Abstract
Peptaibols are a group of small peptides having a high α-aminoisobutyric acid (Aib) content and produced by filamentous fungi, especially by the members of the genus Trichoderma (anamorph Hypocrea). These antibiotics are economically important for their anti-microbial and anti-cancer properties as well as ability to induce systemic resistance in plants against microbial invasion. In this study we present sequences of two classes (11-residue and 14-residue) of peptaibols produced by the biocontrol fungus Trichoderma virens. Of the 35 11-residue peptaibols sequenced, 18 are hitherto not described, and all the 53 14-residue sequences described by us here are new. We have also identified a peptaibol synthetase (non-ribosomal peptide synthetase, NRPS) with 14 complete modules in the genome of this fungus and disruption of this single gene (designated as tex2) resulted in the loss of both the classes of peptaibols. We, thus present here an unprecedented case where a single NRPS encodes for two classes of peptaibols. The new peptaibols identified here could have applications as therapeutic agents for the management of human and plant health.
Keywords: Antibiotics, Antimicrobial Peptides, Fungi, Genetics, Metabolism, NRPS, Non-ribosomal Peptides, Peptaibols, Therapeutics, Trichoderma
Introduction
Microbial resistance to classical antibiotics is a serious medical challenge, and new antimicrobial, membrane-active peptides have been proposed as alternatives to treat infections (1). Peptaibols (a subgroup of peptaibiotics) (2) are peptides characterized by short linear chain lengths (≤20 residues), C-terminal alcohol residues, and high levels of non-standard amino acids, principally α-aminoisobutyric acid (Aib),2 isovaline (Iva), and the imino acid hydroxyproline (Hyp). Naturally occurring peptaibols have been isolated from soil fungi (mostly from the genus Trichoderma (teleomorph Hypocrea) and related genera), and often exhibit antibacterial, antifungal and anti-cancer properties (3–9). The amphipathic nature of peptaibols allows them to self-associate into oligomeric ion-channel assemblies which span the width of lipid bi-layer membranes. Their antibiotic functions arise from their membrane insertion and pore-forming abilities (10). Some of these peptides have also been shown to inhibit the replication of enveloped viruses such as influenza A virus, vesicular stomatitis, and human immunodeficiency virus (11). In addition, some possess anticancer activities or promote wound healing (12). All these properties make these peptides good candidates as new therapeutic agents (13). Recently it has been shown for the first time that peptaibols are regulators involved in both apoptosis and autophagy, suggesting that the class of peptaibols might also serve as potential suppressors of tumor cells (13).
Trichoderma spp., the major producers of peptaibols, are a unique group of filamentous fungi that are omnipresent in soil as highly competitive saprophytes, as well as rhizosphere and root colonizers. They are capable of protecting crops from root-invading fungi, parasitizing mushrooms and other fungi, and in some instances being opportunistic human pathogens (14, 15). Thus, these fungi occupy important roles in agriculture, medicine as well as industry (16, 17).
Within the genus Trichoderma/Hypocrea, so far peptaibol subfamilies 1, 4, 5 and 9 have been described (18). Subfamily 1 (SF1) comprises about half of the known structures and includes peptides ranging from 18 to 20 residues in length. All of these peptides have partial sequence identities or similarities. Subfamily 4 (SF4) is comprised of peptides of 11–14 residues, also sharing sequence similarities, but having no sequence relationship to SF1. Subfamilies 5 and 9 (SF5 and -9) have only a few members and comprise peptides with 11 or 6 and 7 residues, respectively, again with no sequence similarities to the other subfamilies (19). An extensive survey of 28 Trichoderma/Hypocrea species for peptaibol production by intact-cell MALDI-TOF mass spectrometry (ICMS) revealed that peptaibols are produced by all strains with some producing up to five peptide families of different sizes (19). In addition to terrestrial strains, peptaibols have also been isolated from marine strains of Trichoderma (7, 20, 22, 23).
The physiological role of peptaibols in Trichoderma is largely unknown as most of the work has been directed toward isolation of new peptaibols from a pharmaceutical perspective. The role of peptaibols in sporulation and mycoparasitism has been hypothesized (24), and their involvement in induced disease resistance in plants has been demonstrated (18, 25–27). Recently, a microheterogenous mixture of 20-residue peptaibols isolated from an endophytic biocontrol strain of Trichoderma citrinoviride has been demonstrated to have antifungal activities against several important forest tree pathogens, thus implicating their role in biocontrol (28). The 18-residue peptaibols from T. harzianum have been shown to exhibit mammalian cytotoxicity (29).
Peptaibols are synthesized by large multidomain enzymes known as non-ribosomal peptide synthetases (NRPSs) that assemble compounds from a remarkable range of precursors (including nonproteinogenic amino acids and hydroxy or carboxyl acids), which can be N-methylated, acylated, reduced, or epimerized (30, 31). NRPSs have a modular structure in which each module is a semiautonomous unit that recognizes, activates, and modifies a single residue of the final peptide (30). Each module can be further partitioned into distinct adenylation (A), thiolation (T), and condensation (C) domains, which together represent a minimal repeating unit of a NRPS (32). The first genetic evidence of the involvement of an NRPS in peptaibol synthesis came from our studies of Tex1, where we demonstrated that an 18-module NRPS synthesizes the largest peptaibol produced by T. virens (26, 33, 34). However, so far, there have been no genetic studies on the synthesis of the shorter peptaibols, even though presence of these peptaibols are reported in many Trichoderma/Hypocrea spp (19). In this study, we present many new sequences of the 11- and 14-residue peptaibols of T. virens, and for the first time, provide genetic evidence that a single 14-module NRPS is responsible for the synthesis of these two classes of peptaibols.
EXPERIMENTAL PROCEDURES
Strains and Culture Conditions
T. virens strain Gv29-8 (hereafter described as Gv29-8), the genome of which has been sequenced recently by US-DOE-JGI, was used in this study. This strain is rhizosphere competent, exhibits mycoparasitism and also induces defense responses in plants (35). Fungal strains were maintained routinely on potato dextrose agar (PDA) or Vogel's mimimal medium supplemented with 1.5% sucrose (VMS) and stored as glycerol stocks at −80 °C for maintaining genetic stability. For selection following transformation, PDA containing the antibiotic hygromycin (PDAH) was used.
Peptaibol Profiling by MALDI-TOF
For peptaibol profiling, Gv29-8 was inoculated as a spore suspension (105 conidia per 90-mm plate) on cellophane membrane overlying VMS agar and grown for 3 days at 28 °C in an incubator. The sporulating cultures were harvested with a sterile spatula, frozen in liquid nitrogen, and lyophilized. Dried cells (5–10 mg) were suspended in 80 μl of water and gently mixed (brief low speed vortex). The resulting cell suspension was pelleted by centrifugation, and about 20 μl of the supernatant was removed and bound to a C18ZipTip from Millipore (Billerica, MA). After a single wash with water, 0.1% trifluoroacetic acid (TFA), peptaibols were eluted from the reversed phase matrix directly onto the MALDI plate for analysis with matrix solution containing α-cyano-4-hydroxy cinnamic acid in 50% acetonitrile, 50% water, 0.1% TFA. The samples were analyzed using a Voyager DE Pro MALDI-TOF MS. The ions were accelerated with a voltage of 20 kV in reflector mode with 80–120 ns delayed extraction (UMKC, School of Biological Sciences, Kansas City, MO).
Peptaibol Sequencing
Gv29-8 was inoculated onto a Petri dish (10-cm diameter) containing DCA medium (Dextrose Casein Agar) and grown for 7 days at 27 °C under fluorescent lights in 14/12 h day:night regime. Cultures were harvested and mycelia and conidia were scraped from the agar surface and steeped twice in dichloromethane/methanol (1:2 then 2:1, v/v) for 1 h at room temperature. The combined extracts were washed and evaporated to provide a crude extract. The crude extract was fractionated by vacuum liquid chromatography (VLC) on Chromabond adsorbant (OH) 2 Diol (Macherey-Nagel, Düren, Germany) with dichloromethane/ethanol mixtures (98:2; 90:10; 85:15, v/v). Peptaibols were eluted in dichloromethane/ethanol 90:10 (v/v) mixture and purified by HPLC on a preparative reverse column (Inerstil ODS-3, 5 μm, 250 mm × 10 mm, Interchim, Montluçon, France) with methanol/water (85:15, v/v) as the mobile phase. Mobile phase was delivered by a Residueck-Hitachi l-6000 pump at a constant flow rate of 5 ml/min and detection was performed at 230 nm with a LCD Milton Roy UV detector (Pont St Pierre, France).
Peptaibol MS analyses were carried out using a mass spectrometer (LCQ, Finnigan, Thermo Separation Products, San Jose, CA) equipped with an electrospray ionization source (ESI) and an ion trap analyzer (IT). Mass analyses were conducted in positive mode with a capillary temperature of 160 °C. All data were acquired and analyzed by LCQ Xcalibur software (Thermo Separation Products). Total current ion mass spectra (full scan mode) were acquired between m/z 150 and m/z 2000. Charge state and isotopic distribution were analyzed by a narrow-scan range mode (Zoomscan mode). HPLC subfractions were infused as methanolic solutions (5 μg/ml) directly into the ESI probe with a 500-μl micrometrically automated syringe (Hamilton, Massy, France) at a flow rate of 3 μl/min. Methanol of HPLC quality grade was purchased from Baker (Deventer, Holland). Microheterogenous mixtures of peptaibols were sequenced using consecutive fragmentation in ESI-MSn, according to the method described previously (20, 23).
Identification of the Peptaibol Synthetases in the Genome
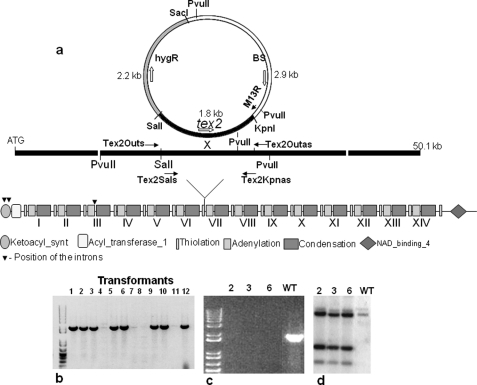
Using the TBALSTN algorithm with T. virens Tex1 (GenBankTM Acc. No. AAM78457) sequence as input, two additional NRPSs with high homology to Tex1 were identified in Gv29-8 genome. The 14-module gene was named as tex2 (protein ID 10003) and the 7-module as tex3 (protein ID 69362).
Disruption of the 14-Module NRPS-encoding Gene tex2
The tex2 gene was disrupted by single crossover homologous integration using hygromycin resistance as the selectable marker; this strategy was earlier used for disruption of a MAP kinase gene in T. virens (36). A primer pair Tex2Sals (GGC TGG GGT CGA CTA TAT AAA ACC) and Tex2Kpnas (TCA TTT GTT GGT ACC GGC TGA) were designed to amplify a 1.8-kb fragment from the genomic sequence (SalI is present in the sequence, KpnI was introduced), digested with this pair of enzymes and ligated to pre-digested pAT-BS harboring the hygromycin resistance cassette. The construct thus designed disrupts module 7 of the tex2 gene. The resulting plasmids containing the hygromycin resistance cassette and a 1.8-kb genomic fragment of the tex2 gene was used for protoplast transformation as described earlier (37). The transformants that developed on PDAH were serially transferred to PDAH (three times), back to PDA (once), and finally to PDAH. DNA was extracted from the transformants that retained their resistance under these conditions of transfer, and subjected to specific PCR amplification that detects homologous recombination using the primer pairs Tex2Outs (AACTAGCTGCGACTGGAATTGTTG) and M13 reverse primer from the vector. The positive strains generating PCR products consistent with an homologous integration event were further purified by single-spore isolation and were subjected to Southern hybridization to determine the copy number. Three transformants with a single copy homologous integration were further analyzed for purity (loss of wild type gene) by PCR using Tex2Outs and Tex2Outas (CAACAAGAGGGTAATTTAGATAAT TTT GAAG) primers. All the three mutants (hence after referred to as tex2–2, tex2–3, tex2–6) were subjected to MALDI-TOF analysis as described above.
Adenylation Domain Alignment and Substrate Prediction
The adenylation domain sequence was determined according to Ref. 38. Adenylation domain amino acid sequences were trimmed to include the selectivity conferring residues as defined by Stachelhaus et al. (39) and aligned using ClustalW 2.0.3. Adenylation substrate prediction was determined using NRPSpredictor (40).
RESULTS
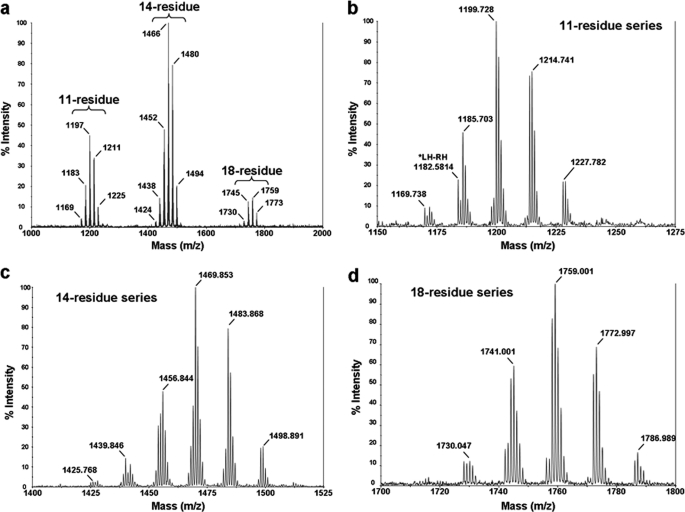
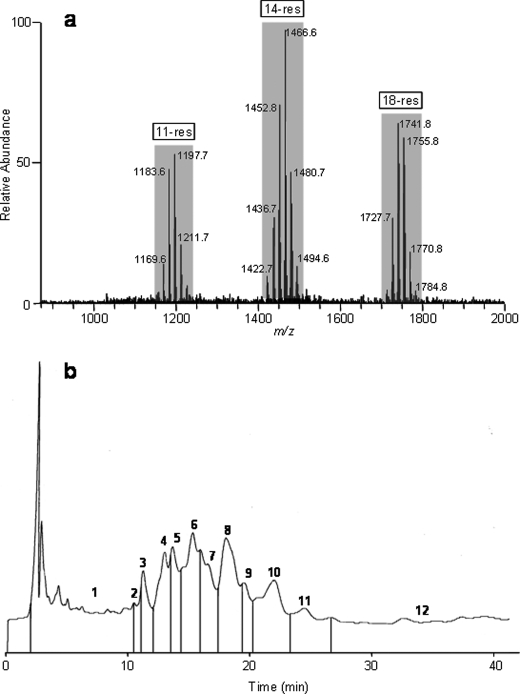
After extraction of the fungal biomass and a consecutive vacuum liquid chromatography, the semi-purified extract consisted of the dichloromethane/ethanol 90:10 (v/v) fraction, which contained a complex mixture of 11, 14, and 18-residue peptaibols (Fig. 1). Additionally, in LC-MS, we have observed the presence of 2 minor products, which depicted [M+Na]+ ions at m/z 689 and 703 from methanolic extract of T. virens; these could correspond to the 7-residue peptaibols (data not presented). In ESI-MS infusion mode, peptaibols appeared as singly charged [M+Na]+ sodium adduct ions ranging from m/z 1150 to 1230, 1400 to 1500, and 1700 to 1800 for 11, 14, and 18-residue peptides, respectively (Fig. 2a). Submitted to a reverse phase preparative HPLC, the semi-purified extract gave 12 HPLC subfractions of microheterogeneous mixtures of isoforms (Fig. 2b).
FIGURE 1.
MALDI-TOF profile of Trichoderma virens Gv29-8 peptaibols (a), 11-residue series (b), 14-residue series (c), and 18-residue series (d).
FIGURE 2.
ESI-MS full-scan mass spectrum (a) and HPLC chromatogram (b) of peptaibols from T. virens Gv29-8.
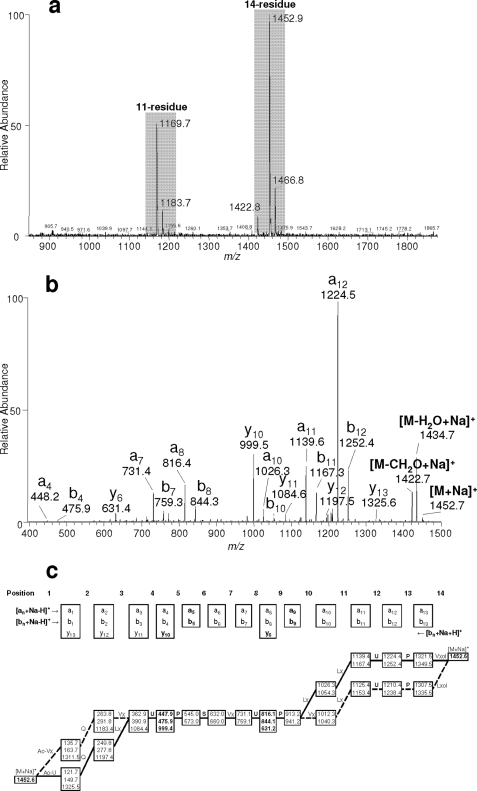
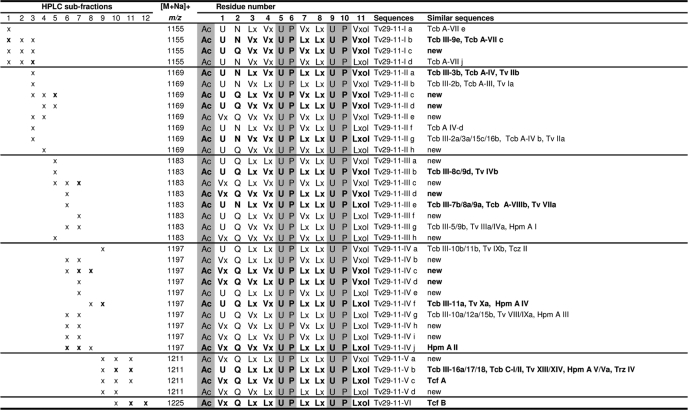
All HPLC subfractions were analyzed in neutral conditions in ESI-MS infusion mode as shown in Fig. 3a for subfraction 5. For each subfraction, resolution of the microheterogeneous composition of observed ions was done according to the method described by Refs. 20, 23 using consecutive fragmentations in ESI-MSn by identifying an, bn, and yn diagnostic fragment ions (Fig. 3b), and realizing a filiation graph for each microheterogenous mixture as presented in Fig. 3c for the 14-residue peptaibol displaying [M+Na]+ at m/z 1452. Sequences of 11- and 14-residue peptaibols identified by ESI-MSn from Gv 29-8 are listed in Tables 1 and 2, respectively. For each sequence the corresponding HPLC subfraction number is indicated. Major sequences for each observed [M+Na]+ ion are written in bold. The 11-residue peptaibols corresponded to a complex mixture of microheterogeneous sequences diplaying [M+Na]+ ions at m/z 1155, 1169, 1183, 1197, 1211, and 1225 with prominent masses of Na+ adducts at m/z 1183 and 1197 (Fig. 2a). Thirty five sequences of 11-residue peptaibols were identified from HPLC subfractions 1–12 (Table 1). They belong to the same peptide family based on the sequence model Ac-XXX-XXX-XXX-XXX-Aib-Pro-XXX-XXX-Aib-Pro-XXXol, where Ac is acetyl-, Aib is α-aminoisobutyric acid, ol indicates reduction to the amino-alcohol, and XXX corresponds to variable amino acids. Seventeen sequences were new and the eighteen others were similar to already known 11-residue peptaibols such as trichobrachins (Trichoderma longibrachiatum) (20, 41), trichofumins (Trichoderma sp. HKI 0276) (42), trichorovins (T. harzianum) (43), trichorozins (T. harzianum) (44), and hypomurocins, formerly originated from Hypocrea muroiana (45), which was recently demonstrated to be H. atroviridis (46).
FIGURE 3.
Full MS spectrum of subfraction 5 (mass range from m/z 800 to 1950) (a). MS2 fragmentation spectrum of the singly charged [M+Na]+ ion at m/z 1452 (b). Filiation graph of the fragment ions (m/z) obtained by successive fragmentations in ESI-MSn of the [M+Na]+ ion at m/z 1452 (c). Major breakings are figured by bold lines, leading to major sequences, and minor ones by dotted lines. Sequences are given in standard single-letter code (Ac, acetyl-, U, Aib, Vx, Val/Iva, Lx, Leu/Ile, and ol represents the C-terminal amino-alcohol).
TABLE 1.
Sequences of 11-residue peptaibols produced by T. virens Gv29-8
Sequences are given in standard single-letter code (Ac, acetyl-; U, Aib; Vx, Val/Iva; Lx, Leu/Ile; and “ol” represents the C-terminal amino-alcohol). Abbreviations: Tcf, trichofumin; Tv, trichorovin; Tcb, trichobrachin; Tcz, trichorozin; Hpm, hypomurocin.
TABLE 2.
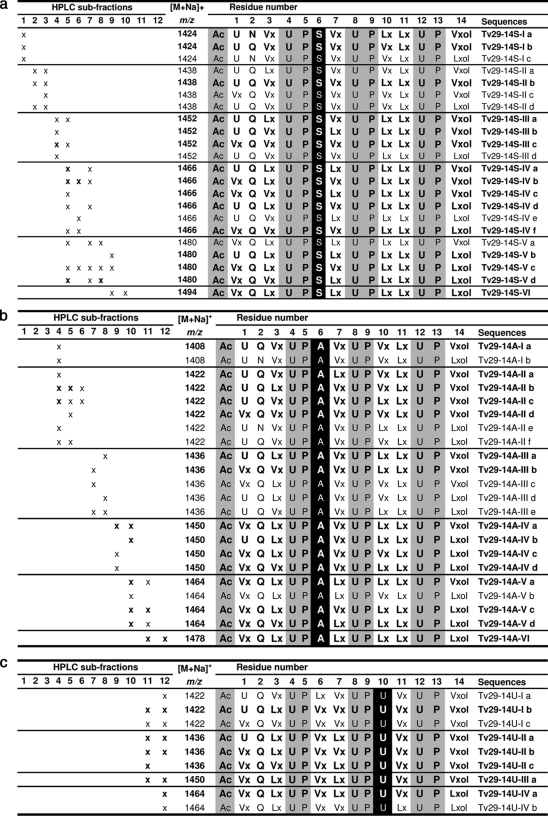
Sequences of 14-residue peptaibols produced by T. virens Gv29-8
Ser-6-containing peptaibols (a) Ala6-containing peptaibols (b) Aib10-containing peptaibols (c). Sequences are given in standard single-letter code (Ac, acetyl-; U, Aib; Vx, Val/Iva; Lx, Leu/Ile; and “ol” represents the C-terminal amino-alcohol).
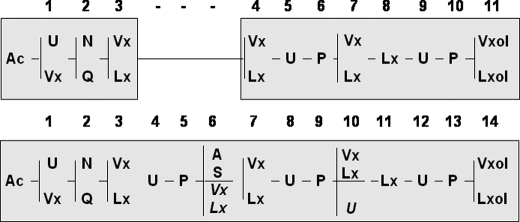
Three series of 14-residue peptaibols were identified in Gv29-8. Each was characterized by a peculiar residue in position 6 or 10. The major one, observed from HPLC subfractions 1 to 10 displayed [M+Na]+ ions at m/z 1424, 1438, 1452, 1466, 1480, and 1494 with prominent masses of Na+ adducts at m/z 1452 and 1466 (Fig. 2a). This series belongs to the same peptide family as harzianins HC isolated from cultures of T. harzianum, which are characterized by a Ser in position 6 (47). Twenty-two new sequences were identified based on the same sequence pattern Ac-XXX-XXX-XXX-Aib-Pro-Ser-XXX-Aib-Pro-XXX-XXX-Aib-Pro-XXXol (Table 2a). The other 14-residue peptaibol series were less predominant than the previous Ser-6 series. They were eluted from HPLC subfractions 4–12. This series displayed [M+Na]+ ions at m/z 1408, 1422, 1436, 1450, 1464, and 1478 with prominent masses of Na+ adducts at m/z 1436 and 1450. Two different microheterogeneous sequence patterns were observed. The most abundant one corresponded to the already known series of harzianins HC containing an Ala in position 6 Ac-XXX-XXX-XXX-Aib-Pro-Ala-XXX-Aib-Pro-XXX-XXX-Aib-Pro-XXXol (47). Twenty-two new sequences have been identified based on this pattern (Table 2b). The second one was a new group of 14-residue peptaibols. In this unprecedented series, Ser/Ala6 is exchanged with a variable VXX/LXX6 and the variable VXX/LXX10 is replaced by a constant Aib10, leading to the pattern Ac-XXX-XXX-XXX-Aib-Pro-XXX-XXX-Aib-Pro-Aib-XXX-Aib-Pro-XXXol expressed in nine sequences (Table 2c). A comparison of the 11-residue and 14-residue peptaibols revealed an interesting fact- the amino acids at positions 4, 5, and 6 are absent in the 11-residue peptaibols (Fig. 4 and supplemental Table S1).
FIGURE 4.
Sequence pattern alignments of 11- and 14-residue peptaibols produced by T. virens Gv29-8. Sequences are given in standard single-letter code (Ac, acetyl-, U, Aib, Vx, Val/Iva, Lx, Leu/Ile, and ol represents the C-terminal amino-alcohol). Numbers above each shaded box correspond to the proposed module order.
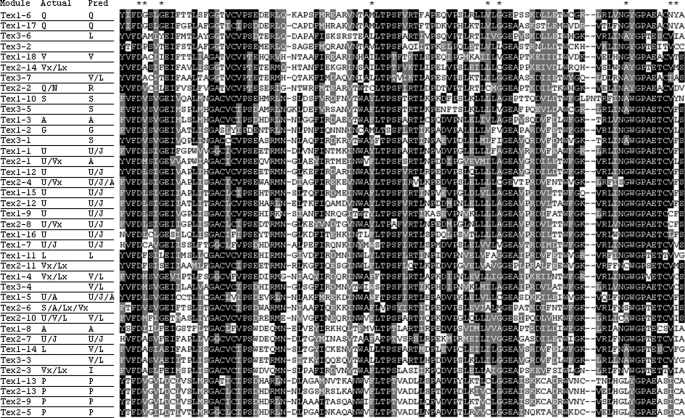
A survey of the Gv29-8 genome indicated the presence of three NRPS genes with high homology to Tex1: the earlier described tex1 (Protein ID 66940), a gene coding for an NRPS of 14 linear modules (tex2, protein ID 10003), and another one with 7 modules (tex3, Protein ID 69362). Significantly, there is no unique NRPS with 11-modules in the genome. The Tex2 NRPS is encoded by a single ORF of 50,096 bp, and interrupted by 3 introns of 85,245, and 248 bp (Fig. 5a). The protein has a β-ketoacyl synthase domain at the N-terminal and 14 complete modules of thiolation-adenylation-condensation domains in a linear fashion. A phylogenetic analysis of all the adenylation domains of both the 18-module NRPSs (Tex1) and 14-module NRPSs from three species of Trichoderma (T. reesei, T. virens, and T. atroviride), along with other NRPSs from Trichoderma spp. and other fungi indicated that the tex2 belongs to the peptaibol class of fungal NRPSs (data not presented). An alignment of the primary sequences between the adenylation domain core motifs A4 and A5 from modules of three NRPSs from Gv29-8 shows strong similarity between all domains (Fig. 6). As is apparent, all but the first and second adenylation domains of Tex-2 agree with the predicted substrate using the Stachelhaus model (39) The extended Rausch code (40), however, predicts the specificity for the first adenylation domain (U/V) as G/A/V/L/I/U/J and is also good at predicting the possible specificities of the most of the other adenylation domains (supplemental Table S2). Neither model accurately predicts the observed specificity of the second adenylation domain (Q/N). As the substrate specificity for more adenylation domains is determined, the accuracy of the current models is certain to increase. A BLASTP search in the NCBI data base indicated that Tex2 is highly homologous to the Tex1 sequence of T. virens and a putative aerothricin synthase of an unknown fungus (query coverage 99 and 93%, respectively, e-value 0.0).
FIGURE 5.
Modular structure and strategy for obtaining loss-of function mutants in the peptaibol synthetase Tex2. Construct used for obtaining loss-of-function mutants (a). The strategy disrupts module 7. Screening of transformants for homologous integration by PCR is shown. The presence of an amplicon using the primer pair Tex2Outs and M13 reverse indicates homologous integration (b). PCR to confirm gene disruption is shown. Note the absence of amplicon in case of the three mutants with the primers Tex2Outs and Tex2Outas; the band is present in the wild type (c). Southern hybridization to confirm the single copy homologous integration is shown (d). The genomic DNA was digested with PvuII and hybridized with the same genomic fragment that was used for making the disruption construct.
FIGURE 6.
A ClustalW alignment of the adenylation domains of three peptaibol-encoding NRPSs of Gv29-8. Identical and similar residues are shaded black and gray, respectively. The actual amino acids activated by each module as well as the predicted substrates are shown. The selectivity conferring residues at positions 235, 236, 239, 278, 299, 301, 322, 330, and 331 (39) are indicated by asterisks.
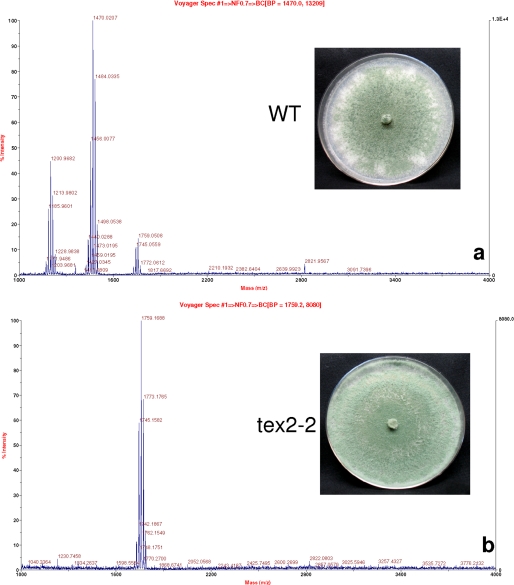
Using the single crossover homologous integration strategy, earlier used successfully to disrupt T. virens MAP kinase (36), we obtained 8 independent transformants showing homologous integration (Fig. 5b). Three (designated as tex2-2, tex2-3, and tex2-6) were selected for further studies. PCR amplification of the putative mutants confirmed the lack of the wild type copy in each of the mutants (Fig. 5c), and the single copy number was additionally confirmed by Southern hybridization (Fig. 5d). There was no major difference between the wild type and the mutants in terms of colony characteristics, except that the mutants exhibited a 12–15% higher radial growth rate. All the mutants lacked both the 11- and 14-residue peptaibols as evidenced from the MALDI-TOF profile (Fig. 7, mutant tex2-2). Because disruption of a single gene resulted in loss of two classes of peptaibols, we further investigated the possible mechanisms. As there is no intron predicted at the end of module 6 (Fig. 5a), alternative splicing is ruled out as a mechanism. We also excluded the possibility of alternative splicing across intron1/2 and intron 3 based on RT-PCR analysis (data not presented).
FIGURE 7.
Peptaibol profile of WT (a) and a tex2 mutant (b). Note that both the 11-residue and 14-residue peptaibols are absent in the mutant. Inset: growth of WT and tex2 mutant on VMS agar after 5 days of incubation.
DISCUSSION
Trichoderma spp. are the major sources of peptaibols/peptaibiotics, synthesized by multi-modular mega-enzymes known as non-ribosomal peptide synthetases. These membrane-channel forming peptides have potential applications as antibiotics, antiviral, and anticancer agents, in addition to their use as inducers of plant defense. In this study, we isolated and characterized a microheterogeneous peptaibol mixture composed of 11 and 14 amino acid residues from the strain Gv29-8. The general structure of both groups of peptaibols is presented in Fig. 4. 18-residue peptaibols also produced by Gv29-8 and belonging to the trichorzin TVB group (TVBI, II, and IV) have been sequenced in a previous study (33). According to their sequence pattern, 11-residue peptaibols isolated from Gv29-8 belong to the major and most widely distributed series of 11-residue peptaibols produced by Trichoderma genus for which more than 100 sequences have been published in the literature. A high microheterogeneity in the 11-residue peptaibol mixture produced by T. virens was observed, composed of 35 analogous sequences, including 18 new, and differing from each other by residue substitutions in positions 1, 2, 3, 4, 7, and 11. These observations are in agreement with previous studies which related high microheterogeneity of similar 11-residue peptaibol trichobrachins isolated from T. longibrachiatum and T. parceramosum (20, 41).
Fewer 14-residue peptaibol sequences have been described in comparison to 11-residue sequences. Eleven sequences have been published corresponding to the harzianin HC group isolated from the phylogenetically undefined T. harzianum strains M-903614 and M-903603 (47). In the present study, numerous 14-residue sequences belonging to harzianins HC group have been observed from Gv29-8, displaying similar general structure with variable residues in positions 1, 2, 3, 6, 7, 10, and 14. The previously known series of harzianins HC differing by the constant residue 6, which can be Ala or Ser, have been detected in Gv29-8. Interestingly all of these microheterogeneous sequences identified correspond to new sequences of the harzianins HC group. Another different series displaying a constant Aib in position 10 and VXX/LXX in position 6 is observed for the first time. It is the highest number of analogous sequences so far observed for 14-residue peptaibols, and the first report of sequences identified from a T. virens strain.
Even though to date there are over 900 peptaibol sequences available (2), the genetics of their synthesis is poorly understood, barring a few examples such as Tex1 (26, 33, 34), and a partial peptaibol synthetase gene in T. harzianum CECT 2413 (48). Recent genome sequencing revealed that in addition to Tex1, an 18-module peptaibol synthetase, the T. virens genome harbors two genes with high homology with Tex1, a 14-module (Tex2) and a 7-module (Tex3). These three genes are phylogenetically closely related to each other (Fig. 6).3 However, the peptaibol profiling using MALDI-TOF MS/MS revealed the presence of an 18-residue, a 14-residue and an 11-residue peptaibol (Fig. 1). Depending upon the strain, Trichoderma spp. produce either a 11-residue peptaibol or a 14-residue peptaibol, or both (19). Although, we initially reported that the deletion of Tex1 resulted in the loss of all three (18, 14, and 11-residue) peptaibols, it was subsequently confirmed by us and another group that Tex1 codes for only the 18-residue peptaibol (26, 33, 34). The synthesis of the 11-residue peptaibol has thus been a mystery as there is no 11-module NRPS gene in the Trichoderma genomes. In the present study, we, for the first time, present genetic evidence that disruption of a single NRPS gene results in the loss of two peptaibols, both 14-residue and the 11-residue. Comparison of the 11- and 14-residue patterns allows the observation of sequence similarities between the two groups of peptaibols (Fig. 4 and supplemental Table S1). As already suggested, both types of peptaibols could originate from a single peptide synthetase. Based on the available sequence comparison of 11-and 14-residue peptaibols, Neuhof et al. (19) predicted that SF4 11- (group 6) and 14-residue (groups 8, 9, and 11) compounds could be produced by a single peptaibol synthetase by deletion of modules 3–5 (Leu3-Pro4-Aib5). Nevertheless, taking into account the alignments of the sequences, it seems that in Gv29-8, 11-residue peptaibols could be derived from 14-residue peptaibols by internal deletion of the residues in positions 4, 5, and 6 or could be synthesized by an enzyme that is lacking the modules for incorporation of residues 4, 5, and 6. Because the 11-residue peptaibol is essentially the 14-residue peptaibol without the amino acids at positions 4, 5, and 6, it seems possible that deletion of these three domains results in the formation of 11-residue from the 14-residue template. Additionally, both forms are acylated at the N terminus and hence functionally utilize the AC and KS domains at the 5′-end of the tex2 gene. Thus, there is no possibility that the 11-module NRPS can arise by an N-terminal truncation of the 14-module enzyme. Disruption of the 7-module NRPS Tex3 did not affect the production of 18-, 14-, or 11-residue peptaibols (data not presented). There are two possibilities by which the 14-module gene could be responsible for synthesis of both 11-residue and 14-residue peptaibols, an alternative splicing of the gene per se or skipping of these three modules by the 14-module NRPS during synthesis. Possibility of an alternative splicing event can be ruled out as there are no introns corresponding to these three modules (Fig. 5a). The only possibility left is skipping of the three modules during biosynthesis of the 11-residue peptaibols. Module skipping during peptaibol synthesis has not been demonstrated yet, but this phenomenon operates during biosynthesis of many types of non-ribosomal peptides giving rise to enhanced chemodiversity (21, 49–54). Synthesis of two groups of compounds (e.g. 11-residue and 14-residue peptaibols) by a single mega-enzyme could have ecological advantage for soil/rhizosphere fungi, like T. virens, as these ecological niches are characterized by intense microbial compounds and the peptaibols are known to be anti-microbial.
In addition to the genetics, we report here the sequences of 35 (of which 18 are new) 11-residue and 53 (all new) 14-residue peptaibols in T. virens 29-8. These new peptaibol sequences described here could be a source of novel medicinally and agriculturally important therapeutic agents for management of human and plant health.
Supplementary Material
This project was supported by the National Research Initiative Competitive Grants Program Grant 2008-35319-04470 (to C. M. K.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2.
- Aib
- α-aminoisobutyric acid
- NRPS
- non-ribosomal peptide synthetase
- Hyp
- hydroxyproline
- Iva
- isovaline.
REFERENCES
- 1. Duclohier H. (2007) Chem. Biodivers. 4, 1023–1026 [DOI] [PubMed] [Google Scholar]
- 2. Degenkolb T., Brückner H. (2008) Chem. Biodivers. 5, 1817–1843 [DOI] [PubMed] [Google Scholar]
- 3. Daniel J. F., Filho E. R. (2007) Nat. Prod. Rep. 24, 1128–1141 [DOI] [PubMed] [Google Scholar]
- 4. Degenkolb T., von Döhren H., Nielsen K. F., Samuels G. J., Brückner H. (2008) Chem. Biodivers. 5, 671–680 [DOI] [PubMed] [Google Scholar]
- 5. Degenkolb T., Gams W., Brückner H. (2008) Chem. Biodivers. 5, 693–706 [DOI] [PubMed] [Google Scholar]
- 6. Kubicek C. P., Komoń-Zelazowska M., Sándor E., Druzhinina I. S. (2007) Chem. Biodivers. 4, 1068–1082 [DOI] [PubMed] [Google Scholar]
- 7. Pruksakorn P., Arai M., Kotoku N., Vilchèze C., Baughn A. D., Moodley P., Jacobs W. R., Jr., Kobayashi M. (2010) Bioorg. Med. Chem. Lett. 20, 3658–3663 [DOI] [PubMed] [Google Scholar]
- 8. Sang Y., Blecha F. (2008) Animal Health Res. Rev. 9, 227–235 [DOI] [PubMed] [Google Scholar]
- 9. Chutrakul C., Alcocer M., Bailey K., Peberdy J. F. (2008) Chem. Biodivers. 5, 1694–1706 [DOI] [PubMed] [Google Scholar]
- 10. Sansom M. S. (1993) Quart. Rev. Biophys. 26, 365–421 [DOI] [PubMed] [Google Scholar]
- 11. Degenkolb T., Berg A., Gams W., Schlegel B., Gräfe U. (2003) J Pept. Sci. 9, 666–678 [DOI] [PubMed] [Google Scholar]
- 12. Marahiel M. A. (2009) J Pept. Sci. 15, 799–807 [DOI] [PubMed] [Google Scholar]
- 13. Shi M., Wang H. N., Xie S. T., Luo Y., Sun C. Y., Chen X. L., Zhang Y. Z. (2010) Mol. Cancer 9, 26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mukherjee P. K., Nautiyal C. S., Mukhopadhyay A. N. (2008) in Molecular Mechanisms of Plant and Microbe Coexistence (Soil Biology Series) (Nautiyal C. S., Dion P. eds) Springer, Netherlands [Google Scholar]
- 15. Verma M., Brar S. K., Tyagi R. D., Surampalli R. Y., Surampalli J., Valéro J. R. (2007). Biochem. Eng. J 37, 1–20 [Google Scholar]
- 16. Brotman Y., Kapuganti J. G., Viterbo A. (2010) Curr. Biol. 20, R390–391 [DOI] [PubMed] [Google Scholar]
- 17. Schuster A., Schmoll M. (2010) Appl. Microbiol. Biotechnol. 87, 787–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Szekeres A., Leitgeb B., Kredics L., Antal Z., Hatvani L., Manczinger L., Vágvölgyi C. (2005) Acta. Microbiol. Immunol. Hung. 52, 137–168 [DOI] [PubMed] [Google Scholar]
- 19. Neuhof T., Dieckmann R., Druzhinina I. S., Kubicek C. P., von Döhren H. (2007) Microbiology. 153, 3417–3437 [DOI] [PubMed] [Google Scholar]
- 20. Ruiz N., Wielgosz-Collin G., Poirier L., Grovel O., Petit K. E., Mohamed-Benkada M., Robiou du Pont T. R., Bisset J., Vérité P., Barnathan G., Pouchus Y. F. (2007) Peptides 28, 1351–1358 [DOI] [PubMed] [Google Scholar]
- 21. Wei M., Wang S., Shang G. (2010) Curr. Org. Chem. 14, 1433–1446 [Google Scholar]
- 22. Ren J., Xue C., Tian L., Xu M., Chen J., Deng Z., Proksch P., Lin W. (2009) J Nat. Prod. 72, 1036–1044 [DOI] [PubMed] [Google Scholar]
- 23. Mohamed-Benkada M., Montagu M., Biard J. F., Mondeguer F., Vérité P., Dalgalarrondo M., Bissett J., Pouchus Y. F. (2006) Rapid Commun Mass Spectrom 20, 1176–1180 [DOI] [PubMed] [Google Scholar]
- 24. Komon-Zelazowska M., Neuhof T., Dieckmann R., von Döhren H., Herrera-Estrella A., Kubicek C. P., Druzhinina I. S. (2007) Eukaryot. Cell 6, 2332–2342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Engelberth J., Koch T., Kühnemann F., Boland W. (2000) Angew. Chem. Int. Ed. Engl. 39, 1860–1862 [DOI] [PubMed] [Google Scholar]
- 26. Viterbo A., Wiest A., Brotman Y., Chet I., Kenerley C. M. (2007) Mol. Plant Pathol. 8, 737–746 [DOI] [PubMed] [Google Scholar]
- 27. Yun B., Yoo I., Kim Y., Kim Y., Lee S., Kim K., Yeo W. (2000) Tetrahedron Lett. 41, 1429–1431 [Google Scholar]
- 28. Maddau L., Cabras A., Franceschini A., Linaldeddu B. T., Crobu S., Roggio T., Pagnozzi D. (2009) Microbiology. 155, 3371–3381 [DOI] [PubMed] [Google Scholar]
- 29. Peltola J., Ritieni A., Mikkola R., Grigoriev P. A., Pócsfalvi G., Andersson M. A., Salkinoja-Salonen M. S. (2004) Appl. Environ. Microbiol. 70, 4996–5004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Marahiel M. A., Stachelhaus T., Mootz H. D. (1997) Chem. Rev. 97, 2651–2674 [DOI] [PubMed] [Google Scholar]
- 31. Zocher R., Keller U. (1997) Adv. Microb. Physiol. 38, 85–131 [DOI] [PubMed] [Google Scholar]
- 32. Stein T., Vater J., Kruft V., Otto A., Wittmann-Liebold B., Franke P., Panico M., McDowell R., Morris H. R. (1996) J. Biol. Chem. 271, 15428–15435 [DOI] [PubMed] [Google Scholar]
- 33. Wiest A., Grzegorski D., Xu B. W., Goulard C., Rebuffat S., Ebbole D. J., Bodo B., Kenerley C. (2002) J. Biol. Chem. 277, 20862–20868 [DOI] [PubMed] [Google Scholar]
- 34. Wei X., Yang F., Straney D. C. (2005) Can. J Microbiol. 51, 423–429 [DOI] [PubMed] [Google Scholar]
- 35. Harman G. E., Howell C. R., Viterbo A., Chet I., Lorito M. (2004) Nat. Rev. Microbiol. 2, 43–56 [DOI] [PubMed] [Google Scholar]
- 36. Mukherjee P. K., Latha J., Hadar R., Horwitz B. A. (2003) Eukaryot. Cell 2, 446–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Thomas M. D., Kenerley C. M. (1989) Curr. Genet. 15, 415–420 [Google Scholar]
- 38. Ansari M. Z., Yadav G., Gokhale R. S., Mohanty D. (2004) Nucleic. Acids Res. 32, 405–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Stachelhaus T., Mootz H. D., Marahiel M. A. (1999) Chem. Biol. 6, 493–505 [DOI] [PubMed] [Google Scholar]
- 40. Rausch C., Weber T., Kohlbacher O., Wohlleben W., Huson D. H. (2005) Nucleic Acids Res. 33, 5799–5808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Krause C., Kirschbaum J., Brückner H. (2007) Chem. Biodivers. 4, 1083–1102 [DOI] [PubMed] [Google Scholar]
- 42. Berg A., Grigoriev P. A., Degenkolb T., Neuhof T., Härtl A., Schlegel B., Gräfe U. (2003) J. Pept. Sci. 9, 810–816 [DOI] [PubMed] [Google Scholar]
- 43. Wada S., Iida A., Akimoto N., Kanai M., Toyama N., Fujita T. (1995) Chem. Pharm. Bull. 43, 910–915 [DOI] [PubMed] [Google Scholar]
- 44. Iida A., Sanekata M., Wada S., Fujita T., Tanaka H., Enoki A., Fuse G., Kanai M., Asami K. (1995) Chem. Pharm. Bull. 43, 392–397 [DOI] [PubMed] [Google Scholar]
- 45. Becker D., Kiess M., Brückner H. (1997) Liebigs. Ann. Recueil. 767–772 [Google Scholar]
- 46. Degenkolb T., Gräfenham T., Nirenberg H. I., Gams W., Brückner H. (2006) J Agric. Food Chem. 54, 7047–7061 [DOI] [PubMed] [Google Scholar]
- 47. Rebuffat S., Goulard C., Bodo B. (1995) J Chem. Soc. Perkin. Trans. I 14, 1849–1855 [Google Scholar]
- 48. Vizcaino J. A., Cardoza R. E., Dubost L., Bodo B., Gutierrez S., Monte E. (2006) Folia Microbiol. 51, 114–120 [DOI] [PubMed] [Google Scholar]
- 49. Bode H. B., Müller R. (2006) J Ind. Microbiol. Biotechnol. 33, 577–588 [DOI] [PubMed] [Google Scholar]
- 50. Weckwerth W., Miyamoto K., Iinuma K., Krause M., Glinski M., Storm T., Bonse G., Kleinkauf H., Zocker R. (2000) J. Biol. Chem. 275, 17909–17915 [DOI] [PubMed] [Google Scholar]
- 51. Wenzel S. C., Müller R. (2005) Curr. Opin. Chem. Biol. 9, 447–458 [DOI] [PubMed] [Google Scholar]
- 52. Wenzel S. C., Kunze B., Höfle G., Silakowski B., Scharfe M., Blöcker H., Müller R. (2005) Chem. Bio. Chem. 6, 375–385 [DOI] [PubMed] [Google Scholar]
- 53. Wenzel S. C., Meiser P., Binz T. M., Mahmud T., Müller R. (2006) Angew. Chem. Int. Ed. Engl. 45, 2296–2301 [DOI] [PubMed] [Google Scholar]
- 54. Lautru S., Deeth R. J., Bailey L. M., Challis G. L. (2005) Nature Chem. Ecol. 1, 265–269 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.