Abstract
Stp1 and Stp2 are two homologous transcription factors activated in response to extracellular amino acid stimuli. Here we show that both ubiquitin-dependent degradation of Stp1 and Stp2 and their intracellular localization are differentially regulated. We have found that the E2 ubiquitin-conjugating enzyme Cdc34 is required for degradation of both full-length and processed Stp1, but not Stp2. We have also found that Grr1, the F-box component of the SCFGrr1 E3 ubiquitin ligase, is the primary factor in degradation of full-length Stp1, whereas both Grr1 and Cdc4 are required for degradation of processed Stp1. Our localization studies showed that full-length Stp1 is localized both in the cytoplasm and at the cell periphery, whereas full-length Stp2 is localized only diffusely in the cytoplasm. We identified two nuclear localization signals of Stp1 and found that the N-terminal domain of Stp1 is required for localization of full-length Stp1 to the cell periphery. We also found that Stp2 is the primary factor involved in basal activation of target gene expression. Our results indicate that the functions of two seemingly redundant transcription factors can be separated by differential degradation and distinct cellular localization.
Keywords: Nuclear Translocation, Signal Transduction, Transcription Factors, Ubiquitin, Yeast, Ptr3, Ssy1, Ssy5, Stp1, Stp2
Introduction
Extracellular amino acid sensing via the SPS amino acid sensing pathway is best characterized in the budding yeast Saccharomyces cerevisiae (1–5). The SPS amino acid sensing pathway includes Ssy1, a plasma membrane-localized sensor for extracellular amino acids, and two downstream factors, Ptr3 and Ssy5 (1–3, 6, 7). Ssy5 is a novel protease, activation of which leads to endoproteolytic processing and activation of two zinc finger transcription factors, Stp1 and Stp2 (8–10). Although protein phosphatase 2A negatively regulates SPS signaling (13), two isoforms of the yeast casein kinase I proteins, Yck1 and Yck2, function as positive regulators of this pathway (8, 11, 12). In addition, the F-box protein in the SCFGrr1 E3 ubiquitin ligase complex, Grr1, is required for SPS-target gene expression (2, 12, 14). The SCF-type E3 ubiquitin ligases, consisting of Skp1, Cdc53, and variable F-box containing proteins such as Cdc4, Grr1, or Met30, are evolutionarily conserved and typically interact with the Cdc34 ubiquitin-conjugating enzyme (E2) (15–17). Furthermore, it has been shown that SPS-target gene expression also requires ubiquitin, other components of the SCFGrr1 complex, as well as Cdc34 (14). The positive regulatory function of Grr1 and Cdc34 in this pathway appears to promote amino acid-induced processing of Stp1 (8). However, Stp1 processing is independent of proteasome function, indicating that Grr1/Cdc34-dependent processing of Stp1 requires ubiquitination but not proteasome-dependent degradation (8, 18).
Ssy5-dependent processing of Stp1 and Stp2 is a key activation step in the regulation of the SPS amino acid sensing pathway. Ssy5 undergoes endoproteolytic processing to generate an N-terminal pro-domain and a C-terminal activity domain, both of which remain associated with each other upon processing (8–10). In cells stimulated with amino acids, Ssy1, Ptr3, Grr1, and Yck1/2-dependent activation of Ssy5 leads to the removal of the N-terminal inhibitory sequence from Stp1 and Stp2 resulting in their nuclear translocation and activation of target gene expression (8–10). The mechanism by which Ssy5 is activated is still unclear. However, it has been proposed that degradation of the pro-domain of Ssy5 leads to activation of the activity domain (9, 19). Nevertheless, it is still unknown how the degradation of the pro-domain of Ssy5 is regulated in response to amino acid signals. Furthermore, Ssy5 has been reported to interact with Ptr3, whose hyperphosphorylation correlates with pathway activation (11, 20). Ptr3 hyperphosphorylation requires Ssy1, Yck1/2, and Grr1. Additionally, a regulatory subunit of the protein phosphatase 2A phosphatase complex, Rts1, negatively regulates SPS signaling, likely by promoting Ptr3 dephosphorylation (11, 13).
Stp1 and Stp2 have been proposed to play redundant roles in the SPS amino acid sensing pathway (18, 21–23). However, a recent publication suggests that Stp1 and Stp2 are derived from a genome duplication event that occurred in a yeast ancestor and functionally diverged during evolution (25). The authors further showed that due to the difference of Stp1 and Stp2 in their N-terminal domains, only Stp2 is processed in response to the stimulation of low levels of amino acids. Stp1 is subject to other types of regulation apart from Ssy5-dependent endoproteolytic processing. It has been shown that Stp1 is rapidly turned over and is phosphorylated (8, 18, 19, 24). Rapamycin treatment induces Stp1 degradation, which has been suggested to result from Stp1 dephosphorylation (24). Although most studies have focused on Stp1, there remains limited understanding of Stp1 degradation with regard to the factors involved. Additionally, very few studies on Stp2 regulation have been reported. Here, we report our findings from a comparative approach to study degradation and intracellular localization of Stp1 and Stp2. We found that different E2 and E3 enzymes are required for degradation of Stp1 and Stp2. We also found that full-length Stp1 and Stp2 are differentially localized. Differential degradation and cellular localization of full-length Stp1 and Stp2 are consistent with the primary role of Stp2 in activation of the SPS amino acid sensing pathway under basal or suboptimal inducing conditions.
EXPERIMENTAL PROCEDURES
Strains and Plasmids
Yeast strains and plasmids used in this study are listed in Tables 1 and 2, respectively. Isogenic strains of PLY126 background (3) were used to generate most of the data in this paper. BY4741 background strains and strains containing cdc34 and cdc53 temperature-sensitive mutations were used for Figs. 2 and 4. Details of plasmid construction are available upon request.
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Source/Ref. |
|---|---|---|
| RBY881 | MATa ura3–52 stp1::kanMX4 AGP1::AGP1-lacZ::kanMX4 | This study |
| RBY903 | MATa ura3–52 stp2::kanMX4 AGP1::AGP1-lacZ::kanMX4 | This study |
| ZLY044 | MATa ura3–52 AGP1::AGP1-lacZ::kanMX4 | 11 |
| ZLY1939 | MATa ura3–52 ssy5::kanMX4 AGP1::AGP1-lacZ::kanMX4 | 11 |
| ZLY1516 | MATa ura3–52 lys2 erg6::kanMX4 [pRS417, CEN-LYS2] | This study |
| ZLY1942 | MATa ura3–52 grr1::kanMX4 AGP1::AGP1-lacZ::kanMX4 | This study |
| MLY43 | MATa ura3–52 leu2::hisG | Joseph Heitman laboratory |
| ZLY827 | MATa ura3–52 stp1::kanMX4 stp2::kanMX4 AGP1::AGP1-lacZ::kanMX4 | This study |
| BY4741 | MATa ura3 leu2 his3 met15 | Yeast genome deletion project |
| BY4741-ubc2 | MATa ura3 leu2 his3 met15 ubc2::kanMX4 | |
| BY4741-ubc4 | MATa ura3 leu2 his3 met15 ubc4::kanMX4 | |
| BY4741-ubc5 | MATa ura3 leu2 his3 met15 ubc5::kanMX4 | |
| BY4741-ubc7 | MATa ura3 leu2 his3 met15 ubc7::kanMX4 | |
| BY4741-ubc8 | MATa ura3 leu2 his3 met15 ubc8::kanMX4 | |
| BY4741-ubc10 | MATa ura3 leu2 his3 met15 ubc10::kanMX4 | |
| BY4741-ubc11 | MATa ura3 leu2 his3 met15 ubc11::kanMX4 | |
| BY4741-ubc12 | MATa ura3 leu2 his3 met15 ubc12::kanMX4 | |
| BY4741-ubc13 | MATa ura3 leu2 his3 met15 ubc13::kanMX4 | |
| MGG15 | MATa ura3 his3 cdc34-2 | 43 |
| MGG12 | MATa ade2 ade3 trp1 ura3 his3 cdc53-1 | 43 |
| YWO1 | MATα his3-Δ200 leu2–3,2–112 lys2–801 trp1-1(am) ura3–52 | 45 |
| YWO23 | MATα his3-Δ200 leu2–3,2–112 lys2–801 trp1-1(am) ura3–52 ubc4::HIS3 ubc5::LEU2 | 46 |
| RJD2125 | MATa his3-Δ200, leu2–3,112, ura3–52 | 47 |
| RJD2152 | MATa his3-Δ200, leu2–3,112, ura3–52 cdc4-1 | 47 |
| ZLY3445 | MATa his3-Δ200, leu2–3,112, ura3–52 cdc4-1 grr1Δ::kanMX4 | This study |
| ZLY1915 | MATa ura3–52 ssy1-Δ13 AGP1::AGP1-lacZ::kanMX4 | 11 |
| ZLY1917 | MATa ura3–52 ptr3-Δ15 AGP1::AGP1-lacZ::kanMX4 | 11 |
| ZLY1939 | MATa ura3–52 ssy5::kanMX4 AGP1::AGP1-lacZ::kanMX4 | 11 |
TABLE 2.
Plasmids used in this study
| Plasmid | Description | Source/Ref. |
|---|---|---|
| pZL1834 | pRS416-STP1-HA, expressing Stp1 with 3× HA tagged at the C terminus | 11 |
| pZL2508 | pRS416-STP2-HA, expressing Stp2 with 3× HA tagged at the C terminus | This study |
| pZL2841 | pRS426-STP1-HA, expressing Stp1with 3× HA tagged at the C terminus | This study |
| pZL2543 | pRS416-GAL1-STP1-HA3-HIS12, expressing Stp1 under control of GAL1 promoter with both a 3× HA and a 12× His tag at the C-terminal end | This study |
| pZL2540 | pRS416-GAL1-STP2-HA3-HIS12, expressing Stp2 under control of GAL1 promoter with both a 3× HA and a 12× His tag at the C-terminal end | This study |
| pZL1553 | pRS415-GAL1-GRR1-myc3, expressing Grr1 under control of GAL1 promoter with 3× myc tag at the C-terminal end | 48 |
| pZL1635 | pRS416-STP1ΔN-HA3, expressing a N-terminal truncation mutant of Stp1 missing the first 97 residues under the native promoter with a 3× HA tag at the C terminus | This study |
| pZL1637 | pRS416-STP2ΔN-HA3, expressing an N-terminal truncation mutant of Stp2 missing the first 105 residues under the native promoter with a 3× HA tag at the C terminus | This study |
| pZL2813 | pRS416-STP1-GFP, expressing C-terminal GFP-tagged Stp1 | This study |
| pZL2849 | pRS416-STP2-GFP, expressing C-terminal GFP-tagged Stp2 | This study |
| pZL2615 | pRS416-STP1(1–125)-GFP, expressing the Stp1(1–125)-GFP fusion | This study |
| pZL2862 | pRS416-STP1(1–159)-GFP, expressing the Stp1(1–159)-GFP fusion | This study |
| pZL2865 | pRS416-STP1(1–273)-GFP, expressing the Stp1(1–273)-GFP fusion | This study |
| pZL2667 | pRS416-STP1(1–299)-GFP, expressing the Stp1(1–299)-GFP fusion | This study |
| pZL2867 | pRS416-STP1(1–331)-GFP, expressing the Stp1(1–331)-GFP fusion | This study |
| pZL2869 | pRS416-STP1(1–376)-GFP, expressing the Stp1(1–376)-GFP fusion | This study |
| pZL2661 | pRS416-STP1(267–519)-GFP, expressing the Stp1(267–519)-GFP fusion | This study |
| pZL2669 | pRS416-STP1(291–519)-GFP, expressing the Stp1(291–519)-GFP fusion | This study |
| pZL2671 | pRS416-STP1(299–519)-GFP, expressing the Stp1(299–519)-GFP fusion | This study |
| pZL2663 | pRS416-STP1(331–519)-GFP, expressing the Stp1(331–519)-GFP fusion | This study |
| pZL2677 | pRS416-STP1(267–376)-GFP, expressing the Stp1(267–376)-GFP fusion | This study |
| pZL2665 | pRS416-STP1(267–331)-GFP, expressing the Stp1(267–331)-GFP fusion | This study |
| pZL2879 | pRS416-STP1(Δ330–336)-GFP, expressing a GFP-tagged Stp1(Δ330–336) truncation construct with residues 330–336 of Stp1 deleted | This study |
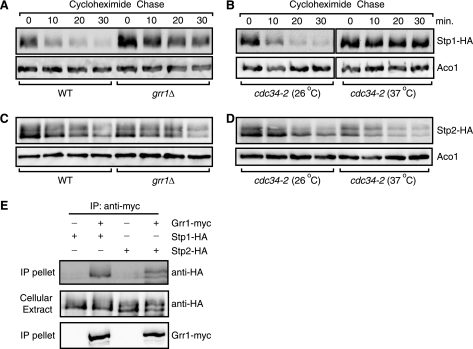
FIGURE 2.
Grr1 is required for the degradation of full-length Stp1, but not full-length Stp2. A, a grr1Δ mutation stabilizes full-length Stp1-HA. Wild-type (ZLY044) and grr1Δ mutant cells (ZLY1942) expressing STP1-HA (pZL1834) were grown in SD medium and stability of Stp1-HA was analyzed. B, a cdc34-2 temperature-sensitive mutation stabilizes full-length Stp1-HA. cdc34-2 mutant cells (MGG15) expressing STP1-HA were cultured in SD medium at 26 or 37 °C and stability of Stp1-HA was analyzed. C, a grr1Δ mutation does not stabilize full-length Stp2-HA. D, a cdc34-2 mutation does not stabilize full-length Stp2-HA. E, Grr1 interacts with the full-length form of Stp1 and Stp2. MLY43 cells expressing HA- or myc-tagged proteins as indicated were subjected to a co-immunoprecipitation assay using anti-myc antibody. Cellular extracts and immunoprecipitates (IP pellet) were immunoblotted with anti-HA and anti-myc antibodies as indicated.
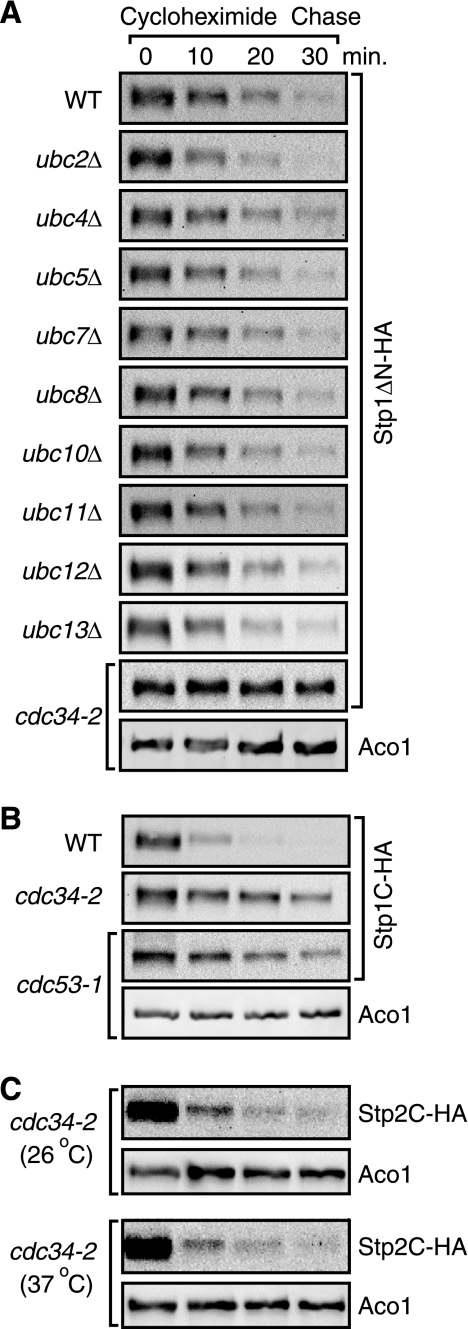
FIGURE 4.
Cdc34 is required degradation of processed Stp1 but not processed Stp2. A, a cdc34-2 temperature-sensitive mutation inhibits degradation of Stp1ΔN-HA. Wild-type (BY4741) and nine isogenic ubcΔ mutant strains, and a cdc34-2 mutant strain (MGG15) expressing STP1ΔN-HA (pZL1635) were grown in SD medium + leucine and stability of Stp1ΔN-HA was determined. B, mutations in CDC34 and CDC53 stabilize natively processed Stp1-HA (Stp1C-HA). Wild-type (ZLY044), cdc34-2 (MGG15), and cdc53-1 (MGG12) mutant strains expressing STP1-HA were analyzed for Stp1C-HA stability. C, a cdc34-2 mutation fails to increase the stability of Stp2C. cdc34-2 (MGG15) mutant cells expressing Stp2-HA were grown in SD medium supplemented with leucine and stability of Stp2C was determined in cells grown at 26 and 37 °C.
Growth Media and Growth Conditions
Yeast cells were grown at 30 °C in SD (0.67% yeast nitrogen base plus 2% dextrose), SRaff (0.67% yeast nitrogen base plus 2% raffinose), or SRaffGal (SRaff plus 2% galactose) media as indicated in the text and figure legends. Temperature-sensitive mutants were grown at 26 and/or 37 °C as indicated in the text. When indicated, leucine (0.02%, 1.5 mm) was added to activate the SPS amino acid sensing pathway. When required, MG132 (50 μm) was added to growth medium to inhibit proteasome activity. When necessary, amino acids, adenine, and/or uracil were added to growth medium at standard concentrations to cover auxotrophic requirements (26).
Yeast Transformation and β-Galactosidase Assays
Yeast transformation and β-galactosidase assays were performed as described (26). Liquid cultures were inoculated with respective yeast strains indicated in the figure legends and cells were grown for at least 6 generations to A600 0.5–0.8 before collection for β-galactosidase activity assays.
Cycloheximide Chase Assay
Cells expressing the indicated HA-tagged proteins were grown in liquid SD medium with or without addition of 0.02% leucine to A600 = 0.6. Cycloheximide was added to a final concentration of 50 μg/ml to initiate the chase. Every 10 or 20 min, 1-ml samples of cell cultures were withdrawn and subjected to trichloroacetic acid precipitation as described below. For protein stability analysis in temperature-sensitive mutants such as cdc4-1, cdc34-2, and cdc53-1, cells were cultured at 26 °C to A600 = 0.4 and shifted to 37 °C for 2–3 h before cycloheximide chase assays were initiated. HA-tagged protein levels were determined by probing Western blots with anti-HA antibody (3F10, Roche Applied Science). As a loading control, Aco1 (mitochondrial aconitase), was probed with polyclonal rabbit anti-Aco1 antibody (27).
Cellular Extract Preparation, Immunoblotting, and Immunoprecipitation
Total cellular protein extracts were prepared by disrupting yeast cells in extraction buffer (1.85 n NaOH, 7.5% β-mercaptoethanol) followed by precipitation with trichloroacetic acid (TCA) as described (28). Co-immunoprecipitation experiments were performed as described (29). Briefly, total cellular extracts were prepared in IP buffer (50 mm Tris-HCl, pH 7.6, 120 mm NaCl, 0.5% Triton X-100, and protease inhibitors). Cell extracts (∼2 mg of proteins) were incubated at 4 °C for 1 h with 8 μg of monoclonal anti-myc antibody (9E10, Roche Applied Science), after which, 30 μl of a 50% slurry of protein G-Sepharose (Roche Applied Science) was added to each sample and the samples were further incubated at 4 °C for 2 h. Immunoprecipitates bound to the Sepharose beads were released by boiling in 1× SDS-PAGE loading buffer after being washed four times with 1 ml of IP buffer. The released immune complexes were analyzed by Western blotting. Myc- and HA-tagged proteins were probed with anti-myc antibody (9E10) and anti-HA antibody (3F10, Roche Applied Science), respectively. Immunoblotting was carried out by incubating blotted nitrocellulose membranes with primary antibody, followed by appropriate horseradish peroxidase-conjugated secondary antibodies. Chemiluminescence images of Western blots were captured using the Bio-Rad Chemi-Doc photodocumentation system (Bio-Rad). For the determination of the half-life of a protein, bands on Western blots were quantified using Bio-Rad QuantityOne software.
Fluorescence Microscopic Analysis of GFP-tagged Proteins
Cells expressing various GFP-tagged proteins as indicated in the text were grown in specified medium to mid-log phase. Cells were concentrated by centrifugation and live cells were observed by fluorescence microscopy using a Nikon Eclipse E800 microscope equipped with an HBO 100 W/2 mercury arc lamp, a Nikon Plan Fluor ×100 objective lens, and a Nikon B-2E/C medium band excitation band pass filter set (excitation light 465–495 nm, emission light 515–555 nm). Digital images were acquired with a Photometrics Coolsnap fx CCD camera and Metamorph Imaging Software (Molecular Devices, Sunnyvale, CA) and processed using ImageJ (National Institutes of Health) and Adobe Photoshop (Mountain View, CA) software.
RESULTS
Full-length Stp2 Is More Stable Than Full-length Stp1
Amino acid treatment leads to Ssy5-dependent processing of Stp1 and Stp2. To analyze post-translational modifications of Stp1 and Stp2, we constructed C-terminal HA-tagged constructs of Stp1 and Stp2, which were found to be able to fully restore amino acid-induced AGP1-lacZ expression to stp1Δ stp2Δ mutant cells, indicating that they are functional (data not shown). The AGP1-lacZ fusion gene has been used extensively as a reporter in this pathway (2, 11, 30, 31). Previously, both full-length and processed Stp1 have been reported to be unstable, with a half-life (t½) of <10 min for full-length Stp1 (18, 24). However, little is known about the stability of Stp2. To that end, C-terminal HA-tagged Stp2 was expressed in wild-type cells from a centromeric plasmid and its stability was determined in cells grown in SD medium without or with leucine supplementation using a cycloheximide chase assay. Full-length Stp2 had an average half-life of 26 ± 0.5 min from two independent experiments (n = 2), and processed Stp2-HA was much more unstable, with a half-life of 5.2 ± 0.3 min (n = 2) (Fig. 1A). An increased turnover rate of processed Stp2-HA over full-length Stp2-HA prompted us to re-examine the turnover rates of full-length and processed Stp1-HA. Consistent with previous reports (18), full-length Stp1 in cells grown without leucine treatment was rapidly degraded with an average half-life of 10 ± 0.7 min (n = 2) (Fig. 1B, lanes 1–4). Processed Stp1-HA in cells exposed to leucine was more unstable, with an average half-life of 5.6 ± 0.8 min (n = 2) (Fig. 1B, lanes 5-8). Together, our data indicate that the protein stabilities of full-length Stp1 and Stp2 are different: full-length Stp1 is rapidly turned over, whereas full-length Stp2 is more stable than full-length Stp1. Our data also indicate that processed Stp1 and Stp2 are more unstable than their full-length counterparts.
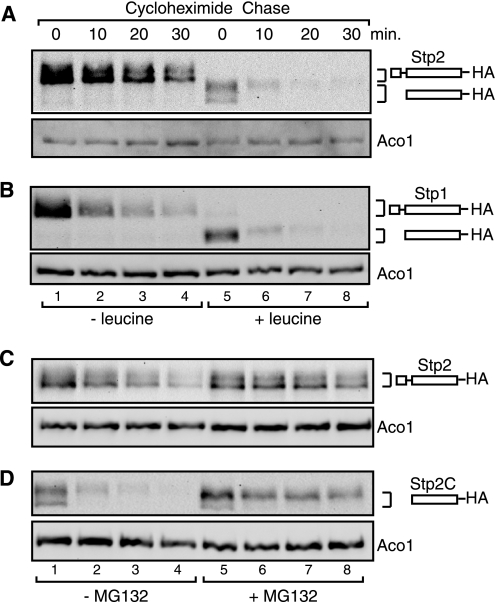
FIGURE 1.
Instability of full-length and processed Stp2 and Stp1. A, full-length Stp2 is more stable than processed Stp2. A cycloheximide chase assay was performed as described under “Experimental Procedures” to determine the stability of full-length and processed Stp2-HA in wild-type cells (ZLY044). Stp2-HA was detected by Western blotting using anti-HA antibody. Full-length and processed Stp2 are indicated diagrammatically to the right of the image. Aco1, mitochondrial aconitase, was included as a loading control. B, both full-length and processed Stp1 are quickly turned over. Stability of full-length and processed Stp1-HA was analyzed as described for panel A. C and D, inhibition of proteasome function by MG132 stabilizes full-length and processed Stp2. erg6Δ mutant cells (ZLY1516) expressing STP2-HA (pZL2508) were grown in SD medium without (panel C) or with (panel D) leucine and treated with or without MG132 (50 μm) as indicated. Stability of full-length Stp2-HA (panel C) and processed Stp2-HA (Stp2C-HA, panel D) was analyzed by cycloheximide chase assay.
Proteasome Function Is Required for Degradation of Full-length and Processed Stp2
It has been shown that proteasome function is required for degradation of full-length and processed Stp1 (19, 24). Therefore, we asked whether proteasome function is also required for turnover of Stp2. To that end, we conducted cycloheximide chase assays on cells treated with or without MG132, an inhibitor of proteasome function. First, an erg6Δ mutant strain was constructed to facilitate uptake of MG132 into cells (32). Next, erg6Δ mutant cells expressing Stp2-HA were grown in SD medium without or with leucine and treated either with DMSO or 50 μm MG132 for 120 min prior to cycloheximide chase initiation. Subsequently, we found that inhibition of proteasome function by MG132 increased the stability of both full-length Stp2-HA (Fig. 1C) and processed Stp2-HA (Fig. 1D), indicating that both full-length and processed Stp2 are subject to proteasome-dependent degradation.
Grr1 Mediates Degradation of Full-length Stp1, but Not Stp2
The factors involved in ubiquitination and subsequent degradation of Stp1 and Stp2 are unknown. Because Grr1, the F-box component of the SCFGrr1 E3 ubiquitin ligase, is required for proteolytic processing of Stp1, we sought to determine the involvement of Grr1 in the regulation of Stp1 and Stp2 stability. Accordingly, turnover of Stp1-HA and Stp2-HA was assessed in grr1Δ mutant cells grown in SD medium. Fig. 2A shows that Stp1-HA was more stable in grr1Δ mutant cells than in wild-type cells grown in SD medium, with half-lives of 9 and 28 min in wild-type and grr1Δ mutant cells, respectively. Because SCFGrr1 usually requires the Cdc34 ubiquitin-conjugating enzyme for substrate ubiquitination, we also assessed turnover of Stp1-HA in cdc34-2 temperature-sensitive mutant cells. We found that a cdc34-2 temperature-sensitive mutation also stabilized full-length Stp1-HA at 37 °C, a non-permissive temperature, but not at 26 °C, a permissive temperature (Fig. 2B), suggesting that Grr1-dependent degradation of Stp1-HA involves Cdc34. In contrast to Grr1-dependent rapid turnover of Stp1-HA, mild turnover of full-length Stp2-HA was not significantly blocked by a grr1Δ mutation (with t½ of 25 and 27 min in wild-type and grr1Δ cells, respectively) (Fig. 2C). Consistently, a cdc34-2 temperature-sensitive mutation also failed to stabilize full-length Stp2-HA at 37 °C (with t½ of 23 and 17 min in cdc34-2 cells grown at 26 and 37 °C, respectively) (Fig. 2D). Altogether, these data uncover a novel role for Grr1: proteasome-dependent degradation of full-length Stp1 and indicates that Grr1 and Cdc34 mediate turnover of full-length Stp1 but not full-length Stp2. Grr1-dependent degradation of Stp1-HA prompted us to determine whether Grr1 interacts with Stp1. We therefore constructed centromeric plasmids encoding either STP1-HA or GRR1-myc under control of the GAL1 promoter and transformed yeast cells with either the STP1-HA construct alone or with both STP1-HA and GRR1-myc constructs. Transformants were cultured in SRaffGal medium to induce expression of Stp1-HA and/or Grr1-myc. Cellular extracts were prepared and subjected to immunoprecipitation using anti-myc antibody. We found that Stp1-HA interacted with Grr1-myc (Fig. 2E). Using a similar procedure, Stp2-HA was also found to interact with Grr1 (Fig. 2E). Because Grr1 is not required for degradation of full-length Stp2, the observed interaction between Stp2 and Grr1 could be due to Grr1 acting as a positive regulator in the processing of Stp2 because Grr1 is required for Ssy5-dependent endoproteolytic processing of Stp2. It is also possible that due to sequence homology with Stp1, Stp2 is able to interact with Grr1, but fails to be ubiquitinated by SCFGrr1 due to sequence divergence. The significance of an interaction between Stp2 and Grr1 at this time is unclear; however, our data suggest that full-length Stp1-HA is a new substrate of Grr1.
Grr1 Is Required for the Turnover of Processed Stp1, but Not Processed Stp2
Because Grr1 mediates the degradation of full-length Stp1, we sought to determine whether Grr1 is involved in the degradation of processed Stp1 and Stp2. Grr1 is essential for Ssy5-dependent endoproteolytic processing of Stp1 and Stp2 and their translocation to the nucleus. Therefore, we could not directly determine stability of processed Stp1 or Stp2 in a grr1Δ mutant. To that end, we constructed plasmids encoding C-terminal HA-tagged, N-terminal truncation mutants of Stp1 (Stp1ΔN-HA) and Stp2 (Stp2ΔN-HA) lacking the first 97 and 105 N-terminal residues, respectively. Consistent with previous findings (18), expression of either Stp1ΔN-HA or Stp2ΔN-HA led to constitutive activation of AGP1-lacZ expression in stp1Δ stp2Δ double mutant cells (Fig. 3A), indicating that Stp1ΔN-HA and Stp2ΔN-HA are constitutively localized to the nucleus and active.
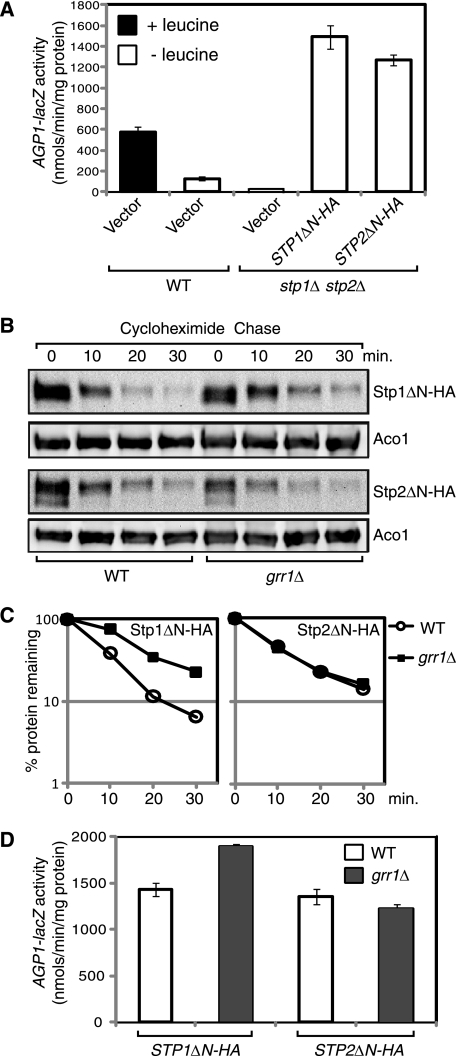
FIGURE 3.
Grr1 mediates degradation of processed Stp1 but not processed Stp2. A, expression of an N-terminal truncation mutant of Stp1 (Stp1ΔN-HA) or Stp2 (Stp2ΔN-HA) leads to constitutive activation of AGP1-lacZ expression. Wild-type (ZLY044) and stp1Δ stp2Δ double deletion mutant cells (ZLY827) carrying control vector pRS416, STP1ΔN-HA (pZL1635), or STP2ΔN-HA (pZL1637) plasmid were grown in SD medium ± leucine and β-galactosidase activities were assayed. B, a grr1Δ mutation partially stabilizes Stp1ΔN-HA, but not Stp2ΔN-HA. Wild-type (ZLY044) and grr1Δ mutant (ZLY1942) cells expressing STP1ΔN-HA or STP2ΔN-HA were grown in SD medium, and cycloheximide chase was carried out as described. C, quantitative analysis of Stp1ΔN-HA and Stp2ΔN-HA levels in panel B. D, stabilization of Stp1ΔN-HA in grr1Δ cells leads to increased AGP1-lacZ expression. Cells described for panel B were grown in SD medium, and β-galactosidase activities were determined. Error bars represent S.E. of results from four independent experiments. The difference in AGP1-lacZ activity due to Stp1ΔN-HA expression in wild-type versus grr1Δ mutant cells is significant (p < 0.0001).
We then investigated whether Grr1 regulates the turnover of Stp1ΔN-HA and Stp2ΔN-HA. Wild-type and grr1Δ mutant cells were transformed with plasmids encoding either STP1ΔN-HA or STP2ΔN-HA and stability of Stp1ΔN-HA and Stp2ΔN-HA was examined. Similar to rapid turnover of natively processed Stp1 and Stp2 (Fig. 1, A and B), Stp1ΔN-HA and Stp2ΔN-HA were also rapidly turned over in wild-type cells (Fig. 3B) and MG132 treatment blocked their rapid turnover in erg6Δ cells (data not shown). In wild-type cells, the turnover of Stp1ΔN-HA was relatively rapid (t½ = 7.4 ± 0.1 min, n = 2) (Fig. 3, B and C). In grr1Δ mutant cells, Stp1ΔN-HA was partially stabilized (t½ = 12.9 ± 0.5 min, n = 2). In contrast, a grr1Δ mutation did not lead to stabilization of Stp2ΔN-HA, which had an average half-life of 10.1 ± 0.6 min (n = 2) in wild-type and 10.6 ± 0.7 min (n = 2) in grr1Δ mutant cells (Fig. 3, B and C). Together, these data suggest that Grr1 mediates partial degradation of processed Stp1, but not Stp2.
We then assessed whether the role played by Grr1 in regulating turnover of processed Stp1 is of any physiological relevance. Accordingly, we expressed Stp1ΔN-HA in both wild-type and grr1Δ mutant strains carrying an integrated AGP1-lacZ reporter gene. We then assayed AGP1-lacZ expression and consistently observed that β-galactosidase activity in the grr1Δ strain was significantly (∼32%) higher than that in wild-type cells (Fig. 3D). In contrast, a grr1Δ mutation did not lead to increased AGP1-lacZ expression in cells expressing Stp2ΔN-HA (Fig. 3D), which was consistent with our data in Fig. 3, B and C, showing that there was no significant increase in Stp2ΔN-HA stability in grr1Δ strains. Altogether, these data suggest that partial stabilization of Stp1ΔN-HA due to a grr1Δ mutation leads to increased AGP1-lacZ expression.
Cdc34 and Cdc53 Are Required for Degradation of Processed Stp1
A minor role for Grr1 in mediating degradation of processed Stp1 prompted us to identify the E2 ubiquitin-conjugating enzyme responsible for rapid turnover of processed Stp1. The yeast genome contains 13 genes encoding E2 ubiquitin-conjugating enzymes (33). Nine yeast strains from the yeast genome deletion project, each with a single deletion mutation in a nonessential gene encoding one of the following E2 enzymes: Ubc2, Ubc4, Ubc5, Ubc7, Ubc8, Ubc10, Ubc11, Ubc12, Ubc13, and a temperature-sensitive cdc34 mutant strain were analyzed for stability of Stp1ΔN-HA. In the wild-type BY4741 background strain, which was used in the yeast deletion project, we noted that rapid turnover of Stp1ΔN-HA was somewhat reduced. Importantly, none of the mutations in the nine nonessential E2 enzyme-encoding genes led to increased stabilization of Stp1ΔN-HA (Fig. 4A). In contrast, the temperature-sensitive cdc34-2 mutation led to a dramatic increase in stability of Stp1ΔN-HA at 37 °C (Fig. 4A). At 30 °C, instability of Stp1ΔN-HA in the cdc34-2 mutant strain was largely restored (data not shown). Together, our data indicate that Cdc34 is required specifically for proteasome-dependent degradation of Stp1ΔN-HA.
Cdc34 is required for amino acid-induced processing of Stp1 (8). But unlike a grr1Δ mutation, a temperature-sensitive cdc34 mutation allowed us to examine the turnover of natively processed Stp1 (Stp1C-HA). cdc34-2 mutant cells expressing STP1-HA were grown at 26 °C in SD medium supplemented with leucine to allow processing of full-length Stp1-HA into the C-terminal active form, Stp1C-HA. Cultures were then moved to the non-permissive temperature, 37 °C, for 2 h prior to cycloheximide chase initiation, which allowed us to examine stability of Stp1C-HA. We observed that a cdc34-2 mutation at 37 °C led to stabilization of Stp1C-HA (Fig. 4B). Using a similar procedure, we also observed that a temperature-sensitive cdc53-1 mutation also led to increased stability of Stp1C-HA at 37 °C, a non-permissive temperature (Fig. 4B). Altogether, these data indicate that turnover of nuclear-localized, processed Stp1 requires Cdc34 and Cdc53.
In an attempt to identify the E2 ubiquitin-conjugating enzyme responsible for rapid turnover of processed Stp2-HA, we expressed STP2-HA in wild-type and E2 enzyme mutant cells as described for Stp1-HA and examined stability of Stp2C-HA in cells grown in SD medium supplemented with leucine and found that none of the single, nonessential E2 enzyme mutations led to a significant increase in Stp2C-HA stability (data not shown). In contrast to increased stability of Stp1C-HA observed in cdc34-2 mutant cells, a cdc34-2 mutation failed to increase stability of Stp2C-HA at 37 °C (Fig. 4C). Together, our data clearly demonstrate that processed Stp1 and Stp2 require different E2 ubiquitin-conjugating enzymes for their turnover.
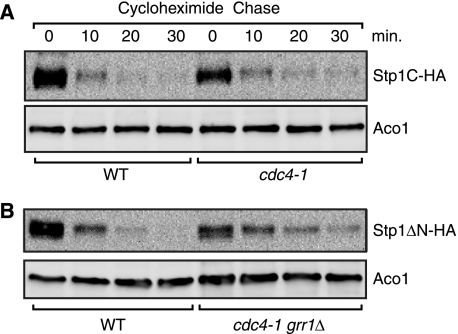
Cdc4 Contributes to Degradation of Processed Stp1
Our observation that a cdc34 mutation had a stronger effect in stabilizing Stp1ΔN than a grr1Δ mutation (compare Figs. 3B and 4A) suggests that other F-box proteins are also involved in Stp1 degradation. To that end, stability of processed Stp1 was analyzed in met30 and cdc4 mutant cells. A met30Δ mutation is lethal, whereas a met4Δ met30Δ double mutant is viable (34). We examined Stp1C stability in met4Δ single and met4Δ met30Δ double mutant cells and found that a met30Δ mutation did not significantly stabilize Stp1C-HA (data not shown). Cdc4 is required for cell viability. Therefore, we used a cdc4-1 temperature-sensitive mutant to determine Stp1C stability. Fig. 5A shows that a cdc4-1 mutation nearly doubled the half-life of Stp1C-HA from 5.8 ± 0.3 min (n = 2) in wild-type cells to 10.4 ± 0.3 min (n = 2) in cdc4-1 mutant cells grown at 37 °C. Because Grr1 is also involved in degradation of processed Stp1 (Fig. 3, B and C), we constructed a cdc4-1 grr1Δ double mutant, examined Stp1ΔN-HA stability, and found that the half-life of Stp1ΔN-HA increased from 5.9 ± 0.3 min (n = 4) in wild-type cells to 14.3 ± 1.4 min (n = 4) in cdc4-1 grr1Δ double mutant cells (Fig. 5B). Grr1 and Cdc4 have been reported to be localized in the nucleus (35, 36). Together, these findings suggest that Cdc4 and Grr1 are both involved in degradation of processed Stp1, which is predominantly localized in the nucleus.
FIGURE 5.
Cdc4 and Grr1 are involved in degradation of processed Stp1. A, a cdc4-1 mutation partially stabilizes natively processed Stp1-HA (Stp1C-HA). Wild-type (RJD2125) and an isogenic cdc4-1 mutant (RJD2152) expressing STP1-HA (pZL1834) were grown at 26 °C in SD medium supplemented with leucine and then transferred to 37 °C for 3 h prior to initiating the cycloheximide chase for Stp1C-HA. B, analysis of Stp1ΔN stability in a cdc4-1 grr1Δ double mutant. Wild-type (RJD2125) and an isogenic cdc4-1 grr1Δ mutant (ZLY3445) expressing STP1ΔN-HA (pZL1635) were grown in SD medium and stability of Stp1ΔN-HA was determined.
Stp1, but Not Stp2, Associates with the Plasma Membrane
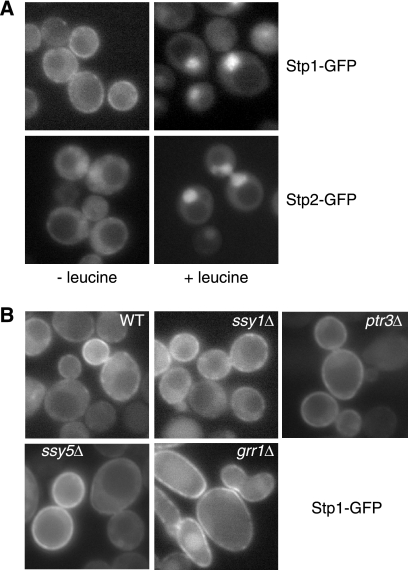
Stp1 has been proposed to associate with the plasma membrane based on results obtained using an Sos recruitment system (18, 37). Furthermore, immunolocalization studies have shown that Stp1 translocates from the cytoplasm into the nucleus following amino acid stimulation, but these studies failed to detect direct plasma membrane localization of Stp1 (18). In a genome-wide yeast protein localization study, GFP-tagged Stp2 was shown to be concentrated in the nucleus in cells grown in SD medium supplemented with amino acids to meet auxotrophic requirements (38); however, no data have been reported on the basal localization of Stp2. We sought to compare the localization of GFP-tagged Stp1 and Stp2 in vivo. Plasmids encoding functional GFP fusions of STP1 and STP2 were constructed and transformed into respective deletion mutant cells. In cells grown in SD medium supplemented with leucine, Stp1-GFP and Stp2-GFP clearly showed nuclear localization (Fig. 6A). In cells without amino acid stimulus, intracellular localization of GFP-tagged Stp1 and Stp2 differed: whereas both showed a diffuse cytoplasmic localization, Stp1-GFP was also localized to the cell periphery (most likely at the plasma membrane), often in a discontinuous manner. Plasma membrane localization of Stp1-GFP is consistent with previous Sos recruitment data (18). Consistent with a low level of Stp2 basal processing in cells grown in SD medium, we found that Stp2-GFP localized weakly in the nucleus in a small percentage of cells (data not shown). Our data not only substantiated plasma membrane localization of Stp1, but revealed that Stp1 and Stp2 are differentially localized outside the nucleus.
FIGURE 6.
Differential localization of Stp1 and Stp2. A, fluorescence microscopic analysis of intracellular localization of Stp1-GFP and Stp2-GFP. stp1Δ mutant cells (RBY881) expressing STP1-GFP (pZL2813) and stp2Δ mutant cells (RBY903) expressing STP2-GFP (pZL2849) were grown in SD medium ± leucine and GFP fluorescence images were captured as described under “Experimental Procedures.” B, plasma membrane localization of Stp1-GFP does not require Ssy1, Ptr3, Ssy5, or Grr1. Wild-type (ZLY044) and isogenic ssy1Δ (ZLY1915), ptr3Δ (ZLY1917), ssy5Δ (ZLY1939), and grr1Δ (ZLY1942) mutant strains expressing STP1-GFP were grown in SD medium and GFP fluorescence images were captured.
The N-terminal domain of Stp1, consisting of the first 125 residues, interacts with Ssy5 (18). Ssy5 has been proposed to form a plasma membrane-localized sensor complex with Ssy1 and Ptr3 (7, 11, 20, 31). It has also been reported that Ssy1 is required for efficient plasma membrane localization of Stp1 using the Sos recruitment system (18). However, direct observation of plasma membrane localization of Stp1-GFP in vivo allowed us to determine whether this association requires the SPS sensor complex. Fig. 6B shows that mutations in SSY1, PTR3, SSY5, or GRR1 did not abolish plasma membrane localization of Stp1-GFP. Together, these data suggest that plasma membrane localization of Stp1 does not require the SPS sensor complex.
Identification of Sequence Elements Required for Plasma Membrane Association of Stp1
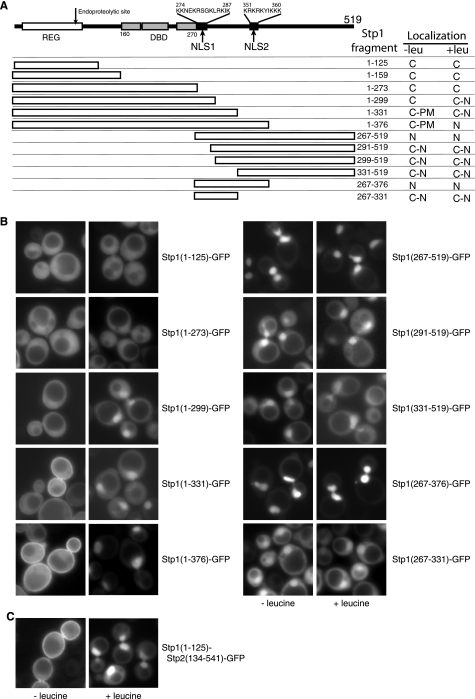
Differential plasma membrane localization of Stp1 and Stp2 prompted us to identify sequence elements required for the plasma membrane association of Stp1. Accordingly, we constructed a series of GFP-tagged Stp1 truncation constructs and examined their cellular localization in stp1Δ mutant cells grown in SD medium without or with leucine (Fig. 7). It has been reported that an N-terminal Stp1 fragment consisting of the first 159 residues associates with the plasma membrane using an Sos recruitment assay (18). Surprisingly, four GFP-tagged N-terminal fragments of Stp1, Stp1(1–125), Stp1(1–159), Stp1(1–273), and Stp1(1–299), failed to localize to the plasma membrane in cells grown in SD medium (Fig. 7B) (data of Stp1(1–159) not shown), suggesting that the N-terminal regulatory domain alone is not sufficient for plasma membrane localization. In contrast to the cytoplasmic localization of Stp1(1–273)-GFP, Stp1(1–299)-GFP showed dual cytoplasmic and nuclear localization in cells treated with leucine, indicating that the sequence comprised of residues 274–299 contains a nuclear localization signal. Examination of residues in that region revealed a Lys/Arg-rich region (residues 274–287), suggestive of a bipartite nuclear localization signal (NLS)2 (39), which we termed NLS1 (Fig. 7A).
FIGURE 7.
Sequence elements of Stp1 required for plasma membrane and nuclear localization. A, diagrammatic representation of Stp1 constructs expressing various fragments of Stp1 with a C-terminal GFP tag. Numbers indicate the Stp1 fragments expressed. The regulatory domain (REG), which confers amino acid-induced proteolytic processing, a DNA binding domain (DBD, residues 160–270), and two nuclear localization signals (NLS1 and NLS2) are indicated. Intracellular localization of C-terminal GFP-tagged Stp1 truncation constructs grown in SD medium without or with leucine from panel B and our unpublished data are summarized to the right of the table (C, cytoplasm; N, nucleus; PM, plasma membrane). B, fluorescence microscopic analysis of GFP-tagged Stp1 truncation constructs. stp1Δ mutant cells expressing various GFP-tagged Stp1 truncation constructs as indicated were grown in SD medium ± leucine. GFP fluorescence images were captured as described. C, the N-terminal domain of Stp1 confers plasma membrane localization to Stp2. stp2Δ mutant cells (RBY903) expressing Stp1(1–125)-Stp2(134–541)-GFP fusion protein were analyzed by fluorescence microscopy.
Compared with cytoplasmic localization of Stp1(1–299)-GFP, Stp1(1–331)-GFP and Stp1(1–376)-GFP exhibited both cytoplasmic and plasma membrane localization in cells without leucine treatment (Fig. 7, A and B), suggesting that residues 300–331 are important for plasma membrane localization of N-terminal fragments of Stp1. However, we still observed plasma membrane localization of a GFP-tagged Stp1 construct lacking residues 300–336, suggesting that residues 300–331 are not essential for plasma membrane localization of Stp1 (data not shown). In cells stimulated with leucine, Stp1(1–331)-GFP exhibited cytoplasmic/nuclear dual localization, whereas Stp1(1–376)-GFP showed strong nuclear localization, suggesting that residues 331–376 contain another NLS, which has been proposed by Andreasson and Ljungdahl (18) and is comprised of residues 351–360. We termed this nuclear localization signal NLS2 (Fig. 7A). In cells stimulated with leucine, Stp1(1–331) and Stp1(1–376) would have ∼100 residues removed from their N terminus (40). The observation that Stp1(1–331)-GFP in cells treated with leucine shows partial cytoplasmic localization, but no plasma membrane localization, suggests that the N-terminal domain removed by Ssy5 in response to amino acid stimuli also contains information required for the plasma membrane localization of Stp1.
Consistent with the critical regulatory role that N-terminal domain removal plays in nuclear translocation of Stp1 (8, 18, 40), GFP fusion constructs of C-terminal fragments of Stp1, Stp1(267–519), Stp1(291–519), and Stp1(331–519), all showed constitutive nuclear localization (Fig. 7B). GFP-tagged Stp1(267–519)-GFP, containing both NLS1 and NLS2, showed a strong nuclear localization, whereas Stp1(291–519)-GFP and Stp1(331–519)-GFP, which only contain NLS2, showed dual cytoplasmic/nuclear localization. A GFP fusion of an internal fragment of Stp1, Stp1(267–376), which contains both NLS1 and NLS2, also showed strong nuclear localization, whereas Stp1(267–331)-GFP, containing only NLS1, showed cytoplasmic/nuclear dual localization (Fig. 7B).
Together, our data suggest that the N-terminal regulatory domain of Stp1, which is removed from Stp1 in response to amino acid stimuli, is required for plasma membrane localization of Stp1. Although they are important for plasma membrane localization of some N-terminal fragments of Stp1, Stp1 residues 300–331 are not essential for plasma membrane localization of full-length Stp1. Our data also indicate the requirement of two nuclear localization signals for optimal nuclear localization of Stp1.
The N-terminal Domain of Stp1 Is Sufficient to Target Stp2 to the Plasma Membrane
To identify the sequence element responsible for differential plasma membrane localization of Stp1 and Stp2, we determined whether the N-terminal domain of Stp1 is able to target Stp2 to the plasma membrane. We replaced the N-terminal domain of Stp2 with the corresponding N-terminal sequence of Stp1 and examined cellular localization of the resultant Stp1(1–125)-Stp2(134–541)-GFP fusion. Stp1(1–125)-Stp2(134–541)-GFP could fully restore leucine-induced activation of AGP1-lacZ expression in stp1Δ stp2Δ mutant cells (data not shown), indicating that it was functional. In cells without leucine treatment, Stp1(1–125)-Stp2(134–541)-GFP exhibited both cytoplasmic and plasma membrane localization (Fig. 7C). In cells treated with leucine, processed Stp1(1–125)-Stp2(134–541)-GFP was localized to the nucleus. Our data demonstrate that differential plasma membrane localization of Stp1 and Stp2 is due to the difference in their N-terminal domains.
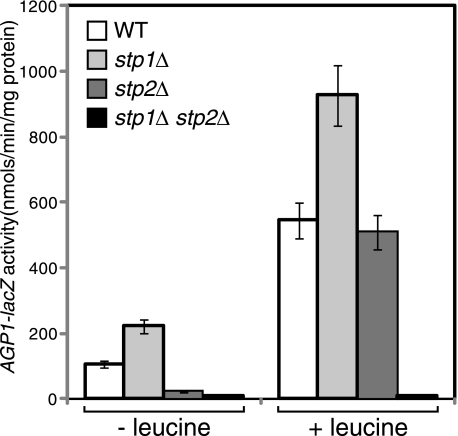
Stp2 Plays a Primary Role in Basal Expression of AGP1-lacZ
Our observation of differential degradation and localization of full-length Stp1 and Stp2 is consistent with a report indicating that Stp1 and Stp2 have functionally diverged (25). It was found that only Stp2 is activated in response to the treatment of low levels of amino acids (25). With our findings that full-length Stp2 was more stable and did not associate with the plasma membrane (Figs. 1A and 6A), we hypothesized that Stp2 may also play a more important role in basal expression of the AGP1-lacZ reporter. To this end, we assayed AGP1-lacZ expression in wild-type, stp1Δ single, stp2Δ single, and stp1Δ stp2Δ double mutant strains under both non-inducing and amino acid-inducing conditions. Consistent with their redundant roles in activating AGP1-lacZ expression (18, 21, 23), an stp1Δ stp2Δ double mutation abolished AGP1-lacZ expression in cells grown in SD medium without or with leucine (Fig. 8). However, we found differential effects of stp1Δ and stp2Δ single mutations on AGP1-lacZ expression under both non-inducing and inducing conditions. Although Stp1 is considered to be a redundant positive regulatory factor in AGP1-lacZ expression, an stp1Δ mutation increased AGP1–1acZ expression by 1.7–2-fold in cells grown in SD medium without or with leucine (Fig. 8), and similar results have been reported in cells stimulated with low levels of phenylalanine (25). In contrast, an stp2Δ mutation reduced basal AGP1-lacZ expression by ∼5-fold in cells grown in SD medium yet had little effect on AGP1-lacZ in cells grown in SD medium with leucine supplementation. Together, these data demonstrate that Stp2 plays a primary role in activating AGP1-lacZ expression under basal conditions.
FIGURE 8.
Stp2 plays the primary role in basal expression of AGP1-lacZ. Wild-type (ZLY044), stp1Δ (RBY881), stp2Δ (RBY903), and stp1Δ stp2Δ double mutant (ZLY827) cells were grown in SD medium ± leucine and AGP1-lacZ activities were determined.
DISCUSSION
Stp1 and Stp2 have been proposed to play a redundant role in activating target gene expression and to be subjected to Ssy5-dependent proteolytic processing in response to amino acid stimuli. A recent study demonstrated that Stp1 and Stp2 have functionally diverged (25). In this study, we show that Stp1 and Stp2 are differentially regulated via several different mechanisms. 1) Full-length Stp1 is a highly unstable protein, whereas full-length Stp2 is relatively more stable. 2) Grr1 and Cdc34 are required for turnover of Stp1, but not Stp2. 3) Full-length Stp1 and Stp2 are differentially localized: latent Stp2 is a cytoplasmic protein, whereas latent Stp1 shows dual localization in the cytoplasm and on the plasma membrane. Differential regulation of Stp1 and Stp2 reported in this work provides a plausible explanation for functional divergence of Stp1 and Stp2.
Consistent with a recent publication (25), we found that Stp1 and Stp2 play differential roles in mediating SPS-target gene expression: Stp2, but not Stp1 is the primary factor that activates target gene expression under basal conditions (Fig. 8). Also consistent with the previous findings that Stp2 is the primary factor that responds to suboptimal amino acid stimuli, we found that in cells grown in SD medium without leucine treatment, only Stp2 was weakly processed into the active form and localized weakly in the nucleus in a small percentage of cells (data not shown). Our data on differential regulation of Stp1 and Stp2 may provide a mechanistic explanation for the primary role that Stp2 plays in the activation of target gene expression under basal or suboptimal induction conditions. In contrast to Stp1, relative stability of full-length Stp2, coupled with its inability to be sequestered at the plasma membrane, probably makes Stp2 more amenable to activation under suboptimal induction conditions. Sequence differences in the N-terminal regulatory domains of Stp1 and Stp2 seem to be responsible for their differential regulation. Wielemans et al. (25) found that the N-terminal domains of Stp1 and Stp2 are responsible for these two proteins being cleaved at different concentrations of amino acids. Likewise, we report that the difference in N-terminal domains of Stp1 and Stp2 is responsible for enabling Stp1 but not Stp2 association with the plasma membrane. It is unclear whether relative stability and/or plasma membrane non-association of full-length Stp2 explains why Stp2 is more easily processed into the active form than Stp1. This should be investigated in the future.
Grr1 plays a positive role in the SPS amino acid sensing pathway by mediating Ssy5-dependent proteolytic processing of Stp1 and Stp2 in response to amino acid stimuli (8, 40). Our data suggest a novel role for Grr1 in this pathway: regulating the degradation of Stp1. Our results contradict a previous study that suggested that Stp2, not Stp1, is a putative Grr1 substrate for degradation (41). Benanti et al. (41) used a proteomics approach to identify Grr1 target proteins by screening for GFP fusion proteins with increased GFP fluorescence due to a grr1Δ mutation and identified Stp2 as one such protein. They likely compared the stability of the processed form of Stp2 in wild-type cells grown in rich medium (YM-1 medium, containing 1% peptone) with the full-length form of Stp2 in grr1Δ deletion mutant cells because Grr1 is absolutely required for Stp2 processing. Therefore, their conclusion that Grr1 is required for Stp2 turnover is likely to be incorrect because it is based on a comparison between relatively stable full-length Stp2 in grr1Δ mutant cells and more rapidly degraded processed Stp2 in wild-type cells. However, our data showing that processed Stp2 is highly unstable and its full-length form is relatively more stable are consistent with their observation that full-length Stp2 in grr1Δ mutant cells is more stable.
Grr1-dependent degradation of processed Stp1 in the nucleus appears to play a negative regulatory role because a grr1Δ mutation significantly increased AGP1-lacZ expression following expression of an N-terminal truncation mutant of Stp1 (Stp1ΔN-HA). Although that effect was relatively weak, our conclusion was strengthened by our observation that the constitutively active, N-terminal truncation construct of Stp2, Stp2ΔN, was not stabilized in grr1Δ mutant cells and did not increase AGP1-lacZ expression in grr1Δ mutant cells compared with wild-type cells. It is likely that Grr1 may also play a negative regulatory role by maintaining a low level of full-length Stp1 in the cytoplasm. Moreover, full-length Stp1 and Stp2 have been reported to “leak” into the nucleus in asi1/2/3Δ mutant cells and activate target gene expression (30, 40, 42). Grr1 is the primary F-box protein required for degradation of full-length Stp1 in the cytoplasm, whereas both Cdc4 and Grr1 contribute to degradation of processed Stp1, which is mostly in the nucleus. All components of the SCFGrr1 E3 ubiquitin ligase complex, Grr1, Cdc53, Skp1, Hrt1, and the associated Cdc34 E2 ubiquitin-conjugating enzyme are located both in the cytoplasm and nucleus (17, 24, 28, 43). Therefore, it is surprising that Grr1 plays a differential role in the degradation of full-length and processed Stp1. Finding a positive role for Grr1 in the processing of Stp1 and Stp2 into their active forms complicated the interpretation of the interaction result we obtained between Grr1 and Stp1/2 by co-immunoprecipitation analysis. It is conceivable that this positive regulatory role of Grr1 entails its interaction with Stp1 and Stp2, which might mask the specific interaction of Grr1 with full-length Stp1 but not Stp2 in its capacity as the E3 ubiquitin ligase for Stp1. However, the differential role that Grr1 plays in regulating two homologous proteins highlights the specificity required for ubiquitin-dependent degradation. Future studies will be directed toward determining how Grr1, Cdc4, and Stp1 degron sequences work together to achieve regulated degradation of Stp1 in the cytoplasm and nucleus. We have detected stabilization of processed Stp2 in ubc4Δ ubc5Δ double mutant cells (data not shown). However, we also found that other proteins were also stabilized in ubc4Δ ubc5Δ mutant cells. Thus, the significance of Stp2C stabilization in such a mutant is unknown. Future work will then be aimed at the isolation of the E2 and E3 ubiquitin ligase responsible for the degradation of processed Stp2 and the identification of the Stp2 degron sequence. This knowledge will help understand differential requirements of E2 and E3 enzymes for degradation of Stp1 and Stp2.
Our GFP fluorescence studies provide direct evidence that Stp1 and Stp2 are differentially localized outside the nucleus. Analyses of GFP-tagged truncation constructs of Stp1 suggest that its N-terminal regulatory domain is the primary determinant in mediating its plasma membrane association. The differential plasma membrane association of Stp1 and Stp2 is due to sequence differences in their N-terminal domains because the N-terminal domain of Stp1 is able to target Stp2 to the plasma membrane. What is the role of plasma membrane association of Stp1? Although it is plausible that plasma membrane association prevents nuclear translocation, additional cytoplasmic retention mechanisms must also exist because Stp1-GFP also localizes diffusely in the cytoplasm in cells lacking amino acid stimulus (Fig. 6). Such mechanisms also exist for Stp2. Thus, plasma membrane association of Stp1, together with its rapid turnover and a hitherto unknown cytoplasmic retention mechanism, may all act in unison to increase the threshold for Stp1 activation. Our data also indicate that there are two NLS in Stp1. Although individual NLSs appear to be sufficient for partial nuclear translocation, when combined, they form a more potent nuclear localization signal. This has also been observed in other proteins such as the mammalian high-mobility group domain transcription factors SRY and SOX9, which carry two NLSs (44). It is unclear why such proteins need two NLSs. It is possible that two NLSs might confer graded nuclear localization if each NLS can be regulated independently. Studies on mechanisms underlying differential processing, degradation, and cellular localization of Stp1 and Stp2 will provide valuable information on the evolution and functional divergence of homologous proteins.
Acknowledgments
We thank Dr. Mark Johnston, Peter Kaiser, Raymond Deshaies, Stefan Jentsch, and Joseph Heitman for yeast strains, Tammy Pracheil for assistance with fluorescence microscopy and manuscript editing, Glenda Castellanos for reporter gene assays, W.M. Keck Foundation for the Keck Facility, Robin Rowe for sequencing, and members of our laboratory for helpful discussions.
This work was supported by the University of New Orleans and Board of Regents of the State of Louisiana Grant LEQSF(2008-11)-RD-A-31.
- NLS
- nuclear localization signal.
REFERENCES
- 1. Didion T., Regenberg B., Jørgensen M. U., Kielland-Brandt M. C., Andersen H. A. (1998) Mol. Microbiol. 27, 643–650 [DOI] [PubMed] [Google Scholar]
- 2. Iraqui I., Vissers S., Bernard F., de Craene J. O., Boles E., Urrestarazu A., André B. (1999) Mol. Cell. Biol. 19, 989–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Klasson H., Fink G. R., Ljungdahl P. O. (1999) Mol. Cell. Biol. 19, 5405–5416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Forsberg H., Gilstring C. F., Zargari A., Martínez P., Ljungdahl P. O. (2001) Mol. Microbiol. 42, 215–228 [DOI] [PubMed] [Google Scholar]
- 5. Poulsen P., Wu B., Gaber R. F., Ottow K., Andersen H. A., Kielland-Brandt M. C. (2005) Biochem. Soc. Trans. 33, 261–264 [DOI] [PubMed] [Google Scholar]
- 6. Forsberg H., Ljungdahl P. O. (2001) Curr. Genet. 40, 91–109 [DOI] [PubMed] [Google Scholar]
- 7. Forsberg H., Ljungdahl P. O. (2001) Mol. Cell. Biol. 21, 814–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Abdel-Sater F., El Bakkoury M., Urrestarazu A., Vissers S., André B. (2004) Mol. Cell. Biol. 24, 9771–9785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Andréasson C., Heessen S., Ljungdahl P. O. (2006) Genes Dev. 20, 1563–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Poulsen P., Lo Leggio L., Kielland-Brandt M. C. (2006) Eukaryot. Cell 5, 601–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liu Z., Thornton J., Spírek M., Butow R. A. (2008) Mol. Cell. Biol. 28, 551–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Spielewoy N., Flick K., Kalashnikova T. I., Walker J. R., Wittenberg C. (2004) Mol. Cell. Biol. 24, 8994–9005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Eckert-Boulet N., Larsson K., Wu B., Poulsen P., Regenberg B., Nielsen J., Kielland-Brandt M. C. (2006) Eukaryot. Cell 5, 174–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bernard F., André B. (2001) FEBS Lett. 496, 81–85 [DOI] [PubMed] [Google Scholar]
- 15. Skowyra D., Craig K. L., Tyers M., Elledge S. J., Harper J. W. (1997) Cell 91, 209–219 [DOI] [PubMed] [Google Scholar]
- 16. Patton E. E., Willems A. R., Sa D., Kuras L., Thomas D., Craig K. L., Tyers M. (1998) Genes Dev. 12, 692–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Seol J. H., Feldman R. M., Zachariae W., Shevchenko A., Correll C. C., Lyapina S., Chi Y., Galova M., Claypool J., Sandmeyer S., Nasmyth K., Deshaies R. J. (1999) Genes Dev. 13, 1614–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Andréasson C., Ljungdahl P. O. (2002) Genes Dev. 16, 3158–3172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pfirrmann T., Heessen S., Omnus D. J., Andréasson C., Ljungdahl P. O. (2010) Mol. Cell Biol. 30, 3299–3309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bernard F., André B. (2001) Mol. Microbiol. 41, 489–502 [DOI] [PubMed] [Google Scholar]
- 21. Abdel-Sater F., Iraqui I., Urrestarazu A., André B. (2004) Genetics 166, 1727–1739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. de Boer M., Nielsen P. S., Bebelman J. P., Heerikhuizen H., Andersen H. A., Planta R. J. (2000) Nucleic Acids Res. 28, 974–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Eckert-Boulet N., Nielsen P. S., Friis C., dos Santos M. M., Nielsen J., Kielland-Brandt M. C., Regenberg B. (2004) Yeast 21, 635–648 [DOI] [PubMed] [Google Scholar]
- 24. Shin C. S., Kim S. Y., Huh W. K. (2009) J. Cell Sci. 122, 2089–2099 [DOI] [PubMed] [Google Scholar]
- 25. Wielemans K., Jean C., Vissers S., André B. (2010) J. Biol. Chem. 285, 855–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Amberg D. C., Burke D. J., Strathern J. N. (2005) Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 27. Chen X. J., Wang X., Butow R. A. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 13738–13743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yaffe M. P., Schatz G. (1984) Proc. Natl. Acad. Sci. U.S.A. 81, 4819–4823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu Z., Sekito T., Spírek M., Thornton J., Butow R. A. (2003) Mol. Cell 12, 401–411 [DOI] [PubMed] [Google Scholar]
- 30. Boban M., Zargari A., Andreasson C., Heessen S., Thyberg J., Ljungdahl P. O. (2006) J. Cell Biol. 173, 695–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Poulsen P., Wu B., Gaber R. F., Kielland-Brandt M. C. (2005) Eukaryot. Cell 4, 1116–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lee D. H., Goldberg A. L. (1998) Mol. Cell. Biol. 18, 30–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hershko A., Ciechanover A. (1998) Annu. Rev. Biochem. 67, 425–479 [DOI] [PubMed] [Google Scholar]
- 34. Patton E. E., Peyraud C., Rouillon A., Surdin-Kerjan Y., Tyers M., Thomas D. (2000) EMBO J. 19, 1613–1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Blondel M., Bach S., Bamps S., Dobbelaere J., Wiget P., Longaretti C., Barral Y., Meijer L., Peter M. (2005) EMBO J. 24, 1440–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Blondel M., Galan J. M., Chi Y., Lafourcade C., Longaretti C., Deshaies R. J., Peter M. (2000) EMBO J. 19, 6085–6097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Aronheim A., Zandi E., Hennemann H., Elledge S. J., Karin M. (1997) Mol. Cell. Biol. 17, 3094–3102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Huh W. K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., Weissman J. S., O'Shea E. K. (2003) Nature 425, 686–691 [DOI] [PubMed] [Google Scholar]
- 39. Mattaj I. W., Englmeier L. (1998) Annu. Rev. Biochem. 67, 265–306 [DOI] [PubMed] [Google Scholar]
- 40. Andréasson C., Ljungdahl P. O. (2004) Mol. Cell. Biol. 24, 7503–7513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Benanti J. A., Cheung S. K., Brady M. C., Toczyski D. P. (2007) Nat. Cell Biol. 9, 1184–1191 [DOI] [PubMed] [Google Scholar]
- 42. Zargari A., Boban M., Heessen S., Andréasson C., Thyberg J., Ljungdahl P. O. (2007) J. Biol. Chem. 282, 594–605 [DOI] [PubMed] [Google Scholar]
- 43. Li F. N., Johnston M. (1997) EMBO J. 16, 5629–5638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Südbeck P., Scherer G. (1997) J. Biol. Chem. 272, 27848–27852 [DOI] [PubMed] [Google Scholar]
- 45. Finley D., Ozkaynak E., Varshavsky A. (1987) Cell 48, 1035–1046 [DOI] [PubMed] [Google Scholar]
- 46. Seufert W., McGrath J. P., Jentsch S. (1990) EMBO J. 9, 4535–4541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lipford J. R., Smith G. T., Chi Y., Deshaies R. J. (2005) Nature 438, 113–116 [DOI] [PubMed] [Google Scholar]
- 48. Liu Z., Spírek M., Thornton J., Butow R. A. (2005) Mol. Biol. Cell 16, 4893–4904 [DOI] [PMC free article] [PubMed] [Google Scholar]