB-agar plates quantified bacterial cfu. ELISAs using commercial kits (Peprotech or eBiosciences) determined the concentration of TNFα and the neutrophil chemokine KC in lavage and lung homogenates. Data shown represent the combined values from homogenates and lung lavage.
Data Analysis
Statistical and graphical analyses of data were performed with Prism software (GraphPad Software). Statistical comparisons were performed with the unpaired, nonparametric Student's t test. Values of p < 0.05 were considered statistically significant. Binding data were analyzed by nonlinear regression of saturation curves fitted to a single site equilibrium binding equation using Prism software to calculate the number of SP-A binding sites/cell (Bmax) and binding affinities (Kd). Analysis of flow cytometric data was accomplished using Cell Quest software (BD Pharmingen). Survival curves were generated by the Kaplan-Meier method, and statistical comparison of survival curves was performed using the Gehan-Breslow-Wilcoxon test.
RESULTS
SP-R210S Binds SP-A-opsonized S. aureus
Previously described COS-1 cells stably transfected with SP-R210S cDNA (25) were used to assess the role of SP-R210S in binding SP-A-opsonized S. aureus bioparticles. The binding of S. aureus in the absence of SP-A was similar in control and SP-R210S cells. Expression of SP-R210S conferred a significant 2.2-fold increase in the attachment of fluorescent S. aureus in the presence of SP-A (Fig. 1A) compared with control cells. Single S. aureus bioparticles were observed on the surface of SP-R210-COS-1 cells in the absence of SP-A, whereas bacterial clusters were observed on the lamellar membrane at the periphery of SP-R210S-COS-1 cells in the presence of SP-A (Fig. 1B). The premonocytic human THP-1 cell line was then used to assess SP-R210-mediated attachment of live S. aureus. Thus, Fig. 1C demonstrates that binding of SP-A-opsonized S. aureus increased 3-fold compared with S. aureus in the absence of SP-A. Importantly, preincubation with neutralizing anti-SP-R210 antibodies (25) blocked the enhanced binding of SP-A-S. aureus complexes. It should be noted that undifferentiated THP-1 cells are poorly phagocytic, although they express opsonic immunoglobulin and complement receptors (38, 39). Immature THP-1 cells selectively express high levels of SP-R210S but not SP-R210L (25). In addition, THP-1 cells do not express the non-opsonic scavenger SR-A (CD204) and mannose receptor, limiting background attachment of unopsonized bacteria (40–42). Therefore, the experiments on Fig. 1C measured attachment rather than uptake of bacteria. These results indicate that SP-R210 is a specific receptor for SP-A-opsonized S. aureus.
FIGURE 1.
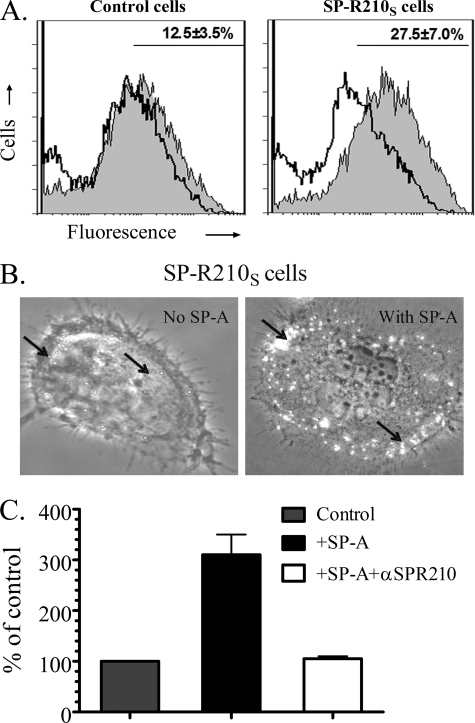
SP-R210S mediates attachment of SP-A-opsonized S. aureus. A, control and SP-R210S-COS-1 cells were incubated with a 50:1 ratio of FITC-labeled S. aureus alone or after preincubation with 20 μg/ml SP-A. Representative histograms show fluorescence of attached non-opsonized (open histograms) and SP-A-opsonized bacteria (gray histograms). The percentage of cells containing SP-A-opsonized bacteria obtained by gating is shown in the graphs. Data are means ± S.D. (n = 4). B, cell-bound bacteria were visualized by partial bright field illumination of fluorescent bacteria under a ×40 phase-contrast lens. Bacteria (arrows) appear as bright dots on control and SP-R210S-COS-1 cells. C, undifferentiated THP-1 cells were incubated with 1.5 × 106 unopsonized or SP-A-opsonized S. aureus for 1 h at 37 °C in opsonization medium. THP-1 cells were treated with 50 μg/ml control or anti-SP-R210n antibodies for 2 h before infection. Washed cells were then lysed, and cell-associated bacterial cfu were enumerated by serial dilution of lysates on TSB-agar. Data are means ± S.D. (error bars) (n = 3).
SP-R210 Suppresses Growth of S. aureus Infection via Secretion of TNFα
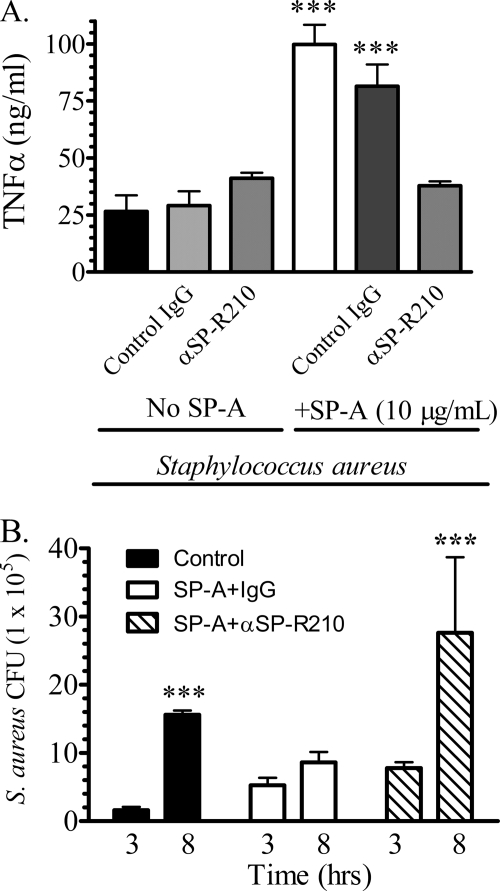
Earlier studies in rat bone marrow-derived macrophages demonstrated that SP-R210 mediates the phagocytosis of SP-A-opsonized Mycobacterium bovis BCG and killing of intracellular bacteria through secretion of TNFα (23, 26). Previous studies determined that TNFα is crucial for clearance of S. aureus in vivo (43). Here, we investigated the link between SP-R210 and S. aureus infection using MBMM. Fig. 2A shows that, consistent with the earlier findings with BCG, MBMM challenged with SP-A-opsonized S. aureus secreted 4-fold more TNFα than cells infected with S. aureus alone (Fig. 2A). However, anti-SP-R210 antibodies but not control IgG blocked the enhanced secretion of TNFα in cells infected with SP-A-opsonized S. aureus (Fig. 2A). Control and anti-SP-R210 antibodies alone did not influence secretion of TNFα in the absence of SP-A. In addition, SP-A alone also did not stimulate secretion of TNFα in uninfected cells (not shown). Quantification of bacterial cfu in macrophage lysates over time (Fig. 2B) demonstrated an 8-fold increase in growth of S. aureus in the absence of SP-A, indicating that macrophages do not kill unopsonized S. aureus despite secretion of TNFα. On the other hand, growth of S. aureus was essentially blocked in the presence of SP-A. Importantly, anti-SP-R210 antibodies blocked not only enhanced secretion of TNFα (Fig. 2A) but also the ability of macrophages to suppress growth of S. aureus (Fig. 2B).
FIGURE 2.
SP-R210 mediates TNFα secretion and control of S. aureus growth in macrophages. Mouse bone marrow-derived macrophages were cultured overnight in 24-well dishes at a density of 500,000 cells/well in DMEM, 10% FCS. Serum-deprived macrophages were then preincubated with 20 μg/ml control or anti-SP-R210 antibodies for 1 h before infection with 5 × 106 unopsonized or SP-A-opsonized S. aureus. A, the concentration of TNFα was measured by ELISA in media collected 3 h after infection. Data are means ± S.D. (error bars) (n = 4). ***, p < 0.005 compared with controls in the absence of SP-A. B, infected macrophages were washed in DMEM, and cell-associated bacterial cfu were quantified after serial dilution of cell lysates on tryptic soy broth-agar plates at 3 and 8 h after infection. Data are means ± S.D. (n = 4). ***, p < 0.005 compared with cfu at 3 h after infection.
SP-A Binding to S. aureus Requires Expression of the Staphylococcal Adhesin Eap
In the course of these studies, binding assays revealed saturable high affinity binding of SP-A both in the absence and presence of calcium, suggesting a protein rather than carbohydrate SP-A ligand on S. aureus. Analysis of binding curves revealed 3443 ± 0.31 SP-A binding sites/cell and an affinity of 3.58 ± 0.31 nm (Fig. 3A) in the absence of calcium. The presence of 1.5 mm Ca2+ increased the number to 12,232 ± 530 sites/cell but lowered the binding affinity to 12.71 ± 0.4 nm, suggesting that calcium modifies SP-A binding capacity and avidity to S. aureus (not shown). Previous studies also found that SP-A does not bind cell wall glycoconjugates peptidoglycan and LTA on Gram-positive bacteria (18). Therefore, ligand blot analysis and MALDI mass spectrometry were used to search for protein ligands on the staphylococcal cell wall (Fig. 3, B and C). Cell wall proteins were extracted with lysostaphin or SDS to obtain anchored and peripheral cell wall proteins, respectively. Ligand blots revealed SP-A-binding proteins only in the SDS-sensitive fraction (Fig. 3B). MALDI fingerprint analysis of trypsin-digested protein in corresponding gel bands identified three staphylococcal proteins: the adhesin Eap, also known as MHCH-like adhesin or Map (44) in 64 and 43 kDa bands, the adhesin Emp (44) in the 43 kDa band, and a leukocidin-like subunit (45) in the 43 kDa band. The leukocidin-like protein is identical to the recently described component S of leukocidin G on the surface of S. aureus (46).
FIGURE 3.
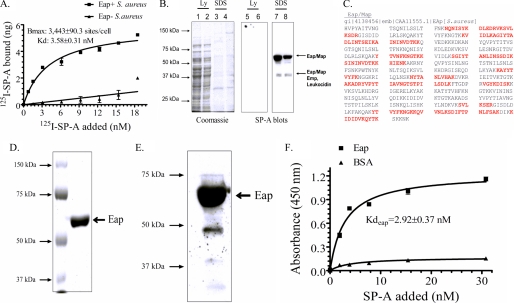
SP-A binding to S. aureus requires expression of the adhesin Eap. A, binding properties of SP-A to Eap+ or Eap− S. aureus Newman were determined by incubating increasing concentrations of 125I-SP-A with 50 × 106 bacteria for 1 h at 37 °C in PBS, 1% BSA without Ca2+. Complexes of bacteria and 125I-SP-A were separated over oil, and bound SP-A was determined using a γ-counter. Data are means ± S.D. (error bars) (n = 4). B, cell wall proteins released from the S. aureus cell wall with either lysostaphin (Ly) (lanes 1, 2, 5, and 6) or 0.4% SDS (lanes 3, 4, 7, and 8) were separated on 10% SDS-polyacrylamide gels. Separated protein was either stained with Coomassie Blue (lanes 1–4) or electrotransferred to nitrocellulose and blotted with biotinylated SP-A (lanes 5–8). MALDI fingerprinting identified the major 64-kDa SP-A-binding protein as the adhesin Eap/Map. The minor 43 kDa band contained peptides from Eap, Emp, and a leukocidin subunit. The results of duplicate experiments are shown. C, the protein sequence of Eap is shown. Identified peptides, shown in red, cover over 50% of the protein. D, native Eap purified from S. aureus Newman (1 μg) was applied on a 10% SDS-polyacrylamide gel and stained with Colloidal Blue. E, purified Eap (1 μg) was visualized by blotting with biotinylated SP-A. F, binding of SP-A to purified Eap was assessed in solid phase assays. Native Eap or BSA adsorbed onto 96-well microtiter plates were incubated with human SP-A (0–20 μg/ml) in blocking buffer for 2 h at 37 °C. Bound SP-A was measured following sequential incubations with rabbit anti-SP-A antibodies and rabbit HRP-conjugated secondary antibody and visualized using 1× tetramethylbenzidine at 450 nm. Data are means ± S.D. (n = 3).
Because Eap was found to be the major component of both 64 and 43 kDa bands, the role of this protein in SP-A binding was pursued further. Identified peptides are shown in red on the Eap protein sequence (Fig. 3C). Purified Eap (Fig. 3D) was used to verify the binding of SP-A in ligand blotting (Fig. 3E) and solid phase assays (Fig. 3F). The binding affinity of SP-A to purified Eap was 2.92 ± 0.37 nm, similar to the binding affinity for SP-A with live S. aureus (Fig. 3A). Importantly, SP-A failed to interact with Eap-deficient S. aureus (Fig. 3A). Binding assays using an inducible Eap-complemented S. aureus strain mAH12(pCXEap) were not feasible due to the low level of Eap induction in this strain (33, 44). However, SP-A was able to detect even low levels of Eap induced in the complemented mAH12(pCXEap) S. aureus strain using the more sensitive ligand blot analysis assay.4 Importantly, SP-A bound full-length recombinant Eap from a different S. aureus strain Mu50 (47) as well; binding of SP-A requires two tandem repeat domains of Eap.4 Eap is specifically expressed by S. aureus and not by other bacterial or staphylococcal species (48, 49), indicating that SP-A is involved in selective recognition of S. aureus via Eap.
Dominant Negative Disruption of SP-R210 Inhibits SP-A Binding and Uptake of SP-A-opsonized S. aureus
In order to study SP-R210 function in more detail, a dominant negative approach was used to disrupt SP-R210 in macrophages. Western blot analysis shows that stable expression of the carboxyl-terminal domain of SP-R210 in Raw264.7 macrophages suppressed expression of endogenous SP-R210 (Fig. 4A, upper band). A cross-reactive 160-kDa protein was not suppressed. The localization of SP-R210 in these dominant negative cells, hereafter called SP-R210(DN) cells, was characterized further. Densitometry indicated a 75% reduction in the expression of SP-R210 in cell lysates of SP-R210(DN) macrophages (not shown). To determine the effect of the SP-R210(DN) mutation in surface localization of SP-R210, cells were surface-biotinylated, and labeled protein was precipitated using streptavidin-Sepharose. Western blot analysis of precipitated protein on Fig. 4B shows that the reduction of SP-R210 expression in SP-R210(DN) cells (Fig. 4A) was accompanied by diminished surface localization of SP-R210 as well. The SP-R210(DN) mutation inhibited surface localization of both the 240-kDa SP-R210L and the 210-kDa SP-R210S isoforms (25). However, SP-R210S is the major isoform in Raw264.7 macrophages. Binding assays demonstrated that the SP-R210(DN) mutation resulted in a partial 50% reduction in SP-A binding sites from 101,811 ± 14,230 sites/cell in control macrophages to 50,732 ± 6891 sites/cell in SP-R210(DN) cells (Fig. 4C). Flow cytometry analysis indicated a 60% reduction in cell surface SP-R210 (not shown). The binding affinity constants were similar, with Kd of 8.8 ± 1.8 and 7.7 ± 1.7 nm in control and SP-R210(DN) cells, respectively.
FIGURE 4.
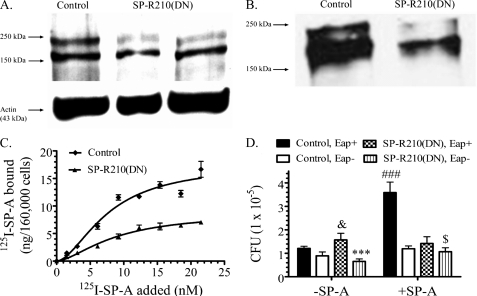
SP-R210 mediates uptake of SP-A-opsonized bacteria. A, dominant negative inhibition of SP-R210 in Raw264.7 macrophages. Western blot analysis assessed expression of SP-R210 in control and SP-R210(DN) macrophages. Proteins (30 μg/lane) were separated on 7% SDS-polyacrylamide gels and electroblotted to nitrocellulose. SP-R210(DN) cell extracts were obtained from two independently derived SP-R210(DN) cells. Probing blots with actin served as loading control. B, control and SP-R210(DN) cells were surface-biotinylated, and extracts were obtained in lysis buffer. Biotinylated protein was precipitated using streptavidin-agarose, and bound protein was separated on 7% SDS-PAGE. Western blot analysis determined the presence of biotinylated SP-R210. A representative blot from two separate experiments is shown. C, binding properties of SP-A to control and SP-R210(DN) cells were measured by incubating cells with increasing concentrations of 125I-SP-A in a 0.1-ml assay volume for 1 h on ice. Bound radioactivity was measured after separation of cells by density centrifugation over oil. Saturation curves were drawn using Prism software. Data shown are means ± S.D. (error bars) (n = 4). D, SP-R210 mediates uptake of SP-A-opsonized Eap+ S. aureus. Control (black and white bars) and SP-R210(DN) (dotted and lined bars) macrophages were infected with a 1:3 ratio of either Eap+ or Eap− S. aureus before or after preincubation with SP-A. After washing, macrophage-associated bacterial cfu were quantified by serial dilution of macrophage lysates. Data shown are means ± S.D. (n = 4 triplicate experiments). ###, p < 0.0001 for SP-A opsonized versus unopsonized Eap+ S. aureus in control macrophages (black bars). ***, p < 0.001 for Eap- versus Eap+ S. aureus infection of SP-R210(DN) macrophages in the absence of SP-A. &, p < 0.05 for Eap+ S. aureus infection between control and SP-R210(DN) cells in the absence of SP-A. $, p < 0.05 for SP-R210(DN) macrophages infected with Eap− S. aureus in the presence versus absence of SP-A.
Simultaneous assays using S. aureus and macrophages deficient in SP-A binding affirmed the specific role of SP-R210 in binding and uptake of SP-A-bound bacteria. Thus, Fig. 4D demonstrates that SP-A enhanced uptake of Eap+ S. aureus almost 4-fold (Fig. 4D, black bars), whereas SP-A did not alter uptake of Eap− S. aureus in control macrophages (Fig. 4D, white bars). Furthermore, SP-A failed to induce uptake of SP-A-opsonized Eap+ S. aureus (Fig. 4E, dotted bars) in SP-R210(DN) cells.
SP-R210(DN) Macrophages Display Enhanced Non-opsonic Uptake of S. aureus
The experiments depicted on Fig. 4D produced two unexpected results. First, SP-A enhanced uptake of the non-SP-A binding Eap− S. aureus by 2-fold in SP-R210(DN) cells (compare stripped bars on Fig. 4D), suggesting that SP-A may have a secondary role in enhancing non-opsonic uptake when the density of SP-R210L is significantly reduced. Second, in the absence of SP-A, the SP-R210(DN) cells displayed a small but consistent 35% increase in uptake of Eap+ S. aureus compared with control macrophages (dotted versus black bars), whereas the uptake of Eap− S. aureus decreased by 2.5-fold compared with Eap+ S. aureus (1.56 ± 0.25 cfu versus 0.66 ± 0.10 cfu; striped versus dotted bars) in the absence of SP-A. Differences were statistically significant. These results indicate compensatory induction of a non-opsonic phagocytic receptor in SP-R210(DN) cells.
The Non-opsonic Scavenger Receptor SR-A Is Induced in SP-R210(DN) Cells
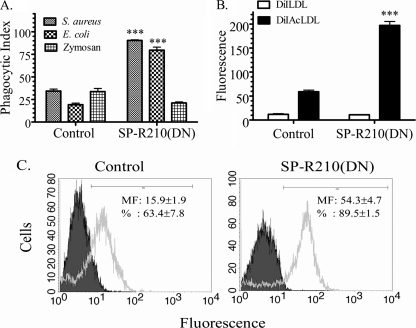
Non-opsonic phagocytosis in SP-R210(DN) cells was probed using commercially available killed fluorescent S. aureus, E. coli, or yeast zymosan bioparticles (Fig. 5A). The experiments depicted in Fig. 5A measured the phagocytic index, an assessment of internalized bacteria (50, 51). Fig. 5A shows a 3–4-fold increase in phagocytosis of S. aureus and E. coli but not zymosan bioparticles by SP-R210(DN) cells. Previous studies identified the scavenger receptor SR-A in phagocytosis of S. aureus and E. coli K-12 bioparticles (52). Enhanced SR-A expression should also lead to increased endocytosis of chemically modified acetylated LDL (53). As shown in Fig. 5B, SP-R210(DN) cells endocytosed 4 times more AcLDL than control cells. The flow cytometry analysis in Fig. 5C shows a 3-fold higher surface expression of SR-A in SP-R210(DN) cells. These results indicate a previously unknown role of SP-R210 in constitutive cross-modulation of the scavenger receptor SR-A. Previous studies showed that SR-A has dual roles in non-opsonic bacterial clearance and dampening of inflammatory responses in vivo (54–57). Based on the above in vitro findings, the following studies investigated in vivo outcomes of S. aureus acute pneumonia in WT and SR-A−/− mice.
FIGURE 5.
Increased expression and function of SR-A in SP-R210(DN) macrophages. A, phagocytic activity of control and SP-R210(DN) macrophages was assessed using FITC-labeled S. aureus, E. coli, or yeast zymosan bioparticles. Control and SP-R210(DN) were incubated with a 10:1 ratio of the indicated bioparticles for 30 min at 37 °C. Data are expressed as phagocytic index, representing the percentage of cells containing internalized bioparticles. Data are means ± S.D. (error bars) (n = 12). ***, p < 0.001 compared with control macrophages. B, scavenger receptor activity of control and SP-R210(DN) was assessed using fluorescently labeled acetylated LDL (DiAcLDL) as an endocytic ligand of scavenger receptors. Native fluorescent LDL (DiLDL) was used as control. Macrophages were incubated with 0.8 μg/ml native fluorescent LDL or fluorescently labeled acetylated LDL for 30 min at 37 °C. Uptake of LDLs was analyzed by flow cytometry. Data are means ± S.D. (n = 8). ***, p < 0.001 compared with control macrophages. C, surface expression of the scavenger receptor SR-A in control and SP-R210(DN) cells was assessed using a monoclonal rat anti-mouse SR-A antibody clone 2F8. Grey-shaded and unshaded histograms show staining with isotype control and anti-SR-A antibodies, respectively. Linear gating was used to calculate fluorescent intensity, and the percentage of SR-A-positive cells in control and SP-R210(DN) cell cultures is shown in histogram insets. Data are means ± S.D. (n = 8).
WT Mice Are Susceptible to Infection with Eap− S. aureus
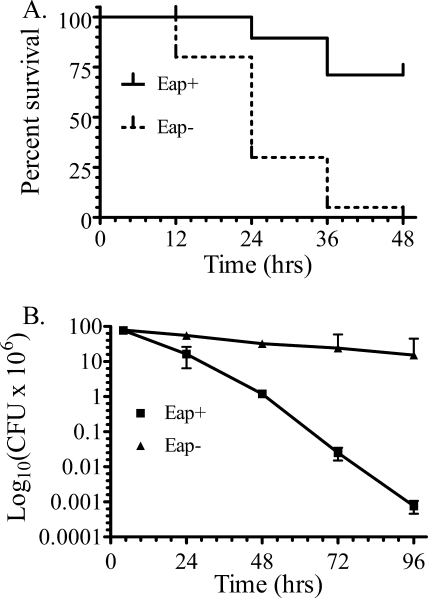
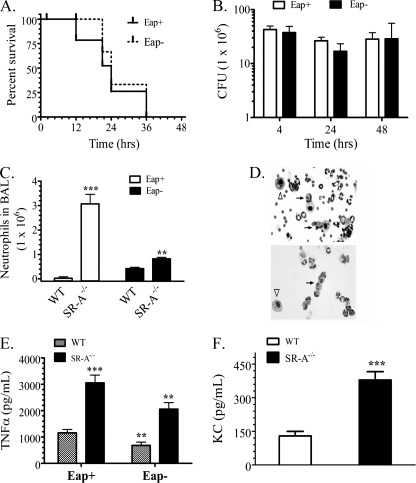
Although SP-R210-deficient mice are not yet available, the Eap+ and Eap− S. aureus strains modeled SP-A opsonization and hence SP-R210 function in vivo. This is predicated from a range of in vitro data in the present and previous work (23) that firmly place SP-R210 as an opsonic receptor for SP-A-bacterial complexes. An intranasal model of S. aureus pneumonia was used to determine survival and bacterial clearance of the SP-A-binding Eap+ or the non-SP-A-binding Eap− S. aureus strains. The reported LD50 for intranasal infection with S. aureus Newman was determined to be 4 × 108 cfu (37). Here, mouse survival was monitored after infection with 3 × 108 cfu of Eap+ or Eap− S. aureus Newman (Fig. 6A) or 75% of the reported LD50. Mice infected with Eap− S. aureus succumbed to the infection with a mean survival of 24 h, whereas mice infected with the parental Eap+ S. aureus had a median survival of >48 h. There were no additional deaths of surviving Eap+ S. aureus-infected mice after 2 weeks of observation (not shown). Pulmonary bacterial clearance was then monitored in lung homogenates from mice infected with a lower 0.5 LD50 dose of Eap+ or Eap− S. aureus. However, the SP-A-resistant Eap− S. aureus strain was cleared slowly with a half-life of 40.3 h, whereas the Eap+ S. aureus was cleared 4 times faster with a half-life of 9.5 h. The lungs of Eap+ and Eap− S. aureus-infected mice retained 0.0001 and 19% of cfu compared with the 4 h time point 96 h after infection (Fig. 6B).
FIGURE 6.
Survival and pulmonary bacterial clearance of mice infected with Eap+ or Eap−S. aureus. A, survival of 6–8-week-old C57BL/6 male mice was monitored at 12-h intervals after intranasal infection with 300 × 106 cfu of Eap+ or Eap− S. aureus Newman. Data shown are from a total of n = 40 mice/group in three independent experiments. Survival curves were generated using the Kaplan-Meier method. The mean survival of mice infected with Eap− S. aureus was 24 h. Differences in survival were statistically significant with p < 0.0001. Statistical analysis of survival curves was performed using the Gehan-Breslow-Wilcoxon test tool using Prism software. B, pulmonary clearance of intranasal infection of mice with 200 × 106 cfu of Eap+ and Eap− S. aureus was monitored in lung homogenates at the indicated time intervals. Data shown are means ± S.E. (error bars) (n = 7–9 at the 4, 24, and 72 h time points, n = 4 at the 96 h time point).
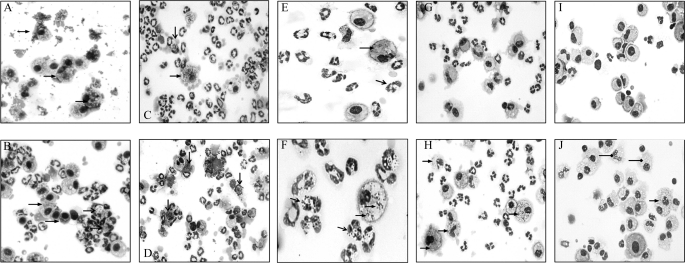
Differential staining of cells in lung lavage over time (Fig. 7) revealed several notable differences in the handling of infections with Eap+ and Eap− S. aureus. First, alveolar macrophages were the dominant cell type 4 h after infection with Eap+ S. aureus (Fig. 7A), whereas the lavage of Eap− S. aureus was already infiltrated with neutrophils (Fig. 7B, open head arrows). Macrophages from mice infected with Eap+ S. aureus contained internalized and aggregated bacteria on the cell surface (Fig. 7A, closed head arrows), consistent with SP-A-mediated agglutination and phagocytosis of the Eap+ strain at the early stage of infection. In contrast, Eap- S. aureus had been taken up by both macrophages (Fig. 7B, closed head arrows) and neutrophils. However, neutrophils were engorged with Eap− staphylococci 4 h (Fig. 7B, open head arrows) and 24 h (Fig. 7D, open head arrows) after infection, indicating that Eap− S. aureus were phagocytosed but not killed by neutrophils. The outcome of this deficiency was persistent infection of neutrophils and macrophages at 48 h (Fig. 7F), 72 h (Fig. 7H), and up to 96 h (Fig. 7J) after infection with Eap− S. aureus, consistent with the slow clearance of the Eap− strain from the lungs (Fig. 6B). In addition, the cells in 96-h lavage from surviving mice infected with Eap− S. aureus were vacuolated, suggesting that persistent Eap− S. aureus elaborated cytotoxic factors inside cells (Fig. 7J, diamond head arrows). In contrast, the Eap+ S. aureus infection was essentially cleared after 48 h; macrophages with oval- or kidney-shaped nuclei without detectable bacteria repopulated the lavage, whereas neutrophils declined 48–96 h after infection (Fig. 7, E, G, and I). It is noteworthy that heavily infected neutrophils were closely associated with macrophages (Fig. 7, B and D). Previous studies have demonstrated that normal host responses involve recruitment of neutrophils that kill bacteria but undergo apoptosis in the process. Apoptotic neutrophils are then ingested by macrophages, limiting neutrophilic injury to host cells and tissues (58). Here, it is possible that abnormal neutrophil turnover resulted in superinfection and lysis of neutrophils with Eap− S. aureus, causing persistent infection of macrophages. The present findings indicate that SP-A binding to S. aureus via Eap drives initial recognition and clearance by alveolar macrophages, mediating subsequent resolution of acute S. aureus infection by macrophages and neutrophils.
FIGURE 7.
Cytospin analysis of alveolar lavage from mice infected with EAP+ and Eap−S. aureus. Bronchoalveolar lavage was collected at 4 h (A and B), 24 h (C and D), 48 h (E and F), 72 h (G and H), and 96 h (I and J) after intranasal infection of mice with 200 × 106 cfu Eap+ (A, C, E, G, and I, left panels) or Eap− (B, D, F, H, and J, right panels) S. aureus. Cells were deposited onto glass slides by cytospin centrifugation and stained with HEMA-3 to evaluate inflammatory cells and cell-associated bacteria. Images in E and F were captured at ×40 magnification. All other images were photographed at ×20 magnification. Open and closed head arrows (A–F, H, and J) point to infected neutrophils and macrophages, respectively. The diamond head arrows (J) indicate vacuolated cells.
Inflammatory Responses in WT Mice Infected with Eap+ and Eap− S. aureus
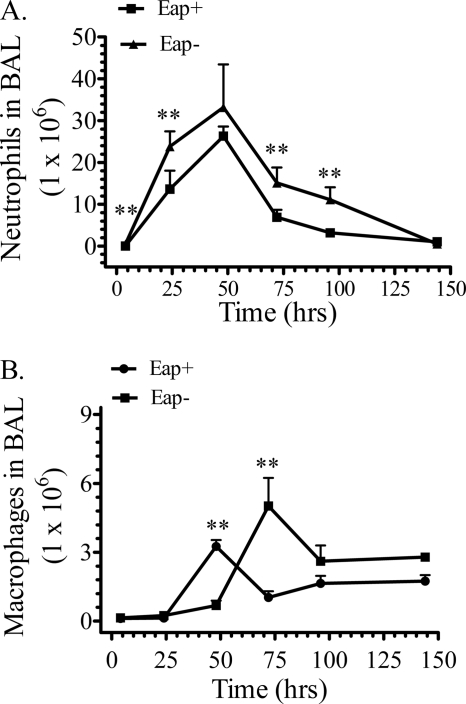
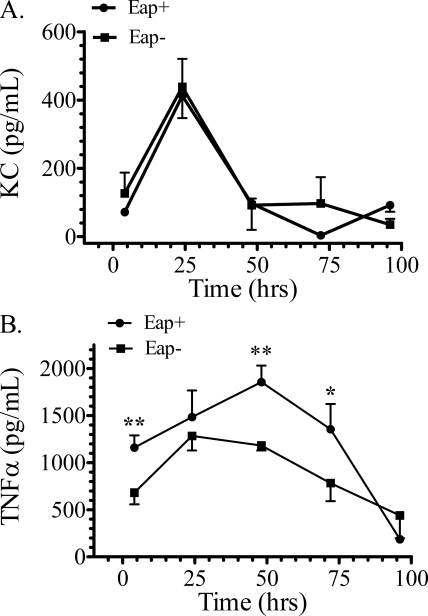
Quantitative analysis of inflammatory cells and mediators measured differences in lung inflammatory responses to Eap+ and Eap− S. aureus infections. The number of neutrophils peaked at 48 h in both infections (Fig. 8A) but was persistently higher in mice infected with Eap− S. aureus. Differences were statistically significant at the early and late time points after infection (Fig. 8A). Consistent with the results in Fig. 7, A and B, the number of neutrophils 4 h after infection with Eap+ or Eap− S. aureus was 0.25 ± 0.06 × 105 and 4.23 ± 0.55 × 105, respectively. However, the levels of the neutrophil chemokine KC (Fig. 9A) were not significantly different, indicating normal KC production in response to both infections in WT mice. KC was not detectable in uninfected mice. However, the present (Fig. 2A) and previous studies (23, 26) showed that SP-A-opsonized bacteria induced secretion of TNFα in macrophages. TNFα is crucial for orchestrating recruitment of neutrophils and resolution of inflammation (59, 60), whereas TNFα primes neutrophils to kill S. aureus infection in vivo (61). Here, TNFα levels peaked 48 h after infection with Eap+ S. aureus (Fig. 9B), whereas TNFα peaked at least 24 h earlier in Eap− S. aureus infection. However, the TNFα levels in Eap− infection were always lower than the Eap+ infection at any time point and indeed 50% lower at the earlier time points (Fig. 9B), indicating that inflammatory responses are regulated differently in the absence of SP-A opsonization. Thus, although both neutrophils and exudate macrophages peaked 48 h after Eap+ S. aureus infection (Fig. 8, A and B), peak recruitment of macrophages in response to Eap− S. aureus was delayed to 72 h in relation to Eap+ S. aureus (Fig. 8B), whereas neutrophil recruitment was not delayed. These findings support the conclusion that SP-A opsonization mediates coordinated recruitment of inflammatory cells and clearance of S. aureus through modulation of TNFα levels by alveolar macrophages.
FIGURE 8.
Recruitment of neutrophils and macrophages in mice infected with Eap+ and Eap−S. aureus. Cellular infiltrates were evaluated in bronchoalveolar lavage (BAL) after intranasal infection of WT mice with 200 × 106 cfu Eap+ or Eap− S. aureus. Total cell numbers were counted using a hemacytometer. Cell types were identified by differential staining following cytospin centrifugation of lavaged cells. Counting of neutrophils (A) and macrophages (B) in 5–10 microscopic fields determined the percentage of each cell type in bronchoalveolar lavage. The percentage of each cell type was multiplied by the total number of cells in bronchoalveolar lavage to obtain the number of neutrophils and macrophages. Data shown are means ± S.E. (n = 7–9 mice at the 4, 24, and 72 h time points; n = 4–6 mice at the 96 h time point). **, p < 0.03 indicates significant differences in neutrophil and macrophage numbers between Eap+ and Eap− S. aureus-infected mice at the indicated time points.
FIGURE 9.
Levels of KC and TNFα in WT mice infected with Eap+ or Eap−S. aureus. The concentration of KC (A) and TNFα (B) was determined by ELISA in lung homogenates after infection of mice with 200 × 106 cfu Eap+ or Eap− S. aureus. Data shown are means ± S.E. (n = 7–9 mice at the 4, 24, and 72 h time points; n = 4–6 mice at the 96 h time point). Statistical differences in TNFα levels at each time point are p < 0.01 (**) at the 4 and 48 h time points and p < 0.05 (*) at the 72 h time point.
Expression of SR-A Is Required for Effective Clearance and Control of Neutrophilic Inflammation in Eap+ S. aureus Infection
Disruption of SP-R210 in macrophages (Figs. 4 and 5) resulted in enhanced expression and function of the scavenger receptor SR-A. Previous studies reported independent roles for SR-A in clearance of S. aureus and control of chemokine-driven neutrophil recruitment in the peritoneal cavity (55, 56). The role of SR-A in pulmonary clearance of S. aureus is not known. In the context of the present study, we considered that modulation of SP-R210 levels during clearance of SP-A-opsonized S. aureus may coordinate inflammatory responses through SR-A. Therefore, the role of SR-A was assessed using SR-A-deficient mice. It is noteworthy that the numbers of initial cfu in both WT (Fig. 6B) and SR-A−/− (Fig. 10B) lungs 4 h after infection were similar among S. aureus strains, indicating that humoral clearance of S. aureus in the very early phase of infection is not dependent on Eap. Previous studies showed that initial killing of S. aureus and other bacteria occurs through non-phagocytic mechanisms (62–64), whereas the subsequent phagocytic phase of the infection is slower, with an estimated half-life of 10.8 h for S. aureus survival in macrophages (62). This rate of clearance is similar to the half-life of Eap+ S. aureus measured in the present study in WT mice in vivo (Fig. 6B). Interestingly, the number of initial cfu at the 4 h time point was 42.7 ± 7.0 × 106 cfu for Eap+ and 37.5 ± 11.5 × 106 cfu for Eap− S. aureus. The number in SR-A−/− mice was 50% lower than in WT mouse lungs, containing 95.7 ± 6.8 × 106 cfu for Eap+ and 85.7 ± 33.8 × 106 cfu for Eap− S. aureus, suggesting increased early humoral host defense in SR-A−/− mice. However, Fig. 10A shows that, unlike WT mice (Fig. 6A), lack of SR-A impaired the ability of mice to survive sublethal infection with Eap+ S. aureus. Most SR-A−/− mice infected with a sublethal dose of S. aureus strains died 4 days after infection (not shown). Importantly, phagocytic clearance of Eap+ S. aureus by SR-A−/− lungs was attenuated, as indicated by similar bacterial burden for both Eap+ and Eap− S. aureus 4–48 h after infection (Fig. 10B). Both strains declined about 2-fold between 4 and 24 h in SR-A−/− mice (Fig. 10B), compared with 6- and 1.6-fold declines in Eap+ and Eap− S. aureus cfu WT mice (Fig. 6B), respectively. Although WT mice cleared Eap− S. aureus slowly compared with the Eap+ strain after 24 h, both S. aureus strains proliferated between 24 and 48 h after infection in SR-A−/− mice. These results indicate decreased capacity of SR-A−/− mice to control Eap+ S. aureus infection in the lung.
FIGURE 10.
Assessment of pneumonia in SR-A−/− mice infected with Eap+ and Eap−S. aureus. A, survival of 6–8-week-old SR-A−/− mice was monitored at 12-h intervals after infection with 300 × 106 cfu of Eap+ or Eap− S. aureus Newman. Data shown are from a total of n = 20 mice/group from two independent experiments. Survival curves were generated using the Kaplan-Meier method. B, pulmonary clearance after infection with 200 × 106 cfu of Eap+ or Eap− S. aureus was monitored in lung homogenates at the indicated time intervals. Data shown are mean ± S.E. (error bars) (n = 6–8 at the 4, 24, and 48 h time points). C, the number of neutrophils in lavage 4 h after infection was counted as described in the legend to Fig. 8. Data shown are means ± S.E. (error bars) (n = 8) in two independent experiments. D, cytospin analysis of lung lavage 4 h after infection with the indicated S. aureus strains. E, the concentration of TNFα was measured in lung homogenates 4 h after infection with the indicated S. aureus strains. Data shown are mean ± S.E. (n = 8) in two independent experiments per group. F, the concentration of KC was measured in lung homogenates 4 h after infection with Eap+ S. aureus. Data are mean ± S.E. (n = 8) in two independent experiments.
Furthermore, assessment of inflammatory responses shows that a lack of SR-A was associated with accelerated influx of neutrophils. The number of neutrophils increased 30-fold more in SR-A−/− mice than in WT mice 4 h after infection with Eap+ S. aureus (Fig. 10C). Interestingly, the number of neutrophils in Eap− S. aureus infection increased only 2-fold more in SR-A−/− mice than in WT mice (Fig. 10C). However, cytospin analysis revealed necrotic neutrophils engorged with bacteria 4 h after infection with both Eap+ and Eap− strains (Fig. 10D, black arrows), suggesting decreased killing of bacteria by neutrophils. This deficiency in bacterial killing was selectively observed in WT mice infected with Eap− S. aureus (Fig. 7, B and D) above but was a prominent early feature of the infection in SR-A−/− mice. The Eap− S. aureus infection was particularly cytotoxic (Fig. 10D, bottom, black arrows). However, the Eap+ strain produced a similar overwhelming infection in neutrophils but was more evident 24 h after infection (not shown). Neutrophil destruction was reflected by a 2–3-fold lower number of neutrophils in SR-A−/− mice compared with WT mice counted 24 h after infection with both S. aureus strains (not shown). Sparse macrophages were either uninfected or filled with bacteria (Fig. 10D, white arrowheads), suggesting impaired clearance of bacteria by these cells as well. In contrast to WT mice (Fig. 9), SR-A−/− mice displayed enhanced secretion of inflammatory mediators. Secretion of TNFα was significantly higher in both Eap+ and Eap− S. aureus infection (Fig. 10E). In addition, the levels of KC, measured in the Eap+ S. aureus infection were significantly higher in SR-A−/− mice compared with WT mice (Fig. 10F). These results indicate that SR-A−/− mice lost the ability to both control inflammation and clear SP-A-opsonized Eap+ S. aureus.
Differential Expression of the SP-R210S Isoform Is Associated with Increased Differentiation of Alveolar Macrophages in SR-A−/− Mice
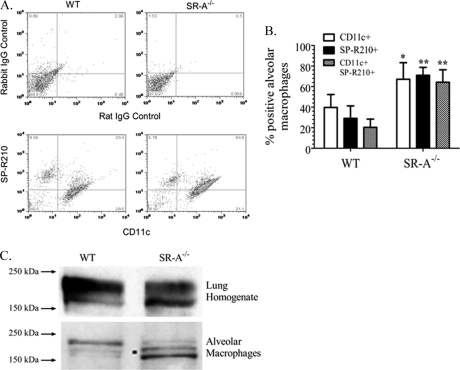
Given the above findings (Figs. 4B and 10), the following experiments determined the effect of SR-A-deficiency on expression of SP-R210. Alveolar macrophages from WT and SR-A−/− mice were stained with antibody to CD11c, a marker for terminal alveolar macrophage differentiation (65, 66), along with anti-SP-R210 antibodies. Representative two-color histograms in Fig. 11A and the combined data from several experiments on Fig. 11B show that about 50% of CD11c+ alveolar macrophages also express SP-R210 in WT alveolar macrophages. Remaining SP-R210 expression was found in CD11c− alveolar macrophages. The expression level of SP-R210 was lower in both alveolar macrophage subpopulations in SR-A−/− mice. The mean fluorescence intensity of SP-R210 was 104.3 ± 9.7 and 88.7 ± 14.3 in WT SP-R210+CD11c− and SP-R210+CD11c+ cells, respectively. In comparison, the mean fluorescent intensity of SP-R210 in SR-A−/− mice was 49.2 ± 2.6 and 32.3 ± 1.1 in SP-R210+CD11c− and SP-R210+CD11c+ alveolar macrophage populations, respectively. However, the percentage of SP-R210+ alveolar macrophages in SR-A−/− mice increased 3-fold to over 60% of alveolar macrophages with more than 95% of SP-R210+ alveolar macrophages also expressing CD11c. Interestingly, the percentage of CD11c+ alveolar macrophages increased significantly, indicating that more SR-A−/− alveolar macrophages reach terminal differentiation. Flow cytometry analysis does not distinguish expression of SP-R210 receptor isoforms. Therefore, Western blot analysis assessed expression of SP-R210 isoforms in whole lung and alveolar macrophage extracts. Fig. 10C demonstrates that the loss of SR-A is associated with an altered expression profile of SP-R210 isoforms. SP-R210L is the most abundant isoform in whole lung extracts from both WT and SR-A−/− lung extracts. Expression of the SP-R210L isoform was reduced in SR-A−/− lung extracts, whereas expression of SP-R210S increased (Fig. 11C, top). However, SP-R210L is the main isoform expressed in WT alveolar macrophages, whereas SR-A−/− alveolar macrophages express mainly SP-R210S (Fig. 11C, bottom). In this regard, the expression profile of SP-R210 isoforms in WT alveolar macrophages differs from that observed in macrophages derived from extrapulmonary sites; SP-R210S rather than SP-R210L is the major isoform in peripheral bone marrow-derived macrophages (25, 29), human monocyte-derived macrophages (27), and peritoneal Raw264.7 macrophages (Fig. 4B). A third 160-kDa species recognized by the affinity-purified antibody also increased in SR-A−/− macrophages (Fig. 11C). This species is similar in molecular weight to an intracellular form in Raw264.7 cells (Fig. 4A) and was also detected in human monocyte-derived macrophages (27). These results indicate that expression of SR-A is linked to differential expression of SP-R210 isoforms; alveolar macrophages expressing SP-R210L in WT mice were replaced with alveolar macrophages that express SP-R210S in SR-A−/− mice. As shown earlier (Fig. 1), SP-R210S mediates attachment but not internalization of SP-A-opsonized S. aureus. In addition, loss of SP-R210L and partial inhibition of SP-R210S in SP-R210(DN) cells blocked the uptake of SP-A-opsonized S. aureus (Fig. 4D). Taken together, the results of the present study support the conclusion that SP-R210L is crucial for internalization and clearance of SP-A-opsonized S. aureus by alveolar macrophages, with SR-A having a regulatory role modulating expression and inflammatory functions of SP-R210.
FIGURE 11.
Differential expression of SP-R210 isoforms in WT and SR-A−/− mice. Flow cytometric analysis determined expression of CD11c and SP-R210 on alveolar macrophages from WT and SR-A−/− mice. A, representative dual color histograms following staining of alveolar macrophages with either isotype-matched IgG in upper panels or anti-CD11c and anti-SP-R210 antibodies in lower panels. B, combined flow cytometric data were expressed as a percentage of single or double positive cells for the indicated markers in WT or SR-A−/− alveolar macrophages. Data are means ± S.E. (error bars) (n = 6 independent experiments on pooled alveolar macrophages from 5 mice/genotype). *, p < 0.05; **, p < 0.01 SR-A−/− compared with WT mice. C, lung or alveolar macrophage extracts were prepared in lysis buffer, separated on 7% SDS-PAGE, and evaluated by Western blot analysis using anti-SP-R210 antibodies. Lanes were loaded with 20 μg of protein. Blots are representative of two separate experiments.
DISCUSSION
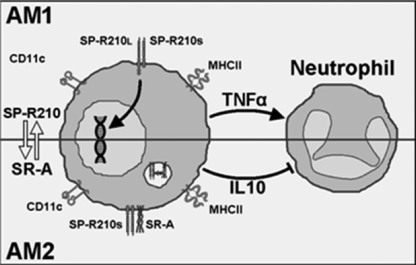
The interaction of surfactant protein A and alveolar macrophages is the first line of communication between the humoral and innate immune systems in the lower respiratory tract. The goal of the current study was to elucidate the role of SP-A and its receptor SP-R210 in opsonization and clearance of S. aureus both in vitro and in vivo. Previous in vitro work showed that SP-R210 orchestrates macrophage activation and phagocytosis of SP-A-opsonized M. bovis BCG (23, 26). The results of the present study not only corroborate earlier findings but also define new roles of SP-A and SP-R210 in linking bacterial opsonization and clearance with appropriate host responses to acute infection with S. aureus. The first advance of the current study was the identification of Eap as the SP-A ligand on S. aureus. The second advance was the identification of a previously unknown cross-modulation between expression of SP-R210 isoforms SP-R210L and SP-R210S and the scavenger receptor SR-A, helping to balance inflammatory responses during SP-A-mediated clearance of S. aureus in vivo. The outcome of dual deficiency in SP-R210L and SR-A in SR-A−/− alveolar macrophages was impaired clearance of the SP-A-opsonized Eap+ S. aureus along with an accelerated inflammatory response attracting neutrophils that were not able to clear the infection. The present findings support the model in which SP-A-opsonized bacteria encounter SP-R210L on alveolar macrophages mediating initial clearance and appropriate priming of the inflammatory response through secretion of TNFα. In this model, counterregulatory interactions between SP-R210S and SR-A control the timing, intensity, and duration of the inflammatory response during infection facilitating clearance and resolution of S. aureus pneumonia in vivo (Fig. 12).
FIGURE 12.
Proposed interaction of SP-R210 and SR-A in alveolar macrophages. Resting alveolar macrophages AM1 express SP-R210L. Ligation of SP-R210L with SP-A-opsonized bacteria in AM1 macrophages induces macrophage activation and secretion of TNFα, timing recruitment and activation of neutrophils. Polarization of AM1 to AM2 macrophages results in expression of SP-R210S and SR-A, coordinating phagocytosis via an SP-R210S·SR-A complex with regulation of the inflammatory response.
SP-A is the principal lung opsonin that associates with S. aureus within 30 min after intranasal infection in mice (67, 68). The present and previous studies demonstrate that two members of the collectin family of proteins, SP-A and MBL, are crucial for eradication of acute infections with S. aureus. Mice deficient in MBL were highly susceptible to systemic S. aureus infection (69). MBL limits hematogenous spread of the bacteria to lung and other tissues. MBL binding to S. aureus cell wall glycoconjugates activates the lectin pathway of complement, enhancing recruitment and clearance of S. aureus through macrophages and neutrophils (69, 70). In the lung, however, MBL appears later in inflammatory fluid from the periphery, associating with S. aureus 6 h after infection (67, 68). Unlike MBL, SP-A does not bind cell wall glycoconjugates, such as LTA or peptidoglycan, on S. aureus. Accordingly, the present studies identified Eap (44) as a critical protein ligand for SP-A binding to S. aureus. Importantly, Eap is necessary for the in vivo clearance of acute S. aureus infection; the absence of Eap presents a defect in SP-A-mediated opsonization consistent with the reduced clearance and high morbidity of WT mice exposed to Eap- S. aureus. In addition to SP-A, lung surfactant contains SP-D, another member of the collectin family of proteins. Both MBL and SP-D share similar binding specificities for cell wall glycoconjugates LTA and peptidoglycan (18, 71). However, MBL and SP-D did not protect mice against acute Eap− S. aureus infection. These results demonstrate that SP-A plays a primary non-redundant role in opsonic clearance of acute S. aureus infection in the lung.
In addition, the present findings indicate that Eap is targeted by both opsonic SP-R210 and non-opsonic SR-A receptors on alveolar macrophages. The alteration in non-opsonic properties of the SP-R210(DN) cells was observed as a small but consistent 30–40% increase in the uptake of Eap+ S. aureus. This assay measured total uptake rather than phagocytic internalization; it was not feasible to distinguish internalized from surface-attached live bacteria. However, flow cytometry using killed fluorescent bacteria, which preferentially interact with SR-A, measured a clear 3–4-fold increase in phagocytic index following quenching of surface-attached bacteria. A significant 2–3-fold reduction in uptake of Eap− S. aureus was measured in the SR-A-enriched SP-R210(DN) macrophages, suggesting that SR-A binds Eap. Recent studies expanded the ligand repertoire of SR-A ligands to surface proteins of Neisseria meningitides (72). Mutation in N. meningitides SR-A-binding protein NMB0667 was lethal in WT mice. It remains to be established whether Eap is also recognized by SR-A.
The adhesin Eap is unique to S. aureus (48, 49). Binding of SP-A to Eap represents a novel mechanism for SP-A recognition of virulent S. aureus. The ability of SP-A to interact with an adhesin widely expressed among diverse S. aureus strains (49) may aid in rapid recognition and clearance of dangerous S. aureus by alveolar macrophages in the distal lung. In contrast, recognition of S. aureus by MBL and SP-D could be limited by common modifications of cell wall glycoconjugates. For example, the acylation status of lipoteichoic acid may abrogate binding of MBL to different S. aureus strains (73). There are two types of S. aureus adhesins: the SERAMs and the MSCRAMMs (74–76). The SERAMs are soluble proteins that reattach to the cell wall upon secretion, whereas MSCRAMMs are covalently anchored on the cell wall. Eap is a member of the SERAM group of staphylococcal adhesins. S. aureus Eap was initially identified as a binding protein for matrix and plasma proteins, interactions that result in tissue invasion and immune modulation (33, 77). Eap contributes to abscess formation, invasion, and persistence of S. aureus in host tissues (78, 79). On the other hand, previous studies showed that deletion of the major MSCRAMM adhesin FnBP (80) enhanced rather than impaired virulence of S. aureus in the lung, although FnBP-deficient S. aureus was less virulent in peripheral tissues. However, FnBP was deleted in the laboratory strain 8325-4, which also does not express Eap (81). The clinical strain S. aureus Newman used here expresses high levels of Eap but lacks surface expression of FnBP (82). It remains to be determined whether modulation of Eap in the host represents a novel invasion strategy for different methicillin-resistant S. aureus strains causing health care- and community-acquired pneumonia. The present results indicate that SP-A binding through Eap is crucial in limiting invasion of S. aureus in the lung.
In this study, we show that recognition of SP-A-opsonized S. aureus by SP-R210 results in enhanced macrophage uptake and clearance of S. aureus. SP-A was not able to induce uptake in the absence of either SP-R210 or Eap, which disabled opsonic recognition of S. aureus. Furthermore, only WT mice infected with the SP-A-targetable S. aureus could elaborate bacterial clearance, accurate timing, and coordination of the inflammatory response in vivo. Agglutinated S. aureus was initially presented on the surface of alveolar macrophages in association with moderate secretion of TNFα that increased gradually until 48 h, coinciding with peak neutrophil recruitment and eradication of over 90% of the bacterial inoculum by this time point. In contrast, mice infected with Eap− S. aureus that is not recognized by SP-A displayed impaired killing and persistent infection in association with elaboration of low but unsustainable levels of TNFα, more intense neutrophilic inflammation, and delayed repopulation of the lung with macrophages. Previous studies reported that SP-A mediates both clearance and restoration of alveolar immune homeostasis in mice infected with Mycoplasma pneumoniae (83). However, the relative importance of SP-A binding to macrophages or bacteria has not been clear because SP-A may have indirect activities on macrophages. The present results indicate that SP-A mediates optimal presentation of S. aureus to alveolar macrophages through its capacity to act as an opsonin, resulting in appropriate priming and modulation of the inflammatory response in the lung.
Furthermore, the present findings support the conclusion that SP-A coordinates innate host defense through two differentially expressed isoforms of SP-R210, SP-R210L and SP-R210S, in collaboration with the scavenger receptor SR-A on the macrophage surface. We show that SP-A-opsonized S. aureus encounters alveolar macrophages that express mainly SP-R210L in WT mice but SP-R210S in SR-A−/− mice. The first symptom associated with the dual alveolar macrophage deficiency in SR-A and SP-R210L was the inability of SR-A−/− mice to survive sublethal infection with the SP-A-opsonized Eap+ S. aureus. The SR-A−/− mice developed an untrained inflammatory response characterized by an extraordinary number of neutrophils rushing into the alveolar space within 4 h rather than over an extended 48-h window observed in WT mice. Accelerated influx of neutrophils at the early stage of infection in SR-A−/− mice was associated with a marked 3–4-fold increase in secretion of both TNFα and the neutrophil chemokine KC compared with WT mice. As indicated earlier, the levels of TNFα were moderately increased by 2-fold in Eap+ compared with Eap− S. aureus infection in WT mice at 4 h, but KC was not affected. In addition, SP-R210S mediated uptake of SP-A-opsonized Eap+ S. aureus along with enhanced secretion of TNFα in bone marrow-derived macrophages. Because SR-A−/− alveolar macrophages express SP-R210S similar to bone marrow-derived macrophages, the SP-R210S isoform was probably responsible for the stronger TNFα response in the absence of SR-A. On the other hand, the present findings show that SR-A controls neutrophil recruitment through secretion of the chemokine KC. An earlier study on sterile peritonitis revealed that SR-A regulates the rate of neutrophil chemotaxis through down-modulation of neutrophil chemokines, including KC (55). Inappropriate levels of KC during infection, however, may counteract TNFα priming of neutrophil bactericidal activities. In this regard, KC and other CXC chemokines are known to potentiate intracellular survival of S. aureus by diverting S. aureus to macropinosomes rather than phagolysosomes (84, 85). Here, cytospin analysis of inflammatory cells in the lavage of SR-A−/− mice showed that neutrophils contained large numbers of S. aureus, indicating defective killing of internalized bacteria. Increased relative concentration of KC and TNFα in WT mice infected with Eap− S. aureus may have contributed to the intracellular persistence of the pathogen as well. Therefore, SP-R210-regulated expression of SR-A is probably critical for the development of effective innate immunity against S. aureus infection in the lung. In support of this hypothesis, previous studies showed that SP-A induced surface localization of SR-A in alveolar but not peritoneal macrophages during infection with Streptococcus pneumoniae (86). In the present work, SP-R210(DN) mutant macrophages compensated with induction of non-opsonic phagocytosis through SR-A, indicating constitutive cross-modulation between one or both isoforms of SP-R210 with SR-A. Conversely, SR-A−/− alveolar macrophages had a reciprocal increase in the number of macrophages expressing SP-R210S. However, the expression level of SP-R210S was more than 50% lower than SP-R210L in WT alveolar macrophages, indicating reduced ability of the shorter SP-R210S isoform to localize to the cell surface in the absence of SP-R210L and SR-A. In this manner, SR-A−/− mice are deficient in both opsonic SP-R210-mediated and non-opsonic SR-A-mediated clearance of S. aureus. The mechanism behind the constitutive cross-modulation between SP-R210 and SR-A is not yet known but could be related to the ability of SR-A to enhance macrophage adhesion. Adhesion modulates differentiation, intercellular communication, and inflammatory responses in macrophages (87, 88). Here, SR-A−/− alveolar macrophages display a state of increased differentiation, as indicated by the higher expression of CD11c with more alveolar macrophages having a SP-R210+CD11c+ phenotype compared with WT mice. On the other hand, treatment of macrophages with surfactant lipids or SP-A was shown to induce SR-A expression (89), indicating that local host factors control basal levels of SR-A on alveolar macrophages. Several signaling pathways were shown to regulate expression and activation of SR-A involving arachidonic acid metabolites of iPLA2 and 12/15 lipoxygenase (87). Such lipid mediators contribute to initiation and resolution of inflammation (90). In addition, SR-A is involved in combinatorial feedback inhibition of proinflammatory signaling mediated by Toll-like receptors (91). In this regard, anti-inflammatory activities of SP-A that were previously attributed to direct binding of SP-A to Toll-like receptors (92, 93) may also involve indirect regulation of SR-A via SP-R210. It is noteworthy that TLR2 (Toll-like receptor 2) contributes to intracellular survival of S. aureus in macrophages (94). TLR2 activation that was not properly controlled by the SP-R210/SR-A pathway could be the mechanism behind the intracellular persistence of Eap− S. aureus in alveolar macrophages of WT mice. Together, these results indicate that homeostatic conditions modulate cross-talk influencing relative expression levels of SP-R210L, SP-R210S, and SR-A in alveolar macrophages. Upon infection, recognition of SP-A-opsonized S. aureus by SP-R210L is a critical early host response through which SP-R210S and SR-A are engaged to facilitate clearance of infection and the development of beneficial inflammatory responses.
The molecular aspects of SP-R210-mediated phagocytosis were beyond the scope of the present work. However, SP-R210, as a surface isoform of unconventional myosin 18A (Myo18A) (25), represents a novel phagocytic receptor. Reorganization of the actin cytoskeleton is essential for engulfment of surface-bound particles. The likely downstream trigger for SP-R210-induced phagocytosis is non-muscle myosin IIA (MyoIIA). MyoIIA is essential for phagocytic internalization via complement and IgG receptors in macrophages (95). SP-R210 co-precipitated with MyoIIA in pull-down assays with either anti-SP-R210 antibodies or immobilized SP-A (25). Expression of SP-R210 in COS-1 cells conferred attachment but not internalization of SP-A-opsonized bacteria; COS-1 cells do not express MyoIIA or SR-A. Antibodies to SP-R210 neutralized attachment of SP-A-opsonized bacteria in immature THP-1 cells; phagocytic internalization is not functional in undifferentiated THP-1 cells (38, 39). Undifferentiated THP-1 cells express high levels of SP-R210S but lack expression of SP-R210L, SR-A, and MyoIIA. In other cells, Myo18A isoforms link indirectly with the MyoIIA cytoskeleton in different subcellular compartments. Myo18A controls retrograde membrane flow through formation of a tripartite complex between the PDZ domain of Myo18A large isoform with a novel protein LRP35a and a Cdc42-related kinase called MRCK in motile cells. Phosphorylation of Myo18A in this system results in activation of MyoIIA and remodeling of the actin cytoskeleton in lamellipodia (96). Myo18A associates with the actin cytoskeleton underlying the Golgi apparatus via GOLPH3, a phosphatidylinositol 4-phosphate-binding protein (97). This interaction modulates formation of vesicles and structure of the trans-Golgi network. Besides Golgi, phosphatidylinositol 4-phosphate that is also enriched at the plasma membrane modulates macrophage phagocytosis via IgG receptors (98, 99) and may have a role in cytoskeleton reorganization during SP-R210-mediated phagocytosis. It remains to be determined whether physical association between SP-R210 isoforms, SR-A, and MyoIIA mediates phagocytosis of SP-A-opsonized bacteria.
Collectively, the present findings support the model in which binding of SP-A-opsonized bacteria to SP-R210L in alveolar macrophages (AM1) is essential for initial uptake and priming of the proinflammatory response through secretion of TNFα, mediating recruitment and activation of neutrophils (Fig. 12). In this model, ligation of SP-R210L (depicted as a heterodimer of L and S isoforms in Fig. 12) results in alveolar macrophage polarization, inducing expression of SP-R210S and SR-A (AM2). Binding of SP-A to SP-R210S induces phagocytosis and anti-inflammatory mediators via association with SR-A (e.g. IL-10 in Fig. 12), enhancing bacterial killing and resolution of the infection. Based on previous findings, SP-R210S (27) and SR-A (100) may coordinate secretion of IL-10, TGFβ, and hydrogen peroxide in AM2. Importantly, it is proposed that temporal control of inflammatory responses via SP-R210S and SR-A contributes to proper recruitment and activation of neutrophils, facilitating eradication of S. aureus infection in the lung. Both IL-10 and TGFβ would contribute to resolution of inflammation and restoration of alveolar homeostasis during clearance of apoptotic neutrophils via SR-A (101, 102). On the other hand, moderate levels of hydrogen peroxide would suppress inflammation through inactivation of NF-κB (103, 104) but enhance bacterial killing through activation of NADPH oxidase (105) during the resolution phase of the disease.
Acknowledgments
We thank Dr. Anna Kurdowska for consultation with KC ELISA, Dr. Jeffrey A. Whitsett for providing SP-A polyclonal antibodies, Dr. Bruce Trapnell for alveolar proteinosis fluid, Dr. Triantafyllos Chavakis for providing purified native Eap from S. aureus Newman, and Dr. Mark A. L. Atkinson for critical reading of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants HL068127 (to Z. C. C.), ES11008 (to L. K.), and AI071028 (to B. V. G.). This work was also supported by the Potts Memorial Foundation (to Z. C. C.), the Juvenile Diabetes Research Foundation (to Z. C. C.), and University of Texas Health Science Center at Tyler research council grants (to Z. S.-C.).
Z. Sever-Chroneos and Z. C. Chroneos, unpublished data.
- SP-A
- surfactant protein A
- Eap
- extracellular adhesion protein
- SR-A
- scavenger receptor class A
- SP-R210
- surfactant protein A receptor 210 identical to myosin 18A
- SP-D
- surfactant protein D
- MBL
- mannose-binding lectin
- BCG
- bacillus Calmette-Guerin
- TSB
- tryptic soy broth
- MBMM
- mouse bone marrow-derived macrophage(s)
- DN
- dominant negative
- KC
- keratinocyte chemokine
- Myo18A
- myosin 18A identical to SP-R210
- MyoIIA
- myosin IIA
- SERAM
- secretable expanded repertoire adhesive molecule
- MSCRAMM
- microbial surface component recognizing adhesive matrix molecules
- FnBP
- fibronectin-binding protein
- LTA
- lipoteichoic acid
- DiI
- 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine
- PE
- phycoerythrin
- AcLDL
- acetylated LDL.
REFERENCES
- 1. Defres S., Marwick C., Nathwani D. (2009) Eur. Respir. J. 34, 1470–1476 [DOI] [PubMed] [Google Scholar]
- 2. Murray R. J., Robinson J. O., White J. N., Hughes F., Coombs G. W., Pearson J. C., Tan H. L., Chidlow G., Williams S., Christiansen K. J., Smith D. W. (2010) PLoS One 5, e8705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sinha B., Fraunholz M. (2010) Int. J. Med. Microbiol. 300, 170–175 [DOI] [PubMed] [Google Scholar]
- 4. Foster T. J. (2009) Vet. Dermatol. 20, 456–470 [DOI] [PubMed] [Google Scholar]
- 5. Floros J., Wang G., Mikerov A. N. (2009) Crit. Rev. Eukaryot. Gene Expr. 19, 125–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gupta G., Surolia A. (2007) BioEssays 29, 452–464 [DOI] [PubMed] [Google Scholar]
- 7. Chroneos Z. C., Sever-Chroneos Z., Shepherd V. L. (2010) Cell Physiol. Biochem. 25, 13–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cañadas O., García-Verdugo I., Keough K. M., Casals C. (2008) Biophys. J. 95, 3287–3294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. García-Verdugo I., Cañadas O., Taneva S. G., Keough K. M., Casals C. (2007) Biophys. J. 93, 3529–3540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chimote G., Banerjee R. (2008) J. Colloid Interface Sci. 328, 288–298 [DOI] [PubMed] [Google Scholar]
- 11. Wang Z., Schwab U., Rhoades E., Chess P. R., Russell D. G., Notter R. H. (2008) Tuberculosis 88, 178–186 [DOI] [PubMed] [Google Scholar]
- 12. Kannan T. R., Provenzano D., Wright J. R., Baseman J. B. (2005) Infect. Immun. 73, 2828–2834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Piboonpocanun S., Chiba H., Mitsuzawa H., Martin W., Murphy R. C., Harbeck R. J., Voelker D. R. (2005) J. Biol. Chem. 280, 9–17 [DOI] [PubMed] [Google Scholar]
- 14. Ragas A., Roussel L., Puzo G., Rivière M. (2007) J. Biol. Chem. 282, 5133–5142 [DOI] [PubMed] [Google Scholar]
- 15. McNeely T. B., Coonrod J. D. (1993) J. Infect Dis. 167, 91–97 [DOI] [PubMed] [Google Scholar]
- 16. Manz-Keinke H., Plattner H., Schlepper-Schäfer J. (1992) Eur. J. Cell Biol. 57, 95–100 [PubMed] [Google Scholar]
- 17. van Iwaarden F., Welmers B., Verhoef J., Haagsman H. P., van Golde L. M. (1990) Am. J. Respir. Cell Mol. Biol. 2, 91–98 [DOI] [PubMed] [Google Scholar]
- 18. van de Wetering J. K., van Eijk M., van Golde L. M., Hartung T., van Strijp J. A., Batenburg J. J. (2001) J. Infect Dis. 184, 1143–1151 [DOI] [PubMed] [Google Scholar]
- 19. Gil M., McCormack F. X., Levine A. M. (2009) J. Biol. Chem. 284, 7495–7504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mikerov A. N., Umstead T. M., Gan X., Huang W., Guo X., Wang G., Phelps D. S., Floros J. (2008) Am. J. Physiol. Lung Cell Mol. Physiol. 294, L121–L130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Crowther J. E., Kutala V. K., Kuppusamy P., Ferguson J. S., Beharka A. A., Zweier J. L., McCormack F. X., Schlesinger L. S. (2004) J. Immunol. 172, 6866–6874 [DOI] [PubMed] [Google Scholar]
- 22. Kudo K., Sano H., Takahashi H., Kuronuma K., Yokota S., Fujii N., Shimada K., Yano I., Kumazawa Y., Voelker D. R., Abe S., Kuroki Y. (2004) J. Immunol. 172, 7592–7602 [DOI] [PubMed] [Google Scholar]
- 23. Weikert L. F., Edwards K., Chroneos Z. C., Hager C., Hoffman L., Shepherd V. L. (1997) Am. J. Physiol. 272, L989–L995 [DOI] [PubMed] [Google Scholar]
- 24. Tenner A. J., Robinson S. L., Borchelt J., Wright J. R. (1989) J. Biol. Chem. 264, 13923–13928 [PubMed] [Google Scholar]
- 25. Yang C. H., Szeliga J., Jordan J., Faske S., Sever-Chroneos Z., Dorsett B., Christian R. E., Settlage R. E., Shabanowitz J., Hunt D. F., Whitsett J. A., Chroneos Z. C. (2005) J. Biol. Chem. 280, 34447–34457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Weikert L. F., Lopez J. P., Abdolrasulnia R., Chroneos Z. C., Shepherd V. L. (2000) Am. J. Physiol. Lung Cell Mol. Physiol. 279, L216–L223 [DOI] [PubMed] [Google Scholar]
- 27. Samten B., Townsend J. C., Sever-Chroneos Z., Pasquinelli V., Barnes P. F., Chroneos Z. C. (2008) J. Leukoc. Biol. 84, 115–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Szeliga J., Jordan J., Yang C. H., Sever-Chroneos Z., Chroneos Z. C. (2005) Anal. Biochem. 346, 179–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chroneos Z. C., Abdolrasulnia R., Whitsett J. A., Rice W. R., Shepherd V. L. (1996) J. Biol. Chem. 271, 16375–16383 [DOI] [PubMed] [Google Scholar]
- 30. Athanasopoulos A. N., Economopoulou M., Orlova V. V., Sobke A., Schneider D., Weber H., Augustin H. G., Eming S. A., Schubert U., Linn T., Nawroth P. P., Hussain M., Hammes H. P., Herrmann M., Preissner K. T., Chavakis T. (2006) Blood 107, 2720–2727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hammel M., Nemecek D., Keightley J. A., Thomas G. J., Jr., Geisbrecht B. V. (2007) Protein Sci. 16, 2605–2617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Xie C., Alcaide P., Geisbrecht B. V., Schneider D., Herrmann M., Preissner K. T., Luscinskas F. W., Chavakis T. (2006) J. Exp. Med. 203, 985–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hussain M., Haggar A., Heilmann C., Peters G., Flock J. I., Herrmann M. (2002) Infect. Immun. 70, 2933–2940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Samten B., Howard S. T., Weis S. E., Wu S., Shams H., Townsend J. C., Safi H., Barnes P. F. (2005) J. Immunol. 174, 6357–6363 [DOI] [PubMed] [Google Scholar]
- 35. Suzuki H., Kurihara Y., Takeya M., Kamada N., Kataoka M., Jishage K., Ueda O., Sakaguchi H., Higashi T., Suzuki T., Takashima Y., Kawabe Y., Cynshi O., Wada Y., Honda M., Kurihara H., Aburatani H., Doi T., Matsumoto A., Azuma S., Noda T., Toyoda Y., Itakura H., Yazaki Y., Kodama T. (1997) Nature 386, 292–296 [DOI] [PubMed] [Google Scholar]
- 36. Zhou H., Imrich A., Kobzik L. (2008) Part. Fibre Toxicol. 5, 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bubeck Wardenburg J., Patel R. J., Schneewind O. (2007) Infect. Immun. 75, 1040–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tsuchiya S., Kobayashi Y., Goto Y., Okumura H., Nakae S., Konno T., Tada K. (1982) Cancer Res. 42, 1530–1536 [PubMed] [Google Scholar]
- 39. Tsuchiya S., Yamabe M., Yamaguchi Y., Kobayashi Y., Konno T., Tada K. (1980) Int. J. Cancer 26, 171–176 [DOI] [PubMed] [Google Scholar]
- 40. Sakamoto H., Aikawa M., Hill C. C., Weiss D., Taylor W. R., Libby P., Lee R. T. (2001) Circulation 104, 109–114 [DOI] [PubMed] [Google Scholar]
- 41. Via D. P., Pons L., Dennison D. K., Fanslow A. E., Bernini F. (1989) J. Lipid Res. 30, 1515–1524 [PubMed] [Google Scholar]
- 42. Rivera-Marrero C. A., Schuyler W., Roser S., Ritzenthaler J. D., Newburn S. A., Roman J. (2002) Am. J. Physiol. Lung Cell Mol. Physiol. 282, L546–L555 [DOI] [PubMed] [Google Scholar]
- 43. Nakane A., Okamoto M., Asano M., Kohanawa M., Minagawa T. (1995) Infect. Immun. 63, 1165–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hussain M., Becker K., von Eiff C., Schrenzel J., Peters G., Herrmann M. (2001) J. Bacteriol. 183, 6778–6786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Inden K., Kaneko J., Miyazato A., Yamamoto N., Mouri S., Shibuya Y., Nakamura K., Aoyagi T., Hatta M., Kunishima H., Hirakata Y., Itoh Y., Kaku M., Kawakami K. (2009) Microbes Infect. 11, 245–253 [DOI] [PubMed] [Google Scholar]
- 46. Ventura C. L., Malachowa N., Hammer C. H., Nardone G. A., Robinson M. A., Kobayashi S. D., DeLeo F. R. (2010) PLoS One 5, e11634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Geisbrecht B. V., Hamaoka B. Y., Perman B., Zemla A., Leahy D. J. (2005) J. Biol. Chem. 280, 17243–17250 [DOI] [PubMed] [Google Scholar]
- 48. Hussain M., Becker K., von Eiff C., Peters G., Herrmann M. (2001) Clin. Diagn. Lab. Immunol. 8, 1271–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hussain M., von Eiff C., Sinha B., Joost I., Herrmann M., Peters G., Becker K. (2008) J. Clin. Microbiol. 46, 470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kim M. K., Huang Z. Y., Hwang P. H., Jones B. A., Sato N., Hunter S., Kim-Han T. H., Worth R. G., Indik Z. K., Schreiber A. D. (2003) Blood 101, 4479–4484 [DOI] [PubMed] [Google Scholar]
- 51. Wright S. D., Licht M. R., Craigmyle L. S., Silverstein S. C. (1984) J. Cell Biol. 99, 336–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Peiser L., Gough P. J., Kodama T., Gordon S. (2000) Infect. Immun. 68, 1953–1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Brown M. S., Basu S. K., Falck J. R., Ho Y. K., Goldstein J. L. (1980) J. Supramol. Struct. 13, 67–81 [DOI] [PubMed] [Google Scholar]
- 54. Areschoug T., Gordon S. (2009) Cell Microbiol. 11, 1160–1169 [DOI] [PubMed] [Google Scholar]
- 55. Cotena A., Gordon S., Platt N. (2004) J. Immunol. 173, 6427–6432 [DOI] [PubMed] [Google Scholar]
- 56. Thomas C. A., Li Y., Kodama T., Suzuki H., Silverstein S. C., El Khoury J. (2000) J. Exp. Med. 191, 147–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Haworth R., Platt N., Keshav S., Hughes D., Darley E., Suzuki H., Kurihara Y., Kodama T., Gordon S. (1997) J. Exp. Med. 186, 1431–1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kobayashi S. D., DeLeo F. R. (2009) Wiley Interdiscip. Rev. Syst. Biol. Med. 1, 309–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Marino M. W., Dunn A., Grail D., Inglese M., Noguchi Y., Richards E., Jungbluth A., Wada H., Moore M., Williamson B., Basu S., Old L. J. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 8093–8098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hodge-Dufour J., Marino M. W., Horton M. R., Jungbluth A., Burdick M. D., Strieter R. M., Noble P. W., Hunter C. A., Puré E. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 13806–13811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kowanko I. C., Ferrante A., Clemente G., Kumaratilake L. M. (1996) Infect. Immun. 64, 3435–3437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Nugent K. M., Pesanti E. L. (1982) Infect. Immun. 36, 1185–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Coonrod J. D., Lester R. L., Hsu L. C. (1984) J. Clin. Invest. 74, 1269–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wu H., Kuzmenko A., Wan S., Schaffer L., Weiss A., Fisher J. H., Kim K. S., McCormack F. X. (2003) J. Clin. Invest. 111, 1589–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Guth A. M., Janssen W. J., Bosio C. M., Crouch E. C., Henson P. M., Dow S. W. (2009) Am. J. Physiol. Lung Cell Mol. Physiol. 296, L936–L946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. von Garnier C., Filgueira L., Wikstrom M., Smith M., Thomas J. A., Strickland D. H., Holt P. G., Stumbles P. A. (2005) J. Immunol. 175, 1609–1618 [DOI] [PubMed] [Google Scholar]
- 67. Ventura C. L., Higdon R., Hohmann L., Martin D., Kolker E., Liggitt H. D., Skerrett S. J., Rubens C. E. (2008) Infect. Immun. 76, 5862–5872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ventura C. L., Higdon R., Kolker E., Skerrett S. J., Rubens C. E. (2008) Infect. Immun. 76, 888–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Shi L., Takahashi K., Dundee J., Shahroor-Karni S., Thiel S., Jensenius J. C., Gad F., Hamblin M. R., Sastry K. N., Ezekowitz R. A. (2004) J. Exp. Med. 199, 1379–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Neth O., Jack D. L., Johnson M., Klein N. J., Turner M. W. (2002) J. Immunol. 169, 4430–4436 [DOI] [PubMed] [Google Scholar]
- 71. Nadesalingam J., Dodds A. W., Reid K. B., Palaniyar N. (2005) J. Immunol. 175, 1785–1794 [DOI] [PubMed] [Google Scholar]
- 72. Plüddemann A., Hoe J. C., Makepeace K., Moxon E. R., Gordon S. (2009) PLoS Pathog. 5, e1000297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Polotsky V. Y., Fischer W., Ezekowitz R. A., Joiner K. A. (1996) Infect. Immun. 64, 380–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Chavakis T., Wiechmann K., Preissner K. T., Herrmann M. (2005) Thromb. Haemost. 94, 278–285 [DOI] [PubMed] [Google Scholar]
- 75. Clarke S. R., Foster S. J. (2006) Adv. Microb. Physiol. 51, 187–224 [DOI] [PubMed] [Google Scholar]
- 76. Patti J. M., Allen B. L., McGavin M. J., Höök M. (1994) Annu. Rev. Microbiol. 48, 585–617 [DOI] [PubMed] [Google Scholar]
- 77. Harraghy N., Hussain M., Haggar A., Chavakis T., Sinha B., Herrmann M., Flock J. I. (2003) Microbiology 149, 2701–2707 [DOI] [PubMed] [Google Scholar]
- 78. Cheng A. G., Kim H. K., Burts M. L., Krausz T., Schneewind O., Missiakas D. M. (2009) FASEB J. 23, 3393–3404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Joost I., Blass D., Burian M., Goerke C., Wolz C., von Müller L., Becker K., Preissner K., Herrmann M., Bischoff M. (2009) J. Infect. Dis. 199, 1471–1478 [DOI] [PubMed] [Google Scholar]
- 80. McElroy M. C., Cain D. J., Tyrrell C., Foster T. J., Haslett C. (2002) Infect. Immun. 70, 3865–3873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Harraghy N., Kormanec J., Wolz C., Homerova D., Goerke C., Ohlsen K., Qazi S., Hill P., Herrmann M. (2005) Microbiology 151, 1789–1800 [DOI] [PubMed] [Google Scholar]
- 82. Grundmeier M., Hussain M., Becker P., Heilmann C., Peters G., Sinha B. (2004) Infect. Immun. 72, 7155–7163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Ledford J. G., Goto H., Potts E. N., Degan S., Chu H. W., Voelker D. R., Sunday M. E., Cianciolo G. J., Foster W. M., Kraft M., Wright J. R. (2009) J. Immunol. 182, 7818–7827 [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 84. McLoughlin R. M., Lee J. C., Kasper D. L., Tzianabos A. O. (2008) J. Immunol. 181, 1323–1332 [DOI] [PubMed] [Google Scholar]
- 85. Gresham H. D., Lowrance J. H., Caver T. E., Wilson B. S., Cheung A. L., Lindberg F. P. (2000) J. Immunol. 164, 3713–3722 [DOI] [PubMed] [Google Scholar]
- 86. Kuronuma K., Sano H., Kato K., Kudo K., Hyakushima N., Yokota S., Takahashi H., Fujii N., Suzuki H., Kodama T., Abe S., Kuroki Y. (2004) J. Biol. Chem. 279, 21421–21430 [DOI] [PubMed] [Google Scholar]
- 87. Lin Y. L., de Villiers W. J., Garvy B., Post S. R., Nagy T. R., Safadi F. F., Faugere M. C., Wang G., Malluche H. H., Williams J. P. (2007) J. Biol. Chem. 282, 4653–4660 [DOI] [PubMed] [Google Scholar]
- 88. Post S. R., Gass C., Rice S., Nikolic D., Crump H., Post G. R. (2002) J. Lipid Res. 43, 1829–1836 [DOI] [PubMed] [Google Scholar]
- 89. Gille C., Spring B., Bernhard W., Gebhard C., Basile D., Lauber K., Poets C. F., Orlikowsky T. W. (2007) J. Lipid Res. 48, 307–317 [DOI] [PubMed] [Google Scholar]
- 90. Serhan C. N., Chiang N., Van Dyke T. E. (2008) Nat. Rev. Immunol. 8, 349–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Seimon T. A., Obstfeld A., Moore K. J., Golenbock D. T., Tabas I. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 19794–19799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Yamada C., Sano H., Shimizu T., Mitsuzawa H., Nishitani C., Himi T., Kuroki Y. (2006) J. Biol. Chem. 281, 21771–21780 [DOI] [PubMed] [Google Scholar]
- 93. Sato M., Sano H., Iwaki D., Kudo K., Konishi M., Takahashi H., Takahashi T., Imaizumi H., Asai Y., Kuroki Y. (2003) J. Immunol. 171, 417–425 [DOI] [PubMed] [Google Scholar]
- 94. Watanabe I., Ichiki M., Shiratsuchi A., Nakanishi Y. (2007) J. Immunol. 178, 4917–4925 [DOI] [PubMed] [Google Scholar]
- 95. Olazabal I. M., Caron E., May R. C., Schilling K., Knecht D. A., Machesky L. M. (2002) Curr. Biol. 12, 1413–1418 [DOI] [PubMed] [Google Scholar]
- 96. Tan I., Yong J., Dong J. M., Lim L., Leung T. (2008) Cell 135, 123–136 [DOI] [PubMed] [Google Scholar]
- 97. Dippold H. C., Ng M. M., Farber-Katz S. E., Lee S. K., Kerr M. L., Peterman M. C., Sim R., Wiharto P. A., Galbraith K. A., Madhavarapu S., Fuchs G. J., Meerloo T., Farquhar M. G., Zhou H., Field S. J. (2009) Cell 139, 337–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Mao Y. S., Yamaga M., Zhu X., Wei Y., Sun H. Q., Wang J., Yun M., Wang Y., Di Paolo G., Bennett M., Mellman I., Abrams C. S., De Camilli P., Lu C. Y., Yin H. L. (2009) J. Cell Biol. 184, 281–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Pizarro-Cerdá J., Payrastre B., Wang Y. J., Veiga E., Yin H. L., Cossart P. (2007) Cell Microbiol. 9, 2381–2390 [DOI] [PubMed] [Google Scholar]
- 100. Józefowski S., Kobzik L. (2004) J. Leukoc. Biol. 76, 1066–1074 [DOI] [PubMed] [Google Scholar]
- 101. Todt J. C., Hu B., Curtis J. L. (2008) J. Leukoc. Biol. 84, 510–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Medeiros A. I., Serezani C. H., Lee S. P., Peters-Golden M. (2009) J. Exp. Med. 206, 61–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Banerjee S., Zmijewski J. W., Lorne E., Liu G., Sha Y., Abraham E. (2010) J. Biol. Chem. 285, 2665–2675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Zmijewski J. W., Lorne E., Zhao X., Tsuruta Y., Sha Y., Liu G., Abraham E. (2009) Am. J. Respir. Crit. Care Med. 179, 694–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. El Jamali A., Valente A. J., Clark R. A. (2010) Free Radic. Biol. Med. 48, 798–810 [DOI] [PMC free article] [PubMed] [Google Scholar]