Abstract
Pyroglutamate-modified Aβ (AβpE3–42) peptides are gaining considerable attention as potential key players in the pathology of Alzheimer disease (AD) due to their abundance in AD brain, high aggregation propensity, stability, and cellular toxicity. Overexpressing AβpE3–42 induced a severe neuron loss and neurological phenotype in TBA2 mice. In vitro and in vivo experiments have recently proven that the enzyme glutaminyl cyclase (QC) catalyzes the formation of AβpE3–42. The aim of the present work was to analyze the role of QC in an AD mouse model with abundant AβpE3–42 formation. 5XFAD mice were crossed with transgenic mice expressing human QC (hQC) under the control of the Thy1 promoter. 5XFAD/hQC bigenic mice showed significant elevation in TBS, SDS, and formic acid-soluble AβpE3–42 peptides and aggregation in plaques. In 6-month-old 5XFAD/hQC mice, a significant motor and working memory impairment developed compared with 5XFAD. The contribution of endogenous QC was studied by generating 5XFAD/QC-KO mice (mouse QC knock-out). 5XFAD/QC-KO mice showed a significant rescue of the wild-type mice behavioral phenotype, demonstrating the important contribution of endogenous mouse QC and transgenic overexpressed QC. These data clearly demonstrate that QC is crucial for modulating AβpE3–42 levels in vivo and prove on a genetic base the concept that reduction of QC activity is a promising new therapeutic approach for AD.
Keywords: Alzheimer Disease, Amyloid, Brain, Enzymes, Mouse Genetics, Transgenic
Introduction
Alzheimer disease (AD)4 is a progressive neurodegenerative disorder characterized by the presence of extracellular amyloid plaques composed of amyloid-β (Aβ) and intracellular neurofibrillary tangles. The discovery that certain early onset familial forms of AD may be caused by enhanced levels of Aβ peptides has led to the hypothesis that amyloidogenic Aβ is intimately involved in the pathogenic process (1).
Besides full-length Aβ 40 and 42 isoforms starting with an aspartate at position 1, a variety of different N-truncated Aβ peptides have been identified in AD brains. Ragged peptides including phenylalanine at position 4 of Aβ have been reported as early as 1985 by Masters et al. (2). In contrast, no N-terminal sequence could be obtained from cores purified in a SDS-containing buffer, which led to the assumption that the N terminus could be blocked (3, 4).
The presence of AβpE3 (N-terminally truncated Aβ starting with pyroglutamate) in AD brain was subsequently shown using mass spectrometry of purified Aβ peptides, explaining at least partially initial difficulties in sequencing Aβ peptides purified from human brain tissue (5). The authors reported that only 10–15% of the total Aβ isolated by this method begins at position 3 with AβpE3. Saido et al. (6) and others (7) subsequently showed that AβpE3 represents a dominant fraction of Aβ peptides in AD brain.
Overexpression of AβpE3–42 in neurons of TBA2 transgenic mice triggers neuron loss and an associated neurological phenotype (8). N-terminal pE formation can be catalyzed by glutaminyl cyclase (QC) and is pharmacologically inhibited by QC inhibitors, both in vitro (9) and in vivo (10). Moreover, QC expression was found up-regulated in the cortex of patients with AD and correlated with the appearance of pE-modified Aβ. Oral application of a QC inhibitor resulted in reduced AβpE3–42 burden in two different transgenic mouse models of AD as well as in a transgenic Drosophila model. Interestingly, treatment of these mice was accompanied by reductions in Aβx-40/42, diminished plaque formation and gliosis, as well as improved performance in context memory and spatial learning tests (10). Thus, AβpE3–42 reduction is a promising target for therapy of AD. In the current work, the contribution of QC was studied for the first time using genetic means by human QC overexpression and endogenous QC-knock-out in an AD mouse model.
EXPERIMENTAL PROCEDURES
Transgenic and Knock-out Mice
5XFAD (11) mice have been described previously. All mice were backcrossed for more than 10 generations on a C57BL/6J genetic background and housed at a 12-h day/12-h night cycle with free access to food and water. For generation of hQC transgenic mice, an expression vector containing the cDNA of human QC under control of the murine Thy1 promoter sequence was constructed, applying standard molecular biology techniques and verified by sequencing. The transgenic founder was generated on C57BL/6J/CBA background by pronuclear injection (JSW, Graz, Austria). The resulting offspring were further characterized for transgene integration by PCR analysis and after crossing to C57BL/6J wild-type mice for transgene expression by RT-PCR (more than 10 generations). QC knock-out mice (QC-KO) were generated on the basis of a classical homologous recombination approach at Genoway, Lyon. The targeting vector contained the mouse chromosomal QC region ranging from intron 3 to exon 6. This region was modified by insertion of two LoxP sites in intron 3 and 5, respectively. In addition, a neomycin resistance cassette flanked by two flippase recognition targets was inserted immediately upstream of the LoxP in intron 5. After homologous recombination and chimera production, the neomycin selection cassette was removed by breeding with Flp-expressing mice followed by breeding of the pups with Cre-expressing mice for deletion of QC exons 4 and 5. The deletion of exons 4 and 5 causes a frameshift in the QC open reading frame generating a stop codon in exon 6. Successful manipulation was confirmed by PCR and Southern hybridization. Absence of murine QC in 5XFAD/QC-KO comparison with 5XFAD and 5XFAD/hQC was further confirmed by RT-PCR (supplemental Methods and supplemental Fig. 1). Animals were handled according to German guidelines for animal care and studies were approved by the local legal authorities (LAVES). Only female mice were used.
Immunohistochemistry
Mouse tissue was processed as described previously (12). In brief, 4-μm paraffin sections were pretreated with 0.3% H2O2 in PBS to block endogenous peroxidases, and antigen retrieval was achieved by boiling sections in 0.01 m citrate buffer, pH 6.0, followed by a 3-min incubation in 88% formic acid. Primary antibodies were incubated overnight, followed by incubation with biotinylated secondary antibodies (DAKO) before staining was visualized using the ABC method with Vectastain kit (Vector Laboratories) and diaminobenzidine as chromogen. Alternatively, fluorochromated secondary antibodies (anti-mouse Alexa Fluor 594 and anti-rabbit Alexa Fluor 488; Invitrogen) were used for immunofluorescence detection.
Antibodies
Aβ antibodies NT78 (against generic Aβ; Synaptic Systems), 22C11 (APP; Millipore) and 2–48 (against N-terminal AβpE3; Synaptic Systems) (12) were used. Antisera (against QC) were raised against recombinant full-length mouse QC (1301) and have been proven to recognize hQC (13).
ELISA of Aβ Levels in Brain
Frozen brains (n = 4–8 per group) were weighed and subsequently subjected to a sequential Aβ extraction. In a first step, brains were homogenized in TBS (120 mm NaCl, 50 mm Tris, pH 8.0, containing complete protease inhibitor (Roche Applied Science)) using a Dounce homogenizer, sonified, and subsequently centrifuged at 27,000 × g for 20 min at 4 °C. The supernatant was removed and stored at −80 °C. The pellet was dissolved in 2.5 ml of 2% SDS, sonificated, and subsequently centrifuged at 80,000 × g for 1 h at 4 °C. Supernatants were directly frozen at −80 °C. The resulting pellets were again resuspended in 0.5 ml of 70% formic acid, sonified, and neutralized using 1 m Tris. Aliquots of the neutralized formic acid fraction were directly frozen at −80 °C. SDS lysates were diluted at least 10-fold for determination of Aβx-42 and AβpE3 using ELISA. All dilutions were carried out using EIA buffer (IBL Co.). The neutralized formic acid fraction and the TBS fraction were applied directly or after dilution using EIA buffer. ELISA measurements were performed in triplicate and according to the protocol of the manufacturer (IBL Co.; catalog nos. JP27716 and JP27711). Samples were run in triplicate.
Quantification of Plaque Load
Extracellular Aβ load was evaluated in mouse brain using an Olympus BX-51 microscope equipped with an Olympus DP-50 camera and the ImageJ software (V1.41; National Institutes of Health). Serial images of 40× magnification (hippocampus) and 100× (cortex) were captured on six sections/animal (n = 5/group), which were at least 30 μm afar from each other. Using ImageJ the pictures were binarized to 16-bit black and white images, and a fixed intensity threshold was applied defining the DAB staining.
Behavioral Testing
Spontaneous alternation rates were assessed using Y- and cross-maze as described previously (11, 14). The alternation percentage was calculated as the percentage of actual alternations to the total number of arm entries. Balance and general motor function were assessed using the balance beam task. A 1-cm dowel beam is attached to two support columns 44 cm above a padded surface. At either end of the 50-cm long beam, a 9 × 15-cm escape platform is attached. The animal is placed on the center of the beam and released. Each animal is given three trials during a single day of testing. The time the animal remained on the beam is recorded and the resulting latencies to fall of all three trials are averaged. If an animal remains on the beam for whole 60-s trial or escapes to one of the platforms, the maximum time of 60 s is recorded (14). For the string suspension test the animals are permitted to grasp the string by their forepaws and are released. A rating system from 0 to 5 is used during the single 60-s trial to assess each animal's performance in this task: 0 = unable to remain on the string; 1 = hangs only by fore- or hindpaws; 2 = as for 1, but attempts to climb onto string; 3 = sits on string and is able to hold balance; 4 = four paws and tail around string with lateral movement; 5 = escape. The following numbers of animals were analyzed in this task (14). The following number of female mice was used at the age of 6 months: 5XFAD, 11; 5XFAD/hQC, 8; hQC, 11; 5XFAD/QC-KO, 4; QC-KO, 6; wild type, 12.
Statistical Analysis
Statistical differences were evaluated using one-way ANOVA followed by Bonferroni post hoc test or unpaired t test as indicated. All data are given as means ± S.E. All statistics were calculated using GraphPad Prism version 5.00 software.
RESULTS
Expression and Distribution of hQC in the Brain of hQC and 5XFAD/hQC Mice
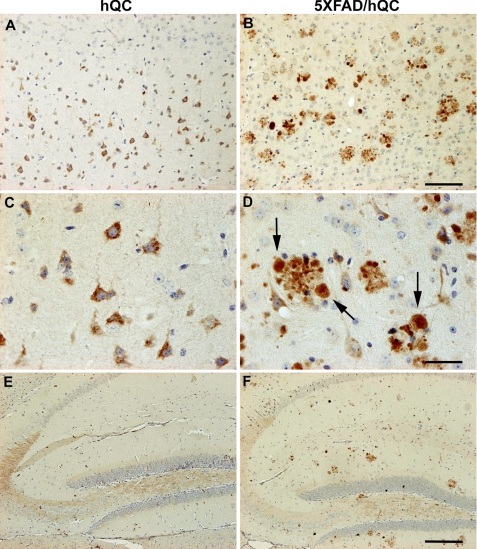
To study human QC overexpression in hQC transgenic mice (hQC and 5XFAD/hQC), the rabbit polyclonal antiserum 1301 recognizing human QC was used to detect the transgene hQC expression in different brain regions (Fig. 1). Because the expression of the hQC trangene is driven by the Thy1 promoter, abundant pyramidal neurons expressing hQC were detected in various brain regions of hQC and 5XFAD/hQC mice including the cortex (Fig. 1, A–D), the hippocampus (Fig. 1, E and F), the midbrain and the cerebellum (data not shown). Notably, a massive staining of hQC was observed in plaque-associated dystrophic neurites in 5XFAD/hQC mice (Fig. 1D). Abundant hQC immunoreactivity was detected in mossy fibers of hQC and 5XFAD/hQC transgenic mice (Fig. 1, E and F).
FIGURE 1.
Immunohistochemical staining of hQC in hQC and 5XFAD/hQC mice. Expression of human transgenic QC was detected in pyramidal neurons in the cortex of hQC (A and C) and 5XFAD/hQC mice and in plaque-associated dystrophic neurites (arrows) of 5XFAD/hQC mice (B and D). In addition, hQC staining was detected in mossy fibers of the hippocampal formation of hQC and 5XFAD/hQC mice (E and F). Scale bars, 100 μm (A and B), 50 μm (C and D), 200 μm (E and F).
Co-localization of APP and QC in Neurons and the Neuritic Component of Plaques
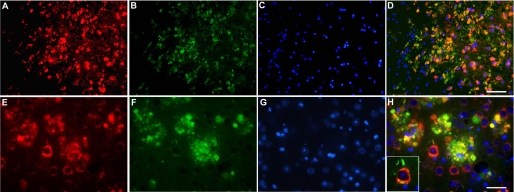
Double immunofluorescence demonstrated co-localization of hQC and APP in the same cellular compartments in the brain of 5XFAD/hQC mice (Fig. 2). APP markedly labels dystrophic neurites around plaques and shows abundant co-localization with hQC suggesting that hQC is axonally transported like APP (Fig. 2).
FIGURE 2.
Transgene human QC is co-localized with APP. Double immunostaining in the cortex of 5XFAD/hQC mice using antibodies against APP (red; A and E), QC (green; B and F) and DAPI (blue; C and G). APP and QC showed co-localization in dystrophic neurites of plaques and in the somatodendritic compartment of pyramidal neurons in the merged images (yellow; D, H, inset in H). Scale bars, 50 μm (A–D), 20 μm (E–H).
Effect of hQC Overexpression and QC Knock-out on Plaque Load in the Frontal Cortex of 6-Month-old 5XFAD Mice
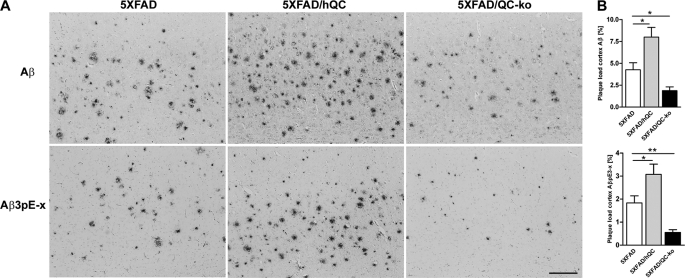
The plaque load for total Aβ (NT78) was significantly higher in 5XFAD/hQC compared with 5XFAD mice (5XFAD/hQC, 7.99 ± 1.1%; and 5XFAD, 4.27 ± 0.79%). In 5XFAD/QC-KO mice, the levels were significantly reduced (1.88 ± 0.43%). The plaque load for AβpE3 showed the same effect. 5XFAD/hQC (3.08 ± 0.44%) had significantly elevated levels compared with 5XFAD mice (1.83 ± 0.31%) and reduced in 5XFAD/QC-KO mice (0.55 ± 0.12%) (Fig. 3).
FIGURE 3.
Effect of hQC overexpression and QC knock-out on plaque load in 5XFAD mice. A, plaque staining in the cortex using antibodies against generic Aβ (NT78) and pyroglutamate-modified Aβ (2–48) in 5XFAD, 5XFAD/hQC, and 5XFAD/QC-KO mice. B, quantification of plaque load demonstrating significantly elevated Aβ and AβpE3 levels in 5XFAD/hQC and significantly reduced levels in 5XFAD/QC-KO mouse brain. Scale bar, 200 μm. *, p < 0.05; **, p < 0.01. Error bars, S.E.
Effect of hQC Overexpression and QC Knock-out on Aβx-42 and AβpE3–42 Levels of 6-Month-old 5XFAD Mice
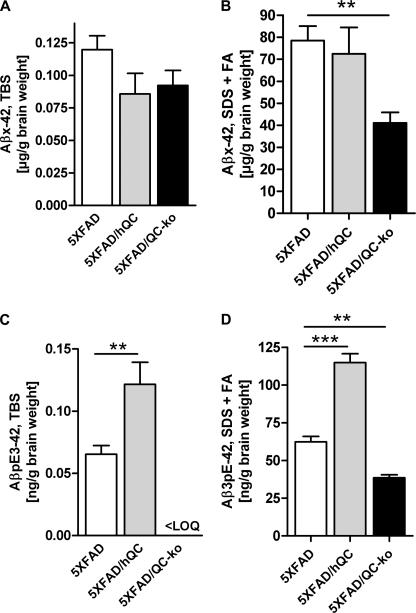
Protein quantification of Aβx-42 (in micrograms/gram of brain weight) and AβpE3–42 (in nanograms/gram of brain weight) levels in brain lysates of 6-month-old 5XFAD, 5XFAD/hQC, and 5XFAD/QC-KO mice revealed significant differences (Fig. 4). In the SDS+FA-soluble fraction there was a significant 48% reduction (p < 0.01) of Aβx-42 levels in 5XFAD/QC-KO mice (41.07 ± 4.79) compared with 5XFAD (78.52 ± 6.54). There was, however, no difference of Aβx-42 levels in the TBS-soluble fraction in 5XFAD/hQC (0.09 ± 0.02) compared with 5XFAD (0.12 ± 0.01) and 5XFAD/QC-KO mice (0.09 ± 0.01). The effects were more pronounced on AβpE3–42 levels. There was an 86% elevation (p < 0.01) of AβpE3–42 levels in the TBS-soluble fraction in 5XFAD/hQC (0.12 ± 0.02) compared with 5XFAD (0.07 ± 0.01) and undetectable levels in 5XFAD/QC-KO mice. In the SDS+FA fraction an 84% elevation (p < 0.001) of AβpE3–42 levels was found in 5XFAD/hQC (114.9 ± 5.89) compared with 5XFAD (62.29 ± 3.66) and significantly reduced levels (−38%) in 5XFAD/QC-KO mice (38.59 ± 1.93, p < 0.01, compared with 5XFAD) (Fig. 4). Despite the differing Aβx-42 and AβpE3–42 levels in 5XFAD, 5XFAD/hQC, and 5XFAD/QC-KO mice, expression levels of transgenic human APP and PS1 were unchanged (supplemental Fig. 2).
FIGURE 4.
Effect of hQC overexpression and QC knock-out on Aβ levels in 5XFAD mice. Quantification of Aβx-42 and AβpE3–42 using ELISA showed significant changes in TBS, SDS, and formic acid (FA) fractions in 5XFAD, 5XFAD/hQC, and 5XFAD/QC-KO mouse brain. SDS and FA fractions were pooled for quantification. Aβx-42 levels were significantly reduced in the SDS+FA fraction of 5XFAD/QC-KO mice. AβpE3–42 levels were significantly elevated in all fractions in 5XFAD/hQC mice. Although in the TBS fraction of 5XFAD/QC-KO mice the levels of AβpE3–42 were below the limit of quantitation, in the SDS+FA fractions of 5XFAD/QC-KO mice the levels of AβpE3–42 were significantly reduced. **, p < 0.01; ***, p < 0.001. LOQ, limit of quantitation. Error bars, S.E.
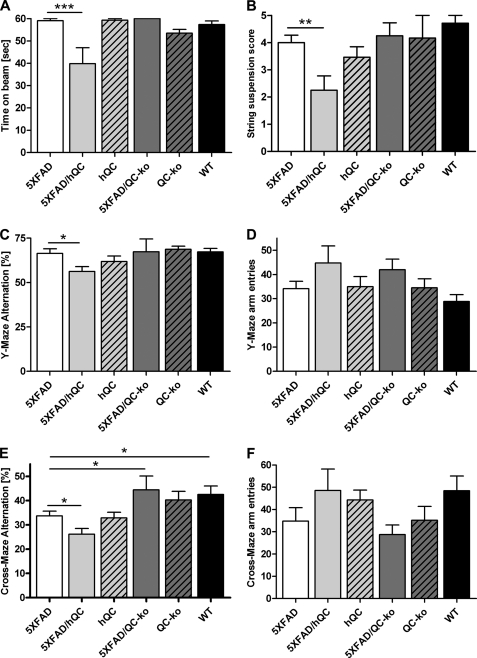
Effect of hQC Overexpression and QC Knock-out on Behavioral Performance in 5XFAD Mice
Motor coordination was assessed by using balance beam and string suspension tasks (Fig. 5, A and B). In both tasks the 5XFAD/hQC performed significantly worse that 5XFAD (p < 0.001 and p < 0.01, respectively). Working memory was assessed using Y- and cross-maze alternation tasks. Analysis in the Y-maze revealed a significantly reduced alternation frequency in 5XFAD/hQC compared with 5XFAD mice (p < 0.05). The number of arm entries during the test period was not different between the groups (Fig. 5, C and D). Assessment using the more complex cross-maze task demonstrated again a significantly reduced alternation frequency in 5XFAD/hQC compared with 5XFAD mice (p < 0.05), and 5XFAD versus wild-type mice (p < 0.05). The latter finding corroborated previous results (14). Moreover, the working memory deficit of 5XFAD mice was rescued in 5XFAD/QC-KO mice (p < 0.05) showing alternation frequencies indistinguishable from wild-type mice. The number of arm entries during the test period was not different among all groups (Fig. 5, E and F).
FIGURE 5.
Effect of hQC overexpression and QC knock-out on behavioral performance in 5XFAD mice. A and B, 5XFAD/hQC mice showed a significantly reduced motor performance in balance beam (A) and string suspension task (B) compared with 5XFAD mice. C, in addition, working memory deficits were detected in 5XFAD/hQC compared with 5XFAD mice using Y- and cross-maze (E). Interestingly, the 5XFAD/QC-KO mice showed a rescue of working memory deficits with alternation frequencies indistinguishable from wild-type mice. D and F, the number of arm entries in Y- and cross-maze did not differ among the groups. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
DISCUSSION
Schilling et al. have shown that cyclization of glutamate at position 3 of Aβ can be driven enzymatically by QC in vitro (15). In addition, it has been demonstrated that QC inhibition significantly reduced AβpE3 formation in vivo, emphasizing the importance of QC activity during cellular maturation of pyroglutamate-containing peptides. The pharmacological inhibition of QC activity by the QC inhibitor PQ150, which significantly reduced the level of AβpE3 in vitro (16) and in vivo (10), suggests that QC inhibition might serve as a new therapeutic approach. Furthermore, the mean level of AβpE3-IgM autoantibodies was significantly decreased in AD patients compared with healthy controls. In the group of mildly cognitive-impaired patients there was a significant positive correlation between AβpE3-IgM and cognitive decline (17).
Interestingly, APP/PS1KI mice, a model with severe neuron loss in the hippocampus, accumulate a large heterogeneity of N-truncated Aβx-42 isoforms including AβpE3 peptides coinciding with the onset of behavioral deficits (18, 19). More specifically, transgenic mice expressing only AβpE3–42 developed a robust and lethal neurological phenotype accompanied by Purkinje cell loss (TBA2 mouse line) (8).
Saido et al. suggested that hypothetically the removal of N-terminal amino acids 1 and 2 of Aβ might be carried out by amino or dipeptidyl peptidase(s) (6). Aminopeptidase A may be responsible in part for the N-terminal truncation of full-length Aβ peptides (20).
N-truncated AβpE3 peptides have been identified by several groups in AD brains (5, 6, 21–32). N-terminal deletions in general enhance aggregation of β-amyloid peptides in vitro (33). AβpE3 has a higher aggregation propensity (34, 35) and stability (36) and shows an increased toxicity compared with full-length Aβ (37). It has been also suggested that N-truncated Aβ peptides are formed directly by β-secretase and not through a progressive proteolysis of full-length Aβ1–40/42 (38).
APP transgenic mouse models have been reported to show no (23) or low AβpE3 levels (31). Maeda et al. have demonstrated that the localization and abundance of [11C]Pittsburgh compound B autoradiographic signals were closely associated with those of N-terminally truncated and modified AβpE3 deposition in AD and different APP transgenic mouse brains, implying that the detectability of amyloid by [11C]Pittsburgh compound B-positron emission tomography is dependent on the accumulation of specific Aβ subtypes (39). APP/PS1KI (12, 18) and 5XFAD (14) mice harbor abundant Aβ3pE levels. Interestingly, both models develop an age-dependent neuron loss and robust behavioral deficits, like TBA2 mice with only AβpE3–42 expression (8).
The findings of the present work are in good agreement with the previous observations that AβpE3–42 levels correlate with behavioral deficits in transgenic mouse models. Here, we demonstrate for the first time genetic evidence for QC as a major target for AD. Overexpression of human QC is co-localized with APP in the neuritic component of plaques, leading to elevated AβpE3–42 levels as detected by ELISA. This finding is corroborated by an increase in the overall plaque pathology including AβpE3–42 in 5XFAD/hQC mice. Consistently, 5XFAD/hQC mice developed a neurological phenotype demonstrated by learning and memory impairments compared with the 5XFAD mouse model at 6 months of age. In addition, we could also show that knock-out of endogenous QC is sufficient to lower Aβ levels, including AβpE3–42, leading to a concomitant rescue of behavioral deficits in 5XFAD mice. The apparent discrepancies between Aβx-42 and AβpE3–42 are likely because the Aβx-42 peptides are ∼1000 times more abundant than AβpE3–42 (micrograms versus nanograms/g wet weight). Therefore it is unlikely that an hQC-dependent increase in AβpE3–42 levels is reflected in a concomitant increase of Aβx-42 levels in a stoichiometric manner. It is, however, surprising that the level of Aβx-42 is significantly reduced in 5XFAD/QC-KO mice. This might be due to a reduced seeding effect of AβpE3–42 on full-length Aβ, which is therapeutically of interest. It is hypothesized that AβpE3–42 elevation does not necessarily lead to increased aggregation of Aβx-42, which might be due to a saturation effect. In addition, the differences in the plaque load of total Aβ and Aβx-42 levels measured by ELISA might be because plaque load was done in the cortex whereas ELISA was performed in whole brain lysates.
Because AβpE3–42 levels were not completely reduced, we assume that other QC-related enzymes like isoQC are responsible for the residual formation of pyroglutamate in QC-KO mice. QC and isoQC represent very similar proteins, which are both present in the secretory pathway of cells. The functions of QCs and isoQC complement each other, suggesting a pivotal role of pyroglutamate modification for protein and peptide maturation (40). To analyze a possible contribution of isoQC to the remaining QC-like activity in 5XFAD/QC-KO mice, we performed a Western blot analysis using an isoQC antibody. The protein levels of isoQC were unchanged in different brain regions between WT and QC-KO mice. This observation demonstrates that the finding of residual pyroglutamate Aβ levels in 5XFAD/QC-KO mice is likely mediated by isoQC (supplemental Fig. 3). In conclusion, reduction of QC was sufficient to rescue the behavioral impairments in the 5XFAD mouse model suggesting a crucial role of QC as a therapeutic target for AD.
Supplementary Material
Acknowledgments
We thank Petra Tucholla, Katrin Schulz and Eike Scheel for technical support.
This work was supported by the German Federal Department of Education, Science and Technology, Grant 3013185 to a collaborative consortium led by H.-U. D.'s group, including T. A. B.'s team.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Methods and Figs. 1–3.
- AD
- Alzheimer disease
- Aβ
- amyloid-β
- AβpE3
- pyroglutamate Aβ
- APP
- amyloid precursor protein
- QC
- glutaminyl cyclase
- hQC
- human QC
- QC-KO
- QC knock-out.
REFERENCES
- 1. Selkoe D. J. (1998) Trends Cell Biol. 8, 447–453 [DOI] [PubMed] [Google Scholar]
- 2. Masters C. L., Simms G., Weinman N. A., Multhaup G., McDonald B. L., Beyreuther K. (1985) Proc. Natl. Acad. Sci. U.S.A. 82, 4245–4249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Selkoe D. J., Abraham C. R., Podlisny M. B., Duffy L. K. (1986) J. Neurochem. 46, 1820–1834 [DOI] [PubMed] [Google Scholar]
- 4. Gorevic P. D., Goñi F., Pons-Estel B., Alvarez F., Peress N. S., Frangione B. (1986) J. Neuropathol. Exp. Neurol. 45, 647–664 [DOI] [PubMed] [Google Scholar]
- 5. Mori H., Takio K., Ogawara M., Selkoe D. J. (1992) J. Biol. Chem. 267, 17082–17086 [PubMed] [Google Scholar]
- 6. Saido T. C., Iwatsubo T., Mann D. M., Shimada H., Ihara Y., Kawashima S. (1995) Neuron 14, 457–466 [DOI] [PubMed] [Google Scholar]
- 7. Portelius E., Bogdanovic N., Gustavsson M. K., Volkmann I., Brinkmalm G., Zetterberg H., Winblad B., Blennow K. (2010) Acta Neuropathol. 120, 185–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wirths O., Breyhan H., Cynis H., Schilling S., Demuth H. U., Bayer T. A. (2009) Acta Neuropathol. 118, 487–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cynis H., Scheel E., Saido T. C., Schilling S., Demuth H. U. (2008) Biochemistry 47, 7405–7413 [DOI] [PubMed] [Google Scholar]
- 10. Schilling S., Zeitschel U., Hoffmann T., Heiser U., Francke M., Kehlen A., Holzer M., Hutter-Paier B., Prokesch M., Windisch M., Jagla W., Schlenzig D., Lindner C., Rudolph T., Reuter G., Cynis H., Montag D., Demuth H. U., Rossner S. (2008) Nat. Med. 14, 1106–1111 [DOI] [PubMed] [Google Scholar]
- 11. Oakley H., Cole S. L., Logan S., Maus E., Shao P., Craft J., Guillozet-Bongaarts A., Ohno M., Disterhoft J., Van Eldik L., Berry R., Vassar R. (2006) J. Neurosci. 26, 10129–10140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wirths O., Bethge T., Marcello A., Harmeier A., Jawhar S., Lucassen P. J., Multhaup G., Brody D. L., Esparza T., Ingelsson M., Kalimo H., Lannfelt L., Bayer T. A. (2010) J. Neural Transm. 117, 85–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hartlage-Rübsamen M., Staffa K., Waniek A., Wermann M., Hoffmann T., Cynis H., Schilling S., Demuth H. U., Rossner S. (2009) Int. J. Dev. Neurosci. 27, 825–835 [DOI] [PubMed] [Google Scholar]
- 14. Jawhar S., Trawicka A., Jenneckens C., Bayer T. A., Wirths O. (2010) Neurobiol. Aging, doi:10.1016/j.neurobiolaging.2010.05.027 [DOI] [PubMed] [Google Scholar]
- 15. Schilling S., Hoffmann T., Manhart S., Hoffmann M., Demuth H. U. (2004) FEBS Lett. 563, 191–196 [DOI] [PubMed] [Google Scholar]
- 16. Cynis H., Schilling S., Bodnár M., Hoffmann T., Heiser U., Saido T. C., Demuth H. U. (2006) Biochim. Biophys. Acta 1764, 1618–1625 [DOI] [PubMed] [Google Scholar]
- 17. Marcello A., Wirths O., Schneider-Axmann T., Degerman-Gunnarsson M., Lannfelt L., Bayer T. A. (2009) Neurobiol. Aging, doi:10.1016/j.neurobiolaging.2009.08.011 [DOI] [PubMed] [Google Scholar]
- 18. Casas C., Sergeant N., Itier J. M., Blanchard V., Wirths O., van der Kolk N., Vingtdeux V., van de Steeg E., Ret G., Canton T., Drobecq H., Clark A., Bonici B., Delacourte A., Benavides J., Schmitz C., Tremp G., Bayer T. A., Benoit P., Pradier L. (2004) Am. J. Pathol. 165, 1289–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Breyhan H., Wirths O., Duan K., Marcello A., Rettig J., Bayer T. A. (2009) Acta Neuropathol. 117, 677–685 [DOI] [PubMed] [Google Scholar]
- 20. Sevalle J., Amoyel A., Robert P., Fournié-Zaluski M. C., Roques B., Checler F. (2009) J. Neurochem. 109, 248–256 [DOI] [PubMed] [Google Scholar]
- 21. Saido T. C., Yamao-Harigaya W., Iwatsubo T., Kawashima S. (1996) Neurosci. Lett. 215, 173–176 [DOI] [PubMed] [Google Scholar]
- 22. Kuo Y. M., Emmerling M. R., Woods A. S., Cotter R. J., Roher A. E. (1997) Biochem. Biophys. Res. Commun. 237, 188–191 [DOI] [PubMed] [Google Scholar]
- 23. Kuo Y. M., Kokjohn T. A., Beach T. G., Sue L. I., Brune D., Lopez J. C., Kalback W. M., Abramowski D., Sturchler-Pierrat C., Staufenbiel M., Roher A. E. (2001) J. Biol. Chem. 276, 12991–12998 [DOI] [PubMed] [Google Scholar]
- 24. Hosoda R., Saido T. C., Otvos L., Jr., Arai T., Mann D. M., Lee V. M., Trojanowski J. Q., Iwatsubo T. (1998) J. Neuropathol. Exp. Neurol. 57, 1089–1095 [DOI] [PubMed] [Google Scholar]
- 25. Harigaya Y., Saido T. C., Eckman C. B., Prada C. M., Shoji M., Younkin S. G. (2000) Biochem. Biophys. Res. Commun. 276, 422–427 [DOI] [PubMed] [Google Scholar]
- 26. Iwatsubo T., Saido T. C., Mann D. M., Lee V. M., Trojanowski J. Q. (1996) Am. J. Pathol. 149, 1823–1830 [PMC free article] [PubMed] [Google Scholar]
- 27. Miravalle L., Calero M., Takao M., Roher A. E., Ghetti B., Vidal R. (2005) Biochemistry 44, 10810–10821 [DOI] [PubMed] [Google Scholar]
- 28. Piccini A., Russo C., Gliozzi A., Relini A., Vitali A., Borghi R., Giliberto L., Armirotti A., D'Arrigo C., Bachi A., Cattaneo A., Canale C., Torrassa S., Saido T. C., Markesbery W., Gambetti P., Tabaton M. (2005) J. Biol. Chem. 280, 34186–34192 [DOI] [PubMed] [Google Scholar]
- 29. Piccini A., Zanusso G., Borghi R., Noviello C., Monaco S., Russo R., Damonte G., Armirotti A., Gelati M., Giordano R., Zambenedetti P., Russo C., Ghetti B., Tabaton M. (2007) Arch. Neurol. 64, 738–745 [DOI] [PubMed] [Google Scholar]
- 30. Russo C., Saido T. C., DeBusk L. M., Tabaton M., Gambetti P., Teller J. K. (1997) FEBS Lett. 409, 411–416 [DOI] [PubMed] [Google Scholar]
- 31. Güntert A., Döbeli H., Bohrmann B. (2006) Neuroscience 143, 461–475 [DOI] [PubMed] [Google Scholar]
- 32. Tekirian T. L., Saido T. C., Markesbery W. R., Russell M. J., Wekstein D. R., Patel E., Geddes J. W. (1998) J. Neuropathol. Exp. Neurol. 57, 76–94 [DOI] [PubMed] [Google Scholar]
- 33. Pike C. J., Overman M. J., Cotman C. W. (1995) J. Biol. Chem. 270, 23895–23898 [DOI] [PubMed] [Google Scholar]
- 34. He W., Barrow C. J. (1999) Biochemistry 38, 10871–10877 [DOI] [PubMed] [Google Scholar]
- 35. Schilling S., Lauber T., Schaupp M., Manhart S., Scheel E., Böhm G., Demuth H. U. (2006) Biochemistry 45, 12393–12399 [DOI] [PubMed] [Google Scholar]
- 36. Kuo Y. M., Webster S., Emmerling M. R., De Lima N., Roher A. E. (1998) Biochim. Biophys. Acta 1406, 291–298 [DOI] [PubMed] [Google Scholar]
- 37. Russo C., Violani E., Salis S., Venezia V., Dolcini V., Damonte G., Benatti U., D'Arrigo C., Patrone E., Carlo P., Schettini G. (2002) J. Neurochem. 82, 1480–1489 [DOI] [PubMed] [Google Scholar]
- 38. Russo C., Salis S., Dolcini V., Venezia V., Song X. H., Teller J. K., Schettini G. (2001) Neurobiol. Dis. 8, 173–180 [DOI] [PubMed] [Google Scholar]
- 39. Maeda J., Ji B., Irie T., Tomiyama T., Maruyama M., Okauchi T., Staufenbiel M., Iwata N., Ono M., Saido T. C., Suzuki K., Mori H., Higuchi M., Suhara T. (2007) J. Neurosci. 27, 10957–10968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Stephan A., Wermann M., von Bohlen A., Koch B., Cynis H., Demuth H. U., Schilling S. (2009) FEBS J. 276, 6522–6536 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.