Abstract
FtsH-related AAA proteases are conserved membrane-anchored, ATP-dependent molecular machines, which mediate the processing and turnover of soluble and membrane-embedded proteins in eubacteria, mitochondria, and chloroplasts. Homo- and hetero-oligomeric proteolytic complexes exist, which are composed of homologous subunits harboring an ATPase domain of the AAA family and an H41 metallopeptidase domain. Mutations in subunits of mitochondrial m-AAA proteases have been associated with different neurodegenerative disorders in human, raising questions on the functional differences between homo- and hetero-oligomeric AAA proteases. Here, we have analyzed the hetero-oligomeric yeast m-AAA protease composed of homologous Yta10 and Yta12 subunits. We combined genetic and structural approaches to define the molecular determinants for oligomer assembly and to assess functional similarities between Yta10 and Yta12. We demonstrate that replacement of only two amino acid residues within the metallopeptidase domain of Yta12 allows its assembly into homo-oligomeric complexes. To provide a molecular explanation, we determined the 12 Å resolution structure of the intact yeast m-AAA protease with its transmembrane domains by electron cryomicroscopy (cryo-EM) and atomic structure fitting. The full-length m-AAA protease has a bipartite structure and is a hexamer in solution. We found that residues in Yta12, which facilitate homo-oligomerization when mutated, are located at the interface between neighboring protomers in the hexamer ring. Notably, the transmembrane and intermembrane space domains are separated from the main body, creating a passage on the matrix side, which is wide enough to accommodate unfolded but not folded polypeptides. These results suggest a mechanism regarding how proteins are recognized and degraded by m-AAA proteases.
Keywords: ATP-dependent Protease, Chaperone Chaperonin, Electron Microscopy (EM), Membrane Proteins, Protein Structure, Yeast Genetics
Introduction
Energy-dependent proteases form oligomeric ring complexes and harbor conserved ATPase domains of the AAA+ family (1). It is widely accepted that AAA+ machines utilize the energy derived from ATP hydrolysis to thread substrate proteins through a central pore resulting in substrate unfolding. FtsH-related AAA proteases form a distinct membrane-associated group of AAA+ machines, present in eubacteria and in mitochondria and chloroplasts of eukaryotic cells (2, 3). Members of this group feature an N-terminal membrane targeting signal, followed by one or two transmembrane helices, and a canonical AAA domain that is covalently linked to the metallopeptidase domain (3, 4).
X-ray crystal structures of the soluble cytosolic domains of the bacterial AAA protease FtsH (FtsHCyt) bound to ADP and in the absence of nucleotide (apo) have been reported (5–7). Although the AAA ring of the apo-FtsHCyt hexamer was 6-fold symmetric (7), the ADP-bound structures revealed a 2- (6) and 3-fold symmetrical hexamer (5). However, the protease ring was 6-fold symmetric in all three structures. Therefore, it remained unclear what symmetry the full-length protease adopts and which of the stereo-specific interactions between neighboring subunits are found in the intact enzyme. Moreover, the membrane anchoring of the AAA protease subunits raises the intriguing but yet unresolved question as to how substrates have access to the central pore of the AAA ring before translocation into the proteolytic chamber. Notably, it has been reported that deletion of the N-terminal region of FtsH and related AAA protease subunits impairs the degradation of membrane-embedded substrates (8, 9), supporting a key role for the N-terminal transmembrane and intermembrane space domains for protease function. Hence, to fully understand the structure-function relationship, the three-dimensional structure of the intact AAA protease is required.
Yeast possess two FtsH-related AAA proteases, an i-AAA protease and an m-AAA protease present in the mitochondrial inner membrane. i-AAA proteases expose their catalytic side toward the intermembrane space, whereas AAA proteases are active on the matrix side and are composed of homologous Yta10 and Yta12 subunits (10). Unlike the yeast hetero-oligomeric m-AAA protease, bacterial FtsH is a homo-oligomer, and both hetero- (consisting of Paraplegin and AFG3L2 subunits) and homo-oligomeric m-AAA protease isoforms (consisting of AFG3L2 subunits only) exist in human mitochondria and can substitute for the yeast m-AAA protease in vivo (11). Interestingly, mutations that affect only hetero-oligomeric or both hetero- and homo-oligomeric human m-AAA protease complexes are linked to two distinct neurological disorders. Although mutations in the Paraplegin subunit are responsible for hereditary spastic paraplegia (12), it was shown more recently that mutations in AFG3L2 cause a dominant form of hereditary spinocerebellar ataxia (SCA28) (13). These findings raise the question whether homo- and hetero-oligomeric complexes are functionally and mechanistically equivalent. Moreover, the structural determinants for homo- and hetero-oligomerization remained unclear.
Here, we used genetic approaches, together with cryo-EM and atomic structure fitting, to provide molecular insights into the structure of a detergent-solubilized, full-length yeast m-AAA protease variant in the ATP-bound state. Our experiments reveal for the first time the structural organization of the transmembrane and intermembrane space domains and highlight functional similarities between homologous Yta10 and Yta12 subunits. These findings suggest a common mechanism by which homo- and hetero-oligomeric m-AAA proteases recognize and degrade their substrates.
EXPERIMENTAL PROCEDURES
Generation of Yta10 and Yta12 Chimeras
Yta10 and Yta12 variants with the AAA domain of the respective assembly partner have been described previously (14). To replace the N-terminal domain or the C-terminal helical (CH)4 region of Yta12 by those of Yta10, DNA fragments encoding the respective segment of Yta10 were amplified by PCR. Δyta10Δyta12 cells (YKO200) were transformed with the PCR product and a DNA fragment obtained by restriction digest of YCplac111ADH1-YTA12 within the coding sequence of the domain to be replaced. To replace the N-terminal domain or CH of Yta10 by those of Yta12, PCR fragments encoding the respective segment were transformed into Δyta10Δyta12 cells together with the linearized plasmid encoding His-tagged Yta10 (YCplac22ADH1-YTA10−His6). To obtain the Yta12 chimera containing the protease domain (PD) of Yta10, we first generated a Yta12 variant containing the PD and CH of Yta10. The resulting construct was linearized and transformed into Δyta10Δyta12 cells together with a PCR fragment encoding the CH of Yta12. A similar approach was used to generate the Yta10 chimera containing the PD of Yta12. To do this, we first generated a Yta10 chimera containing the PD and CH of Yta12. The obtained plasmid was digested and transformed into Δyta10Δyta12 cells together with a PCR fragment encoding the CH of Yta10. Transformants were selected on SC-LEU (YCplac111 constructs) or SC-TRP (YCplac22 constructs) plates containing glucose.
Genetic Screening Procedures
To isolate Yta12 mutant variants capable of homo-oligomerization, a 0.8-kb DNA fragment encoding the PD and flanking regions of Yta12 (residues 502–765) was amplified by error-prone PCR in the presence of 0.1 mm Mn2+. Δyta10Δyta12 cells (YKO200) were transformed with the PCR fragment and an 8.6-kb DNA fragment obtained by restriction digest of YCplac111ADH1-YTA12 by XhoI (which had been introduced by site-directed mutagenesis at 1,557 bp of the ORF) and BsmI. Transformants were selected on SC-LEU plates containing glucose and were replica-plated on SC-LEU plates containing glycerol as the sole carbon source for selection. Plasmids were recovered from growing colonies and mutations in Yta12 were identified by DNA sequencing. Single or multiple mutations were introduced into YCplac111ADH1-YTA12 by site-directed mutagenesis and tested for their suppressive activity.
Purification of the m-AAA Protease
Mitochondria were resuspended in 50 mm Tris-HCl, pH 7.2, 150 mm NaCl, 1 mm phenylmethylsulfonyl fluoride, EDTA-free protease inhibitor mixture (Roche Applied Science), 10% glycerol, 50 mm imidazole, 3% n-octyl-β-d-glucopyranoside (β-OG) at a protein concentration of 5 mg/ml. Following incubation for 20 min at 4 °C, insoluble material was removed by centrifugation, and the supernatant was applied to a 1-ml Ni2+-nitrilotriacetic acid agarose-affinity column (GE Healthcare). The column was washed with 50 mm Tris-HCl, pH 7.2, 150 mm NaCl, 1 mm phenylmethylsulfonyl fluoride, 10% glycerol, and 1% β-OG containing an increasingly higher concentration of imidazole (130, 150, and 180 mm). Bound complexes were eluted in 50 mm Tris-HCl, pH 7.2, 150 mm NaCl, 10% glycerol, 500 mm imidazole, and 1% β-OG. The eluate was concentrated and further purified by size exclusion chromatography on a Superose 6 column (GE Healthcare) that was pre-equilibrated in 50 mm Tris-HCl, pH 7.2, 150 mm NaCl, 10% glycerol, and 1% β-OG. After purification, Yta10 and Yta12 were present at roughly equimolar concentrations within assembled complexes, independent of which subunit had been overexpressed (14).
Cryo-EM Specimen Preparation
Purified, detergent-solubilized m-AAA protease was diluted to a final concentration of 0.3 mg/ml in 50 mm Tris-HCl, pH 7.2, 150 mm NaCl, 0.5% glycerol, 1% β-OG, 10 mm MgCl2, and 5 mm ATP. The hetero-oligomeric assembly was stabilized with 0.05% glutaraldehyde, followed by incubation for 60 min on ice. A 2-μl sample was applied to commercial holey-film copper grids that were coated with a continuous carbon film prior to use. Excess solution was blotted, and the grid was flash-frozen in liquid ethane using the Vitrobot (FEI).
Cryo-EM Data Collection
Grids were imaged in low dose mode (∼17 e−/Å2) at −180 °C using a JEOL JEM-2010F transmission electron microscope operated at 200 keV. Images were taken at an effective magnification of 82,800 with defocus ranging from 1.8 to 6.5 μm and recorded using a 4 × 4 k CCD camera. This resulted in a sampling rate of 1.81 Å per pixel at the specimen level.
Image Processing and Three-dimensional Reconstruction
All image processing and refinements were performed using the EMAN software suite (15). Particles were selected semi-automatically using the boxer routine. The contrast transfer function of the microscope was determined using ctfit. Reference-free two-dimensional class averages were prepared using refine2d.py. Top-down views from these class averages indicate a 6-fold symmetrical hexamer (supplemental Fig. S1). Therefore, C6 symmetry was applied throughout the reconstruction and refinement. An initial model was generated from the reference-free class averages using the startcsym routine and was refined using projection-based particle classification. Each refinement cycle includes particle alignment, classification, averaging, and reconstruction of a new model and was carried out iteratively until the model converged. This procedure was repeated for several rounds with a decreasing angular step size. The final reconstruction included 32,311 of 37,882 particles. The resolution of the final reconstruction was 12 Å based on the 0.5 criterion of the Fourier shell correlation (supplemental Fig. S2).
Atomic Structure Fit
To generate an atomic model for the m-AAA protease, we manually fitted the x-ray structure of the Thermatoga maritima apo-FtsHCyt hexamer (PDB code 3KDS (7)) into our cryo-EM density. Next, to improve the envelope fit, we replaced the AAA domain of apo-FtsHCyt (7) with that of the nucleotide-bound structure (6) by superimposing the small helical domain from each structure. Because the pore loop-2 (residues 393–404 in Yta10 and 453–463 in Yta12) is disordered in the T. maritima FtsHCyt crystal structure, the loop was modeled according to the conformation seen in the x-ray structure of Thermus thermophilus FtsHCyt (PDB code 2DHR (5)), followed by manually adjusting the position of the AAA domains. The fitted hexamer was then energy-minimized using CNS (16) to release minor side chain clashes. Finally, the program foldhunter (17) was used to further improve the rigid body fit of the hexamer. The final correlation coefficient between the cryo-EM density of the main body and the fitted hexamer model was 0.86.
RESULTS
Determinants for Hetero-oligomeric Assembly of m-AAA Protease Subunits
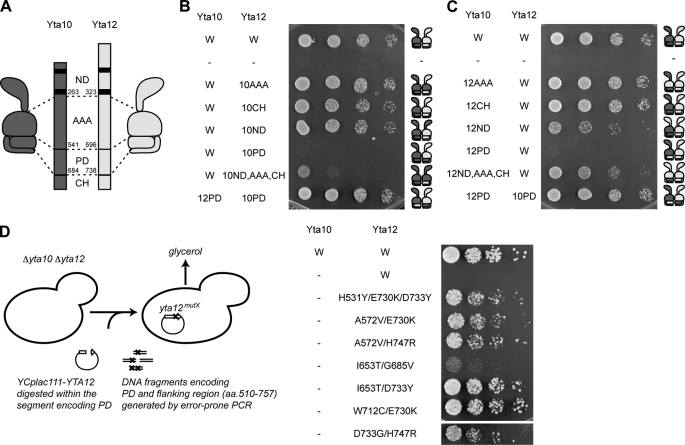
To assess the functional similarity between subunits of the hetero-oligomeric yeast m-AAA protease, we screened for amino acid segments that drive hetero-oligomerization. In the first step, we constructed a series of chimeric m-AAA protease subunits, carrying domain(s) from the respective assembly partner (Fig. 1A). These chimeras were co-expressed with their wild-type counterpart in Δyta10Δyta12 cells and analyzed for their ability to suppress the respiratory deficiency of cells lacking m-AAA proteases (Fig. 1, B and C). Respiratory growth indicates assembly of the chimera into functional hetero-oligomeric complexes because expression of the chimera in the absence of the wild-type assembly partner did not suppress the respiratory deficiency of Δyta10Δyta12 cells (data not shown). Exchange of AAA domains between Yta10 and Yta12 did not interfere with respiratory growth (Fig. 1, B and C), demonstrating that yeast m-AAA proteases with only one type of AAA domain are functionally active. Similarly, m-AAA protease complexes harboring the N-terminal domain or CH from only one type of subunit restored respiratory growth in Δyta10Δyta12 cells (Fig. 1, B and C). In contrast, replacement of the homologous PDs with that of the other subunit did not allow respiratory growth (Fig. 1, B and C), indicating that PDs from both subunits are required for functional assembly.
FIGURE 1.
Protease domains confer hetero-oligomeric assembly of the m-AAA protease. A, domain structure of the yeast m-AAA protease subunits Yta10 (dark gray) and Yta12 (light gray). ND, N-terminal domain; AAA, AAA domain. B, respiratory growth of Δyta10Δyta12 cells upon co-expression of Yta10 and chimeras of Yta12. C, respiratory growth of Δyta10Δyta12 cells upon co-expression of Yta12 and chimeras of Yta10. D, forward genetic screen for Yta12 variants allowing respiratory growth in the absence of Yta10. Screening procedures (left panel) and respiratory growth of Δyta10Δyta12 cells upon expression of the identified Yta12 variants (right panel) are shown.
Yta12 Variants Capable of Homo-oligomerization
Because the PD is a highly conserved domain in Yta10, Yta12, and other FtsH-related AAA protease subunits (supplemental Fig. S3), we reasoned that only minor differences in amino acid sequence between Yta10 and Yta12 subunits may facilitate hetero-oligomer assembly. To identify residues involved in oligomerization, we generated Yta12 PD mutants by PCR-based mutagenesis and screened for Yta12 variants that were able to form active homo-oligomeric complexes (Fig. 1D). As anticipated, expression of Yta12 wild-type did not restore the respiratory growth of Δyta10Δyta12 cells. Using genetic screening, we identified seven independent Yta12 variants harboring a mutant PD, which allowed respiratory growth when expressed in Δyta10Δyta12 cells (Fig. 1D). As all variants carried multiple mutations, in most cases two mutations, we generated Yta12 variants harboring only individual mutations by site-directed mutagenesis. However, none of these variants allowed respiratory growth of Δyta10Δyta12 cells, suggesting that synergistic effects of at least two mutations are required to promote homo-oligomerization of Yta12 (supplemental Fig. S4). These experiments revealed that only minor differences between the amino acid sequence of Yta10 and Yta12 drive hetero-oligomerization of the m-AAA protease.
Cryo-EM Structure of the Intact m-AAA Protease
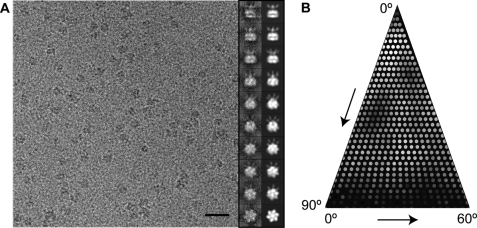
To provide a spatial framework for our identified Yta12 mutations in context of the full-length protease, we determined the cryo-EM structure of the intact yeast m-AAA protease. To do so, we expressed and purified an ATP hydrolysis-deficient m-AAA protease variant (Trap) carrying a mutation of the conserved glutamate residue within the Walker B motif in both subunits (Yta10E388Q and His6-Yta12E448Q) (14). This Walker B mutant effectively traps ATP and keeps m-AAA protease subunits in one nucleotide conformation. Examination of a frozen hydrated specimen by cryo-EM showed that the Trap-ATP hexamer particles were well distributed and displayed a wide range of orientations in vitreous ice (Fig. 2). Top-down views from reference-free two-dimensional class averages (Fig. 3A, top, and supplemental Fig. S1) indicate that the intact m-AAA protease is 6-fold symmetric (Fig. 3). This is somewhat surprising considering the fact that the yeast m-AAA protease is a hetero-oligomer and may be attributed to the resolution limit of our data. It is noteworthy that we examined the structure of a detergent-solubilized m-AAA protease, which might differ from the native structure. Our attempts to perform an asymmetric reconstruction or obtaining a cryo-EM reconstruction using only 3-fold symmetry averaging were unsuccessful, despite the large number of particles available (data not shown).
FIGURE 2.
Cryo-EM analysis of an intact yeast m-AAA protease. A, representative area of a digital micrograph of the Trap-ATP hexamer embedded in vitreous ice. Selected projection views (right column) and their corresponding class averages (left column) are also shown. Scale bar, 300 Å. B, distribution of particle orientations over an asymmetric unit used in the three-dimensional reconstruction. Brighter dots indicate a larger number of particles.
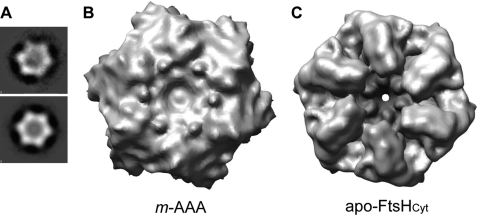
FIGURE 3.
Comparison of the hexameric structures of the m-AAA protease and apo-FtsHCyt. A, top-down view of a reference-free two-dimensional class average of the m-AAA protease without symmetry (top) and with C6 symmetry applied (bottom). B, 12 Å resolution cryo-EM reconstruction of the m-AAA protease in the ATP-bound state depicted as isosurface representation. For clarity, only the matrix domain is shown. C, mass density of the apo-FtsHCyt hexamer calculated at 12 Å resolution from the atomic coordinates of the x-ray structure (PDB code 3KDS (7)).
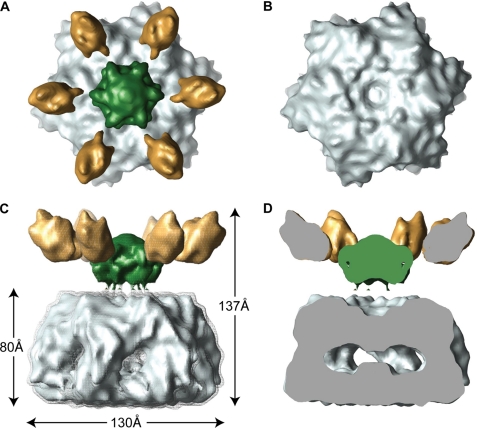
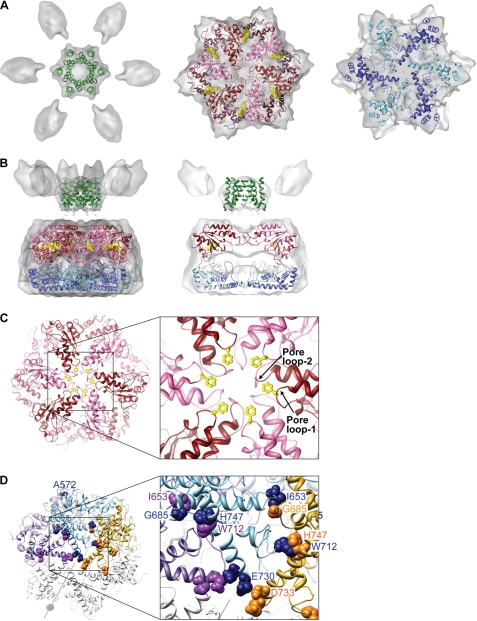
Our 12 Å resolution cryo-EM reconstruction shows that the yeast m-AAA protease has a bipartite structure with a height of 137 Å and a diameter of 130 Å in its widest dimension (Fig. 4). The lower main body is similar in overall dimensions to the apo-FtsHCyt hexamer (Fig. 4, C and D) (7). However, in our reconstruction, the protease ring is somewhat wider, presumably because of the additional 23 and 33 amino acid residues present at the C termini of Yta10 and Yta12 subunits. Moreover, there are six ∼25 Å wide lateral openings in the main body, which may function as exit holes allowing cleaved peptides to leave the m-AAA protease. The upper, funnel-shaped density sits on top of the main hexamer body and consists of a larger central mass and six smaller mass densities that splay out from the 6-fold symmetry axis (Fig. 4, A, C, and D, green and gold mass densities). The additional mass was not present in the x-ray structures of FtsHCyt and therefore can be attributed to the 12 transmembrane spanning helices and six intermembrane space domains of the intact m-AAA protease. Notably, the central mass is separated from the main body by about 13 Å but is connected when contoured at a lower threshold (Fig. 4C).
FIGURE 4.
Cryo-EM structure of an intact yeast m-AAA protease. The structure of the Trap-ATP hexamer is shown as isosurface representation with the transmembrane domain colored green, the intermembrane space domain gold, and the matrix domain gray. Because the transmembrane and intermembrane space domains appear to be more flexible than the main body, we split the cryo-EM map into two parts and applied different contour levels. The contour level of the solid isosurface representation was chosen to cover 91 kDa for the transmembrane segments and intermembrane space domains and 436 kDa for the matrix domains. A, top-down view; B, top-down view of the main body only. C, side view; D, vertical central section. The meshed density is contoured at a lower threshold to show the connectivity between the transmembrane segments and the AAA ring.
Atomic Structure Fit of an Intact m-AAA Protease
To generate an atomic model for the m-AAA protease, we fitted the x-ray structure of apo-FtsHCyt from T. maritima (PDB 3KDS (7)) into our cryo-EM reconstruction (Fig. 5, A and B). The central mass and six smaller mass densities observed in the upper part of the bipartite structure are in good agreement with the calculated mass for the 12 transmembrane-spanning helices (30 kDa) and six intermembrane space domains (6 × 10 kDa). We therefore modeled the arrangement of transmembrane helices according to a pair of transmembrane-spanning helices (residues 114–136 and 150–172) of similar length and topology present in the x-ray structure of photoactivated rhodopsin (PDB 2I37 (18)). No structural information is available for the intermembrane space domains, which were not fitted.
FIGURE 5.
Atomic structure fit. The cryo-EM reconstruction of the Trap-ATP hexamer is shown as a semitransparent surface with the fitted structure of the hetero-oligomeric m-AAA protease docked in. The protease is depicted as ribbon diagram with the transmembrane domain colored green, the AAA red/pink, and the PD blue/sky blue. The dark and light hues represent the Yta10 (red/blue) and Yta12 subunits (pink/sky blue) of the hetero-oligomer. A, top-down views. B, side view (left) and vertical central section (right) of the fitted transmembrane domain, AAA ring, and protease ring. Bound ATP molecules are depicted as CPK models. C, top-down view of the fitted AAA ring. The inset shows an enlarged view of the pore loops and surrounding structural elements. The critical Phe-361 (Yta10) and Phe-421 (Yta12) are depicted as ball-and-stick models. D, model of an Yta12 homo-hexamer. For clarity, three neighboring subunits are colored purple, sky blue, and yellow. Yta12 mutations that allow respiratory growth in the absence of Yta10 are shown as CPK models.
Although the x-ray structure of the apo-FtsHCyt hexamer was almost compatible with our envelope fit, the atomic coordinates of the AAA domains required some adjustment. Our cryo-EM map supports the notion that the relative orientation of the large α/β and the smaller helical domain is different in the ATP-bound state, and it involves a hinge motion that alters the angle between the two domains. We therefore replaced the AAA domain of apo-FtsHCyt (7) with that of the nucleotide-bound structure (6), which resulted in a better envelope fit.
Atomic Model for the Hetero-oligomeric m-AAA Protease Hexamer
To examine the inter-subunit interface, we generated homology models for the structurally related Yta10 and Yta12 subunits with SWISS-MODEL (19) using the fitted homo-hexamer structure as template. However, at this resolution, it was not possible to distinguish between Yta10 and Yta12 subunits because of their high structural homology and the C6 symmetry applied during image reconstruction. We therefore assigned subunits according to our previous biochemical and genetic results that were most consistent with an alternating arrangement of Yta10 and Yta12 subunits (14).
In our fitted structure, the 12 transmembrane helices of the Yta10 and Yta12 subunits form a tight bundle of helices that presumably anchor the m-AAA protease to the mitochondrial inner membrane (Fig. 5, A and B). The six copies of the ATPase domain account for the upper half of the main body, whereas the protease ring composed of PD and CH fits snuggly into the lower half of the mass density (Fig. 5B). In our fitted structure, the AAA domain and PD are in a staggered configuration with the large α/β domain sitting on top of the PD of the neighboring subunit and the small helical domain on top of the PD of the same protomer. This domain arrangement places the large α/β domain of the Yta10 and Yta12 subunits near the central pore and the smaller helical domains close to the inner rim of the main body. When viewed top down from the membrane-facing side, the pore loop-1 and -2 of alternating Yta10 and Yta12 subunits line the central pore that traverses the AAA ring (Fig. 5C). The arrangement of Yta10 and Yta12 pore loops is reminiscent of other AAA+ machines (7, 20–25) and can explain the role of the critical (Phe/Tyr)-Val-Gly motif (residues 361–363 in Yta10 and 421–423 in Yta12) in substrate translocation and processing via handover of unfolded polypeptides from the ATPase to the protease domains (26).
Unlike the AAA domains, the structure of the protease ring is fully compatible with that of the x-ray structures of T. maritima (6, 7) and T. thermophilus FtsHCyt (5). The latter supports the notion that the protease ring is required for hexamer assembly but does not undergo conformational changes in solution, which are driven by ATP binding and hydrolysis.
Residues Critical for Functional Assembly Are Located at the Protomer Interface
To determine the structural basis for homo-oligomeric assembly of the Yta12 variants, we mapped our identified Yta12 mutations onto the fitted structure of the hetero-oligomeric m-AAA protease (Fig. 5D). Strikingly, we found that all mutated amino acid residues are located at the interface between two neighboring protomers in the hexameric ring complex and are often in close proximity to each other. Notably, most of these amino acids are not conserved among m-AAA protease subunits, suggesting that the nature of those amino acids likely determines the assembly status of m-AAA protease isoenzymes. It is therefore conceivable that alterations of amino acid residues at the PD interface during evolution gave rise to homo- and hetero-oligomeric m-AAA protease complexes and fine-tuned their specific cellular activities. Interestingly, mutations in Yta12, which allow homo-oligomerization, cluster within the same region of the PD as disease-causing mutations in SCA28 patients (13), suggesting that these mutations may also affect assembly of human m-AAA proteases.
DISCUSSION
Here, we have presented the 12 Å resolution cryo-EM structure of a hetero-oligomeric m-AAA protease, revealing for the first time the structural organization of the intact enzyme. Our combined structural and genetic findings highlight the similarities between different types of m-AAA protease subunits and demonstrate that only minor differences in amino acid residue composition of the PDs confer homo- or hetero-oligomeric assemblies of m-AAA protease subunits. Our cryo-EM analysis indicates that the intact m-AAA protease is hexameric and has a bipartite structure consisting of a funnel-shaped upper density, corresponding to the N-terminal transmembrane and intermembrane space domains, and a more rigid lower body consisting of the AAA, PD, and CH domains, which is similar to the x-ray structure of the isolated 6-fold symmetric apo-FtsHCyt hexamer (7) but differs in detail. Notably, the relative orientation of the large α/β and the small helical domain is different, consistent with a closing of the nucleotide-binding cleft upon ATP binding. The x-ray structure of the protease ring (5–7), on the other hand, is fully compatible with our cryo-EM map. The PD likely drives the correct assembly of the complex, but our work also suggests that both the transmembrane and AAA domains contribute toward oligomerization by stabilizing the hexamer assembly through stereo-specific interactions with domains from neighboring subunits.
Within the resolution limit of our cryo-EM analysis, the intact m-AAA protease in the ATP-bound state forms a 6-fold symmetrical hexamer (Fig. 3A and supplemental Fig. S1). However symmetry breaks are apparent in some of the class images before application of C6 symmetry (supplemental Fig. S1A). This might reflect variations in the nucleotide occupancy of AAA domains within a ring, which are expected from the observation of a coordinated ATP hydrolysis between neighboring subunits (14). This heterogeneity and asymmetry may mask minor differences between Yta10 and Yta12, preventing the successful application of C3 symmetry. which is predicted from the alternating architecture.
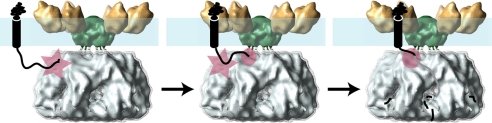
The cryo-EM structure of the intact m-AAA protease with its transmembrane domain suggests a mechanistic model for protein degradation. Unlike other ring-forming AAA+ machines, m-AAA proteases and other FtsH-like proteases are membrane-bound, raising the question how soluble and membrane-integral protein substrates enter the protease and are degraded. It is possible that the transmembrane segment of m-AAA proteases could function as a gated pore allowing membrane proteins to pass through the mitochondrial inner membrane. However, our three-dimensional structure of the intact m-AAA protease suggests another mechanism. It revealed a 13 Å wide gap between the main hexamer body and the N-terminal transmembrane segment, which is large enough to accommodate unfolded but not folded polypeptides. After initial substrate contact on the distal surface of the AAA domain (27), this gap might enable a guided transfer of the unfolded polypeptide to the central pore of the AAA ring (Fig. 6). Notably, the minimal length of ∼20 amino acid residues of an unfolded polypeptide segment on the matrix side required for substrate degradation (28) closely matches the distance between the initial substrate binding site and the central pore loops of the AAA ring. Once the unfolded segment is bound to the central pore loops, the substrate is threaded in an ATP hydrolysis-driven manner into the proteolytic chamber of the m-AAA protease, where the unfolded substrate is degraded, and the cleaved peptides are released through nearby lateral openings (Fig. 6).
FIGURE 6.
Proposed mechanism of membrane-integral protein dislocation and degradation by the m-AAA protease. The m-AAA protease hexamer is shown as described in Fig. 4. The mitochondrial inner membrane (blue) and a membrane-integral protein substrate (black) are shown schematically with the initial (star) and secondary contact sites (circle) on the m-AAA protease depicted by red symbols.
Supplementary Material
Acknowledgments
We thank S. J. Ludtke for help with EMAN, and members of the Tsai and Langer laboratories for discussions. We are grateful to W. Chiu for access to the cryo-EM facilities of the National Center for Macromolecular Imaging (NCRR P41RR002250).
This work was supported in part by National Institutes of Health Grant R01-AI076239, by Welch Foundation Grant Q-1530, Department of Defense, and American Cancer Society (to F. T. F. T.), by a grant from the American Heart Association Texas Affiliate (to S. L.), and by Deutsche Forschungsgemeinschaft and European Research Council grants and German-Israeli-Project DIP Grant F.5.1. (to T. L.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4 and additional references.
The cryo-EM map of the m-AAA protease hexamer has been deposited under EMDB code EMD-1712.
- CH
- C-terminal helical region
- PD
- protease domain
- β-OG
- n-octyl-β-d-glucopyranoside
- PDB
- Protein Data Bank.
REFERENCES
- 1. Erzberger J. P., Berger J. M. (2006) Annu. Rev. Biophys. Biomol. Struct. 35, 93–114 [DOI] [PubMed] [Google Scholar]
- 2. Ito K., Akiyama Y. (2005) Annu. Rev. Microbiol. 59, 211–231 [DOI] [PubMed] [Google Scholar]
- 3. Koppen M., Langer T. (2007) Crit. Rev. Biochem. Mol. Biol. 42, 221–242 [DOI] [PubMed] [Google Scholar]
- 4. Tatsuta T., Langer T. (2009) Res. Microbiol. 160, 711–717 [DOI] [PubMed] [Google Scholar]
- 5. Suno R., Niwa H., Tsuchiya D., Zhang X., Yoshida M., Morikawa K. (2006) Mol. Cell 22, 575–585 [DOI] [PubMed] [Google Scholar]
- 6. Bieniossek C., Schalch T., Bumann M., Meister M., Meier R., Baumann U. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 3066–3071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bieniossek C., Niederhauser B., Baumann U. M. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 21579–21584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Akiyama Y., Ito K. (2000) EMBO J. 19, 3888–3895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Korbel D., Wurth S., Käser M., Langer T. (2004) EMBO Rep. 5, 698–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Arlt H., Tauer R., Feldmann H., Neupert W., Langer T. (1996) Cell 85, 875–885 [DOI] [PubMed] [Google Scholar]
- 11. Koppen M., Metodiev M. D., Casari G., Rugarli E. I., Langer T. (2007) Mol. Cell. Biol. 27, 758–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Casari G., De Fusco M., Ciarmatori S., Zeviani M., Mora M., Fernandez P., De Michele G., Filla A., Cocozza S., Marconi R., Dürr A., Fontaine B., Ballabio A. (1998) Cell 93, 973–983 [DOI] [PubMed] [Google Scholar]
- 13. Di Bella D., Lazzaro F., Brusco A., Plumari M., Battaglia G., Pastore A., Finardi A., Cagnoli C., Tempia F., Frontali M., Veneziano L., Sacco T., Boda E., Brussino A., Bonn F., Castellotti B., Baratta S., Mariotti C., Gellera C., Fracasso V., Magri S., Langer T., Plevani P., Di Donato S., Muzi-Falconi M., Taroni F. (2010) Nat. Genet. 42, 313–321 [DOI] [PubMed] [Google Scholar]
- 14. Augustin S., Gerdes F., Lee S., Tsai F. T., Langer T., Tatsuta T. (2009) Mol. Cell 35, 574–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ludtke S. J., Baldwin P. R., Chiu W. (1999) J. Struct. Biol. 128, 82–97 [DOI] [PubMed] [Google Scholar]
- 16. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 17. Jiang W., Baker M. L., Ludtke S. J., Chiu W. (2001) J. Mol. Biol. 308, 1033–1044 [DOI] [PubMed] [Google Scholar]
- 18. Salom D., Lodowski D. T., Stenkamp R. E., Le Trong I., Golczak M., Jastrzebska B., Harris T., Ballesteros J. A., Palczewski K. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 16123–16128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schwede T., Kopp J., Guex N., Peitsch M. C. (2003) Nucleic Acids Res. 31, 3381–3385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang J., Song J. J., Franklin M. C., Kamtekar S., Im Y. J., Rho S. H., Seong I. S., Lee C. S., Chung C. H., Eom S. H. (2001) Structure 9, 177–184 [DOI] [PubMed] [Google Scholar]
- 21. Siddiqui S. M., Sauer R. T., Baker T. A. (2004) Genes Dev. 18, 369–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Weibezahn J., Tessarz P., Schlieker C., Zahn R., Maglica Z., Lee S., Zentgraf H., Weber-Ban E. U., Dougan D. A., Tsai F. T., Mogk A., Bukau B. (2004) Cell 119, 653–665 [DOI] [PubMed] [Google Scholar]
- 23. Park E., Rho Y. M., Koh O. J., Ahn S. W., Seong I. S., Song J. J., Bang O., Seol J. H., Wang J., Eom S. H., Chung C. H. (2005) J. Biol. Chem. 280, 22892–22898 [DOI] [PubMed] [Google Scholar]
- 24. Hinnerwisch J., Fenton W. A., Furtak K. J., Farr G. W., Horwich A. L. (2005) Cell 121, 1029–1041 [DOI] [PubMed] [Google Scholar]
- 25. Lee S., Choi J. M., Tsai F. T. (2007) Mol. Cell 25, 261–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tatsuta T., Augustin S., Nolden M., Friedrichs B., Langer T. (2007) EMBO J. 26, 325–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Graef M., Seewald G., Langer T. (2007) Mol. Cell. Biol. 27, 2476–2485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Leonhard K., Guiard B., Pellecchia G., Tzagoloff A., Neupert W., Langer T. (2000) Mol. Cell 5, 629–638 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.