Abstract
Rev-erbβ is a heme-binding nuclear hormone receptor that represses a broad spectrum of target genes involved in regulating metabolism, the circadian cycle, and proinflammatory responses. Here, we demonstrate that a thiol-disulfide redox switch controls the interaction between heme and the ligand-binding domain of Rev-erbβ. The reduced dithiol state of Rev-erbβ binds heme 5-fold more tightly than the oxidized disulfide state. By means of site-directed mutagenesis and by UV-visible and EPR spectroscopy, we also show that the ferric heme of reduced (dithiol) Rev-erbβ can undergo a redox-triggered switch from imidazole/thiol ligation (via His-568 and Cys-384, based on a prior crystal structure) to His/neutral residue ligation upon oxidation to the disulfide form. On the other hand, we find that change in the redox state of iron has no effect on heme binding to the ligand-binding domain of the protein. The low dissociation constant for the complex between Fe3+- or Fe2+-heme and the reduced dithiol state of the protein (Kd = ∼20 nm) is in the range of the intracellular free heme concentration. We also determined that the Fe2+-heme bound to the ligand-binding domain of Rev-erbβ has high affinity for CO (Kd = 60 nm), which replaces one of the internal ligands when bound. We suggest that this thiol-disulfide redox switch is one mechanism by which oxidative stress is linked to circadian and/or metabolic imbalance. Heme dissociation from Rev-erbβ has been shown to derepress the expression of target genes in response to changes in intracellular redox conditions. We propose that oxidative stress leads to oxidation of cysteine(s), thus releasing heme from Rev-erbβ and altering its transcriptional activity.
Keywords: Disulfide, Electron Paramagnetic Resonance (EPR), Heme, Metabolic Regulation, Thiol
Introduction
Nuclear receptors regulate gene expression in response to small molecules, controlling key eukaryotic events such as development, differentiation, reproduction, and metabolic homeostasis (1). All nuclear receptors possess a highly conserved N-terminal DNA-binding domain and a less conserved C-terminal ligand-binding domain (LBD),2 which recognizes small molecules (2). The LBD in nuclear receptors is typically coupled to transcriptional activation through an activation function-2 domain, which promotes recruitment of a coactivator (1). Rev-erbβ (also known as RVR and BD73) and Rev-erbα are unique members of the superfamily of nuclear receptors because they lack an activation function-2 domain (3, 4).
Rev-erbs are transcriptional repressors that are present in the nucleus and bind as a monomer to the Rev responsive element or as a dimer to a Rev-RE direct repeat, RevDR-2 (5, 6). Rev-erbs had been considered to be an orphan nuclear receptor until the Drosophila ortholog of human Rev-erb (E75) (7) and then Rev-erb α (8, 9) and Rev-erbβ (8) were recently demonstrated to bind heme at their LBDs. Heme binding promotes recruitment of nuclear corepressor and histone deacetylase (HDAC) (8, 10, 11). Rev-erbα recruits HDAC3, whereas Rev-erbβ recruits HDAC1 (3, 8, 9). In turn, the Rev-erb-nuclear corepressor-HDAC complex facilitates the repression of target genes involved in glucose, lipid, and bile acid metabolism, as well as in regulating the circadian cycle and the proinflammatory response (3, 4, 10). Therefore, Rev-erbα and Rev-erbβ have been proposed to link metabolism and inflammation to the circadian clock (5, 12–14). Because heme controls the activity of Rev-erb, understanding the regulation of heme binding is of special significance.
Tip60 and ZNHIT-1 are involved in derepression of Rev-erbβ-regulated genes (e.g. apoCIII) (15, 16); however, what facilitates the derepression of Rev-erbα regulated genes is not known. In addition to its activity as a repressor, Rev-erbβ can also act as a transcriptional activator (17).
Several recent studies have focused on the relationship between heme and Rev-erbs. Heme has long been implicated to play a key role in maintaining the circadian rhythm via binding to different circadian proteins such as NPAS2 and mPer2 (18, 19). Correspondingly, heme shows a circadian pattern of expression (20). Rev-erbβ also has been recently shown to bind heme, and the x-ray crystal structure reveals Cys-384 and His-568 as axial ligands for the ferric heme (11), similar to the Drosophila homolog, E75 (21). A ligand switch appears to occur upon reduction of the Fe3+-heme to Fe2+ with His-568 proposed to remain as one of the Fe2+ ligands (22). Raman and magnetic CD data indicated the presence of one or two neutral ligands in the Fe2+ state (22). An important attribute of heme-dependent proteins is their ability to bind gaseous signaling molecules (CO, NO, etc.) (23, 24). Like some other heme-dependent transcriptional regulators, Rev-erbβ appears to bind diatomic (CO/NO) gases (11); however, the affinity has not been measured. Based on Raman and magnetic CD studies, it has been proposed that NO/CO replaces Cys-384 as an axial ligand and that a neutral ligand serves as the sixth ligand in the (CO/NO)-bound form of Rev-erbβ (11, 22).
Here, we show that a thiol-disulfide redox switch regulates binding of heme to the LBD of Rev-erbβ. Heme binds more tightly to the reduced dithiol state of Rev-erbβ, forming a complex with a dissociation constant that is similar to the low nanomolar intracellular concentration of free heme. Mass spectrometric and mutational analyses reveal that formation of the disulfide bond between Cys-384 and Cys-374 upon oxidation lowers the affinity for heme by ∼5-fold. UV-visible and EPR spectroscopic studies with the purified protein and with Escherichia coli cells expressing the Rev-erbβ LBD indicate that oxidation of Rev-erbβ also triggers a ligand switch converting His/Cys ligated heme to another six-coordinated heme ligated via His and an unknown neutral residue. Based on EPR spectra, the neutral ligand could be another His, Met, or Lys. Increasing the pH above 10 also converts the His/neutral residue ligation state of the oxidized protein to His/Cys ligation. Our results indicate that the thiol/disulfide redox switch (but not the ligand switch) regulates heme affinity. Thus, the present study demonstrates a mechanism by which the redox status of Rev-erbβ can alter heme binding and may facilitate its ability to regulate the cyclic expression of metabolic and proinflammatory genes in response to changes in intracellular redox conditions.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
In all experiments, the LBD (residues 247–579) of human Rev-erbβ was used. The wild-type Rev-erbβ LBD clone was generously provided by Thomas P. Burris (Scripps Research Institute, Jupiter, FL). The protein was expressed in a pET-46Ek/LIC vector as an N-terminal His6-tagged protein. All of the cysteines were kept intact in the protein unless otherwise specified. E. coli cells were grown without any heme supplement, unless otherwise stated. The protein was purified using a nickel-nitrilotriacetic acid column (Qiagen) and was then dialyzed against a buffer containing 20 mm Tris-HCl, 300 mm NaCl, and 10% glycerol, pH 8.0 (Buffer A). No significant amount of heme remained bound to the protein after overnight dialysis as was confirmed by the pyridine hemochrome assay (25).
Site-directed Mutagenesis of Rev-erbβ LBD
Site-directed mutagenesis was performed as described previously (26), and all of the clones were sequenced at the DNA Sequencing Core Facility (University of Michigan, Ann Arbor, MI).
Reduction and Oxidation of Rev-erbβ LBD
Throughout, we refer to the disulfide and dithiol states of the Rev-erbβ LBD when we describe the “oxidized” and “reduced” protein; the redox state of the heme, i.e. Fe3+ or Fe2+, is stated explicitly. To generate the fully reduced or fully oxidized form of the protein, the following methods were employed. To obtain the fully reduced protein, the Rev-erbβ LBD was first incubated with a 30-fold molar excess of tris(2-carboxyethyl)phosphine (TCEP) for 30 min on ice, and then TCEP was removed on a Bio-Gel P-6 (Bio-Rad) column. Therefore, no TECP was present during the experiments with the reduced protein. The number of free thiols in the reduced Rev-erbβ LBD was quantified by an assay as described earlier (27–29). The TCEP treatment, incubation, chromatography, and assay were carried out inside an anaerobic chamber (Vacuum Atmospheres Co., Hawthorne, CA). Then the protein was transferred to anaerobically closed vials/cuvettes or EPR tubes, thus maintaining the reducing environment.
To fully oxidize the protein, the reduced Rev-erbβ LBD was treated with 400 μm diamide; alternatively, the air-oxidized protein was used. Complete oxidation of thiols was verified by the 5,5′-dithio-bis(2-nitrobenzoic acid) assay before performing experiments with the oxidized protein.
Heme Binding Analysis
Heme titrations were performed by difference spectroscopy in a double beam spectrometer using 0.2–0.5 μm protein, as described earlier (29), except that the buffer contained Buffer B (20 mm Tris-HCl, 300 mm NaCl, and 10% glycerol, 3% Me2SO, pH 8.0). In these titrations, the blank cuvette contained buffer, the sample cuvette had both protein and buffer, and heme from a 50 μm stock solution was added to both cuvettes. Because heme solutions are prone to dimerization and oligomerization, special precautions, as described earlier (29), were taken in preparation of the stock heme solutions, e.g. by including Me2SO, which has been shown to prevent heme aggregation (30). Briefly, hemin (Fe3+-heme) stocks were prepared fresh for each use by dissolving hemin (Sigma) in 0.1 m NaOH and 5% Me2SO, filtering with a 0.2-μm syringe filter (Amicon, Beverly, MA) and diluting to 50 μm by adding an aliquot into a solution of 20 mm Tris-HCl, pH 8.0, 300 mm NaCl, 5% Me2SO, and 10% glycerol. To prepare Fe2+-heme, the Fe3+ in the 50 μm hemin stock solution was reduced by adding 2.5 mm sodium dithionite in the anaerobic chamber (Vacuum Atmospheres Co., Hawthorne, CA) and was handled anaerobically in vials and cuvettes that were sealed with rubber serum stoppers.
To determine the binding parameters, the data obtained from the heme (21) titrations were plotted and fit to an equation describing a single binding site (Equation 1), which is quadratic and is used when the Kd value is similar to the concentration of protein, because it accounts for the amount of bound ligand in the solution (29). Here, ΔA refers to the absorbance difference at 416, 424, and 428 nm between the sample (containing protein and added heme) and reference (containing only protein) cuvettes; Δϵ is defined as the difference in extinction coefficient between free and bound heme, and EL is the concentration of the heme-protein complex. EL was calculated from Equation 2, where E0 refers to the total protein concentration, L0 refers to the total heme concentration, and Kd refers to the dissociation constant. Thus, the lines shown in the heme titrations are all theoretical fits of the data (the symbols) to these equations.
Surprisingly, for Rev-erbβ LBD, the apparent Kd value seemed to depend on the protein concentration. When the titration was performed at increasing concentrations of the oxidized protein (from 0.4 to 5 μm), the Kd value increased from 0.12 to 2 μm, the latter value being similar to values reported earlier (see above). Likewise, variation in concentration of reduced protein from 0.3 to 5 μm resulted in an increase in the Kd from 23 nm to 0.9 μm. It is likely that aggregation of Rev-erbβ at higher protein concentrations is responsible for this nonideal behavior in heme-binding experiments. It is also possible that this behavior results in part from aggregation of heme (31); however, we took special precautions (preparing heme solutions and performing titrations in the presence of Me2SO (30)) to minimize heme dimerization and aggregation. Correspondingly, native gel electrophoresis experiments with the oxidized LBD show high molecular weight species that are not present with the reduced protein or with reduced protein that has been freshly treated with diamide. When the concentration of the Rev-erbβ LBD was kept below 1 μm, Kd values in the 0.20 μm range for the oxidized protein and in the 23 nm range for the reduced protein were consistently observed. Thus, the high Kd values in the 2–6 μm range are likely to be artifactual. Furthermore, free heme concentrations in the cell are below 0.1 μm (32–34), and free heme levels above 1 μm are toxic (34). Thus, we continued our experiments using protein concentrations below 1 μm.
EPR Spectroscopy
EPR spectra were recorded at 15 K on an X-band Bruker EMX spectrometer (Bruker Biospin Corp., Billerica, MA) containing an Oxford ITC4 temperature controller, a Hewlett-Packard model 5340 automatic frequency counter, and a Bruker gaussmeter. All of the EPR samples were prepared in Buffer A (see above). The samples were frozen with liquid nitrogen before the experiment.
Whole Cell EPR Spectroscopy
For in vivo detection of the Fe3+-heme complex with Rev-erb, whole cell EPR experiments were performed. E. coli cells expressing the Rev-erbβ LBD were grown overnight at 25 °C after induction with 1 mm isopropyl β-d-thiogalactopyranoside, and 10 ml of culture was centrifuged at 2977 × g for 10 min. The pellet obtained was resuspended in 100 μl of Buffer B, transferred to an EPR tube, and frozen in liquid nitrogen before obtaining the EPR spectrum.
Alkylation of Cysteines
Samples of oxidized and reduced (with 10 mm TCEP) Rev-erbβ LBD (2 mg/ml) were subjected to a two-step alkylation protocol, as has been described previously (29). Briefly, samples were first treated with iodoacetamide in a denaturing buffer and then with 4-vinyl pyridine after reduction with 10 mm TCEP.
Mass Spectrometric Analysis
Alkylated Rev-erbβ LBD samples were digested with trypsin and analyzed by mass spectrometry in the Protein Structure Facility at the University of Michigan, Ann Arbor. LC-MS/MS was performed on a NanoAqcuity/Qtof Premier Instrument (Waters Inc., Milford, MA) (see supplemental methods for details).
Quantification of Redox States of the Cysteines in Rev-erbβ LBD Using the ICAT Technique
OxICAT, a novel mass spectrometric method that couples thiol trapping with the ICAT technique to quantify oxidative thiol modifications, was performed. For trapping the redox states of purified Rev-erbβ LBD, 100 μg of oxidized Rev-erbβ LBD was applied to a two-step alkylation procedure with light/heavy ICAT reagents as described previously (35, 36). The alkylated protein was then digested with trypsin, and the cysteine-containing peptides, which are linked to biotin, were enriched on a cation exchange cartridge followed by an avidin affinity cartridge (Applied Biosystems, Foster City, CA). The biotin tag, which was conjugated to the cysteine-containing peptides, was removed afterward, and then samples were analyzed by nano-liquid chromatography/tandem mass spectrometry at the Michigan Proteome Consortium to quantify the amounts of reduced (dithiol) protein containing the light ICAT and oxidized (disulfide) protein containing the heavy ICAT adduct. For determining the redox state of cysteines in Rev-erbβ LBD, different peptides were analyzed containing the cysteine(s) of interest. As expected, the high resolution mass spectra consist of envelopes of multiple peaks reflecting the presence of 0.018% 2H, 1.11% 13C, 0.45% 15N, and 0.20% 18O in each peptide. The m/z ratios for the oxidized form of these peptides are 9 (for peptides containing single cysteine) and 18 (for the peptide with two cysteines) mass units larger than those for the reduced protein (37).
Protein Preparation for Spectroscopic Analysis at Different pH Values
The protein was first dialyzed in 50 mm acetic acid, 50 mm MES, and 100 mm triethanolamine, pH 8.0 buffer (38), and then the pH was adjusted with 10 n NaOH before performing UV-visible and EPR spectroscopy (as described above) at the desired pH.
CO Binding Assay
Rev-erbβ LBD samples were prepared anaerobically, treated with sodium dithionite (final concentration, 2.5 mm), and transferred anaerobically to the reaction cuvette. Aliquots of CO were added to the assay mixture from an anaerobic stock solution consisting of Buffer A that had been bubbled with a gas mixture of N2/CO (75%/25%, providing a CO concentration of 0.25 mm). Gas cylinders (25%) were purchased from Metro Welding Supply Corporation (Detroit, MI). The final concentration of CO in the CO-saturated buffer was confirmed by titration with myoglobin, whose concentration was determined based on its published extinction coefficient (121 mm−1 cm−1) (39).
RESULTS
Thiol-disulfide Redox Regulation of Heme Binding to Rev-erbβ
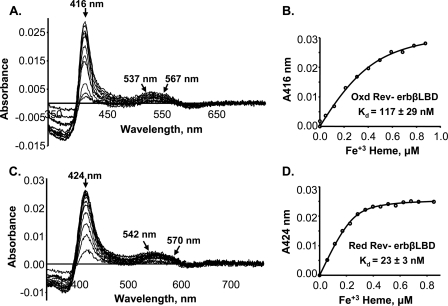
Heme binding studies were performed with the oxidized and reduced forms (assessed by the 5,5′-dithio-bis(2-nitrobenzoic acid) assay as described under “Experimental Procedures”) of the purified LBD of Rev-erbβ and were monitored by difference UV-visible spectroscopy. All of the heme titrations were performed in Buffer A (see “Experimental Procedures” for details), unless stated otherwise. When the oxidized Rev-erbβ LBD was titrated with Fe3+-heme, the difference spectra exhibited absorbance changes in the Soret region at 416 nm (Fig. 1A) and in the alpha (567 nm) and beta (537 nm) bands. Titration of reduced Rev-erbβ LBD (after TCEP removal) with Fe3+-heme resulted in shifts in the Soret peak to 424 nm, and the alpha and beta peaks were at 570 and 542 nm (Fig. 1C), respectively, which is characteristic of His/Cys axial ligands (21, 40). The UV-visible spectra of the oxidized and reduced protein-heme complexes are shown in supplemental Fig. S1 and show similar peak shifts as observed in the difference spectra. The oxidized protein with bound Fe3+-heme has its Soret band at 415 nm, whereas two long wavelength peaks are at 566 and 534 nm. However, the TCEP-reduced protein containing bound Fe3+-heme exhibits a Soret peak at 424 nm and alpha and beta peaks at 571 and 541 nm, respectively. These spectral changes between the oxidized and reduced protein in the Fe3+-heme state indicate that a ligand switch is associated with oxidation/reduction of the Rev-erbβ LBD (this conclusion is verified by the mutagenesis and EPR studies described below).
FIGURE 1.
Redox-dependent binding of Fe3+-heme to Rev-erbβ LBD. Difference absorption spectra and titration curves of oxidized (0. 4 μm) (A and B, respectively) and reduced (0.3 μm) (C and D, respectively) Rev-erbβ LBD are shown. The titrations were performed in Buffer B (see “Experimental Procedures”). Titration of reduced Rev-erbβ LBD with Fe3+-heme was performed anaerobically to avoid thiol-oxidation during the experiment. The lines in B and D were generated from fits to Equation 1.
By plotting the change in absorbance at 416 nm (for the oxidized LBD) or at 424 nm (for the reduced protein) versus the concentration of added Fe3+-heme, typical hyperbolic profiles were obtained, indicating saturation binding. When the data for the oxidized protein were fit to Equation 1, the Kd value was 117 nm (Fig. 1B), significantly lower than the values of ∼2 μm (8) or ∼6 μm (11) reported earlier. Furthermore, the reduced protein binds Fe3+-heme even more tightly, with a Kd value of 23 nm (Fig. 1D). Because the crystal structure identified His-568 and Cys-384 as heme ligands, we performed heme titrations of the H568R, H568A, and C384A variants. The heme binding affinities of these variants, both in oxidized and reduced states, were significantly lower than those of the wild-type protein (see Table 3). The Kd for the complex between heme and the H468R variant is ∼20-fold higher than that for the H568A mutant, suggesting that this substitution many have an effect on the active site other than simply removal of the heme ligand.
TABLE 3.
Redox-dependent heme binding properties of wild type and mutants
The peaks from optical absorption spectra are given in nm.
| Oxidized protein-Fe3+ heme |
Reduced protein-Fe3+ heme |
Reduced protein-Fe2+ heme |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Soret | α | β | Kd | Soret | α | β | Kd | Soret | α | β | Kd | |
| nm | nm | nm | ||||||||||
| WT | 416 | 567 | 537 | 117 ± 29 | 424 | 570 | 542 | 23 ± 3 | 428 | 560 | 530 | 16 ± 4 |
| C374S | 416 | 567 | 537 | 12 ± 4 | 424 | 570 | 542 | 13 ± 6 | ||||
| C355S/C374S | 416 | 567 | 537 | 17 ± 5 | 424 | 570 | 542 | 24 ± 4 | ||||
| C384A | 415 | 567 | 537 | 1010 ± 110 | 420 | Broad peak at 550 | Broad peak at 550 | 46 ± 14 | 428 | 560 | 530 | 209 ± 30 |
| H568R | 416 | Broad peak at 550 | Broad peak at 550 | 13090 ± 780 | 424 | Broad peak at 550 | Broad peak at 550 | 175 ± 44 | 431 | 563 | 536 | 1520 ± 490 |
| H568A | 416 | Broad peak at 550 | Broad peak at 550 | 713 ± 135 | 420 | Broad peak at 550 | Broad peak at 550 | 2291 ± 580 | 427 | 560 | 530 | 199 ± 54 |
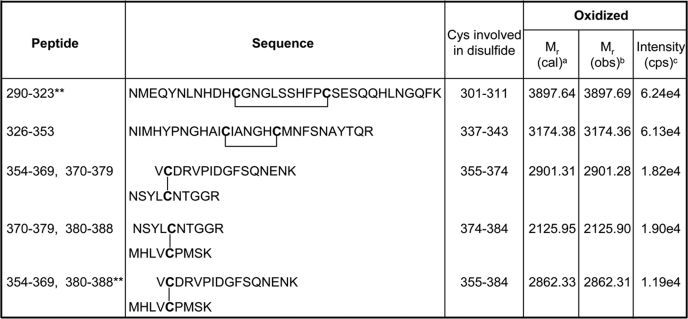
Because the heme binding assays demonstrated that oxidation of the protein weakens affinity for heme, we used LC-MS/MS analyses (see supplemental methods) to determine whether Cys-384 forms a disulfide bond and (if so) to which residue it is linked, as well as to evaluate the redox status of all seven cysteines present in the LBD. Initially, we performed the LC-MS/MS analysis of the trypsin-digested oxidized protein and searched for the peptides carrying disulfide bonds. The result of this analysis is described in Table 1, which compares the predicted molecular mass of each peptide (based on sequence) to the experimentally determined mass. The intensity values given in the table do not measure the absolute quantity of a peptide but quantify the relative abundance under similar LC-MS/MS conditions. Intact disulfide bonds were found between Cys-337 and Cys-343, between Cys-374 and Cys-384, and between Cys-355 and Cys-374 in their respective peptides. Two other disulfides, Cys-301 and Cys-311 and Cys-355 and Cys-384 were also detected but with very low ionization intensity. The disulfide bonds between Cys-301 and Cys-311 and Cys-337 and Cys-343 were present in single peptides. Because there are trypsin cleavage sites between the interlinked cysteine residues, the other three disulfide bonds connected two peptides. Nevertheless, all of the detected disulfides appear to be intramolecular, because no dimeric form of the protein was observed in nonreducing SDS-PAGE analyses of the oxidized protein.
TABLE 1.
Disulfide bond analysis after mass spectrometry of unmodified air-oxidized Rev-erbβ
a Relative molecular mass based on the matched peptide sequence.
b Observed m/z factored by z.
c Counts per second.
** These peptides had very low ionization intensity.
LC-MS/MS was also performed on tryptic-digested products of the oxidized and reduced Rev-erbβ LBD that had been subjected to the two-step alkylation with iodoacetamide and vinyl pyridine, as described previously (41). This analysis further confirmed the involvement of all of the above mentioned cysteines in disulfide bonds, in the oxidized state of the protein (Table 2). In the reduced protein, all of the Cys residues were modified by iodoacetamide, confirming that all Cys residues are in the thiol(ate) state in the reduced protein. In the oxidized protein, Cys-301 and Cys-311 from peptide 290–323 and Cys-337 and Cys-343 from peptide 326–353 showed only 4-vinyl pyridine modifications, indicating that these cysteines were involved in the disulfide bond in the oxidized protein. However, peptides containing the other three cysteine residues (Cys-355, Cys-374, and Cys-384) were detected with both iodoacetamide and 4-vinyl pyridine modifications in different fractions. This might result from an incomplete oxidation of the cysteines. Regardless, the peptide containing 4-vinyl pyridine-modified Cys-384 (peptide 380–388) was almost 50 times more abundant than that containing iodoacetamide modification. Moreover, the peptides containing Cys-374–Cys-384 was 1.6-fold more abundant than the peptides containing Cys-355–Cys-384. Thus, based on the above two mass spectrometric analyses, we hypothesize that Cys-384 may form a disulfide with either Cys-374 or Cys-355.
TABLE 2.
LC-MS/MS results of oxidized and reduced Rev-erbβ LBD after two-step alkylation
cps, counts per second; ND, peptide not detected; IAM, idoacetamide; 4VP, 4-vinylpyridine.
| Peptide | Sequence | Oxidized |
Reduced |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| No.a |
Mr |
Intensity (cps) | No.a |
Mr |
Intensity (cps) | ||||||
| IAM | 4VP | Calculatedb | Observedc | IAM | 4VP | Calculatedb | Observedc | ||||
| 290–323 | NMEQYNLNHDHCGNGLSSHFPCSESQQHLNGQFK | 0 | 2 | 4322.90 | 4322.93 | 1.75e4 | ND | ND | ND | ND | ND |
| 326–353 | NIMHYPNGHAICIANGHCMNFSNAYTQR | 0 | 2 | 3386.55 | 3386.51 | 8.54e4 | 2 | 0 | 3290.44 | 3290.44 | 2.09e4 |
| 354–369 | VCDRVPIDGFSQNENK | 0 | 1 | 1924.91 | 1924.91 | 5.15e4 | 1 | 0 | 1876.86 | 1876.86 | 1.55e4 |
| 1 | 0 | 1876.87 | 1876.87 | 4.10e3 | |||||||
| 370–379 | NSYLCNTGGR | 0 | 1 | 1188.53 | 1188.54 | 4.69e2 | 1 | 0 | 1140.50 | 1140.50 | 4.07e2 |
| 1 | 0 | 1140.50 | 1140.50 | 6.46e2 | |||||||
| 380–388 | MHLVCPMSK | 0 | 1 | 1149.55 | 1149.51 | 1.14e5 | 1 | 0 | 1101.51 | 1101.50 | 8.84e5 |
| 1 | 0 | 1101.51 | 1101.51 | 2.27e3 | |||||||
a Number of cysteine residues modified.
b Relative molecular mass based on the matched peptide sequence.
c Observed m/z factored by z.
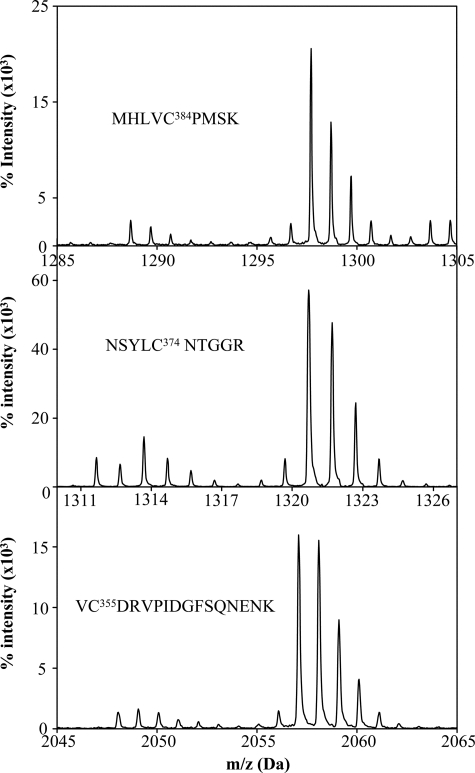
To test the above hypothesis about Cys-384, we used the oxICAT method to quantify the redox state of cysteines in the oxidized protein. As described under “Experimental Procedures,” we treated Rev-erbβ LBD with the light ICAT reagent, reduced all of the existing oxidative thiol modifications, and alkylated all newly reduced thiols with the heavy ICAT. Thus, a peptide containing a free Cys thiol(ate) will have an m/z value that is 9 units smaller than one containing a Cys that is engaged in a disulfide bond (37). Mass spectral analysis of the affinity-purified tryptic peptide harboring Cys-384 (MHLVC384PMSK) reveals peaks at 1288.69 and 1297.65 (+9), with the major peak at 1297.65, resulting from labeling with one heavy ICAT molecule (Fig. 2). Similarly, peptides containing Cys-374 (NSYLC374NTGGR) and Cys-355 (VC355DRVPIDGFSQNENK) showed major peaks at 1320.64 (+9) and 2057.02 (+9) from the addition of one heavy ICAT molecule (Fig. 2). Thus, the majority of the oxidized protein contains oxidized forms of Cys-384 (89%), Cys-374 (87%), and Cys-355 (92%). In fact, all of the LBD peptides containing Cys residues showed major peaks with an increase in their mass by 9 or 18 Da, indicating that all of the cysteines in the oxidized state of the LBD are in the oxidized form (data not shown).
FIGURE 2.
Oxidation of cysteines in oxidized Rev-erbβ LBD. OxICAT analysis of the redox state of purified and oxidized Rev-erbβ LBD reveals incorporation of heavy ICAT (13C) in Cys-384 (top panel), Cys-374 (middle panel), and Cys355 (bottom panel) in their respective peptides, which demonstrates that these cysteines are oxidized upon oxidation of the protein.
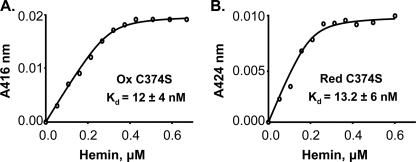
To determine which disulfides are involved in regulating heme binding, we generated C374S and C355S/C374S variants. Interestingly, mutation of Cys-374 to Ser was sufficient for the loss of redox-dependent heme binding (Table 3). Both C374S and C355S/C374S variants exhibited similar affinity for heme in both oxidized and reduced states. The C374S variant has binding constants of 12 and 13 nm for the oxidized and reduced states, respectively (Fig. 3), whereas the C355S/C374S variant has binding constants of 13 and 24 nm for the oxidized and reduced proteins, respectively (supplemental Fig. S2). Altogether, mass spectrometric and heme binding analyses demonstrate that disulfide bond formation between Cys-384 and Cys-374 is responsible for lower heme affinity of the oxidized Rev-erbβ LBD as compared with the reduced Rev-erbβ LBD.
FIGURE 3.
Loss of redox dependence of binding of Fe3+-heme to the C374S variant. Fe3+-heme titration curves of oxidized (A, 0. 3 μm) and reduced (B, 0.2 μm) C374S variants. The titrations were performed in Buffer B (see “Experimental Procedures”). Titration of reduced Rev-erbβ LBD variant with Fe3+-heme was performed anaerobically to avoid thiol-oxidation during the experiment. The lines were generated from fits to Equation 1.
A Ligand Switch Occurs upon Changing the Thiol Redox State
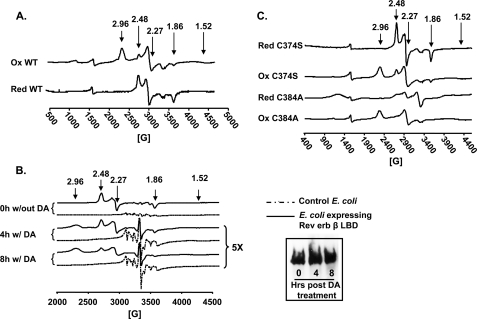
To further examine the nature of the ligand switch in the Rev-erbβ LBD predicted by the UV-visible absorption measurements (described above), X-band EPR experiments were performed of the Fe3+-heme-bound oxidized and reduced proteins (Fig. 4). The EPR spectrum of the anaerobically prepared complex of the reduced protein and Fe3+-heme manifests a rhombic spectrum with g values of 2.49, 2.27, and 1.86. These g values are consistent with His and Cys axial ligation to a low spin six-coordinate heme (Fig. 4A) (22). On the other hand, the predominant EPR spectrum of the complex between the oxidized protein and heme has g values (2.96, 2.27, and 1.52) that are characteristic of a low spin six-coordinate Fe3+-heme that is ligated by His and a neutral residue. The minor component (∼30%) consists of His/Cys ligation (g values of 2.49, 2.27, and 1.87) (Fig. 4A). The g values (2.96, 2.27, and 1.52) of the major species are fairly close to those assigned to a bis-His ligated heme (42); however, because the EPR spectra of His/Lys- or His/Met-coordinated heme are similar, the possibility of His/Met or His/Lys ligation cannot be excluded (43). When Cys-384 was mutated to Ala, as expected, the EPR spectrum of the oxidized C384A-heme complex exhibited pure His/neutral residue ligation (g values at 2.96, 2.27, and 1.52), with no detectable g = 2.48 peak. On the other hand, EPR experiments of the reduced C384A-heme complex (Fig. 4C) did reveal low amounts of a low spin EPR spectrum characteristic of His/Cys ligation (which might be provided from another unknown Cys) along with a g = 2.0 species of unknown origin.
FIGURE 4.
Ligand switching associated with a change in redox state of Rev-erbβ LBD. A, EPR spectroscopic analysis of heme complexes with the reduced (Red) and oxidized (Ox) proteins (complexes were prepared at a ratio of 1:1.5 of Fe3+-heme:LBD). B, EPR analysis of E. coli cells overexpressing Rev-erbβ LBD (without or with oxidation by 30 mm diamide (DA) for 4 and 8 h). Inset, Western blot analysis of the EPR samples from the diamide treatment at 0, 4, and 8 h using anti-pentahistidine antibody. C, EPR analysis of C384A-heme and C374S-heme complexes as in A. Note that the in vitro experiments were performed in Buffer B. The minor peaks at g values of 2.0 and 1.99 are from the buffer.
Assuming that the EPR spectrum with g values at 2.96, 2.27, and 1.5 originates from bis/His ligation, we assessed whether His-381, which is near Cys-384, might be the ligand that replaces Cys-384 in this ligand switch. However, as shown in supplemental Fig. S3, the EPR spectrum of the H381A variant is nearly identical to that of the wild-type protein, with no diminution in the g = 2.96 peak. To ensure that this His/neutral residue ligation is not due to an artifact resulting from the binding of one of the His residues in the His tag, we removed this tag by proteolytic cleavage with enterokinase and performed EPR experiments on the oxidized protein. The EPR spectrum of the oxidized tagless variant is identical to that of the wild-type protein (supplemental Fig. S3), demonstrating that the His tag does not affect the ligation state of the heme.
To further examine whether the LBD binds heme in vivo and whether the Cys-to-unknown neutral residue ligand switch occurs in growing cells, we performed whole cell EPR analyses on E. coli cells overexpressing the Rev-erbβ LBD. One might criticize the use of a bacterial system for these studies; however, use of the oxICAT methodology revealed that the redox state of human heme oxygenase-2 under normoxic growth conditions, as well as the response to oxidative and reductive conditions, was similar whether the protein was expressed in E. coli or in the human embryonic kidney cell line (HEK293) (44). Even though the E. coli cells were grown without heme precursor, the cells were amber-colored (supplemental Fig. S5C), indicating that overexpression of the human Rev-erbβ LBD induces heightened production of heme in E. coli. The whole cell EPR spectrum exhibited a single rhombic species with g values (g = 2.48, 2.27, and 1.86) identical to those obtained for the purified reduced LBD-Fe3+-heme complex (Fig. 4B, 0 h), confirming that heme binds to the LBD through His/Cys ligation. This spectrum is absent when cells lacking the overexpression construct are similarly treated. When cells were treated with 30 mm diamide, a partial switch from His/Cys to His/neutral residue ligation was observed. Initially, only the set of g values (2.48, 2.27, and 1.86) characteristic of His/Cys ligation was detected; then, after 30 min, the spectrum of a His/neutral residue-ligated species appeared (g = 2.96, 2.27, and 1.52). By 8 h, there was an equal mixture of the EPR spectra from the His/Cys- and His/neutral residue-ligated species (Fig. 4B and supplemental Fig. S4). The amount of Rev-erbβ LBD was unchanged throughout the experiment (Fig. 4B, inset). Thus, treatment of the cells with oxidants caused a Cys-to-neutral residue ligand switch, as observed for the purified protein. For control experiments, when either the H568R, H568A, or C384A variants were overexpressed, the cells did not turn red (supplemental Fig. S5C), and the whole cell EPR spectra revealed a pronounced g = 4.3 spectrum, without detectable amounts of low spin Fe3+-heme (supplemental Fig. S5A), even when the growth medium was supplemented with δ-amino levulinic acid (data not shown). Control E. coli cells not expressing Rev-erbβ also had the same peak (g = 4.3), although with lower intensity. The g = 4.3 EPR spectrum, which is characteristic of “extraneous” iron, probably derives from other iron proteins in the cell. Expression levels of the variants are similar to those of the wild-type Rev-erbβ LBD (supplemental Fig. S5B).
Because the mass spectrometric results (above) demonstrated a disulfide linkage between Cys-384 and Cys-374, we hypothesized that mutating Cys-374 would enable free Cys-384 to ligate heme in the oxidized state of the protein. Thus, EPR experiments of the heme-bound C374S variant were performed. To our surprise, the variant showed an EPR spectrum that was similar to that of the wild-type protein, both in the reduced and oxidized states, although the oxidized C374S variant exhibits slightly more Cys/His ligation (43%) than in the corresponding wild-type protein (29%) (Fig. 4C). The EPR spectra of the C355S/C374S variant are similar to those of C374S (data not shown).
Overall, mutation of Cys-374 resulted in the loss of the redox dependence of heme binding, but not redox-dependent ligand switching. This indicates that the oxidation of Cys-384 (resulting in a disulfide bond with Cys-374) is not solely responsible for the ligand switch. Perhaps oxidation of Cys-384 induces a conformational shift that moves Cys-384 away from the heme. This hypothesis is supported by the electronic absorption spectra (supplemental Fig. S6) of the oxidized wild-type, C384A, C374S, and C355S/C374S proteins. Similar absorption spectra of wild-type and cysteine variants indicate similar heme ligation states (i.e. His/neutral residue) in the oxidized forms of these variants and the wild-type protein (supplemental Fig. S6).
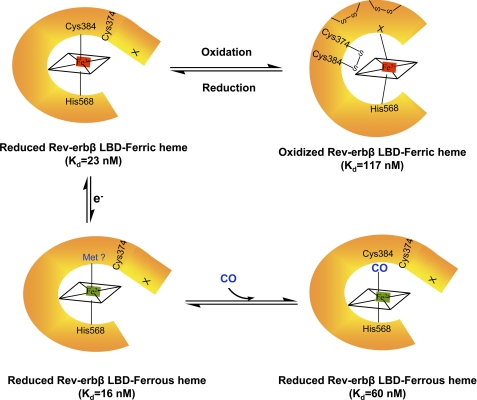
However, unlike the wild-type protein, the oxidized and reduced states of the C374S variant bind heme with similar affinities. Therefore, it is the redox switch, not the ligand switch, that alters the heme binding affinity. It is plausible that a conformational change in the LBD, resulting from oxidation of Cys-384, might block the heme-binding site, thus decreasing affinity of the oxidized protein for heme (see Fig. 7).
FIGURE 7.
Model for redox modulation of heme binding and heme ligand switching in Rev-erbβ. Ferric heme is ligated via Cys-384 and His-568 in the ligand-binding domain of the thiol-reduced form of Rev-erbβ. Disulfides, formed after the oxidation of the protein, change the conformation of the protein, which results in His/neutral residue ligation (His/His or His/Met or His/Lys) of the heme. Formation of the disulfide bond between Cys-374 and Cys-384 interferes with heme binding. The LBD can bind ferrous heme with similar affinity to the ferric heme. In this state, Cys-384 appears to be replaced by a neutral ligand, suggested to be Met (45, 46). CO binds tightly to the ferrous state of Rev-erbβ, replacing the sulfur ligation from Cys-384.
pH-dependent Ligand Switch in the Oxidized Rev-erbβ LBD
Apart from the redox-dependent ligand switch, the oxidized protein shows a pH-dependent switch from His/neutral residue to His/Cys ligation. At increasing pH values, the Soret peak of the wild-type oxidized protein showed a red shift from 416 to 424 nm along with a significant decrease in intensity from pH 8 to 10 (supplemental Fig. S7A), and the color of the complex changed from dark red to green over the pH range from pH 8 to 10. The titrations could not be performed at lower pH values, because the protein precipitated below pH 7. Similar observations were made for the C374S variant (supplemental Fig. S7C). Similarly, EPR spectroscopic results indicate a ligand switch from His/neutral residue to His/Cys ligation above pH 10 for wild-type LBD (supplemental Fig. S7B) and for the C374S (supplemental Fig. S7D) variant. It is plausible that a base-induced cleavage of the Cys-374–Cys-384 disulfide bond occurs at pH 10, which releases Cys-384 for ligation to the heme.
As the pH is increased, a minor amount of high spin heme (g = ∼6.2) is observed, as seen earlier with cytochrome b561 (42). In addition, small amounts of a low spin species with a g value at 2.68 appears, which could result from His/water ligation (29). Furthermore, a peak at g = 4.3 is observed that is likely to reflect heme decomposition.
No Effect of Change in Iron Redox State on Heme Binding Affinity
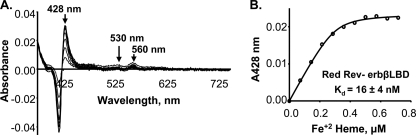
Pardee et al. (11) recently showed that Rev-erbβ is capable of binding both Fe3+- and Fe2+-heme. We performed Fe2+-heme titrations to determine whether the reduced protein prefers Fe3+- versus Fe2+-heme. All of the titrations with Fe2+-heme were performed in anaerobically closed vials in the presence of 2.5 mm sodium dithionite to prevent oxidation of the Fe2+-heme to Fe3+. The absorption spectrum of Fe2+-heme-reduced Rev-erbβ LBD complex is shown in supplemental Fig. S1. The spectrum has a Soret band at 428 nm and alpha and beta bands at 560 and 530 nm, respectively, indicating that the Cys ligand is released from the Fe2+-heme (45, 46). Similarly, the difference spectrum of the complex between the reduced Rev-erbβ LBD and Fe2+-heme exhibited a Soret peak at 428 nm along with the alpha (560 nm) and beta (530 nm) bands (Fig. 5A). Titration of the reduced Rev-erbβ LBD with Fe2+-heme resulted in a sharply saturating binding isotherm (Fig. 5B), which, when fit to the quadratic equation (Equation 1), gave a Kd value in the low nanomolar range (Kd = 15.8 ± 4.1 nm). This Kd value is similar to that obtained in the titration of the reduced form of the Rev-erbβ LBD with Fe3+-heme (above). Thus, the reduced protein exhibits similar high affinity for Fe2+- and Fe3+-heme. Because dithionite, which is used to reduce the heme, also reduces the disulfide groups in the LBD, it is not possible to determine the Kd of the oxidized protein for Fe2+-heme.
FIGURE 5.
Similar affinity of reduced Rev-erbβ LBD toward Fe3+- and Fe2+-heme. The Rev-erbβ LBD (0. 3 μm) was anaerobically titrated with Fe2+-heme in Buffer B in the presence of 2.5 mm dithionite. The Soret peak for Fe2+-heme is at 428 nm and exhibits sharp alpha and beta peaks at 560 and 530 nm, respectively (A). The Kd values of the complexes of Fe3+- and Fe2+-heme with reduced Rev-erbβ LBD are 23 ± 2.7 nm (Fig. 1D) and 15.8 ± 4.1 nm (Fig. 5B), respectively. The line in B was generated from fits to Equation 1.
Magnetic circular dichroism and resonance Raman spectroscopic experiments have revealed that the Fe2+-heme bound to Rev-erbβ can exist in a five-coordinate environment with a single neutral ligand as well as a six-coordinate state with two neutral ligands (22). His-568 has been postulated to remain as an axial ligand in the Fe2+ state. To further examine the ligand environment of Fe2+-heme, we determined the affinity of the H568R variant for Fe2+-heme. Substitution of His-568 for Arg severely diminished the affinity of the Rev-erbβ LBD for Fe2+ heme (Kd = 1.52 ± 0.49 μm). On the other hand, mutation of Cys-384 to Ala had a more modest effect on the affinity (Kd = 210 ± 30 nm) for Fe2+-heme (Table 3). These observations lend further support for the conclusion that in the Fe2+ state, His-568 remains as an axial ligand, whereas Cys-384 ligation might be lost. Dissociation of a thiolate anionic ligand upon reduction of Fe3+-heme is a fairly common phenomenon (47).
Tight CO Binding to Rev-erbβ LBD
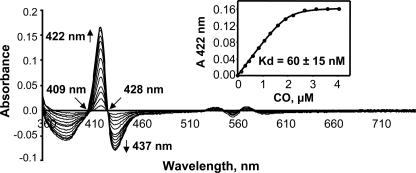
CO is an important signaling molecule and has been shown to interact in vitro with the Fe2+ heme-Rev-erbβ complex, probably by replacing Cys as an axial ligand. However, it was stated that no significant interaction was measured by in vivo experiments (although details were not provided) (11). To quantify the interaction between CO and the LBD, we performed CO titrations. The difference spectra are shown in Fig. 6, and the real spectra are presented in supplemental Fig. S8. The addition of CO to the Rev-erbβ LBD resulted in a peak shift from 428 to 422 nm. The data obtained from the titration were subjected to quadratic analysis (Equation 1), which showed very high affinity of the LBD for CO (Kd = 60 ± 15 nm).
FIGURE 6.
High affinity of CO toward Fe2+ heme- Rev-erbβ LBD complex. Difference spectra following the binding of CO to the Fe2+-heme complex with the Rev-erbβ LBD. Inset, fitting the binding isotherm for the CO titration yielded a Kd value of 60 ± 15 nm. The CO titration was performed anaerobically in Buffer B in the presence of 2.5 mm dithionite with 2 μm Fe2+-heme-LBD complex, as described under “Experimental Procedures.” The line shown in the inset was generated from a fit to Equation 1.
DISCUSSION
Rev-erbα and Rev-erbβ are heme-binding nuclear receptors that are highly expressed in the brain, liver, adipose tissue, and skeletal muscle. The Rev-erbs exhibit circadian patterns of expression and play important roles in controlling the expression of genes constituting the primary transcriptional-translational feedback loop that regulates the vertebrate circadian clock (48, 49). This primary negative feedback loop involves a highly conserved suite of proteins (CLOCK, the CLOCK paralog NPAS2, BMAL1, Per1, Per2, Cry1, and Cry2) that are expressed within the suprachiasmatic nucleus of the hypothalamus, the central circadian pacemaker in humans, as well as in peripheral tissues that are subject to circadian modulation. In this primary feedback loop, the CLOCK-BMAL1 heterodimer serves as a positive acting transcription factor that activates expression of the Cry and Per proteins, which feed back to repress expression of BMAL1. In a secondary negative feedback loop, Rev-erb represses the expression of BMAL1 by binding to two ROR response elements in the BMAL1 promoter, yet expression of Rev-erb is activated by the CLOCK-BMAL1 heterodimer, which binds to E-boxes within the Rev-erb promoter (14, 50, 51). Transcriptional regulation through the primary and secondary circadian loops, coupled to post-translational modification and degradation of circadian clock proteins (52, 53), synchronizes to the daily light-dark cycle a wide range of metabolic and behavioral processes, including the sleep-wake cycle, feeding behavior, body temperature, and blood hormone levels (49).
Heme, which has been shown to regulate processes as diverse as enzyme and ion channel activity, DNA binding, signal transduction, and protein complex assembly (54), is linked to the circadian cycle at several levels. Like Rev-erb, heme exhibits circadian patterns of expression, at least in part because of circadian regulation of δ-amino levulinic acid synthase, the rate-limiting enzyme in heme synthesis (20). Heme also modulates the activity of multiple nuclear receptors such as Hap1, HRI kinase, elF2α (54), and several key players involved in the circadian rhythm, including Rev-erbα and Rev-erbβ (8, 10, 19, 55). Heme binding to Rev-erb is required for efficient nuclear corepressor recruitment; thus, alterations in intracellular heme levels modulate the expression of BMAL1 in a manner suggesting that heme stabilizes the Rev-erb-nuclear corepressor complex (14).
The intracellular redox state has also been proposed to entrain the circadian oscillator by modulating the ability of the CLOCK-BMAL1 and NPAS-BMAL1 heterodimers to bind DNA (48). One mechanism of redox regulation of gene expression is by thiol-based redox modification of nuclear receptors (56). For example, upon treatment with thiol-oxidizing reagents, the ligand binding activity of the glucocorticoid receptor is severely impaired but is restored by overexpression of thioredoxin (56, 57).
One mechanism by which proteins link redox- and heme-dependent regulation is by thiol-disulfide regulation of heme binding. For example, human heme oxygenase-2 and the BK channel contain redox switches that undergo reversible thiol-disulfide interconversion, with different heme binding affinities in the oxidized disulfide state and the reduced state (29, 44, 58). The heme regulatory motif consists of a conserved Cys-Pro core sequence that is usually flanked at the N terminus by basic amino acids and at the C terminus by a hydrophobic residue (59–61). Given that the LBD of Rev-erbβ contains an heme regulatory motif (HLVC384PMSK), in which Cys-384 is a heme ligand (11), we hypothesized that Cys-384 may be involved in a ligand switch in which oxidation of this residue would abolish (or reduce) binding of Fe3+-heme. Because heme binding to Rev-erb regulates the expression of target genes (8, 10, 11), such a redox and/or ligand switch could help explain how intracellular redox state and heme metabolism can control the circadian cycle.
Our combined mutagenesis, spectroscopic (EPR and UV-visible), and mass spectrometric experiments provide novel insights into redox- and ligand-based changes in the LBD and lend support to the conclusions based on previous resonance Raman and crystallographic experiments that Cys-384 and His-568 can serve as the axial ligands to the heme in Rev-erbβ (11, 22). Based on heme titrations, the reduced state of the Rev-erbβ LBD binds Fe3+-heme (Kd = 23 nm) with ∼5-fold greater affinity than does the oxidized protein (Kd = 117 nm). This change in heme affinity appears to be associated with the reduction of the disulfide state of Cys-384 (linked to Cys-374) to the free thiolate state. With such a redox-coupled ligand switch, the Cys residue would only be available for heme ligation under reducing conditions within the cell. Mutagenesis studies further indicate that Cys-384 is redox active, because the EPR spectra of the oxidized C384A variant, which has only His/neutral residue ligation to the heme, are similar to those of the diamide-oxidized wild-type Rev-erb β LBD. It is likely that His-568 remains as one of the heme ligands in the oxidized protein, because mutation of His-568 to either Ala or Arg markedly lowered heme binding affinity (Table 3). To attempt to identify the other neutral residue that binds heme in the oxidized state, we performed EPR studies of the residue that seemed most likely to be involved in the ligand switch, e.g. His-381, based on the crystal structure of the protein (11). However, the H381A variant exhibits an EPR spectrum that superimposes with that of the wild-type protein. Thus, the identity of the other neutral residue that can bind heme in the oxidized state of the protein is unknown.
The heme titrations quantitatively demonstrate that the reduced state of the purified LBD tightly binds heme at the low nanomolar concentrations of free heme present within the cell, and the in vivo EPR and mutagenesis experiments confirm that the LBD tightly binds heme within growing E. coli cells. We used E. coli cells for these studies because obtaining sufficient numbers of mammalian cells for such an experiment is prohibitive, and at least from a redox perspective, the intracellular redox potential of E. coli is approximately −250 mV (62, 63), which is within the range of the ambient redox potential of mammalian cells, from −170 to −325 mV (64, 65). Furthermore, the ratios of oxidized/reduced thiols are similar whether heme oxygenase-2 is expressed under different redox conditions in mammalian or E. coli cells (44).
That the LBD binds heme with high affinity is also indicated by observation of the amber color of E. coli cells that are overexpressing the LBD in the absence or presence of the heme precursor, δ-amino levulinic acid. Similarly, when cytochrome b and NPAS2 are overexpressed, the bacterial cells exhibit the characteristic amber color of heme (55, 66). Earlier studies on heme binding to the Rev-erbβ LBD have yielded Kd values of 2 and 6 μm (8, 11), which are problematic because these values are much higher than the physiological concentration of free heme (<100 nm) (67). Based on a 2 μm Kd value, at 100 nm free heme, ∼95% of the Rev-erb population would be in the apo form. However, based on the published UV-visible spectra (including Soret peaks for the Fe3+-heme-bound 415–420 nm) (8, 9, 11, 22), the previous studies were performed with a mixture of oxidized and reduced LBD. Our results also indicate that, in addition to the potential problems associated with working with mixed redox states of the LBD, the very high Kd values reported in prior studies result from protein aggregation at the high concentrations used in those experiments. Here, we show heme binding to the Rev-erbβ LBD at physiologically relevant heme concentrations and that heme affinity is redox-regulated.
The EPR spectrum of the LBD-bound heme is easily observed in E. coli cells, and the g values, which are fingerprints for the heme ligation state, can be readily assigned. Thus, the mutagenesis and in vivo EPR experiments demonstrate that the switch between His/neutral residue and His/Cys coordination environments observed for the purified oxidized and reduced proteins also occurs within cells. It appears likely that the coordination environment is governed by the ambient intracellular redox potential.
The studies described here also explain the discrepancy noted by De Rosny et al. (68), i.e. that although His/Cys ligation was observed in the crystal structure of Rev-erbβ LBD, the Soret peak is unexpectedly at 415 nm. Our results show that the fully oxidized Fe3+-protein exhibits a Soret peak at 416 nm, whereas this band for the reduced ferric protein is observed at 424 nm, which is characteristic of His/Cys ligation (68, 69). Therefore, the crystal structure of the TCEP-incubated protein seems to have trapped one of the two stable states of the protein, i.e. the reduced state, whereas the His/neutral residue state is responsible for the 415-nm band.
A novel aspect of the ligand switch between the His/neutral residue and His/Cys states of the Fe3+-heme is that it is regulated by the change in redox state of thiols in Rev-erbβ. This ligand switch in Rev-erbβ also can be brought about by changes in the redox state of the heme (22), a property that is commonly observed when the iron undergoes redox changes between the Fe3+and Fe2+ states (69–72). In Rev-erbβ, the reduced LBD binds Fe2+- and Fe3+-heme with similar affinity (16 and 23 nm, respectively). Generally the anionic thiolate ligand is replaced upon reduction of the ferric ion (47). Based on the earlier experiments (11), it is possible that Fe2+-heme is ligated via His only; however, the UV-visible spectrum cannot rule out the possibility of His/Met ligation (45, 46). Although the physiological relevance of Fe2+-heme binding is not very well established (73), it is clear that replacement of a strong ligand (like thiolate) by a weak ligand facilitates reduction of the iron and promotes binding of extrinsic axial ligands, like CO or NO (Fig. 7).
We propose that CO and/or NO binding to Rev-erb may help explain the signaling role of CO in the circadian clock (74). Although previous studies have shown that CO can interact with Rev-erbβ (11, 22), the high affinity (60 nm) for CO demonstrated here emphasizes the potential role for CO in regulating the properties of Rev-erbβ. In comparison, NPAS2 (Kd = 1 μm) and a bacterial CO sensor (CooA) (Kd = 11 μm) bind CO with significantly lower affinity (55, 75).
Redox-mediated ligand switching allows adjustment of the activity of heme proteins in response to changes in the ambient redox potential within the cell, which occur during oxidative stress or conversely hypoxia. Oxidative stress, a factor in many important diseases (76–79), is correlated with disruptions in the circadian rhythm (80, 81). Oxidative stress may also play a role in different diseases, such as shift work sleep disorder, originating from imbalance in circadian rhythm (81). Because Rev-erbβ is considered to link the circadian cycle and metabolism (12), we propose that under more reducing intracellular redox conditions, Rev-erbβ (containing the reduced thiolate form of Cys-384) is in its heme-bound state, which is optimal for recruitment of nuclear corepressor and repression of target genes. When subjected to oxidative stress conditions, Cys-384 of Rev-erbβ undergoes oxidation to form a disulfide bridge with Cys-374, also triggering a ligand switch that results in release of bound heme and derepression of target genes. Furthermore, the tight binding of CO offers yet another possible layer of regulating Rev-erb activity. These redox- and ligand-dependent changes could then trigger alterations in circadian patterns and, if not reversed, result in circadian and/or metabolic disorders. The effects of heme, redox, and intrinsic and extrinsic (CO and NO) ligands on RevErb would likely be complemented by those on other heme proteins with key roles in the circadian cycle, such as NPAS2 and Per2 (82, 83).
Supplementary Material
Acknowledgments
We thank Dr. Thomas P. Burris (Scripps Research Institute, Jupiter, FL) for providing the pET-46Ek/LIC::Rev-erbβ LBD construct. We are also thankful to Dr. Henriette A. Remmer and Amber Peariso from University of Michigan Protein Structure Facility for performing the mass spectrometry analyses.
This work was supported, in whole or in part, by National Institutes of Health Grants R21 HL089837 and R01 HL102662.

The on-line version of this article (available at http://www.jbc.org) contains supplemental text and Figs. S1–S8.
- LBD
- ligand-binding domain
- TCEP
- tris(2-carboxyethyl)phosphine
- HDAC
- histone deacetylase
- MES
- 4-morpholineethanesulfonic acid.
REFERENCES
- 1. Bain D. L., Heneghan A. F., Connaghan-Jones K. D., Miura M. T. (2007) Annu. Rev. Physiol. 69, 201–220 [DOI] [PubMed] [Google Scholar]
- 2. Olefsky J. M. (2001) J. Biol. Chem. 276, 36863–36864 [DOI] [PubMed] [Google Scholar]
- 3. Downes M., Burke L. J., Bailey P. J., Muscat G. E. (1996) Nucleic Acids Res. 24, 4379–4386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Retnakaran R., Flock G., Giguère V. (1994) Mol. Endocrinol. 8, 1234–1244 [DOI] [PubMed] [Google Scholar]
- 5. Duez H., Staels B. (2008) FEBS Lett. 582, 19–25 [DOI] [PubMed] [Google Scholar]
- 6. Terenzi H., Cassia R. O., Zakin M. M. (1996) Protein Expr. Purif. 8, 313–318 [DOI] [PubMed] [Google Scholar]
- 7. Reinking J., Lam M. M., Pardee K., Sampson H. M., Liu S., Yang P., Williams S., White W., Lajoie G., Edwards A., Krause H. M. (2005) Cell 122, 195–207 [DOI] [PubMed] [Google Scholar]
- 8. Raghuram S., Stayrook K. R., Huang P., Rogers P. M., Nosie A. K., McClure D. B., Burris L. L., Khorasanizadeh S., Burris T. P., Rastinejad F. (2007) Nat. Struct. Mol. Biol. 14, 1207–1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yin L., Wu N., Curtin J. C., Qatanani M., Szwergold N. R., Reid R. A., Waitt G. M., Parks D. J., Pearce K. H., Wisely G. B., Lazar M. A. (2007) Science 318, 1786–1789 [DOI] [PubMed] [Google Scholar]
- 10. Yin L., Lazar M. A. (2005) Mol. Endocrinol. 19, 1452–1459 [DOI] [PubMed] [Google Scholar]
- 11. Pardee K. I., Xu X., Reinking J., Schuetz A., Dong A., Liu S., Zhang R., Tiefenbach J., Lajoie G., Plotnikov A. N., Botchkarev A., Krause H. M., Edwards A. (2009) PLoS Biol. 7, e43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ramakrishnan S. N., Muscat G. E. (2006) Nucl. Recept. Signal. 4, e009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kovac J., Husse J., Oster H. (2009) Mol. Cells 28, 75–80 [DOI] [PubMed] [Google Scholar]
- 14. Burris T. P. (2008) Mol. Endocrinol. 22, 1509–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang J., Liu N., Liu Z., Li Y., Song C., Yuan H., Li Y. Y., Zhao X., Lu H. (2008) Biochim. Biophys. Acta 1783, 224–236 [DOI] [PubMed] [Google Scholar]
- 16. Wang J., Li Y., Zhang M., Liu Z., Wu C., Yuan H., Li Y. Y., Zhao X., Lu H. (2007) FEBS J. 274, 5370–5381 [DOI] [PubMed] [Google Scholar]
- 17. Ramakrishnan S. N., Lau P., Crowther L. M., Cleasby M. E., Millard S., Leong G. M., Cooney G. J., Muscat G. E. (2009) Biochem. Biophys. Res. Commun. 388, 654–659 [DOI] [PubMed] [Google Scholar]
- 18. Boehning D., Snyder S. H. (2002) Science 298, 2339–2340 [DOI] [PubMed] [Google Scholar]
- 19. Yang J., Kim K. D., Lucas A., Drahos K. E., Santos C. S., Mury S. P., Capelluto D. G., Finkielstein C. V. (2008) Mol. Cell. Biol. 28, 4697–4711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kaasik K., Lee C. C. (2004) Nature 430, 467–471 [DOI] [PubMed] [Google Scholar]
- 21. de Rosny E., de Groot A., Jullian-Binard C., Gaillard J., Borel F., Pebay-Peyroula E., Fontecilla-Camps J. C., Jouve H. M. (2006) Biochemistry 45, 9727–9734 [DOI] [PubMed] [Google Scholar]
- 22. Marvin K. A., Reinking J. L., Lee A. J., Pardee K., Krause H. M., Burstyn J. N. (2009) Biochemistry 48, 7056–7071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ignarro L. J. (2000) Nitric Oxide: Biology and Pathobiology, pp. 386–387, Academic Press, San Diego, CA [Google Scholar]
- 24. Wu L., Wang R. (2005) Pharmacol. Rev. 57, 585–630 [DOI] [PubMed] [Google Scholar]
- 25. Berry E. A., Trumpower B. L. (1987) Anal. Biochem. 161, 1–15 [DOI] [PubMed] [Google Scholar]
- 26. Gupta N., Ragsdale S. W. (2008) J. Biol. Chem. 283, 28721–28728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bulaj G., Kortemme T., Goldenberg D. P. (1998) Biochemistry 37, 8965–8972 [DOI] [PubMed] [Google Scholar]
- 28. Ellman G. L. (1958) Arch. Biochem. Biophys. 74, 443–450 [DOI] [PubMed] [Google Scholar]
- 29. Yi L., Ragsdale S. W. (2007) J. Biol. Chem. 282, 21056–21067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kuzelová K., Mrhalová M., Hrkal Z. (1997) Biochim. Biophys. Acta 1336, 497–501 [DOI] [PubMed] [Google Scholar]
- 31. de Villiers K. A., Kaschula C. H., Egan T. J., Marques H. M. (2007) J. Biol. Inorg. Chem. 12, 101–117 [DOI] [PubMed] [Google Scholar]
- 32. Garrick M. D., Scott D., Kulju D., Romano M. A., Dolan K. G., Garrick L. M. (1999) Biochim. Biophys. Acta 1449, 125–136 [DOI] [PubMed] [Google Scholar]
- 33. Liu S. C., Zhai S., Palek J. (1988) Blood 71, 1755–1758 [PubMed] [Google Scholar]
- 34. Sassa S. (2006) J. Clin. Biochem. Nutr. 38, 138–155 [Google Scholar]
- 35. Leichert L. I., Gehrke F., Gudiseva H. V., Blackwell T., Ilbert M., Walker A. K., Strahler J. R., Andrews P. C., Jakob U. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 8197–8202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yang J., Ishimori K., O'Brian M. R. (2005) J. Biol. Chem. 280, 7671–7676 [DOI] [PubMed] [Google Scholar]
- 37. Gygi S. P., Rist B., Gerber S. A., Turecek F., Gelb M. H., Aebersold R. (1999) Nat. Biotechnol. 17, 994–999 [DOI] [PubMed] [Google Scholar]
- 38. Ellis K. J., Morrison J. F. (1982) Methods Enzymol. 87, 405–426 [DOI] [PubMed] [Google Scholar]
- 39. Antonini E., Brunori M. (1971) Hemoglobin and Myoglobin in Their Rections with Ligands, pp. 43–47, North Holland Publishing Co., London [Google Scholar]
- 40. Igarashi J., Kitanishi K., Martinkova M., Murase M., Iizuka A., Shimizu T. (2008) Acta Chim. Slov. 55, 67–74 [Google Scholar]
- 41. Pop S. M., Gupta N., Raza A. S., Ragsdale S. W. (2006) J. Biol. Chem. 281, 26382–26390 [DOI] [PubMed] [Google Scholar]
- 42. Tsubaki M., Nakayama M., Okuyama E., Ichikawa Y., Hori H. (1997) J. Biol. Chem. 272, 23206–23210 [DOI] [PubMed] [Google Scholar]
- 43. Cheesman M. R., Little P. J., Berks B. C. (2001) Biochemistry 40, 10562–10569 [DOI] [PubMed] [Google Scholar]
- 44. Yi L., Jenkins P. M., Leichert L. I., Jakob U., Martens J. R., Ragsdale S. W. (2009) J. Biol. Chem. 284, 20556–20561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ito S., Igarashi J., Shimizu T. (2009) J. Inorg. Biochem. 103, 1380–1385 [DOI] [PubMed] [Google Scholar]
- 46. Kooter I. M., Moguilevsky N., Bollen A., van der Veen L. A., Otto C., Dekker H. L., Wever R. (1999) J. Biol. Chem. 274, 26794–26802 [DOI] [PubMed] [Google Scholar]
- 47. Perera R., Sono M., Sigman J. A., Pfister T. D., Lu Y., Dawson J. H. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 3641–3646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rutter J., Reick M., McKnight S. L. (2002) Annu. Rev. Biochem. 71, 307–331 [DOI] [PubMed] [Google Scholar]
- 49. Takahashi J. S., Hong H. K., Ko C. H., McDearmon E. L. (2008) Nat. Rev. Genet 9, 764–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sato T. K., Panda S., Miraglia L. J., Reyes T. M., Rudic R. D., McNamara P., Naik K. A., FitzGerald G. A., Kay S. A., Hogenesch J. B. (2004) Neuron 43, 527–537 [DOI] [PubMed] [Google Scholar]
- 51. Preitner N., Damiola F., Lopez-Molina L., Zakany J., Duboule D., Albrecht U., Schibler U. (2002) Cell 110, 251–260 [DOI] [PubMed] [Google Scholar]
- 52. Lee C., Etchegaray J. P., Cagampang F. R., Loudon A. S., Reppert S. M. (2001) Cell 107, 855–867 [DOI] [PubMed] [Google Scholar]
- 53. Shirogane T., Jin J., Ang X. L., Harper J. W. (2005) J. Biol. Chem. 280, 26863–26872 [DOI] [PubMed] [Google Scholar]
- 54. Mense S. M., Zhang L. (2006) Cell Res. 16, 681–692 [DOI] [PubMed] [Google Scholar]
- 55. Dioum E. M., Rutter J., Tuckerman J. R., Gonzalez G., Gilles-Gonzalez M. A., McKnight S. L. (2002) Science 298, 2385–2387 [DOI] [PubMed] [Google Scholar]
- 56. Tanaka H., Makino Y., Okamoto K., Iida T., Yoshikawa N., Miura T. (2000) Oncology 59, (Suppl. 1) 13–18 [DOI] [PubMed] [Google Scholar]
- 57. Makino Y., Okamoto K., Yoshikawa N., Aoshima M., Hirota K., Yodoi J., Umesono K., Makino I., Tanaka H. (1996) J. Clin. Invest. 98, 2469–2477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Yi L., Morgan J. T., Ragsdale S. W. (2010) J. Biol. Chem. 285, 20117–20127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zhang L., Guarente L. (1995) EMBO J. 14, 313–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Steiner H., Kispal G., Zollner A., Haid A., Neupert W., Lill R. (1996) J. Biol. Chem. 271, 32605–32611 [DOI] [PubMed] [Google Scholar]
- 61. McCoubrey W. K., Jr., Huang T. J., Maines M. D. (1997) J. Biol. Chem. 272, 12568–12574 [DOI] [PubMed] [Google Scholar]
- 62. Ostergaard H., Henriksen A., Hansen F. G., Winther J. R. (2001) EMBO J. 20, 5853–5862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Taylor M. F., Boylan M. H., Edmondson D. E. (1990) Biochemistry 29, 6911–6918 [DOI] [PubMed] [Google Scholar]
- 64. Dooley C. T., Dore T. M., Hanson G. T., Jackson W. C., Remington S. J., Tsien R. Y. (2004) J. Biol. Chem. 279, 22284–22293 [DOI] [PubMed] [Google Scholar]
- 65. Jones D. P. (2002) Methods Enzymol. 348, 93–112 [DOI] [PubMed] [Google Scholar]
- 66. Rivera M., Walker F. A. (1995) Anal. Biochem. 230, 295–302 [DOI] [PubMed] [Google Scholar]
- 67. Ogawa K., Sun J., Taketani S., Nakajima O., Nishitani C., Sassa S., Hayashi N., Yamamoto M., Shibahara S., Fujita H., Igarashi K. (2001) EMBO J. 20, 2835–2843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. de Rosny E., de Groot A., Jullian-Binard C., Borel F., Suarez C., Le Pape L., Fontecilla-Camps J. C., Jouve H. M. (2008) Biochemistry 47, 13252–13260 [DOI] [PubMed] [Google Scholar]
- 69. Marvin K. A., Kerby R. L., Youn H., Roberts G. P., Burstyn J. N. (2008) Biochemistry 47, 9016–9028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kurokawa H., Lee D. S., Watanabe M., Sagami I., Mikami B., Raman C. S., Shimizu T. (2004) J. Biol. Chem. 279, 20186–20193 [DOI] [PubMed] [Google Scholar]
- 71. Inagaki S., Masuda C., Akaishi T., Nakajima H., Yoshioka S., Ohta T., Pal B., Kitagawa T., Aono S. (2005) J. Biol. Chem. 280, 3269–3274 [DOI] [PubMed] [Google Scholar]
- 72. Ishitsuka Y., Araki Y., Tanaka A., Igarashi J., Ito O., Shimizu T. (2008) Biochemistry 47, 8874–8884 [DOI] [PubMed] [Google Scholar]
- 73. Igarashi J., Murase M., Iizuka A., Pichierri F., Martinkova M., Shimizu T. (2008) J. Biol. Chem. 283, 18782–18791 [DOI] [PubMed] [Google Scholar]
- 74. Artinian L. R., Ding J. M., Gillette M. U. (2001) Exp. Neurol. 171, 293–300 [DOI] [PubMed] [Google Scholar]
- 75. Yamashita T., Hoashi Y., Watanabe K., Tomisugi Y., Ishikawa Y., Uno T. (2004) J. Biol. Chem. 279, 21394–21400 [DOI] [PubMed] [Google Scholar]
- 76. Uttara B., Singh A. V., Zamboni P., Mahajan R. T. (2009) Curr. Neuropharmacol. 7, 65–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Watanabe K., Thandavarayan R. A., Gurusamy N., Zhang S., Muslin A. J., Suzuki K., Tachikawa H., Kodama M., Aizawa Y. (2009) Acta Physiol. Hung. 96, 277–287 [DOI] [PubMed] [Google Scholar]
- 78. Shi P., Gal J., Kwinter D. M., Liu X., Zhu H. (2010) Biochim. Biophys. Acta 1802, 45–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Elswaifi S. F., Palmieri J. R., Hockey K. S., Rzigalinski B. A. (2009) Infect. Disord. Drug Targets 9, 445–452 [DOI] [PubMed] [Google Scholar]
- 80. Hardeland R., Coto-Montes A., Poeggeler B. (2003) Chronobiol. Int. 20, 921–962 [DOI] [PubMed] [Google Scholar]
- 81. Sharifian A., Farahani S., Pasalar P., Gharavi M., Aminian O. (2005) J. Circadian. Rhythms. 3, 15–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Kitanishi K., Igarashi J., Hayasaka K., Hikage N., Saiful I., Yamauchi S., Uchida T., Ishimori K., Shimizu T. (2008) Biochemistry 47, 6157–6168 [DOI] [PubMed] [Google Scholar]
- 83. Uchida T., Sato E., Sato A., Sagami I., Shimizu T., Kitagawa T. (2005) J. Biol. Chem. 280, 21358–21368 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.