Abstract
Hepcidin is a liver-derived hormone with a key role in iron homeostasis. In addition to iron, it is regulated by inflammation and hypoxia, although mechanisms of hypoxic regulation remain unclear. In hepatocytes, hepcidin is induced by bone morphogenetic proteins (BMPs) through a receptor complex requiring hemojuvelin (HJV) as a co-receptor. Type II transmembrane serine proteinase (TMPRSS6) antagonizes hepcidin induction by BMPs by cleaving HJV from the cell membrane. Inactivating mutations in TMPRSS6 lead to elevated hepcidin levels and consequent iron deficiency anemia. Here we demonstrate that TMPRSS6 is up-regulated in hepatic cell lines by hypoxia and by other activators of hypoxia-inducible factor (HIF). We show that TMPRSS6 expression is regulated by both HIF-1α and HIF-2α. This HIF-dependent up-regulation of TMPRSS6 increases membrane HJV shedding and decreases hepcidin promoter responsiveness to BMP signaling in hepatocytes. Our results reveal a potential role for TMPRSS6 in hepcidin regulation by hypoxia and provide a new molecular link between oxygen sensing and iron homeostasis.
Keywords: Bone Morphogenetic Protein (BMP), Hepatocyte, Hypoxia, Iron Metabolism, Metabolic Regulation, BMP Signaling, HIF, Hemojuvelin, TMPRSS6, Hypoxia
Introduction
The main physiological regulator of systemic iron homeostasis is the hormone hepcidin. Synthesized in the liver, this 25-amino acid peptide binds to and internalizes the ferroportin iron exporter, which in turn blocks iron uptake from duodenal enterocytes and iron reutilization by the reticuloendothelial system (1, 2).
Hepcidin expression in the liver is subject to control by a number of signaling pathways including interleukin-6 (IL-6)-stimulated Janus kinase/signal transducers and activators of transcription (JAK/STAT) signaling, probably contributing to the anemia of chronic inflammation (3). In addition, hepcidin is induced by the bone morphogenetic proteins, BMP3 2, 4, 6, and 9 that signal through son of mothers against decapentaplegic (SMAD) binding to bone morphogenetic response elements (BREs) in the hepcidin promoter (4, 5). Activity of the BMP receptor complex BMRI/II is regulated by transferrin levels through the non-classical MHC class I molecule HFE. When transferrin levels are low, HFE interacts on the cell surface with transferrin receptor 1 (TfR1). When transferrin levels are high, its binding to TfR1 releases HFE to stimulate hepcidin production through the BMPRI/II complex (6, 7).
Activity of the BMPRI/II complex is further modulated by interaction with hemojuvelin (HJV), a glycosylphosphatidylinositol (GPI)-anchored protein that acts as co-receptor for BMPs (8). Inactivating mutations in either HFE or HJV lead to reduced levels of hepcidin, increased iron absorption and hemochromatosis (9, 10). HJV is subject to post-translational processing by the trans-Golgi associated enzyme furin, that releases a 42-kDa soluble component (sHJV) into the extracellular space (11), which then acts as a “decoy-receptor” competing with membrane HJV (mHJV) for the BMP ligand (12).
Recently, a second HJV-processing enzyme, transmembrane serine proteinase TMPRSS6 (also known as matriptase-2), has emerged as a key physiological regulator of hepcidin (13, 14). Expressed predominantly in the liver, TMPRSS6 antagonizes BMP signaling in hepatocytes by cleaving membrane-bound HJV (15). Both targeted knockdown of TMPRSS6 in mouse models and random inactivation in human disease lead to inappropriately high levels of hepcidin and consequent iron-deficiency, underscoring the importance of TMPRSS6 in the regulation of hepcidin (11, 15–21).
In addition to sensing iron levels, hepcidin production also responds to hypoxia (3, 22). Mice subjected to hypobaric hypoxia exhibit reduced hepcidin levels (22, 23) but this hypoxic regulation may be due in part to iron utilization resulting from increased erythropoiesis driven by hypoxically stimulated erythropoietin (24). More direct effects are suggested by the finding that in some cell types under certain conditions hypoxia may down-regulate hepcidin mRNA in cell culture (22, 25), although these effects are difficult to reproduce under all culture conditions (26).
Cellular homeostatic responses to altered oxygen are chiefly orchestrated through the hypoxia-inducible factor (HIF) transcriptional pathway, which regulates the expression of many hundreds of genes, including several with known roles in iron homeostasis (27, 28). Activity of the HIF transcription factor is controlled through stability of its α-subunits and its ability to recruit the p300 transcriptional co-activator by oxygen-dependent, enzyme-mediated hydroxylation of key residues that in turn requires iron as a cofactor and 2-oxoglutarate as a co-substrate (29, 30). Inhibitory binding of HIF to the hepcidin promoter has been proposed as a mechanism of hypoxic down-regulation of the gene in mice (25). However, down-regulation of gene expression by direct HIF binding is uncommon (28, 31), and the HIF-binding site identified in mice is not conserved in humans. Furthermore, other laboratories have not observed a direct effect of HIF on the human hepcidin promoter (26).
Indirect transcriptional regulation of hepcidin by HIF may be mediated in part by furin, which is itself transcriptionally regulated in response to iron oxygen and HIF (11, 32). We explored the possibility that hypoxia signaling through the HIF transcriptional cascade might impinge on the expression of additional regulatory proteins upstream of hepcidin.
In this study, we interrogate the role of the HJV-processing enzyme, TMPRSS6, in hepcidin regulation by hypoxia. We show that TMPRSS6 is a HIF-regulated gene that responds to hypoxia and other activators of the HIF cascade. This activation is sufficient to alter levels of membrane HJV and to regulate activity of the hepcidin promoter in response to SMAD signaling. These findings provide a new mechanism through which hypoxia impinges upon hepcidin regulation, and highlight a novel link between oxygen sensing and iron homeostasis.
EXPERIMENTAL PROCEDURES
Cells and Cell Culture
HJV-Hep3B cells were made by infecting Hep3B cells with an IRES pQCXIX Retroviral Vector encoding human HJV cDNA and GFP 3′ to the CMV promoter. The hepatocellular carcinoma cell line Hep3B was cultured at 37 °C in Dulbecco modified Eagle's medium (α-modification) supplemented with 10% fetal calf serum, 2.5 mm l-glutamine, 50 units/ml penicillin, and 50 μg/ml streptomycin (Sigma-Aldrich). Cells were seeded at 105 cells/ml for all experiments.
Hypoxia Mimetics
The cell-permeable 2-oxoglutarate analog dimethyloxalyl glycine (DMOG) was purchased from Frontier Scientific (Logan, UT) and dissolved in water to a stock concentration of 0.5 m. The cell-permeable iron chelator 2,2′-bipyridyl (BIP) was purchased from Sigma Aldrich and dissolved in DMSO to a stock concentration of 25 mm. Cells were seeded in normoxia for 24 h before treatment.
RNA Extraction and cDNA Synthesis
Adherent cells were washed twice with phosphate-buffered saline (PBS), lysed in TRIzol (Sigma-Aldrich), and mRNA was then extracted by phase separation. Equal amounts of the mRNA template were used for cDNA synthesis by Moloney murine leukemia virus reverse transcriptase (Invitrogen, Carlsbad, CA).
Quantitative Real-time PCR
Gene expression was measured by real-time quantitative PCR using TaqMan® Gene Expression Assays on a StepOne thermocycler (both Applied Biosystems). Expression was normalized to the constitutively expressed gene cyclophilin and calculated using the ΔCT method.
Antibodies for Immunoblotting
Rabbit polyclonal anti-TMPRSS6 (Abcam, ab56180, at 1:500), rabbit polyclonal anti-CA9 antibody (Abcam, ab15086, at 1:1000), mouse anti-HIF-1α (BD Transduction Laboratories, clone 54, at 1:1000), mouse anti-HIF-2α(33) at 1:4, and mouse polyclonal anti-HJV antibody (Abnova H00148738, at 1:1000) were used, with the appropriate secondary antibodies, for the detection of TMPRSS6, CA9, HIF-1α, HIF-2α, and HJV proteins, respectively.
RNA Interference
Hep3B cells were transfected in serum-free medium with small interfering RNA (siRNA) oligonucleotides (Dharmacon RNA Technologies, Lafayette, CO) at a final concentration of 40 nm, using Oligofectamine transfection reagents (Invitrogen). Cells were transfected on days 1 and 2 and harvested on day 3. Drosophila HIF siRNA was used as an irrelevant control siRNA in all transfections.
Luciferase Vectors
Human hepcidin promoter constructs wtBRE and Hepc.prom-2.4kb were generated by inserting the 140-bp and 2.4-kb regions upstream of the Hamp transcriptional start site, respectively, into a pGL3 luciferase reporter vector, containing a constitutive SV40 enhancer and no promoter (Promega). The BRE and STAT3 sites (shown in italics) in the 140-bp segment were mutated by site-directed mutagenesis using the following pairs of primers (mutations are underlined): wtBRE construct: Forward-GCCTTTTCGGCGCCACCACCTTCTTGGCCGTGAGAC, Reverse-GTCTCACGGCCAAGAAGGTGGTGGCGCCGAAAAGGC, mutBRE construct: Forward-GCCTTTTCGGTGCCACCACCTTCTTGGCCGTGAGAC, Reverse-GTCT-CACGGCCAAGAAGGTGGTGGCACCGAAAAGGC.
Combined Luciferase and siRNA Transfection
1 × 105 cells were seeded in 24-well plates in serum-free medium 24 h before transfection. Each well was transfected with 50 ng of the relevant luciferase reporter vector, 50 ng of the control β-galactosidase reporter vector and 20 pmol of the relevant siRNA oligonucleotide (40 nm final concentration), using DharmaFECT Duo transfection reagent (Dharmacon). Cells were then cultured for a further 48 h in normoxia or 0.5% hypoxia. Relative luciferase activity was measured using the Promega Luciferase kit according to the manufacturer's instructions. To correct for transfection efficiency, all luciferase readouts were normalized to the β-galactosidase signal for the corresponding well, assayed by ortho-nitrophenyl-β-galactoside substrate assay.
Flow Cytometry
Hep3B cells were detached using 10 mm EDTA in PBS, then stained for 45 min with 5 μg/ml mouse monoclonal antibody to human HJV (Abcam ab54431). After washing to remove unbound primary antibody with PBS, cells were then stained for a further 30 min with phycoerythrin-conjugated goat anti-mouse IgG2b antibody (Invitrogen). Cells were then analyzed by flow cytometry using BD FACSCanto II instrument and FACSDiva software.
Isolation of Membrane Fractions for HJV Immunoblot
5 × 105 HJV-Hep3B cells were seeded in 6 cm dishes. Cells were detached using 10 mm EDTA in PBS, washed twice in ice-cold PBS, and membrane fractions were isolated using Subcellular Protein Fractionation kit (ThermoScientific). Equal amounts of protein from each fraction were loaded for immunoblotting.
Statistical Analysis
Data are mean plus or minus S.E. of three independent biological replicates. Analysis of mRNA data were performed on the normalized threshold cycles (ΔCT) before antilog transformation. One-way ANOVA with Dunnett's post-test and two-way ANOVA with Bonferroni's post-test corrections were performed using GraphPad Prism version 4.00 for Windows, GraphPad Software, San Diego. A value of p less than 0.01 was deemed significant.
RESULTS
Regulation of TMPRSS6 Expression by Oxygen Levels, Iron Availability, and by a 2-Oxoglutarate Analog
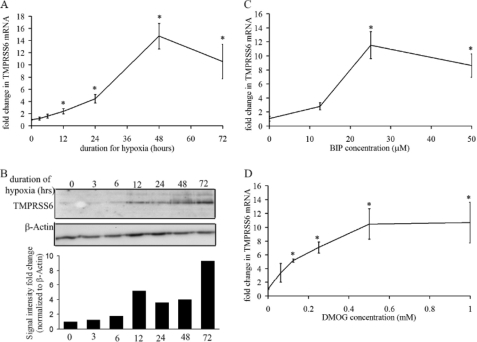
To determine the role of TMPRSS6 in the hepcidin response to hypoxia, we first examined whether TMPRSS6 itself was regulated by hypoxia and its mimetics. Hep3B cells were cultured under normoxia (21% O2) or hypoxia (0.5% O2), harvested at the designated time points and assayed for mRNA by quantitative real-time PCR and for protein by immunoblot. Real-time PCR showed a significant increase in TMPRSS6 mRNA under hypoxia (Fig. 1A). Similarly, immunoblot analysis of Hep3B lysates revealed increased TMPRSS6 protein levels from 12 h of hypoxia, with continued increase at 24, 48, and 72 h of hypoxia (Fig. 1B).
FIGURE 1.
TMPRSS6 is up-regulated by hypoxia, BIP, and DMOG. Hep3B cells were cultured under normoxia (21% O2) or hypoxia (0.5% O2) and harvested at the designated time points for mRNA and protein analyses. A, quantitative real-time PCR showing TMPRSS6 mRNA expression (*, p < 0.0001 by one-way ANOVA, with p < 0.01, at 12 h and beyond using Dunnett's post-test). B, immunoblot analysis of TMPRSS6 protein levels, following indicated duration of hypoxic incubation. TMPRSS6 signal intensity was quantitated by densitometry, normalized to β-actin signal intensity and plotted for the indicated hypoxic time points. C and D, quantitative real-time PCR showing TMPRSS6 mRNA expression after 16 h treatment with increasing concentrations of BIP and DMOG respectively (*, p < 0.0001 by one-way ANOVA with p < 0.01 at all doses using Dunnett's post-test). mRNA expression levels are normalized to cyclophilin. All data points are averages of three independent biological replicates.
Having observed induction of both TMPRSS6 mRNA and protein by hypoxia, we next determined whether other activators of HIF, the major transcriptional pathway regulating gene expression in response to hypoxia, could mimic this hypoxic response. The iron chelator bipyridyl (BIP) activates HIF by inhibiting the activity of HIF hydroxylases. We observed a dose-dependent increase in TMPRSS6 mRNA in Hep3B cells after exposure to BIP (Fig. 1C) indicating that TMPRSS6 levels are iron- as well as oxygen-sensitive. Similarly, the 2-oxoglutarate analog, dimethyloxalyl glycine (DMOG), also leads to induction of the HIF pathway through inhibition of HIF hydroxylase activity. An analogous dose-dependent increase in TMPRSS6 mRNA was observed in Hep3B cells after exposure to DMOG. Similar effects were observed in HepG2 cells (data not shown).
TMPRSS6 Up-regulation by Hypoxia, BIP, and DMOG Is Dependent on HIF-1α and HIF-2α
Taken together, these results indicate that TMPRSS6 levels are regulated in an oxygen, iron, and 2-oxoglutarate-dependent manner, suggesting that they might be regulated, at least in part, by the HIF transcriptional pathway. Therefore, we next examined the role of HIF-1α and HIF-2α in the regulation of TMPRSS6 mRNA by hypoxia, iron chelation, and DMOG.
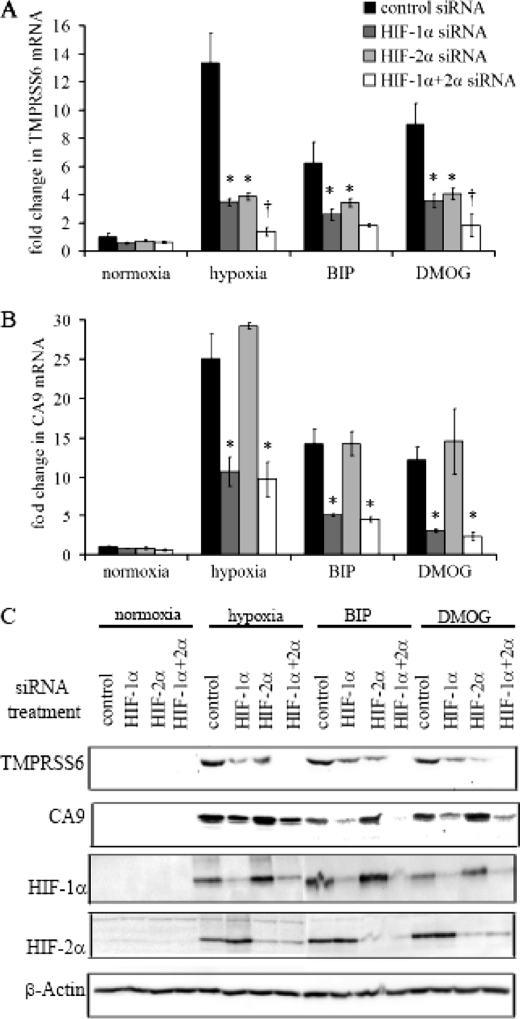
Hep3B cells were transfected with control siRNA, HIF-1α siRNA, HIF-2α siRNA, or combined HIF-1α & -2α siRNA. Cells were then incubated in normoxia or hypoxia (0.5% O2 for 48 h), or in the presence of 25 μm BIP or 0.5 mm DMOG for 16 h before harvesting. Induction of HIF-1α and HIF-2α by hypoxia, BIP, and DMOG, and knockdown by the respective siRNAs were confirmed by immunoblots (Fig. 2C).
FIGURE 2.
Regulation of TMPRSS6 by hypoxia, BIP, and DMOG is dependent on HIF-1α and HIF-2α. Hep3B cells were transfected with control siRNA, HIF-1α siRNA, HIF-2α siRNA or combined HIF-1α & HIF-2α siRNA followed by incubation either in hypoxia (0.5% O2 for 48 h), or in the presence of 25 μm BIP or 0.5 mm DMOG for 16 h. A and B, quantitative real-time PCR showing TMPRSS6 and CA9 mRNA expression respectively (*, p < 0.0001 by two-way ANOVA with p < 0.01 for each siRNA using Bonferroni's post-test analysis, †, p < 0.01 relative to single siRNA treatments). mRNA expression levels are normalized to cyclophilin, and data points are averages of three independent biological replicates. C, immunoblot analysis of TMPRSS6, CA9, HIF-1α, and HIF-2α levels in Hep3B cells. β-Actin levels were measured to control for loading differences.
Quantitative real-time PCR analysis demonstrated attenuation of the TMPRSS6 response to all three stimuli by both HIF-1α siRNA and HIF-2α siRNA. Furthermore, combined HIF-1α and -2α siRNA abrogated these responses still further (Fig. 2A). By contrast, up-regulation of the HIF-target gene, carbonic anhydrase (CA9), by all three stimuli was reversed by HIF-1α siRNA, but not by HIF-2α siRNA, with combined siRNA showing effects comparable to HIF-1α siRNA alone (Fig. 2B). This is consistent with previous observations that CA9 responds to the HIF-1 isoform alone (33), and suggests that TMPRSS6 is a transcriptional target of both HIF isoforms in Hep3B cells. TMPRSS6 and CA9 protein levels were also measured by immunoblot and reflected changes in transcript level (Fig. 2C).
To examine whether TMPRSS6 is a direct transcriptional target of HIF, we performed chromatin immunoprecipitation studies in Hep3B cells, using HIF-1α and HIF-2α antibodies, and tested for enrichment at all HIF-binding motifs within 3 kb of the TMPRSS6 promoter. We found no evidence for HIF binding within this region (data not shown). However, because transcription factor binding may be seen 100 kb or more from a regulated gene, we cannot conclude definitively whether TMPRSS6 is a direct or an indirect target of the HIF transcriptional pathway.
Hypoxia Reduces Membrane HJV (mHJV) Levels through HIF and TMPRSS6
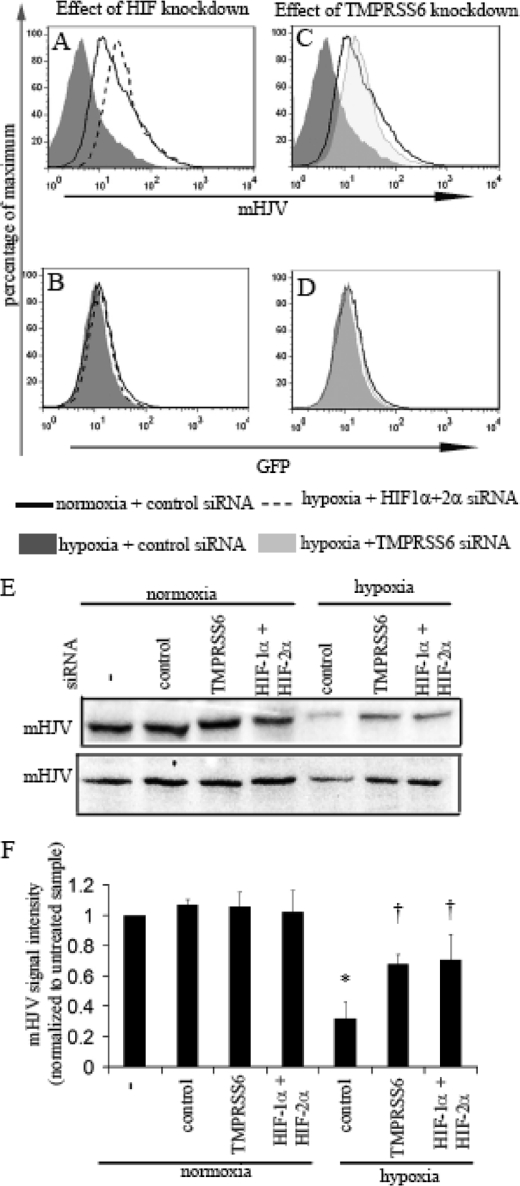
Given the pivotal role of TMPRSS6 in the processing of membrane HJV, and its consequent effects on the regulation of BMP signaling to the hepcidin promoter, we next examined whether the increased TMPRSS6 expression observed during hypoxia was capable of reducing membrane HJV levels. Although HJV mRNA is detectable in WT-Hep3B cells, cell surface protein expression is too low to quantify reliably by flow cytometry (data not shown). Instead, we analyzed HJV-Hep3B cells, in which a lentiviral vector containing a CMV promoter drives the bi-cistronic expression of HJV and GFP cDNAs, separated by an internal ribosomal entry site (IRES).
Using flow cytometry and an antibody to the extracellular domain of HJV, we first measured the effects of hypoxia on membrane HJV levels in HJV-Hep3B. Hypoxia (0.5% O2 for 48 h) resulted in a significant decrease in membrane HJV levels relative to cells cultured in normoxia (Fig. 3A). In contrast, GFP levels were not appreciably altered by hypoxia, indicating that activity of the bi-cistronic CMV promoter was not responsible for the decreased membrane HJV levels observed in hypoxia (Fig. 3B). Furthermore, in these overexpression studies we observed no appreciable effect of hypoxia on either the expression levels or on the half-life of total cellular HJV protein (supplemental Fig. S1).
FIGURE 3.
Hypoxia reduces membrane HJV levels through HIF and TMPRSS6. Histogram analysis of membrane HJV (mHJV) (A) and GFP (B) expression in HJV-Hep3B pre-transfected with control siRNA and cultured in normoxia or hypoxia for 48 h (p < 0.01 comparing mean fluorescence intensity for mHJV signal in hypoxia relative to normoxia). The effect of combined HIF-1α and 2α knockdown prior to hypoxia are also shown. Histogram analysis of mHJV (C) and GFP (D) expression in HJV-Hep3B pre-transfected with TMPRSS6 siRNA prior to hypoxia. Results are representative of two independent experiments in HJV-Hep3B cells. E, immunoblot analysis of HJV levels in membrane fractions (mHJV) of HJV-Hep3B cells from two independent experiments. F, mHJV levels were quantitated by densitometry from the two immunoblots, and then normalized to the signal intensity in untreated samples. (*, p < 0.001 relative to normoxic samples transfected with control siRNA, †, p < 0.001 relative to hypoxia samples transfected with control siRNA, both p values calculated using Bonferroni's post-test analysis of one-way ANOVA).
When cells were pretransfected with combined HIF-1α and -2α or TMPRSS6 siRNA prior to hypoxia, the decrease in membrane HJV was abolished (Fig. 3, A and C), without altering GFP protein levels (Fig. 3, B and D). HIF and TMPRSS6 suppression by siRNA were confirmed by immunoblot and by quantitative real-time PCR respectively (data not shown). HJV mRNA measured by real time-PCR was not altered by any of the above treatments (data not shown).
To further confirm the effects of hypoxia, HIF siRNA and TMPRSS6 siRNA on membrane HJV levels, we also performed immunoblot analysis of membrane fractions derived from HJV-Hep3B cells. Membrane HJV levels were decreased by hypoxia, in two independent experiments, and this decrease in membrane HJV was inhibited when cells were transfected with combined HIF-1α and -2α or with TMPRSS6 siRNA prior to hypoxia (Fig. 3, E and F). Taken together, these results indicate that hypoxia reduces membrane HJV levels through a post-transcriptional mechanism that is dependent on HIF-1α, HIF-2α, and TMPRSS6.
Hypoxia Reduces BRE Activity in the Hepcidin Promoter through HIF and TMPRSS6
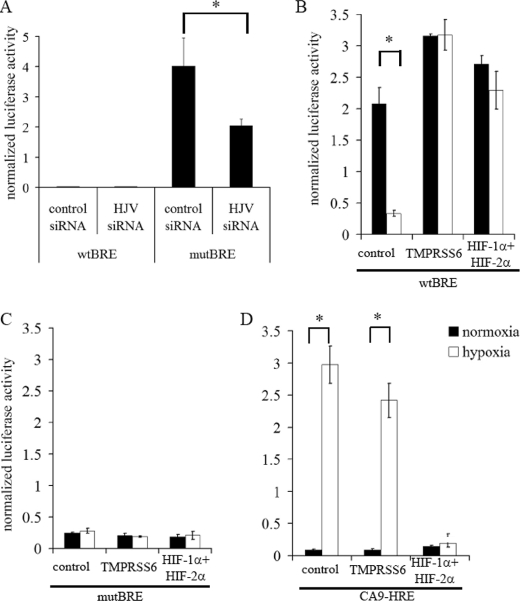
Because membrane HJV is a co-receptor for BMPs, which in turn lead to activation of intracellular SMAD pathways and of BMP response elements (BREs), we next wished to examine whether the hypoxic reduction in membrane HJV might suppress hepcidin production through BREs in its promoter. To test this directly, we used a luciferase reporter gene fused to a 140-bp fragment of DNA containing the proximal BRE from the human hepcidin promoter in HJV-Hep3B cells, and tested its responsiveness to hypoxia, HIF and TMPRSS6 knockdown. Importantly, this region does not contain any HIF-binding RCGTG motifs. However, the proximal BRE is in close proximity to a STAT3-binding site that mediates hepcidin induction by inflammatory cytokines such as IL-6. We therefore mutated this STAT3-binding site to exclude any confounding effects from these pathways. To further confirm that effects were due to the BRE, the wild-type BRE construct (wtBRE) was compared with a mutated BRE containing a known inactivating substitution in the core sequence (mutBRE) (34). A luciferase vector containing the previously established CA9-HRE was used as a control for hypoxia and HIF regulation.
Activity of the wtBRE promoter construct in HJV-Hep3B cells was significantly greater than the mutBRE construct and was suppressible by HJV siRNA, while HJV suppression had no significant effect on activity of the mutBRE reporter (Fig. 4A). Activity of wtBRE, but not mutBRE reporter constructs was significantly reduced by 48 h of incubation in 0.5% hypoxia (Fig. 4, B and C). Suppression of either TMPRSS6 or HIF-α by siRNA abolished the hypoxic regulation of the wtBRE reporter and restored activity to normoxic levels (Fig. 4B), concordant with the observed effects on membrane HJV levels. No effects were seen on the mutBRE reporter (Fig. 4C), while hypoxic regulation of the CA9-HRE reporter was dependent on the presence of HIF, but not TMPRSS6 (Fig. 4D).
FIGURE 4.
Hypoxia reduces BRE activity in the hepcidin promoter through HIF and TMPRSS6. A, HJV-Hep3B cells were co-transfected with one of the luciferase vectors (wtBRE, mutBRE) in combination with control siRNA or HJV siRNA. Levels of luciferase signal were then measured after 48 h (*, p < 0.001 using Bonferroni's post-test analysis of one-way ANOVA). HJV-Hep3B cells were transfected with one of the luciferase vectors wtBRE (B), mutBRE (C), or CA9-HRE (D) in combination with control siRNA, TMPRSS6 siRNA or combined HIF-1α and -2α siRNA. Levels of the luciferase signal were then measured after 48 h of normoxia or hypoxia (0.5% O2) (*, p < 0.001 using Bonferroni's post-test analysis of one-way ANOVA). Luciferase signal was normalized to β-galactosidase signal to correct for transfection efficiency.
Down-regulation of wtBRE activity by HJV knockdown and by hypoxia, and the inhibition of these hypoxic effects by HIF and TMPRSS6 siRNA were also observed in wt-Hep3B cells, without the HJV expressing lentivirus (supplemental Fig. S2), indicating that although we could not detect membrane HJV by flow cytometry in these cells that functional levels of endogenous HJV were present and behaved similarly to the overexpressed protein.
Taken together these results demonstrate that both TMPRSS6 and HIF-α are necessary for the hypoxic regulation of the wtBRE reporter and are consistent with the hypothesis that hypoxia leads to regulation of BRE activity through HIF-dependent activation of TMPRSS6 and consequent reduction in membrane HJV.
We saw no consistent effects of hypoxia on endogenous hepcidin production in WT or HJV-Hep3B cells (data not shown). However, it is known that in vivo effects of iron and oxygen on hepcidin expression are difficult to reproduce in cell culture models (26). Tests with a reporter construct encompassing 2.4-kb of the hepcidin promoter and containing two BRE sequences (supplemental Fig. S3) demonstrated that the effects of hypoxia, TMPRSS6, and HIF siRNA were preserved, suggesting that any putative cis-acting suppressors of hypoxic transcriptional activity at the hepcidin gene locus, either lie outside of this 2.4-kb region or involve processes, such as chromatin modification, that are not represented in transient transfection studies.
DISCUSSION
In this study, we demonstrate HIF-dependent hypoxic regulation of the membrane serine proteinase TMPRSS6. This enzyme cleaves membrane HJV from the cell surface (13) and the hypoxic induction of TMPRSS6 that we observe results in reduced levels of membrane HJV. Membrane HJV acts as a co-receptor for BMP signaling(8) and we demonstrate that hypoxic regulation of TMPRSS6 and membrane HJV leads to altered BMP signaling to a hepcidin-promoter derived BMP response element. Taken together, these findings reveal a novel HIF-dependent pathway leading to the hypoxic regulation of BMP/BRE signaling through the induction of TMPRSS6. Because HIF is also regulated through iron availability (29), this HIF-dependent regulation of BMP/BRE signaling through TMPRSS6 provides another potential mechanism through which iron bioavailability may contribute to the regulation of hepcidin.
Implications for Hepcidin and Iron Regulation
Several lines of evidence, including mouse knock-out models and human disease associated mutations, point to the importance of TMPRSS6 and membrane HJV levels in the transcriptional regulation of hepcidin in response to BMPs in vivo (11, 16–21). While the hypoxic induction of TMPRSS6 resulted in the transcriptional regulation of isolated hepcidin promoter sequences, no effect of hypoxia or TMPRSS6 was seen on endogenous hepcidin production itself. Other workers have also reported that in vivo regulation of hepcidin by hypoxia is not always reflected in tissue culture studies (26). This suggests that additional, as yet incompletely understood, levels of control operate to regulate the transcription of hepcidin in vivo and that not all of these controls are reflected in tissue culture models. Despite these uncertainties, our data indicate that the SMAD/BRE pathway is intact in our model and that signaling from hypoxia to the BRE in the hepcidin promoter, through an HIF- and TMPRSS6-dependent mechanism, is possible.
Cooperation with Furin-mediated Release of Soluble Hemojuvelin
In addition to cleavage by TMPRSS6, HJV is also cleaved by the trans-Golgi associated enzyme furin(11). However, unlike TMPRSS6, furin cleavage does not result in appreciable alteration in the level of membrane HJV(11) (supplemental Fig. S4, A and B). Instead, furin releases a 42-kDa soluble component that acts extracellularly as a “decoy receptor,” inhibiting BMP signaling (11). Transcriptional regulation of furin by the HIF pathway, leading to increased soluble HJV, has also been reported (11, 32). Thus our discovery of a second pathway linking HIF activation to reduced membrane HJV through TMPRSS6, reveals a synergistic mechanism by which hypoxia can alter BMP signaling through cooperative effects on levels of both the co-receptor and the decoy receptor (Fig. 5). However, the relative contribution of each limb of this mechanism to the overall regulation of BMP/BRE signaling is difficult to establish. In our model both TMPRSS6 and furin siRNA altered hypoxic signaling to the BRE (supplemental Fig. S4, C and D) confirming that both pathways can regulate BMP signaling. Importantly, in mouse models and in human disease, furin is not able to compensate for the effects of TMPRSS6 loss on hepcidin production, suggesting that in this aspect of BMP signaling, at least, TMPRSS6 is non-redundant.
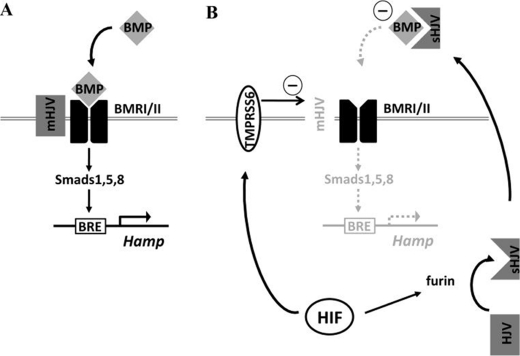
FIGURE 5.
Schematic overview. A, in normoxia BMP binds its receptor, activating intracellular SMAD phosphorylation pathways that lead to activation of the hepcidin promoter through its BRE. B, in hypoxia, HIF-dependent transcription of TMPRSS6 leads to cleavage and shedding of the BMPR co-receptor, HJV, from the cell membrane leading to reduced BMP/BRE signaling. In addition, cleavage of HJV in the Golgi apparatus by furin leads to increased secretion of soluble HJV, which acts as a “decoy receptor” that synergizes with reduced co-receptor levels to further reduce signaling to the BRE.
Wider Signaling Implications
As well as defining a novel link between hypoxic signaling and hepcidin, the regulation of TMPRSS6 and SMAD/BRE signaling by hypoxia and HIF has potential implications for other pathways. Although under normal conditions, TMPRSS6 expression is largely limited to hepatocytes (35), membrane HJV may act as a co-receptor for several BMPs, including those involved in differentiation, apoptosis, cell-cycling, and embryonic development (36, 37). Regulation of membrane HJV by the HIF/TMPRSS6 pathway may therefore influence these other functions. Furthermore, when compared with other serine proteases, TMPRSS6 exhibits a degree of promiscuity (38), raising the possibility that TMPRSS6 may also signal through protease action on substrates other than HJV, including fibronectin, fibrinogen, and type I collagen (35).
Because TMPRSS6 and other members of the type II transmembrane serine protease family, are known to regulate invasion and metastasis of prostate and breast tumors (39, 40), this raises the possibility that HIF-dependent activation of TMPRSS6 plays a role in tumor invasion and/or metastasis, contributing to the well recognized association between HIF expression and poor prognosis in cancer (41).
In summary, these data reveal a new hypoxia-responsive pathway leading to the modulation of BMP/BRE signaling through induction of TMPRSS6. This provides a novel molecular mechanism linking oxygen sensing by HIF to the regulation of iron homeostasis by hepcidin and opens the possibility of the HIF pathway affecting additional diverse processes via TMPRSS6.
Supplementary Material

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
- BMP
- bone morphogenetic protein
- TMPRSS6
- type II transmembrane serine proteinase 6
- HIF
- hypoxia-inducible factor
- DMOG
- dimethyloxalyl glycine
- BIP
- bipyridyl
- BRE
- BMP response element
- HRE
- hypoxia response element
- SMAD
- signal through son of mothers against decapentaplegic
- HJV
- hemojuvelin
- mHJV
- membrane hemojuvelin
- CA9
- carbonic anhydrase IX.
REFERENCES
- 1. Nemeth E., Tuttle M. S., Powelson J., Vaughn M. B., Donovan A., Ward D. M., Ganz T., Kaplan J. (2004) Science 306, 2090–2093 [DOI] [PubMed] [Google Scholar]
- 2. Yamaji S., Sharp P., Ramesh B., Srai S. K. (2004) Blood 104, 2178–2180 [DOI] [PubMed] [Google Scholar]
- 3. Nemeth E., Rivera S., Gabayan V., Keller C., Taudorf S., Pedersen B. K., Ganz T. (2004) J. Clin. Investig. 113, 1271–1276 [DOI] [PubMed] [Google Scholar]
- 4. Truksa J., Peng H., Lee P., Beutler E. (2006) Proc. Natl. Acad. Sci. U. S. A. 103, 10289–10293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Andriopoulos B., Jr., Corradini E., Xia Y., Faasse S. A., Chen S., Grgurevic L., Knutson M. D., Pietrangelo A., Vukicevic S., Lin H. Y., Babitt J. L. (2009) Nature Genet. 41, 482–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Giannetti A. M., Björkman P. J. (2004) J. Biol. Chem. 279, 25866–25875 [DOI] [PubMed] [Google Scholar]
- 7. Schmidt P. J., Toran P. T., Giannetti A. M., Bjorkman P. J., Andrews N. C. (2008) Cell Metabol. 7, 205–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Babitt J. L., Huang F. W., Wrighting D. M., Xia Y., Sidis Y., Samad T. A., Campagna J. A., Chung R. T., Schneyer A. L., Woolf C. J., Andrews N. C., Lin H. Y. (2006) Nat. Genet. 38, 531–539 [DOI] [PubMed] [Google Scholar]
- 9. Feder J. N., Gnirke A., Thomas W., Tsuchihashi Z., Ruddy D. A., Basava A., Dormishian F., Domingo R., Jr., Ellis M. C., Fullan A., Hinton L. M., Jones N. L., Kimmel B. E., Kronmal G. S., Lauer P., Lee V. K., Loeb D. B., Mapa F. A., McClelland E., Meyer N. C., Mintier G. A., Moeller N., Moore T., Morikang E., Prass C. E., Quintana L., Starnes S. M., Schatzman R. C., Brunke K. J., Drayna D. T., Risch N. J., Bacon B. R., Wolff R. K. (1996) Nat. Genet. 13, 399–408 [DOI] [PubMed] [Google Scholar]
- 10. Papanikolaou G., Samuels M. E., Ludwig E. H., MacDonald M. L., Franchini P. L., Dubé M. P., Andres L., MacFarlane J., Sakellaropoulos N., Politou M., Nemeth E., Thompson J., Risler J. K., Zaborowska C., Babakaiff R., Radomski C. C., Pape T. D., Davidas O., Christakis J., Brissot P., Lockitch G., Ganz T., Hayden M. R., Goldberg Y. P. (2004) Nat. Genet. 36, 77–82 [DOI] [PubMed] [Google Scholar]
- 11. Silvestri L., Pagani A., Camaschella C. (2008) Blood 111, 924–931 [DOI] [PubMed] [Google Scholar]
- 12. Lin L., Goldberg Y. P., Ganz T. (2005) Blood 106, 2884–2889 [DOI] [PubMed] [Google Scholar]
- 13. Silvestri L., Pagani A., Nai A., De Domenico I., Kaplan J., Camaschella C. (2008) Cell Metab. 8, 502–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Folgueras A. R., de Lara F. M., Pendás A. M., Garabaya C., Rodríguez F., Astudillo A., Bernal T., Cabanillas R., López-Otín C., Velasco G. (2008) Blood 112, 2539–2545 [DOI] [PubMed] [Google Scholar]
- 15. Finberg K. E., Whittlesey R. L., Fleming M. D., Andrews N. C. (2010) Blood 115, 3817–3826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Du X., She E., Gelbart T., Truksa J., Lee P., Xia Y., Khovananth K., Mudd S., Mann N., Moresco E. M., Beutler E., Beutler B. (2008) Science 320, 1088–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Silvestri L., Guillem F., Pagani A., Nai A., Oudin C., Silva M., Toutain F., Kannengiesser C., Beaumont C., Camaschella C., Grandchamp B. (2009) Blood 113, 5605–5608 [DOI] [PubMed] [Google Scholar]
- 18. Guillem F., Lawson S., Kannengiesser C., Westerman M., Beaumont C., Grandchamp B. (2008) Blood 112, 2089–2091 [DOI] [PubMed] [Google Scholar]
- 19. Melis M. A., Cau M., Congiu R., Sole G., Barella S., Cao A., Westerman M., Cazzola M., Galanello R. (2008) Haematologica 93, 1473–1479 [DOI] [PubMed] [Google Scholar]
- 20. Beutler E., Van Geet C., te Loo D. M., Gelbart T., Crain K., Truksa J., Lee P. L. (2010) Blood Cells, Mol. Dis. 44, 16–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Finberg K. E., Heeney M. M., Campagna D. R., Aydinok Y., Pearson H. A., Hartman K. R., Mayo M. M., Samuel S. M., Strouse J. J., Markianos K., Andrews N. C., Fleming M. D. (2008) Nat. Genet. 40, 569–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nicolas G., Chauvet C., Viatte L., Danan J. L., Bigard X., Devaux I., Beaumont C., Kahn A., Vaulont S. (2002) J. Clin. Investig. 110, 1037–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Leung P. S., Srai S. K., Mascarenhas M., Churchill L. J., Debnam E. S. (2005) Gut 54, 1391–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pinto J. P., Ribeiro S., Pontes H., Thowfeequ S., Tosh D., Carvalho F., Porto G. (2008) Blood 111, 5727–5733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Peyssonnaux C., Zinkernagel A. S., Schuepbach R. A., Rankin E., Vaulont S., Haase V. H., Nizet V., Johnson R. S. (2007) J. Clin. Investig. 117, 1926–1932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Volke M., Gale D. P., Maegdefrau U., Schley G., Klanke B., Bosserhoff A. K., Maxwell P. H., Eckardt K. U., Warnecke C. (2009) PLoS ONE 4, e7875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wenger R. H., Stiehl D. P., Camenisch G. (2005) Sci. STKE 2005, re12 [DOI] [PubMed] [Google Scholar]
- 28. Mole D. R., Blancher C., Copley R. R., Pollard P. J., Gleadle J. M., Ragoussis J., Ratcliffe P. J. (2009) J. Biol. Chem. 284, 16767–16775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schofield C. J., Ratcliffe P. J. (2004) Nat. Rev. Mol. Cell Biol. 5, 343–354 [DOI] [PubMed] [Google Scholar]
- 30. Kaelin W. G., Jr., Ratcliffe P. J. (2008) Mol. Cell 30, 393–402 [DOI] [PubMed] [Google Scholar]
- 31. Xia X., Lemieux M. E., Li W., Carroll J. S., Brown M., Liu X. S., Kung A. L. (2009) Proc. Natl. Acad. Sci. U. S. A. 106, 4260–4265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. McMahon S., Grondin F., McDonald P. P., Richard D. E., Dubois C. M. (2005) J. Biol. Chem. 280, 6561–6569 [DOI] [PubMed] [Google Scholar]
- 33. Wiesener M. S., Turley H., Allen W. E., Willam C., Eckardt K. U., Talks K. L., Wood S. M., Gatter K. C., Harris A. L., Pugh C. W., Ratcliffe P. J., Maxwell P. H. (1998) Blood 92, 2260–2268 [PubMed] [Google Scholar]
- 34. Island M. L., Jouanolle A. M., Mosser A., Deugnier Y., David V., Brissot P., Loréal O. (2009) Haematologica 94, 720–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Velasco G., Cal S., Quesada V., Sánchez L. M., López-Otín C. (2002) J. Biol. Chem. 277, 37637–37646 [DOI] [PubMed] [Google Scholar]
- 36. Nohe A., Keating E., Knaus P., Petersen N. O. (2004) Cell Signal. 16, 291–299 [DOI] [PubMed] [Google Scholar]
- 37. Xiao Y. T., Xiang L. X., Shao J. Z. (2007) Biochem. Biophys. Res. Commun. 362, 550–553 [DOI] [PubMed] [Google Scholar]
- 38. Béliveau F., Désilets A., Leduc R. (2009) FEBS J. 276, 2213–2226 [DOI] [PubMed] [Google Scholar]
- 39. Parr C., Sanders A. J., Davies G., Martin T., Lane J., Mason M. D., Mansel R. E., Jiang W. G. (2007) Clin. Cancer Res. 13, 3568–3576 [DOI] [PubMed] [Google Scholar]
- 40. Sanders A. J., Parr C., Martin T. A., Lane J., Mason M. D., Jiang W. G. (2008) J. Cell. Physiol. 216, 780–789 [DOI] [PubMed] [Google Scholar]
- 41. Semenza G. L. (2010) Oncogene 29, 629–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.