Abstract
Ceramides, which are membrane sphingolipids and key mediators of cell-stress responses, are generated by a family of (dihydro) ceramide synthases (Lass1–6/CerS1–6). Here, we report that brain development features significant increases in sphingomyelin, sphingosine, and most ceramide species. In contrast, C16:0-ceramide was gradually reduced and CerS6 was down-regulated in mitochondria, thereby implicating CerS6 as a primary ceramide synthase generating C16:0-ceramide. Investigations into the role of CerS6 in mitochondria revealed that ceramide synthase down-regulation is associated with dramatically decreased mitochondrial Ca2+-loading capacity, which could be rescued by addition of ceramide. Selective CerS6 complexing with the inner membrane component of the mitochondrial permeability transition pore was detected by immunoprecipitation. This suggests that CerS6-generated ceramide could prevent mitochondrial permeability transition pore opening, leading to increased Ca2+ accumulation in the mitochondrial matrix. We examined the effect of high CerS6 expression on cell survival in primary oligodendrocyte (OL) precursor cells, which undergo apoptotic cell death during early postnatal brain development. Exposure of OLs to glutamate resulted in apoptosis that was prevented by inhibitors of de novo ceramide biosynthesis, myriocin and fumonisin B1. Knockdown of CerS6 with siRNA reduced glutamate-triggered OL apoptosis, whereas knockdown of CerS5 had no effect: the pro-apoptotic role of CerS6 was not stimulus-specific. Knockdown of CerS6 with siRNA improved cell survival in response to nerve growth factor-induced OL apoptosis. Also, blocking mitochondrial Ca2+ uptake or decreasing Ca2+-dependent protease calpain activity with specific inhibitors prevented OL apoptosis. Finally, knocking down CerS6 decreased calpain activation. Thus, our data suggest a novel role for CerS6 in the regulation of both mitochondrial Ca2+ homeostasis and calpain, which appears to be important in OL apoptosis during brain development.
Keywords: Apoptosis, Bioenergetics, Brain, Calcium, Calpain, Mitochondrial Apoptosis, Neurodevelopment, Sphingolipid
Introduction
Sphingolipids are essential structural components of cellular membranes, playing prominent roles in signal transduction that governs cell proliferation, differentiation, migration, and apoptosis (1). Most sphingolipids are ubiquitous, but complex sphingolipids, including sphingomyelin (SM)2 and glycosphingolipids, are more abundant in the brain and in myelin formed by oligodendrocytes (OLs). The building block of many complex sphingolipids is ceramide, which has numerous cellular signaling functions (2). Ceramides are a family of distinct molecular species characterized by various acyl chains as well as the desaturation and hydroxylation of those chains. Highly hydrophobic ceramides are generated by membrane-associated enzymes and exert their effects proximal to the ceramide generation site, or they require specific transporter proteins to reach their targets in other intracellular compartments (1, 3).
Ceramides are synthesized de novo at the cytosolic side of the endoplasmic reticulum (4, 5), serving as precursors for the biosynthesis of glycosphingolipids and SM in the Golgi (6, 7). Mitochondria are another important intracellular compartment of sphingolipid metabolism (8), and several sphingolipid-metabolizing enzymes were found to be associated with mitochondria, including neutral ceramidase (9), novel neutral sphingomyelinase (10), and (dihydro) ceramide synthase (EC 2.3.1.24), a key enzyme in de novo ceramide synthesis (11, 12). Recently, mitochondrial ceramide engagement in apoptosis has been shown using loss-of-function mutants of ceramide synthase in the germ cell line of Caenorhabditis elegans (13). Specifically, ionizing radiation-induced apoptosis of germ cells was blocked upon inactivation of ceramide synthase, and apoptosis was restored upon microinjection of long-chain ceramide. Radiation-induced increases in ceramide localized to the mitochondria were required for activation of CED-3 caspase and apoptosis.
Each of the 6 mammalian ceramide synthase (CerS, originally known as Lass) genes appears to regulate synthesis of a specific subset of ceramides, and each has a unique substrate specificity for chain-length and/or saturation of fatty acid acyl-CoA. Overexpression of any CerS protein in mammalian cells resulted in increases in a specific subset of ceramide species. CerS1 has high specificity for C18:0-CoA generating C18:0-ceramide (14, 15). CerS2, CerS4, and CerS3 appear to have broader specificity (16, 17). CerS2 or CerS4 mainly synthesizes C20:0-, C22:0-, C24:1-, C24:0-, C26:1-, and C26:0-ceramide, but is unable to synthesize C16:0- or C18:0-ceramide (14, 17). CerS3 generates C18:0-, C20:0-, C22:0-, and C24:0-ceramide (16). It has been shown that CerS5 generates C14:0-, C16:0-, C18:0-, and C18:1-ceramide (14, 18); and CerS6 produces C14:0-, C16:0-, and C18:0-ceramide (14).
Our studies described here were designed to ascertain the functional role of ceramide and CerS6 in mitochondria during postnatal animal brain development. Herein, we report that, contrary to most ceramide species, C16:0-ceramide was down-regulated, as was CerS6 expression, in mitochondria. The data imply that CerS6 could be a primary ceramide synthase, generating C16:0-ceramide in brain mitochondria. Functional analysis revealed a significant decrease in Ca2+-loading capacity in mitochondria from the adult rat brain compared with the postnatal day 10 (P10) brain, and this decrease occurred with lower CerS6 expression and decreased C16:0-ceramide. Exogenously added C16:0-ceramide completely restored the Ca2+-loading capacity of adult mitochondria to that of the young rat brain. Co-immunoprecipitation studies exposed selective CerS6 association with adenine nucleotide translocator (ANT), the mitochondrial permeability transition pore (MPTP) component in the inner mitochondrial membrane. This suggests that CerS6 could generate C16:0-ceramide in close proximity of MPTP and prevent pore opening that results in an increased mitochondrial Ca2+-buffering capacity. Gene knockdown experiments revealed a critical role for CerS6 in promoting OL apoptosis. Thus, knocking down CerS6 enhanced OL survival in response to glutamate- or nerve growth factor-induced apoptosis. Investigation of downstream targets of the CerS6-mediated signaling pathway revealed an important contribution of mitochondrial Ca2+ and calpain in promoting ceramide-dependent apoptosis in OLs. Specifically, OL exposure to inhibitors of mitochondrial Ca2+ uptake or calpain activity enhanced cell survival in response to glutamate and NGF. Knocking down CerS6 reduced calpain activation. These studies identify CerS6 as an important regulator of mitochondrial Ca2+ homeostasis and suggest a pro-apoptotic role in OLs during postnatal brain development.
EXPERIMENTAL PROCEDURES
Animals and Reagents
Female timed-pregnant Sprague-Dawley rats (Charles River Laboratories, Wilmington, MA) were acclimated for 1 week prior to experimentation. Experimental protocols were reviewed and approved by the Institutional Animal Care and Use Committee of Medical University of South Carolina (MUSC), Charleston SC, and followed the National Institutes of Health guidelines for experimental animal use. Cell culture was Dulbecco's modified Eagle's medium (DMEM), fetal bovine serum (FBS), and N2 supplement (from Invitrogen). Complete Mini Protease Inhibitor Mixture was from Roche Applied Science. A new generation pan-caspase inhibitor, Q-VD-OPH, was from BioVision (Mountain View, CA). Calpain inhibitor PD150606 was supplied by Santa Cruz Biotechnology (Santa Cruz, CA). Calpeptin was from EMD Chemicals (Gibbstown, NJ). Nerve growth factor (NGF) was purchased from Neuromics (Edina, MN). All other chemicals were purchased from Sigma.
Antibodies
The following antibodies were used: mouse anti-LASS2/CerS2 (clone 1A6), mouse monoclonal anti-LASS4/CerS4 (clone7D1), rabbit polyclonal anti-LASS5/CerS5 (PAB13440), and mouse monoclonal anti-LASS6/CerS6 (clone 5H7). These antibodies were obtained from Abnova (Taipei, Taiwan). The rabbit polyclonal anti-LASS1/CerS1 antibody was from Sigma Genosys (Woodlands, TX). The specificity of each anti-CerS antibody was verified in knockdown experiments using specific siRNA targeting CerS in OLs. Anti-β-actin mouse monoclonal (A1978) and rabbit polyclonal anti-p75NTR antibodies were purchased from Sigma. Rabbit polyclonal antibodies against voltage-dependent anion channel were supplied by EMD Chemicals (Gibbstown, NJ). Goat polyclonal anti-myelin basic protein (myelin marker) antibody, rabbit polyclonal anti-cyclophilin D, anti-Tom20, and anti-ANT antibodies were obtained from Santa Cruz (Santa Cruz, CA). The rabbit polyclonal anti-LAMP-2 (lysosomal marker), mouse monoclonal anti-α1 subunit of the sodium/potassium ATPase (plasma membrane marker), and the rabbit polyclonal anti-calnexin (ER marker) antibody were purchased from Abcam (Cambridge, MA). Rat monoclonal anti-myelin proteolipid protein antibody was generously provided by Dr. Wendy Macklin (University of Colorado, Denver, CO). Secondary horseradish peroxidase-conjugated antibodies were supplied by Jackson ImmunoResearch Laboratories Inc.
Isolation of Rat Brain Mitochondria
All procedures were performed at 4 °C as described (12). Briefly, tissue was placed immediately in ice-cold isolation medium containing 230 mm mannitol, 70 mm sucrose, 10 mm HEPES, and 1 mm EDTA, pH 7.4. Brain tissue (∼1 g) was homogenized in 10 ml of isolation medium using a Teflon-glass homogenizer. The homogenate was centrifuged at 900 × g for 10 min. The supernatant was then centrifuged at 12,000 × g for 10 min. The pellet was re-suspended in the isolation medium and centrifuged again at 12,000 × g for 10 min. The pellet was re-suspended in 2 ml of 15% Percoll-Plus (GE Healthcare) and placed atop the discontinuous Percoll gradient consisting of a bottom layer of 4 ml of 40% Percoll and a top layer of 4 ml of 23% Percoll. The gradient was spun at 31,000 × g for 15 min in a SW-Ti40 rotor in a Beckman LE80K centrifuge. The fraction at the 23–40% interface, which contained mitochondria, was washed 3 times with isolation medium by centrifugation at 12,000 × g for 10 min. Protein concentration was measured with a bicinchoninic acid assay (Sigma) using bovine serum albumin as a standard. Typically, the contamination of mitochondria with ER was <1% by activity measurements of ER-specific marker enzyme, NADPH-cytochrome c reductase (12).
Mitochondrial Respiratory Chain Activity
Mitochondrial respiration was measured by recording oxygen consumption at 25 °C in a chamber equipped with a Clark-type oxygen electrode (Instech Laboratories, Plymouth Meeting, PA) as previously described (12). Briefly, mitochondria were incubated in the medium containing 125 mm KCl, 10 mm HEPES, 2 mm KH2PO4, 5 mm MgCl2, and 0.5 mg/ml of mitochondrial protein supplemented with either Complex I substrate (mixture of 5 mm glutamate and 5 mm malate) or Complex II substrate (5 mm succinate) in the presence of 1 μm rotenone or Complex IV substrate (1 mm ascorbate in the presence of 250 μm TMPD and 1 μm antimycin). A respiratory control ratio was measured as the oxygen consumption rate in the presence of the substrate and 100 μm ADP (State 3) divided by the rate in the resting state (State 4) in the presence of 2 μg/ml of oligomycin. Uncoupler-stimulated (State 3u) respiration was measured in the presence of 50 nm carbonyl cyanide p-trifluoromethoxyphenylhydrazone.
Measurement of Mitochondrial Ca2+ Load
The Ca2+-loading capacity (CLC) of mitochondria was monitored using a Ca2+-selective electrode (Thermo Scientific/Orion, Rockford, IL) in a medium containing 250 mm sucrose, 10 mm HEPES, and 2 mm KH2PO4, pH 7.4 (adjusted with Tris base). Mitochondria were energized by 10 mm succinate with 1 μm rotenone and pulsed with 100 μm Ca2+ every 1.5 min. The increasing Ca2+ load caused a decline in Ca2+ uptake rates. Maximal CLC was defined as an amount of Ca2+ (per mg of protein) required to decrease the Ca2+ uptake rate by >90%. Simultaneously with CLC measurements, mitochondrial swelling was monitored using a Brinkman probe colorimeter as described (19).
Western Blot
Proteins were analyzed by Western blot as previously described (12, 20). Cells or tissue samples were lysed in a buffer containing 50 mm Tris-HCl, 5 mm EDTA, 150 mm NaCl, 1% Triton X-100, pH 7.4, 1 mm Na3VO4, and 10 mm NaF, supplemented with a protease inhibitor mixture. After 1 h on ice, cell lysates were centrifuged at 15,000 × g for 10 min to remove insoluble material. Protein samples were prepared by boiling lysates in reducing SDS-sample buffer. Proteins were separated by 8–10% SDS-PAGE, blotted to PVDF membrane, blocked with 5% nonfat dry milk in TBS-T buffer (10 mm Tris, 150 mm NaCl, and 0.2% Tween 20, pH 8.0) overnight at 4 °C, and subsequently probed with the appropriate primary antibody. Immunoreactive bands were visualized using a chemiluminescence SuperSignal West Femto substrate (Thermo Scientific).
Cell Culture
Dissociated rat neonatal cortices were cultured on poly-l-lysine-coated flasks as described (20). Briefly, the cerebra of rat pups were dissected and minced to generate a single-cell suspension. Cells were plated into 75-cm2 flasks and grown in DMEM with 10% FBS at 37 °C and 5% CO2. By day 10, mixed glial cultures were obtained, consisting of OLs and microglia growing on an astrocyte monolayer. OLs were purified from mixed glial cell cultures using a shake-off procedure. Cells were shaken initially for 1 h at 100 × g to remove microglia, re-fed, and shaken again for 22–24 h at 37 °C at 200 × g. OLs were collected by centrifugation at 1,200 × g for 4 min. OLs were used immediately for transfections or further culturing. Cell culture plates and cell culture dishes were pre-coated with the 10 μg/ml of fibronectin solutions overnight at 37 °C. All cultures contain less than 2% of GFAP+ astrocytes and non-detectable CD11+ microglia.
siRNA transfection
To down-regulate Lass6/CerS6, siGenome SMARTpool silencing RNAs were obtained from Thermo Scientific/Dharmacon (Rockford, IL). The set consists of 4 siRNAs targeting different regions of the gene to minimize the off-target effects. In addition, silencing RNA targeting Lass5/CerS5 or Lass6/CerS6 were purchased from the Qiagen High Performance GenomeWide siRNA bank (Qiagen, Valencia, CA). The following target sequences were used: Lass6/CerS6, 5′-GAACUGGCGUCCUGACUAG-3′, 5′-GAACACCGGACUUAACUAU-3′, 5′-GGACAGAGGUGCAAGACGC-3′, 5′-CGACACAGGAGUGGACAAA-3′; Lass5/CerS5, 5′-TTCGAGCGATTTATTGCTAAA-3′; Lass6/CerS6, 5′-GGACAGAGGUGCAAGACGC-3′. OLs were transfected with siRNA using the Nucleofector electroporation system (Amaxa Biosystems, Gaithersburg, MD) according to the manufacturer's instructions with efficiencies of >70% as described (20). Cells (6 × 106) were mixed with 100 μl of Nucleofector reagent and 0.5 μl of siRNA in the cuvette of the Amaxa electroporation device. AllStars negative control siRNA (Qiagen, Valencia, CA) was used as a control.
Cell Survival Assay
Cell death was measured using a lactate dehydrogenase-based CytoTox-ONETM Homogeneous Membrane Integrity Assay (Promega, Madison, WI), according to the manufacturer's recommendations. Cell survival was expressed as percent of viable cells based on the measurements of lactate dehydrogenase activity associated with the cells versus the lactate dehydrogenase activity in the medium. The fluorescence of the sample was measured at 590 nm emission with 560 nm excitation in a microplate reader (FLUOstar Optima, BMG LABTECH Inc., Durham, NC).
Caspase Activity Assay
The activities of executioner caspases 3/7 were determined using Apo-One® Homogeneous kit (Promega) according to the manufacturer's instructions. Cleavage of non-fluorescent substrate, Z-DEVD-Rodamine-110 by caspase 3/7 resulted in fluorescent rodamine-110. The fluorescence of the sample was measured at 530 nm emission and 490 nm excitation in the microplate reader FLUOstar Optima.
Calpain Activity Assay
Calpain activity was measured using the SensoLyte520 fluorimetric calpain activity assay kit (AnaSpec, Freemont, CA) according to the manufacturer's instructions. The assay employs a novel internally quenched 5-FAM/OXLTM 520 FRET substrate to increase the sensitivity of the measurements. Calpain cleaved the FRET substrate yielding the release of fluorescent 5-FAM, which was monitored at 520 nm emission and 490 excitation in the microplate reader FLUOstar Optima.
Immunoprecipitation
For immunoprecipitation, cell lysates (500 μg) were precleared in buffer A (0.15 m NaCl, 0.5 mm EDTA, 1% Triton X-100, 10 mm NaF, and 1 mm Na3VO4, protease inhibitor mixture, 0.05 m Tris, pH 7.5, 0.2% BSA) by incubation with appropriate species-specific, IgG-conjugated magnetic beads (Dynabeds, Invitrogen/Dynal, Carlsbad, CA) for 1 h. Antibodies then were added. After incubation at 4 °C overnight with gentle mixing, antibody-antigen complexes were captured with Dynabeads and washed two times with buffer A (without BSA), and then washed twice with Tris-buffered saline, pH 7.5. The immunoprecipitates were eluted by boiling in SDS-sample buffer. As a control, the same immunoprecipitation procedure was performed except for the primary antibody application.
Analysis of Sphingolipids by Tandem Mass Spectrometry
Cells or tissues were lysed in a buffer containing 10 mm Tris and 1% Triton X-100, pH 7.4, for analysis by reverse-phase high pressure liquid chromatography coupled to electrospray ionization followed by separation by MS. Sphingolipid analysis was performed in the Lipidomics Core Facility at MUSC using a Thermo Finnigan TSQ 7000 triple quadrupole mass spectrometer, operating in a multiple reaction monitoring positive-ionization mode (20, 21). The peaks for the target analytes and internal standards were collected and processed with the Xcalibur software system. Calibration curves were constructed by plotting peak area ratios of synthetic standards, representing each target analyte, to the corresponding internal standard. The target analyte peak area ratios from the samples were similarly normalized to their respective internal standard and compared with the calibration curves using a linear regression model. Each sample was normalized to its respective total protein levels.
Statistical Analysis
All experiments and assays were performed three or more times. Typically, there were at least six replicates of each treatment in each assay. Data were collected, and the mean value of the treatment groups and the standard error were calculated. Data were analyzed for statistically significant differences between groups by one-way analysis of variance with a post hoc Tukey test, which adjusts for multiple simultaneous comparisons (SAS version 9.1.3). Statistical significance was ascribed to the data when p < 0.05.
RESULTS
C16:0-ceramide Is Down-regulated in Mitochondria during Postnatal Brain Development
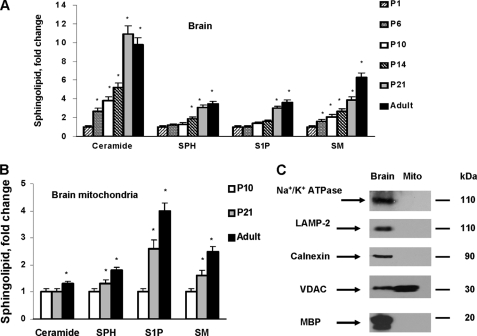
Sphingolipids, including sphingomyelin (SM), sphingosine (SPH), sphingosine 1-phosphate (S1P), and ceramide, were measured in brains of rats at different postnatal ages: postnatal day 1 (P1) through P21 and 6-month-old (adult) rats (Fig. 1A). The data are expressed as fold-increases of P1 brain sphingolipid content (Table 1). Postnatal brain growth was accompanied by profound increases in all sphingolipids (Fig. 1A), which is consistent with the structural role of sphingolipids in cell membranes.
FIGURE 1.
Sphingolipid changes in brain tissue or brain mitochondria during rat development. Sphingolipids were analyzed in total brain tissue lysate (A) or isolated brain mitochondria (B) from rats at various ages. Ceramide, SPH, S1P, or SM content was gradually increased in developing rat brain tissue or mitochondria. The data are expressed as fold-increase in mitochondria content for postnatal day 1 (P1) brains or postnatal day 10 (P10) brains. Data are mean ± S.E., *, p < 0.05, n = 16. Each sample was normalized to its respective total protein levels. C, lack of mitochondrial contamination with various cellular membranes was characterized by Western blot using specific antibodies: anti-α1 subunit Na+/K+-ATPase (plasma membrane marker), anti-calnexin (ER marker), anti-LAMP-2 (lysosomal marker), anti-VDAC (mitochondrial marker), and anti-myelin basic protein (MBP, myelin marker). An equal amount of brain (Brain) or brain mitochondria (Mito) lysate (10 μg) was loaded into the lane.
TABLE 1.
Sphingolipid content of rat brain tissue and brain mitochondria
Total ceramide, SPH, S1P, or SM content (pmol/mg of protein) was determined in the P1 rat brain or mitochondria purified from P10 rat brain. Each sample was normalized to its respective total protein. Values are mean ± S.E., n = 16.
| Sphingolipid | Ceramide | SPH | S1P | SM |
|---|---|---|---|---|
| Brain (P1) | 915.4 ± 24.3 | 15.9 ± 0.6 | 1.3 ± 0.4 | 2,313.6 ± 98.2 |
| Mitochondria (P10) | 979.3 ± 26.8 | 16.4 ± 0.8 | 2.1 ± 0.5 | 4,281.4 ± 121.7 |
Reports suggest that some sphingolipids, including ceramide, exert pleotropic effects on cell signaling pathways and can also mediate a variety of cell responses (22). It has been emphasized that the function of the sphingolipid is determined by its local concentration, which could vary even between the two leaflets of the lipid bilayer in the membrane or between organelles (1, 23). Recently, mitochondria have emerged as an important intracellular compartment of sphingolipid biosynthesis and bioactive sphingolipid-mediated signaling pathways (8). To gain more insight into sphingolipid function, the content of isolated brain mitochondria was analyzed (Fig. 1B). The data are presented as fold-increases of P10 brain mitochondria sphingolipid content (Table 1). Substantial developmental increases in SM, SPH, or S1P occurred, but there were smaller changes in ceramide content compared with brain tissue samples. To rule out possible contamination of the mitochondrial preparation with other cellular membranes as a source of sphingolipids, Western blot was performed with specific antibodies against calnexin (ER marker), LAMP-2 (lysosomal marker), Na+/K+ ATPase (plasma membrane marker), and myelin basic protein (myelin marker) (Fig. 1C).
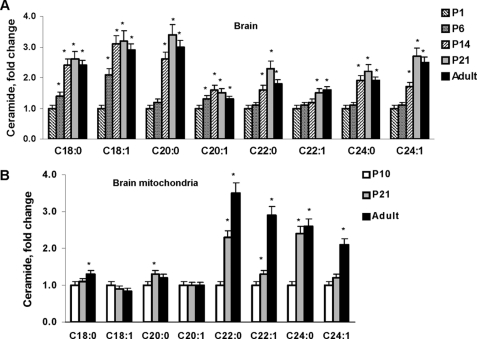
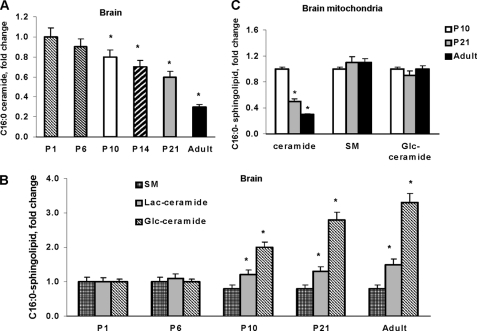
The ceramide species profile of the developing rat brain revealed increased C18:0 -, C18:1-, C20:0-, C20:1-, C22:0-, C22:1-, C24:0-, and C24:1-ceramide (Fig. 2A). In mitochondria, there were substantial increases in very long-chain ceramide species, including C20:0-, C22:0-, C22:1-, C24:0- and C24:1-ceramide. Although C18:0-ceramide or C20:0-ceramide appear only to be moderately increased, there were large mass changes in these ceramides because of their abundance, whereas C18:1- and C20:1-ceramide did not change (Fig. 2B). Data are presented as fold-increases of P1 brain tissue or P10 brain mitochondria sphingolipid content (Table 2). Intriguingly, C16:0-ceramide was decreased by 70% in the adult rat brain compared with the P1 brain (Fig. 3A).
FIGURE 2.
Ceramide species changes in brain tissue or brain mitochondria during rat development. Ceramide species were analyzed in total brain lysate (A) or isolated brain mitochondria (B) from rats at various ages. The data are expressed as fold-increase of ceramide species content of P1 brain tissue or mitochondria from P10 brain. Data are mean ± S.E., *, p < 0.05, n = 16. Each sample was normalized to its respective total protein levels.
TABLE 2.
Ceramide species content in brain tissue and brain mitochondria
Ceramide species content (pmol/mg of protein) was determined in P1 rat brain or mitochondria purified from P10 rat brain. Each sample was normalized to its respective total protein levels. Values are mean ± S.E., n = 16.
| Ceramide | C18:0 | C18:1 | C20:0 | C20:1 | C22:0 | C22:1 | C24:0 | C24:1 |
|---|---|---|---|---|---|---|---|---|
| Brain (P1) | 632.9 ± 13.2 | 501.4 ± 10.9 | 138.1 ± 7.1 | 36.9 ± 2.2 | 25.1 ± 2.5 | 11.8 ± 1.1 | 118.8 ± 7.5 | 140.5 ± 6.9 |
| Mitochondria (P10) | 328.0 ± 9.6 | 241.7 ± 7.3 | 135.3 ± 5.4 | 23.9 ± 1.9 | 3.8 ± 0.5 | 2.1 ± 0.6 | 15.4 ± 0.8 | 16.5 ± 0.9 |
FIGURE 3.
C16:0-containing sphingolipid changes during brain development. A, C16:0-ceramide content was down-regulated during brain development. The data are expressed as fold-decrease of C16:0-ceramide content in P1 brain tissue (203.9 ± 12.5 pmol/mg of protein). B, developmental changes in brain content of complex sphingolipids containing C16:0 moeity. The data are expressed as fold-increase of sphingolipid content of P1 brain tissue that contains: C16:0-sphingomyelin (SM), 779.1 ± 18.9 pmol/mg of protein; C16:0-glucosylceramide (Glc-ceramide), 22.9 ± 1.1 pmol/mg of protein; C16:0-lactosylceramide (Lac-ceramide), 19.3 ± 0.7 pmol/mg of protein. C.C16:0-ceramide content was down-regulated in mitochondria, whereas C16:0-sphingomyelin or C16:0-glucosyl-ceramide was not changed. The content of sphingolipids in mitochondria from P10 brain: C16:0-ceramide, 24.5 ± 0.9 pmol/mg of protein; C16:0-sphingomyelin, 646.4 ± 17.3 pmol/mg of protein; C16:0-glucosyl-ceramide, 20.1 ± 0.8 pmol/mg of protein. Data are mean ± S.E., compared with appropriate control. *, p < 0.05, n = 12. Each sample was normalized to its respective total protein levels.
Ceramide serves as a building block for complex sphingolipids, including SM and glycosphingolipids. Two classes of glycosphingolipids carry galactose or glucose as a first sugar on the ceramide backbone. Glucosyl-ceramide (Glc-ceramide) can be further glycosylated to lactosyl-ceramide (Lac-ceramide), a precursor for gangliosides, which constitute 10–12% of total cell membrane lipids in the animal brain (24). Fig. 3B illustrates the developmental changes in C16:0-ceramide-containing complex sphingolipids in the brain. Glycosphingolipid content (Glc-ceramide and Lac-ceramide) was increased, but C16:0-SM did not change. Further investigation revealed profound down-regulation of C16:0-ceramide in mitochondria, whereas the content of C16:0-SM or C16:0-Glc-ceramide was unchanged during brain development (Fig. 3C). The data suggest that C16:0-ceramide could play a specific functional role in mitochondria.
CerS6 Is Primarily Involved in Producing C16:0-ceramide in Mitochondria
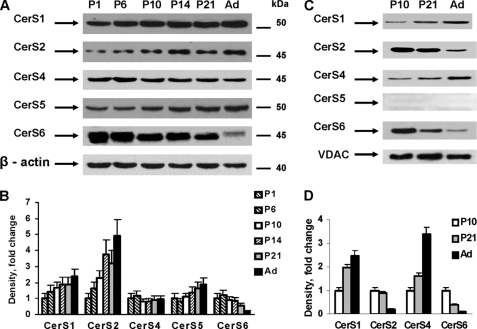
To define the role of ceramide in mitochondria, we focused on enzymes involved in ceramide biosynthesis. We measured protein expression of various (dihydro) ceramide synthase (CerS) isoforms in rat brains at different developmental stages (Fig. 4). In agreement with our finding of increases in C18:0-ceramide content (Fig. 2, A and B), CerS1 protein expression was gradually increased in brain and mitochondria (Fig. 4, A and C, respectively). These data are consistent with previous findings that CerS1 selectively utilizes stearoyl-CoA as acyl donor (14, 15) to generate C18:0-ceramide.
FIGURE 4.
Ceramide synthases are differentially expressed during brain development. Ceramide synthase protein expression was analyzed in brain tissue (A) or in mitochondria (C) by Western blot using specific antibodies. Membranes were stripped and probed with anti-β-actin antibody (brain) or anti-voltage-dependent anion channel (VDAC) antibody (mitochondria) to confirm equal loading of samples. B and D, quantification of the ceramide synthases expression in brain (B) or mitochondria (D). Data are mean ± S.E. (from 4 independent experiments).
The protein expression of CerS2 (Fig. 4A), a ceramide synthase that utilizes very long-chain acyl-CoAs, was up-regulated and this reflected increased levels of very long-chain ceramides, C20:0-, C20:1-, C22:0-, C22:1-, C24:0-, and C24:1-ceramide, during brain development (Fig. 2A). Whereas protein expression of CerS4, a ceramide synthase with a similar acyl-CoA specificity, did not differ significantly with developmental changes (Fig. 4A), we noted differential expression of CerS2 and CerS4 in mitochondria during brain development (Fig. 4B). Specifically, CerS2 protein expression was significantly down-regulated as CerS4 was up-regulated, suggesting that CerS4 regulates very long-chain ceramide levels in brain mitochondria. The data suggest a lack of uniform developmental changes in CerS2 and CerS4 expression in mitochondria compared with cellular levels of these enzymes in the brain. This finding lends support to the idea that mitochondria represent a separate and unique intracellular compartment involved in sphingolipid metabolism (12, 13, 25).
CerS5 and CerS6 have a similar substrate specificity: both ceramide synthases can utilize palmitoyl-CoA to produce C16:0-ceramide (14). Fig. 4A illustrates differential protein expression of CerS5 and CesR6 in the developing brain. Specifically, CerS5 expression was gradually up-regulated during postnatal brain development, whereas CerS6 expression was significantly down-regulated in the adult rat brain. Consistent with previous findings (12), CerS5 was not localized to mitochondria, whereas CerS6 expression was down-regulated in the organelle (Fig. 4B). Decreases in CerS6 expression were associated with reduced C16:0-ceramide in mitochondria during brain development (Fig. 3C). Altogether, the data suggest that CerS6 is a primary ceramide synthase that produces C16:0-ceramide in brain mitochondria.
Ceramide Is Involved in Regulation of Mitochondrial Ca2+ Homeostasis
To investigate whether differential CerS6 expression affected mitochondrial function, respiratory chain activity was measured. Mitochondrial oxygen consumption supported by respiratory chain substrates of Complex I, glutamate and malate, or substrates of Complex II, succinate, or of Complex IV, ascorbate and TMPD, were measured with/without ADP. Respiration rates in the presence of any tested substrate alone (state 2) were similar among mitochondria isolated from P10, P21, and adult (6-month-old) rat brains. There were no significant differences in state 3 (in the presence of ADP) respiration rates among brain mitochondria at various developmental ages, according to measurements of respiration supported by glutamate and malate (109.3 ± 6.2 nAO/min/mg of protein), succinate (128.1 ± 6.9 nAO/min/mg of protein), or ascorbate and TMPD (150.6 ± 7.5 nAO/min/mg of protein). Respiratory control ratios were 6.72 ± 0.31 for mitochondria isolated from P10 or P21 or adult rat brains. These data indicate a lack of change in oxidative phosphorylation parameters despite differential CerS6 expression in brain mitochondria.
In addition to generating ATP, mitochondria also maintain low cytosolic Ca2+ levels (26) by sequestering Ca2+ inside the mitochondrial matrix complexed with phosphate (27, 28). Energized mitochondria take up Ca2+ via the mitochondrial calcium uniporter, which has been recently described as a highly selective, inwardly rectifying channel (29, 30). The mitochondrial calcium uniporter is activated by Ca2+ concentrations greater than 200 nm, and an estimated 10–40 mitochondrial calcium uniporter channels per μm2 are thought to be localized in the inner mitochondrial membrane (29).
Excessive accumulation of Ca2+ in the mitochondrial matrix could trigger opening of MPTP at a high conductance state, which would be accompanied by dissipation of the transmembrane potential and mitochondrial swelling. In brain mitochondria, Ca2+ may also activate a limited permeability state of MPTP opening (31) that only depolarizes mitochondria without causing swelling (32). This depolarization dramatically reduces the driving force for Ca2+ influx via mitochondrial calcium uniporter, thus limiting the mitochondrial ability to sequester Ca2+ (33).
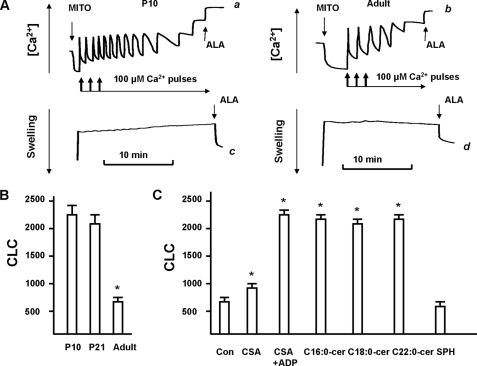
To determine the effect of differential CerS6 expression on the ability of mitochondria to regulate Ca2+, the CLC was measured in mitochondria from brains of rats at different postnatal developmental ages (P10, P21, or 6 months old) (Fig. 5). Pulses of 100 μm Ca2+ were added to mitochondria energized by the substrate of Complex II, succinate, whereas electron transport through Complex I was inhibited by 1 μm rotenone. In line with previous studies (33, 34), sequential Ca2+ additions caused gradual decreases in the Ca2+ uptake rates until virtually complete inhibition of Ca2+ uptake was achieved (Fig. 5A). Notably, mitochondria retained all accumulated Ca2+ and did not swell, which is consistent with the MPTP opening at a low conductance state (31, 33). The addition of the pore-forming peptide alamethicin permitted detection of mitochondrial swelling under these conditions. Quantification of CLC revealed a profoundly reduced ability of adult rat brain mitochondria to retain Ca2+ (Fig. 5B), compared with P10 or P21 brains.
FIGURE 5.
Mitochondrial CLC is decreased in adult rat brain compared with young rat and could be rescued by ceramide. A, sequential Ca2+ pulses (100 μm each) caused gradual decreases in the Ca2+ uptake rates (traces a and b). The arrows indicate the addition of mitochondria (MITO) and Ca2+. No changes in swelling (traces c and d) at the increasing Ca2+ load were detected. Maximal Ca2+ accumulation was followed by addition of 30 μg/ml of pore-forming peptide alamethicin (ALA). Data are representative of four independent experiments. B, quantitative assessment of CLC (nanomole of Ca2+/mg of protein). Data are mean ± S.E., *, p < 0.05, n = 4. C, CLC was measured in mitochondria purified from an adult (6 month old) rat brain in the presence of vehicle control (con) or 1 nmol/mg of protein CSA or 1 mm ADP with 1 nmol/mg of protein CSA, C16:0-ceramide, C18:0-ceramide, C22:0-ceramide, or SPH. Data are mean ± S.E., *, p < 0.05, n = 3.
To investigate whether decreased CLC is dependent on lower CerS6 expression and C16:0-ceramide in mitochondria from adult brains, mitochondria were supplemented with 1 nmol of C16:0-ceramide/mg of protein. Fig. 5C shows that C16:0-ceramide restored the CLC of adult brain mitochondria to that of mitochondria from young rat brains. The data suggest an involvement of CerS6 and C16:0-ceramide in the regulation of mitochondrial Ca2+-buffering capacity.
Next, we explored the underlying mechanism of C16:0-ceramide-mediated increases in CLC of mitochondria from adult brains. Physiologically relevant concentrations of C16:0-ceramide have been shown to prevent mitochondrial swelling in response to high Ca2+ load (35), which is indicative of the MPTP opening at a high conductance state. However, the MPTP ceramide-binding site had low specificity toward long-chain and very long-chain ceramides (35). Consistent with these findings, addition of C18:0-ceramide (Fig. 5C), C22:0-ceramide (Fig. 5C), or C24:0-ceramide (not shown) restored the CLC of adult brain mitochondria to that of mitochondria from young rat brains. In contrast, sphingosine had no effect.
To examine whether ceramide-dependent modulation of MPTP opening is responsible for increased mitochondrial CLC; another inhibitor of MPTP, cyclosporin A (CSA), was tested (Fig. 5C). CSA is a potent inhibitor of MPTP in heart or liver mitochondria, but it had a limited ability to prevent the MPTP opening at a high conductance state in brain mitochondria. It was more effective with ADP addition (32). CSA was moderately effective in enhancing the CLC of adult brain mitochondria, but CSA with ADP restored CLC to that of mitochondria from young rat brains (Fig. 5C). This supports the concept that Ca2+ could induce MPTP opening at a low conductance state that results in no swelling and dissipation of transmembrane potential, the driving force for Ca2+ uptake (31, 33). Therefore, inhibitors of MPTP opening at a high conductance state can block MPTP at a low conductance state resulting in increased mitochondrial CLC.
These data suggest an involvement of CerS6 and C16:0-ceramide in the regulation of Ca2+ homeostasis in brain mitochondria. Thus, lower CerS6 expression and C16:0-ceramide content were associated with reduced mitochondrial CLC in adult brain mitochondria, whereas exogenous C16:0-ceramide restored CLC to that of young brain mitochondria. Despite their ability to increase CLC in adult mitochondria, C18:0-, C22:0-, and C24:0-ceramide seem unlikely candidates for regulating CLC, because there were inverse correlations between their levels and CLC in mitochondria during brain development. In contrast to C16:0-ceramide, these three ceramide species were more abundant in mitochondria from adult brains with lower CLC compared with mitochondria from young animal brains (characterized by reduced content of these ceramides and higher CLC).
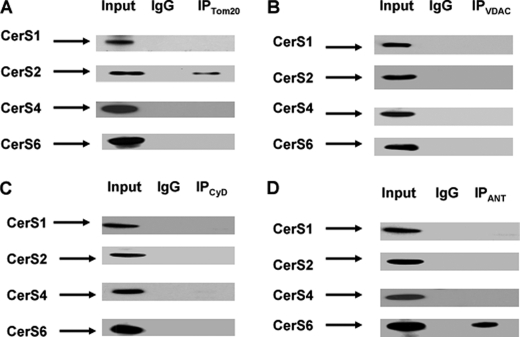
Highly hydrophobic ceramide appears to be segregated within the membrane with its generating enzyme and require specific transporter proteins to reach other membrane compartments (1). This suggests that CerS6 may produce C16:0-ceramide proximal to the MPTP ceramide-binding site in the inner membrane, whereas CerSs generating C18:0-, C22:0-, and C24:0-ceramide could be localized to other intra-mitochondrial compartments. To examine the intra-mitochondrial localization of CerS6, co-immunoprecipitations were performed using antibodies against protein components of MPTP: outer mitochondrial membrane resident protein voltage-dependent anion channel, inner mitochondrial membrane resident ANT, and matrix protein cyclophilin D. These studies reveal a selective CerS6 association with ANT, the inner membrane component of MPTP (Fig. 6). In contrast, CerS2 associated with the outer membrane resident protein Tom20, a receptor of the protein import complex. The data suggest CerS6/ceramide could regulate the MPTP activity and mitochondrial Ca2+ homeostasis.
FIGURE 6.
CerS6 is associated with ANT, whereas CerS2 is associated with protein import complex. Mitochondria were purified from the brain of an adult rat. Mitochondria lysate (10 μg) was loaded into the lane (Input). Complexing with each CerS was detected by immunoprecipitation using antibodies against Tom20, a receptor of the outer mitochondrial membrane protein import complex (A); voltage-dependent anion channel (VDAC), outer mitochondrial membrane component of MPTP (B); cyclophilin D (CyD), matrix protein-regulator of MPTP (C); or ANT, the inner mitochondrial membrane component of MPTP (D). As a control, the same immunoprecipitation procedure was performed except for primary antibody application (IgG).
CerS6 and Ceramide Generation Are Required for OL Apoptosis in Response to Glutamate
To further investigate the role of CerS6 during brain development, our studies were limited to OLs. OL precursor cells were purified from mixed glial cultures, plated onto fibronectin-coated plates, and cultured for 21 days in medium supplemented with N2, which is known to promote OL differentiation. In culture, OL precursor cells successfully progressed through the lineage that was manifested by increased expression of an OL-specific differentiation marker, myelin proteolipid protein (Fig. 7A). Similar to the mixed cell population in the brain (Fig. 4A), CerS6 protein expression was down-regulated during OL differentiation, whereas CerS5 expression was up-regulated (Fig. 7A). During normal brain development OL precursor cells are greatly overproduced and the final OL cell number is adjusted to the number of axons requiring myelination by increased OL apoptosis (36). It has been emphasized that neuron-derived factors, including the neurotransmitter glutamate, could control OL responses (36, 37).
FIGURE 7.
Glutamate-induced OL death is ceramide-dependent. A, CerS5 and CerS6 protein expression was analyzed in OLs during cell differentiation in culture by Western blot. Proteolipid protein is a marker for OL differentiation, and β-actin was used for normalization purposes. B, quantification of the ceramide synthases expression in cultured OLs during differentiation. C, OLs were exposed to glutamate with/without 20 μm fumonisin B1 (FB1) or 1 μm myriocin (MR) or 200 μm cystine, glutamate antiporter inhibitor, and cell death was quantified 24 h later. Data are mean ± S.E., *, p < 0.05, n = 12. D, OLs were exposed to 6 mm glutamate (Glut) with/without 20 μm FB1 or 1 μm myriocin and ceramide content was analyzed 24 h later. Data are mean ± S.E., *, p < 0.05, n = 12.
To ascertain the role of CerS6 in apoptosis, OL precursor cell response to glutamate was examined. CerS6 expression in OL precursor cells is depicted in Fig. 7A at day 1. Glutamate can damage OLs via excitotoxicity, which is caused by sustained activation of ionotropic glutamate receptors, and by receptor-independent mechanisms, secondary to glutamate uptake (38, 39). Glutamate enters the cell via bidirectional cystine/glutamate antiporter that results in reduced cytosolic cystine leading to glutathione depletion and cell death (40, 41). Indeed, OL exposure to glutamate reduced cell survival (Fig. 7C). As expected, blocking glutamate uptake with 200 μm cystine protected OLs against high glutamate concentrations (above 1 mm) indicating the involvement of the cystine/glutamate antiporter.
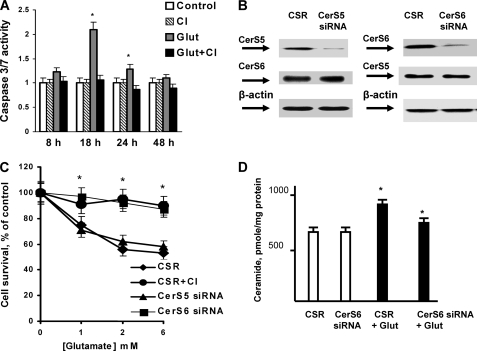
To elucidate whether glutamate-induced OL death is mediated by CerS and ceramide, a specific inhibitor of CerS activity, fumonisin B1 (FB1), and an inhibitor of ceramide biosynthesis, myriocin, were employed. CerS produces ceramide via two pathways: by de novo ceramide biosynthesis and the recycling or salvage pathway (1, 8, 42). If blocking CerS activity with FB1 enhanced OL survival, it would indicate CerS activity is required for glutamate-induced OL death. If blocking ceramide biosynthesis with myriocin increased OL survival, the involvement of recycling or the salvage pathway could be ruled out (42). In fact, both inhibitors protected OL from glutamate toxicity, suggesting that glutamate triggers activation of CerS via the de novo ceramide biosynthetic pathway leading to OL death (Fig. 7C). Furthermore, sphingolipid analysis revealed an about 3-fold increase in ceramide content after OL treatment with glutamate that was abolished by FB1 or myriocin (Fig. 7D). The data suggest that ceramide is an essential mediator of glutamate-induced OL death.
Glutamate-induced OL death appears to be mediated by ceramide-dependent apoptotic mechanisms. Fig. 8A shows that glutamate-triggered activation of executioner caspases 3/7 peaked at 18 h after treatment with glutamate. A new generation pan-caspase inhibitor, Q-VD-OPH blocked caspase activation (Fig. 8A). These studies suggest that glutamate-induced OL apoptosis is dependent on activation of ceramide synthase that participates in de novo ceramide biosynthesis.
FIGURE 8.
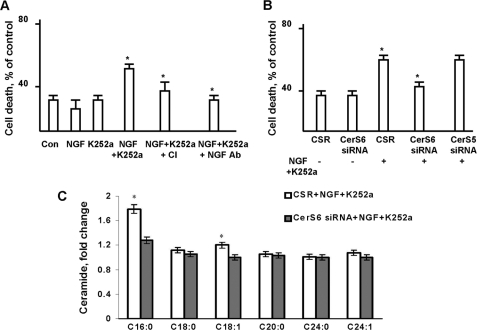
CerS6 knockdown decreased ceramide and enhanced cell survival. A, OLs were exposed to glutamate (Glut) with/without 20 μm pan-caspase inhibitor (CI), and caspase 3/7 activity was measured. Data are mean ± S.E., *, p < 0.05, n = 12. B, OLs were transfected with 1 nm CerS6-specific siRNA or 1 nm CerS5-specific siRNA or control siRNA (CSR) and cultured for 24 h. CerS5 or CerS6 protein expression was confirmed by Western blot. C, OLs were transfected with 1 nm CerS6-specific siRNA or 1 nm CerS5-specific siRNA or control siRNA (CSR) and cultured for 24 h. Then, cells were treated with glutamate with/without 20 μm pan-caspase inhibitor (CI). Cell survival was measured 24 h later. Data are mean ± S.E., *, p < 0.05, n = 12. D, OLs were transfected with 1 nm CerS6-specific siRNA or 1 nm CerS5-specific siRNA or control siRNA (CSR) and cultured for 24 h. Then, cells were treated with 6 mm glutamate (Glut), and ceramide was analyzed 24 h later. Data are mean ± S.E., *, p < 0.05, n = 18.
To identify the ceramide synthase promoting apoptotic mechanisms in response to glutamate, CerS6 or CerS5 were knocked down using siRNA. OLs were transfected with 1 nmol of siRNA targeting CerS6 or CerS5 or control siRNA (CSR), and cultured for 48 h. Western blot analysis indicated ∼80% knockdown of CerS6 or CerS5 expression (Fig. 8B). Importantly, there was no interference between effects of CerS5 and CerS6 siRNA. Thus, siRNA targeting CerS5 did not affect the protein expression of CerS6, and vice versa. It should be mentioned that CerS6 siRNA at 10 nmol and higher could decrease CerS5 protein expression (not shown). Knocking down CerS6 protected OLs from glutamate toxicity, whereas knocking down CerS5 had no effect on OL survival (Fig. 8C).
Pan-caspase inhibitor prevented glutamate-induced OL death, thereby confirming the involvement of apoptotic mechanisms. Furthermore, knockdown of CerS6 impeded the increase in ceramide after OL exposure to glutamate (Fig. 8D).
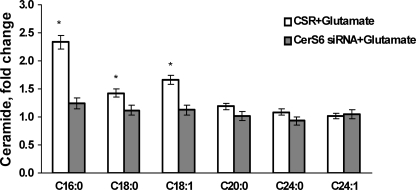
Analysis of the ceramide species revealed the most dramatic increase in C16:0-ceramide (up to 2.3-fold) and smaller increases in C18:0- and C18:1-ceramide (up to 1.4- and 1.7-fold, respectively) after OL treatment with glutamate (Fig. 9). Knocking down CerS6 attenuated increases in ceramide species in response to glutamate. The data are expressed as fold-increases of OL ceramide content shown in Table 3. The results of these studies support a critical role for mitochondrial CerS6-generated C16:0-ceramide in glutamate-induced apoptosis and suggest the additional pro-apoptotic input of C18:0- and C18:1-ceramide, which could also be generated by CerS6. To verify these results, OLs were transfected with another siRNA targeting CerS6 (see “Experimental Procedures”) that yielded similar OL protection from glutamate toxicity (not shown). These data suggest that CerS6 is required for glutamate-induced OL apoptosis.
FIGURE 9.
CerS6 knockdown effect on ceramide species profile in response to OL treatment with glutamate. OLs were transfected with 1 nm CerS6-specific siRNA or control siRNA (CSR) and cultured for 24 h. Then, cells were treated with 6 mm glutamate, and ceramide species was analyzed 24 h later. Data are mean ± S.E., *, p < 0.05, n = 18.
TABLE 3.
Ceramide species content of OLs transfected with CSR or CerS6 siRNA
Ceramide species were determined in OL cultures transfected with 1 nmol of siRNA targeting CerS6 or 1 nmol of control siRNA and cultured for 24 h. Each sample was normalized to its respective total protein levels. Values are mean ± S.E., n = 18.
| Ceramide | C16:0 | C18:0 | C18:1 | C20:0 | C24:0 | C24:1 |
|---|---|---|---|---|---|---|
| CSR | 89.7 ± 6.8 | 179.2 ± 10.6 | 5.9 ± 0.8 | 18.1 ± 1.3 | 192.0 ± 11.3 | 221.1 ± 8.4 |
| CerS6 siRNA | 80.6 ± 5.2 | 177.8 ± 9.7 | 4.9 ± 1.1 | 20.9 ± 1.1 | 205.9 ± 10.8 | 218.3 ± 9.7 |
CerS6 Is Necessary for NGF-induced OL Apoptosis
To determine whether the CerS6 requirement is specific to glutamate-induced OL apoptosis, we utilized another apoptotic stimulus, NGF. Apoptotic cell death is an essential feature of normal brain development, and it is controlled in part by a wide array of neurotrophic factors, including NGF, which binds and activates both the p75 neurotrophin receptor (p75NTR) and the receptor tyrosine kinase (TrkA) to dictate specific cell responses. Thus, blocking p75NTR function with antibodies resulted in hypomyelination of peripheral nerves, whereas blocking TrkA signaling with K252a yielded increased myelination, suggesting that myelination is under mutual control of p75NTR and TrkA signaling (43). Whereas TrkA receptors have a well defined trophic function, p75NTR exerts activities ranging from trophism to apoptosis. OLs have been shown to undergo p75NTR-dependent apoptosis in vitro (44) mediated by Rac GTPase activity (45), JNK phosphorylation, and caspase activation (46).
Fig. 10A shows that treatment of OLs with 1 mm NGF or 20 μm of the specific TrkA inhibitor K252a alone did not affect cell survival. In contrast, OL exposure to 1 mm NGF in the presence of 20 μm K252a significantly increased OL death that was completely prevented by anti-p75NTR antiserum, indicating the involvement of p75NTR. As expected, the pan-caspase inhibitor protected OL from NGF-induced toxicity.
To learn whether CerS6 is an important molecular determinant in NGF-induced pro-apoptotic signaling, CerS6 was knocked down using siRNA. CerS6 knockdown protected OLs from NGF-induced apoptosis, whereas knocking down CerS5 had no effect on OL survival (Fig. 10B). Furthermore, knocking down CerS6 prevented increases in C16:0- or C18:1-ceramide in response to NGF + K252a treatment (Fig. 10C). Data are expressed as fold-increases of OL ceramide species content (Table 3). Total ceramide did not increase after OLs exposure to NGF with K252a (not shown). The results of these studies indicate CerS6 involvement in promoting NGF-initiated apoptotic signaling in OLs. Collectively, the data suggest CerS6 activation and ceramide generation are important for OL apoptosis regardless of the apoptotic stimuli.
FIGURE 10.
CerS6 knockdown protected OLs against NGF-induced apoptosis. A, OLs were exposed to 1 mm NGF with/without TrkA receptor inhibitor, 50 μm K252a, or 1 μg/ml of anti-NGF receptor antibody, and cell death was measured 24 h later. B, OLs were transfected with 1 nm CerS6-specific siRNA or 1 nm CerS5-specific siRNA or control siRNA (CSR) and cultured for 24 h. Then, cells were treated with 1 mm NGF, 50 μm K252a, and cell survival was measured 24 h later. Data are mean ± S.E., *, p < 0.05, n = 12. C, OLs were transfected with 1 nm CerS6-specific siRNA control siRNA (CSR) and cultured for 24 h. Then, cells were treated with 1 mm NGF, 50 μm K252a, and ceramide species content was measured 24 h later. Data are mean ± S.E., *, p < 0.05, n = 12.
CerS6 Promotes OL Apoptosis by Increasing Ca2+ Influx in Mitochondria and Calpain Activation
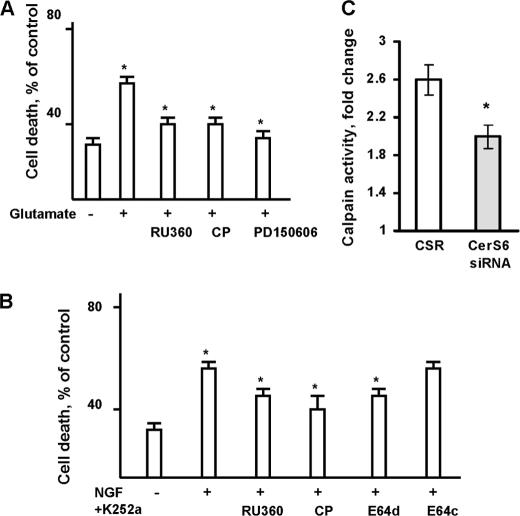
To identify downstream targets of CerS6-mediated pro-apoptotic signaling, we investigated the possibility that CerS6-dependent disturbance of mitochondrial Ca2+ homeostasis could be crucial for OL apoptosis. Having shown the ceramide-induced increases in mitochondrial CLC in isolated brain mitochondria, we focused our studies on the role of mitochondrial Ca2+ accumulation in promoting OL apoptosis using a specific inhibitor of the mitochondrial uniporter channel, RU360 (IC50 = 2 nm) (29). Blocking mitochondrial Ca2+ uptake with 1 μm RU360 significantly enhanced OL survival in response to glutamate or NGF (Fig. 11, A or B, respectively). This suggests that glutamate or NGF triggers activation of CerS6 and generation of ceramide in OL mitochondria leading to MPTP closure, increased Ca2+ influx via the uniporter channel, and reduced cell survival.
FIGURE 11.
Glutamate- or NGF-induced OL apoptosis involves disturbance of mitochondrial Ca2+ and calpain activation. A, OLs were exposed to glutamate with/without the mitochondrial Ca2+-uniporter inhibitor, 1 μm RU360, or calpain inhibitors, 1 μm calpeptin or 1 μm PD150606. Cell death was assessed 24 h later. Data are mean ± S.E., *, p < 0.05, n = 16. B, OLs were exposed to 1 mm NGF plus 20 μm K252a with/without 1 μm RU360 or 1 μm calpeptin or 1 μm E64d or 1 μm E64c, and cell death was measured 24 h later. Data are mean ± S.E., *, p < 0.05, n = 12. C, OLs were transfected with 1 nm CerS6-specific siRNA or control siRNA (CSR) and cultured for 24 h. Then, cells were treated with glutamate for 24 h and calpain activity was measured. Data are mean ± S.E., *, p < 0.05, n = 6.
Increased Ca2+ accumulation in OL mitochondria could result in activation of a Ca2+-dependent cysteine protease calpain (EC 3.4.22.17). Calpains are a 15-member family of non-lysosomal enzymes that degrade diverse proteins via limited proteolysis. Although calpains are mainly cytosolic enzymes, recent studies have shown that calpains 1, 2, and 10 exist in mitochondria and participate in the cleavage of aspartate aminotransferase, apoptosis-inducing factor, respiratory chain Complex I subunits, and the MPTP (47–49). Calpains exhibit different Ca2+ sensitivities in vitro, Kd = 25 μm Ca2+ for calpain 1, and Kd = 750 μm Ca2+ for calpain 2 (50). Mounting evidence supports mitochondrial calpain involvement in both caspase-dependent and -independent pathways of apoptotic cell death (51). Specifically, calpain has been shown to truncate apoptosis-inducing factor, a caspase-independent death effector, and to induce its release from the mitochondria (48, 51).
To elucidate whether calpain activation is required for OL apoptosis, structurally unrelated cell-permeable specific inhibitors of calpain activity were used. Peptide derivative (benziloxycarbonyl-Leu-nLeu-H) calpeptin (52) competes for the active site of calpain, whereas epoxysuccinyl peptide (E64d) covalently and irreversibly binds to a critical sulfhydryl group in the active site of the enzyme (53). In contrast, an α-mercaptoacrylic acid derivative, PD150606, is a selective nonpeptide uncompetitive inhibitor of calpain activity (Ki = 0.21 μm for calpain 1, and 0.37 μm for calpain 2) (54). OLs were exposed to glutamate with/without 1 μm calpeptin or 1 μm PD150606, and cell death was assessed 24 h later. Both inhibitors of calpain activity enhanced OL survival in response to glutamate (Fig. 11A). These data agree with previous reports of calpain involvement in pro-apoptotic signaling triggered by glutamate in motor neurons (55). Similarly, OLs were protected by 1 μm calpeptin or 1 μm E64c against NGF receptor-induced cell death (Fig. 11B). Whereas E64c, a cell-impermeable analog of E64d, had no effect. Study results suggest that glutamate- or NGF receptor-triggered pro-apoptotic signaling leads to a disturbance of mitochondrial Ca2+ homeostasis and activation of calpain in OLs.
To learn whether calpain is a downstream target in the CerS6-mediated pro-apoptotic signaling pathway, calpain activity was measured in OLs with down-regulated CerS6. Fig. 11C shows that glutamate increased (∼2.6 fold) calpain activity in OLs transfected with non-targeting siRNA (CSR). Glutamate-induced calpain activation was partly attenuated by knocking down CerS6. The data indicate CerS6 involvement in regulation of calpain activity in OLs. Altogether, the results of these studies suggest that CerS6-ceramide-mediated signaling increases mitochondrial Ca2+ load and calpain activity to promote apoptosis in OLs.
DISCUSSION
The present studies are unique in establishing a novel pro-apoptotic signaling pathway mediated by CerS6 in OLs. We have shown that an apoptotic stimulus triggers activation of CerS6 and generation of ceramide, thereby disturbing mitochondrial Ca2+ homeostasis and calpain activation, which results in OL death. Furthermore, CerS6 is down-regulated during postnatal brain development and appears to generate C16:0-ceramide in brain mitochondria. This is the first demonstration of an essential role of CerS6 in the neural cell apoptosis.
Apoptosis is important during brain development, eliminating excess cells and ensuring the establishment of a proper synaptic connection network. In contrast to OLs, neuronal apoptosis has been extensively studied, and two waves of neuronal cell death have been described. The first wave consists of a large number of dividing neurons being eliminated during a peak of neurogenesis at mid-embryogenesis due to competition for a limited supply of neurotrophic factors and intracellular processes (56). The second wave consists of differentiated neurons dying while migrating toward their target location or while connecting to target cells during the early postnatal period (57).
To provide electrical insulation and maximize their conduction velocity, the axonal tracts in the central nervous system are myelinated by OLs in early postnatal life. The myelin biogenesis is coordinated by neuronal signals that control OL proliferation, differentiation, and survival (36, 58). OLs are greatly overproduced and the cell number is adjusted to the number and length of axons requiring myelination (36). Only the OLs that manage to ensheath the axon survive, whereas those that fail degenerate (36). Given the significance of neuron-derived factors in regulation of OL survival, the characterization of a ceramide-mediated apoptotic pathway triggered by glutamate and nerve growth factor is a valuable contribution to our understanding of sphingolipid signaling in OL apoptosis during brain development.
Our studies identify CerS6 as a novel determinant of the pro-apoptotic signaling cascade in OLs. Here, we show that OL exposure to high concentrations of glutamate activates de novo ceramide biosynthesis, leading to cell death. Blocking ceramide production with a specific inhibitor of de novo ceramide biosynthesis or an inhibitor of ceramide synthase enhanced OL survival (Fig. 7, D and C). Gene knockdown experiments suggested involvement of CerS6, but not CerS5, in glutamate-induced OL death (Fig. 8, B–D). Further investigations revealed that CerS6-mediated OL death involves apoptotic mechanisms through activation of functionally similar executioner caspases 3 and 7 (Fig. 8A). Based on the knock-out mice studies, caspases 3 and 7 have been implicated as key mediators of apoptotic events downstream of mitochondria (59). Caspase activation requires the release of pro-apoptotic proteins from mitochondria due to mitochondrial dysfunction and loss of integrity. Indeed, glutamate-induced OL apoptosis appears to involve a disturbance in mitochondrial Ca2+ homeostasis and activation of the Ca2+-dependent protease, calpain (Fig. 11, A and C). Knocking down CerS6 reduced calpain activation in response to glutamate, suggesting that calpain is a downstream target of CerS6.
Furthermore, these studies show that a CerS6/ceramide-mediated pro-apoptotic signaling pathway is essential for p75NTR-induced OL apoptosis (Fig. 10). Whereas p75NTR-induced responses in OLs are certainly understudied, ceramide participation in p75NTR-initiated signaling activities, ranging from growth and differentiation to apoptosis, is well established in neurons (60, 61). Thus, stimulation of p75NTR activates neutral and/or acid sphingomyelinases in the vicinity of the receptor in the plasma membrane that results in SM hydrolysis and ceramide generation. It has been emphasized that transient ceramide production upon sphingomyelinase activation takes place within 1–5 min and mainly serves the membrane structure, thereby facilitating the clustering of the death receptors localized in lipid rafts and promoting apoptosis (62, 63). Our studies point to an important pro-apoptotic role of ceramide generated by CerS6 in mitochondria, and they agree with the concept that endogenous ceramide production should be considered in its topological context (1, 63).
Mitochondria are being appreciated as vital intracellular compartments for ceramide metabolism. Mitochondria have been shown to contain a variety of sphingolipids, including SM and ceramide (64, 65). Although several enzyme activities involved in ceramide metabolism have been shown in mitochondria, the nature of ceramide biosynthesis enzymes in this organelle is still a matter of debate (17).
Ceramide synthase activity was first detected (66, 67) and partially purified from a bovine brain mitochondria-enriched fraction (68). Mitochondrial enzymes had ∼2-fold higher specific ceramide synthase activity than the ceramide synthase from the ER. The mitochondrial enzyme had a pH optimum ∼7.5 and maximal catalytic efficiency with C16:0- or C18:0-acyl-CoA (68). Purification of ceramide synthase from bovine liver mitochondria yielded two major protein bands: 62 and 72 kDa (69). Detailed analysis of ceramide synthase activity in highly purified mitochondria by Bionda et al. (11) essentially confirmed previous findings. Thus, ceramide synthase activity was shown in rat liver mitochondria and in the subcompartment of the ER that is closely associated with mitochondria. Further submitochondrial investigation of ceramide synthase activity revealed enzyme localization to both outer and inner mitochondrial membranes (11).
Our studies describing CerS1, CerS2, CerS4, and CerS6, in purified brain mitochondria support the idea that several ceramide synthesizing enzymes could be localized to the mitochondria and/or to ER fragments tethered to the outer mitochondrial membrane (25, 70). The results of our studies suggest that CerS6 could be localized to the inner mitochondrial membrane proximal to the MPTP, whereas CerS1, CerS2, and CerS4 are likely to be found in the outer mitochondrial membrane. The additional source of ceramide in mitochondria is a reverse reaction of a neutral ceramidase, e.g. formation of ceramide as a result of condensation of palmitate and sphingosine (71). On the basis of molecular cloning and confocal microscopy data, this activity was ascribed to mitochondria (9), and it was demonstrated in purified mitochondria (11). A recent report suggests that ceramide could be generated by novel mitochondrial neutral sphingomyelinase hydrolyzing SM (10). Continued research efforts are required to better understand the mechanisms of mitochondrial ceramide generation and utilization along with its influence on mitochondrial functions.
Our studies provide further support for the concept of distinct roles of ceramide species in cell metabolism. As expected, sphingolipids and most ceramide species were increased in mitochondria during postnatal brain growth and development (Figs. 1 and 2). In contrast, C16:0-ceramide was severely reduced concomitantly with the down-regulation of CerS6 expression (Figs. 3 and 4). Although overexpressed in mammalian cells, CerS6 could generate C14:0-, C16:0-, and C18:0-ceramide (14); in brain tissue, CerS6 appears to specifically regulate C16:0-ceramide content in brain mitochondria during organ development. Increasing evidence suggests that the fatty acid chain of ceramide is an important characteristic of the biological effect mediated by the individual ceramide species. Generation of C18:0-ceramide, and not C16:0-ceramide, has been shown to repress the human telomerase reverse transcriptase promoter in lung carcinoma cells (72). Activation of acid sphingomyelinase in the salvage pathway brought about a selective accumulation of C16:0-ceramide (20, 73) due to the involvement of CerS5 (73). Another study revealed a specific role for dihydro-C16:0-ceramide in the adaptive cardiac tissue response to hypoxia (74). Although certain ceramide species could have different effects on biophysical properties of the membrane lipid bilayer (75), it remains unclear how ceramides containing different fatty acids exert their effects upon cell physiology. These studies suggest that regulated expression of specific CerS in intracellular compartments in conjunction with availability of certain fatty acyl-CoA species could be an important mechanism for controlling the fatty acid composition of ceramides and their biological effects on cell metabolism.
In the present study, we investigated the effect of differential CerS6 protein expression on mitochondrial functions in brain mitochondria from young (high CerS6 expression) or adult (low CerS6 expression) rat brains (Fig. 5). Assessment of respiratory chain enzyme activities revealed no changes in oxidative phosphorylation parameters between brain mitochondria from animals at different ages. Consistent with previous reports (76), mitochondria from young animal brains were characterized by higher CLC compared with brain mitochondria from adult rats (Fig. 5). Remarkably, long-chain or very long-chain ceramide addition to brain mitochondria from adult rats enhanced their ability to retain Ca2+ such that the mitochondria were similar to mitochondria from young animal brains. Ceramide-mediated blockade of MPTP opening seems to be the underlying mechanism of the increased CLC. Further studies reveal selective association of CerS6 with ANT the inner mitochondrial membrane component of MPTP thereby linking CerS6 to the regulation of MPTP activity (Fig. 6). These studies suggest a novel role for CerS6/ceramide in governing Ca2+ homeostasis in brain mitochondria.
The results from our study implicate CerS6 as an upstream regulator of calpain activity in the cellular response to apoptotic stimuli in OLs (Fig. 11). Calpains are part of a broad family of intracellular cysteine proteases that are independent from caspases. Typically, calpains function as key regulators in cytoskeletal remodeling through their substrates, including the microtubule-associated proteins neurofilament, Tau, and actin (77). Conversely, calpain activation was found to increase as intracellular Ca2+ increased during oxidative stress, leading to induction of apoptotic pathways (55, 78).
Whereas calpains are mainly cytosolic proteins, numerous reports exist of mitochondrial calpain-like activity (51). Cytosolic contamination of mitochondrial preparations has been a concern, and a few studies report the presence of calpain in the inner membrane fraction (47). Mitochondrial calpains are thought to facilitate apoptosis-inducing factor release from the intra-membrane space, inducing caspase-independent apoptosis, but direct experimental evidence has been elusive (79). Relevant to this study is the ability of mitochondrial calpain to modulate the activity of the MPTP, and the subsequent release of proteins initiating the caspase-dependent apoptotic cascade. Overexpression of calpain 10 induced mitochondrial fragmentation and swelling, consistent with the MPTP opening at a high conductance state and this altered mitochondrial morphology was blocked by MPTP inhibitors in kidney cells (49).
Conceivably, an apoptotic stimulus triggers a cytosolic Ca2+ influx into the mitochondria in OLs and an activation of mitochondrial CerS6 that then elevates ceramide. Ceramide blocks the MPTP opening at a low conductance state, leading to increased Ca2+ in the mitochondrial matrix. Rising mitochondrial Ca2+ activates calpain 10, which could cleave protein components of the MPTP resulting in the MPTP opening at a high conductance state, swelling, and rupture of the outer mitochondrial membrane and release of cytochrome c to initiate caspase activation. Noteworthy, all 8 calpain 10 splice variants seem to possess a mitochondrial targeting sequence localized to the NH2-terminal 15 amino acids (49). Mitochondrial calpain expression appears to be tissue-specific. Also, it has been shown that smooth muscle and rat liver mitochondria do not contain calpain 10, whereas kidney mitochondria express only calpain 10 and not calpains 1 or 2 (51). Identification of the calpain isoform activated by the CerS6/ceramide-dependent pro-apoptotic pathway in OL mitochondria would be helpful, and these studies are currently underway in our laboratory.
In summary, this study provides experimental evidence that apoptotic stimuli trigger activation of CerS6 and accumulation of ceramide that results in an increased Ca2+ in mitochondrial matrix and activation of calpain in OLs, and the data shed more light on the compartmentalization of sphingolipid metabolism and function in brain.
Acknowledgments
We are very grateful to Drs. Narayan Bhat and Edward Goetzl for stimulating discussions regarding the manuscript. We thank Dr. Wendy Macklin for generously providing anti-proteolipid protein antibody. We thank Dr. Jennifer G. Schnellmann for help with preparation of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant P20 RR 17677 for NCCR COBRE in Lipidomics and Pathobiology (to T. I. G.), Grant AG16583 (to L. M. O.), and Veterans Affairs Rehabilitation Research and Development Merit Awards (to T. I. G. and M. S. K.).
- SM
- sphingomyelin
- CI
- caspase inhibitor
- CerS
- ceramide synthase
- CLC
- Ca2+-loading capacity
- CSR
- control siRNA
- CSA
- cyclosporin A
- FB1
- fumonisin B1
- Glc-ceramide
- glucosyl-ceramide
- Lac-ceramide
- lactosyl-ceramide
- MPTP
- mitochondrial permeability transition pore
- OL
- oligodendrocyte
- p75NTR
- p75 neurotrophin receptor
- SPH
- sphingosine
- S1P
- sphingosine-1-phosphate
- TrkA
- receptor tyrosine kinase
- TMPD
- N,N,N′,N′-tetramethyl-p-phenelenediamine
- ANT
- adenine nucleotide translocator.
REFERENCES
- 1. Hannun Y. A., Obeid L. M. (2008) Nat. Rev. Mol. Cell Biol. 9, 139–150 [DOI] [PubMed] [Google Scholar]
- 2. Kolesnick R. N., Goñi F. M., Alonso A. (2000) J. Cell. Physiol. 184, 285–300 [DOI] [PubMed] [Google Scholar]
- 3. Futerman A. H., Riezman H. (2005) Trends Cell Biol. 15, 312–318 [DOI] [PubMed] [Google Scholar]
- 4. Merrill A. H., Jr. (2002) J. Biol. Chem. 277, 25843–25846 [DOI] [PubMed] [Google Scholar]
- 5. Mandon E. C., Ehses I., Rother J., van Echten G., Sandhoff K. (1992) J. Biol. Chem. 267, 11144–11148 [PubMed] [Google Scholar]
- 6. Kolter T., Proia R. L., Sandhoff K. (2002) J. Biol. Chem. 277, 25859–25862 [DOI] [PubMed] [Google Scholar]
- 7. Futerman A. H., Stieger B., Hubbard A. L., Pagano R. E. (1990) J. Biol. Chem. 265, 8650–8657 [PubMed] [Google Scholar]
- 8. Novgorodov S. A., Gudz T. I. (2009) J. Cardiovasc. Pharmacol. 53, 198–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. El Bawab S., Roddy P., Qian T., Bielawska A., Lemasters J. J., Hannun Y. A. (2000) J. Biol. Chem. 275, 21508–21513 [DOI] [PubMed] [Google Scholar]
- 10. Wu B. X., Rajagopalan V., Roddy P. L., Clarke C. J., Hannun Y. A. (2010) J. Biol. Chem. 285, 17993–18002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bionda C., Portoukalian J., Schmitt D., Rodriguez-Lafrasse C., Ardail D. (2004) Biochem. J. 382, 527–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yu J., Novgorodov S. A., Chudakova D., Zhu H., Bielawska A., Bielawski J., Obeid L. M., Kindy M. S., Gudz T. I. (2007) J. Biol. Chem. 282, 25940–25949 [DOI] [PubMed] [Google Scholar]
- 13. Deng X., Yin X., Allan R., Lu D. D., Maurer C. W., Haimovitz-Friedman A., Fuks Z., Shaham S., Kolesnick R. (2008) Science 322, 110–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mizutani Y., Kihara A., Igarashi Y. (2005) Biochem. J. 390, 263–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Spassieva S., Seo J. G., Jiang J. C., Bielawski J., Alvarez-Vasquez F., Jazwinski S. M., Hannun Y. A., Obeid L. M. (2006) J. Biol. Chem. 281, 33931–33938 [DOI] [PubMed] [Google Scholar]
- 16. Mizutani Y., Kihara A., Igarashi Y. (2006) Biochem. J. 398, 531–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Laviad E. L., Albee L., Pankova-Kholmyansky I., Epstein S., Park H., Merrill A. H., Jr., Futerman A. H. (2008) J. Biol. Chem. 283, 5677–5684 [DOI] [PubMed] [Google Scholar]
- 18. Lahiri S., Futerman A. H. (2005) J. Biol. Chem. 280, 33735–33738 [DOI] [PubMed] [Google Scholar]
- 19. Novgorodov S. A., Gudz T. I., Milgrom Y. M., Brierley G. P. (1992) J. Biol. Chem. 267, 16274–16282 [PubMed] [Google Scholar]
- 20. Chudakova D. A., Zeidan Y. H., Wheeler B. W., Yu J., Novgorodov S. A., Kindy M. S., Hannun Y. A., Gudz T. I. (2008) J. Biol. Chem. 283, 28806–28816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bielawski J., Szulc Z. M., Hannun Y. A., Bielawska A. (2006) Methods 39, 82–91 [DOI] [PubMed] [Google Scholar]
- 22. Hannun Y. A., Obeid L. M. (2002) J. Biol. Chem. 277, 25847–25850 [DOI] [PubMed] [Google Scholar]
- 23. van Meer G. (2005) EMBO J. 24, 3159–3165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Degroote S., Wolthoorn J., van Meer G. (2004) Semin. Cell Dev. Biol. 15, 375–387 [DOI] [PubMed] [Google Scholar]
- 25. Futerman A. H. (2006) Biochim. Biophys. Acta 1758, 1885–1892 [DOI] [PubMed] [Google Scholar]
- 26. Nicholls D. G. (1985) Prog. Brain Res. 63, 97–106 [DOI] [PubMed] [Google Scholar]
- 27. Chalmers S., Nicholls D. G. (2003) J. Biol. Chem. 278, 19062–19070 [DOI] [PubMed] [Google Scholar]
- 28. Kristian T., Pivovarova N. B., Fiskum G., Andrews S. B. (2007) J. Neurochem. 102, 1346–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kirichok Y., Krapivinsky G., Clapham D. E. (2004) Nature 427, 360–364 [DOI] [PubMed] [Google Scholar]
- 30. Hoppe U. C. (2010) FEBS Lett. 584, 1975–1981 [DOI] [PubMed] [Google Scholar]
- 31. Novgorodov S. A., Gudz T. I. (1996) J. Bioenerg. Biomembr. 28, 139–146 [DOI] [PubMed] [Google Scholar]
- 32. Brustovetsky N., Dubinsky J. M. (2000) J. Neurosci. 20, 8229–8237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Brustovetsky N., Dubinsky J. M. (2000) J. Neurosci. 20, 103–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kushnareva Y. E., Wiley S. E., Ward M. W., Andreyev A. Y., Murphy A. N. (2005) J. Biol. Chem. 280, 28894–28902 [DOI] [PubMed] [Google Scholar]
- 35. Novgorodov S. A., Gudz T. I., Obeid L. M. (2008) J. Biol. Chem. 283, 24707–24717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Barres B. A., Hart I. K., Coles H. S., Burne J. F., Voyvodic J. T., Richardson W. D., Raff M. C. (1992) Cell 70, 31–46 [DOI] [PubMed] [Google Scholar]
- 37. Gudz T. I., Komuro H., Macklin W. B. (2006) J. Neuros ci. 26, 2458–2466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Matute C., Domercq M., Sánchez-Gómez M. V. (2006) Glia 53, 212–224 [DOI] [PubMed] [Google Scholar]
- 39. Goldberg M. P., Ransom B. R. (2003) Stroke 34, 330–332 [DOI] [PubMed] [Google Scholar]
- 40. Wang L., Hinoi E., Takemori A., Nakamichi N., Yoneda Y. (2006) J. Biol. Chem. 281, 24553–24565 [DOI] [PubMed] [Google Scholar]
- 41. Iemata M., Takarada T., Hinoi E., Taniura H., Yoneda Y. (2007) J. Cell. Physiol. 213, 721–729 [DOI] [PubMed] [Google Scholar]
- 42. Kitatani K., Idkowiak-Baldys J., Hannun Y. A. (2008) Cell Signal. 20, 1010–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cosgaya J. M., Chan J. R., Shooter E. M. (2002) Science 298, 1245–1248 [DOI] [PubMed] [Google Scholar]
- 44. Casaccia-Bonnefil P., Carter B. D., Dobrowsky R. T., Chao M. V. (1996) Nature 383, 716–719 [DOI] [PubMed] [Google Scholar]
- 45. Harrington A. W., Kim J. Y., Yoon S. O. (2002) J. Neurosci. 22, 156–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gu C., Casaccia-Bonnefil P., Srinivasan A., Chao M. V. (1999) J. Neurosci. 19, 3043–3049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kar P., Chakraborti T., Samanta K., Chakraborti S. (2008) Arch. Biochem. Biophys. 470, 176–186 [DOI] [PubMed] [Google Scholar]
- 48. Ozaki T., Yamashita T., Ishiguro S. (2009) Biochim. Biophys. Acta 1793, 1848–1859 [DOI] [PubMed] [Google Scholar]
- 49. Arrington D. D., Van Vleet T. R., Schnellmann R. G. (2006) Am. J. Physiol. Cell Physiol. 291, C1159–1171 [DOI] [PubMed] [Google Scholar]
- 50. Elce J. S., Hegadorn C., Arthur J. S. (1997) J. Biol. Chem. 272, 11268–11275 [DOI] [PubMed] [Google Scholar]
- 51. Kar P., Samanta K., Shaikh S., Chowdhury A., Chakraborti T., Chakraborti S. (2010) Arch. Biochem. Biophys. 495, 1–7 [DOI] [PubMed] [Google Scholar]
- 52. Tsujinaka T., Kajiwara Y., Kambayashi J., Sakon M., Higuchi N., Tanaka T., Mori T. (1988) Biochem. Biophys. Res. Commun. 153, 1201–1208 [DOI] [PubMed] [Google Scholar]
- 53. Lampi K. J., Kadoya K., Azuma M., David L. L., Shearer T. R. (1992) Toxicol. Appl. Pharmacol. 117, 53–57 [DOI] [PubMed] [Google Scholar]
- 54. Wang K. K., Nath R., Posner A., Raser K. J., Buroker-Kilgore M., Hajimohammadreza I., Probert A. W., Jr., Marcoux F. W., Ye Q., Takano E., Hatanaka M., Maki M., Caner H., Collins J. L., Fergus A., Lee K. S., Lunney E. A., Hays S. J., Yuen P. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 6687–6692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Das A., Sribnick E. A., Wingrave J. M., Del Re A. M., Woodward J. J., Appel S. H., Banik N. L., Ray S. K. (2005) J. Neurosci. Res. 81, 551–562 [DOI] [PubMed] [Google Scholar]
- 56. Meier P., Finch A., Evan G. (2000) Nature 407, 796–801 [DOI] [PubMed] [Google Scholar]
- 57. de la Rosa E. J., de Pablo F. (2000) Trends Neurosci. 23, 454–458 [DOI] [PubMed] [Google Scholar]
- 58. Barres B. A., Raff M. C. (1999) J. Cell Biol. 147, 1123–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lakhani S. A., Masud A., Kuida K., Porter G. A., Jr., Booth C. J., Mehal W. Z., Inayat I., Flavell R. A. (2006) Science 311, 847–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Brann A. B., Scott R., Neuberger Y., Abulafia D., Boldin S., Fainzilber M., Futerman A. H. (1999) J. Neurosci. 19, 8199–8206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Blöchl A., Blöchl R. (2007) J. Neurochem. 102, 289–305 [DOI] [PubMed] [Google Scholar]
- 62. Grassmé H., Cremesti A., Kolesnick R., Gulbins E. (2003) Oncogene 22, 5457–5470 [DOI] [PubMed] [Google Scholar]
- 63. van Blitterswijk W. J., van der Luit A. H., Veldman R. J., Verheij M., Borst J. (2003) Biochem. J. 369, 199–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ardail D., Popa I., Alcantara K., Pons A., Zanetta J. P., Louisot P., Thomas L., Portoukalian J. (2001) FEBS Lett. 488, 160–164 [DOI] [PubMed] [Google Scholar]
- 65. Tserng K. Y., Griffin R. (2003) Anal. Biochem. 323, 84–93 [DOI] [PubMed] [Google Scholar]
- 66. Morell P., Radin N. S. (1970) J. Biol. Chem. 245, 342–350 [PubMed] [Google Scholar]
- 67. Ullman M. D., Radin N. S. (1972) Arch. Biochem. Biophys. 152, 767–777 [DOI] [PubMed] [Google Scholar]
- 68. Shimeno H., Soeda S., Yasukouchi M., Okamura N., Nagamatsu A. (1995) Biol. Pharm. Bull. 18, 1335–1339 [DOI] [PubMed] [Google Scholar]
- 69. Shimeno H., Soeda S., Sakamoto M., Kouchi T., Kowakame T., Kihara T. (1998) Lipids 33, 601–605 [DOI] [PubMed] [Google Scholar]
- 70. Csordás G., Hajnóczky G. (2009) Biochim. Biophys. Acta 1787, 1352–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. El Bawab S., Birbes H., Roddy P., Szulc Z. M., Bielawska A., Hannun Y. A. (2001) J. Biol. Chem. 276, 16758–16766 [DOI] [PubMed] [Google Scholar]
- 72. Wooten-Blanks L. G., Song P., Senkal C. E., Ogretmen B. (2007) FASEB J. 21, 3386–3397 [DOI] [PubMed] [Google Scholar]
- 73. Kitatani K., Idkowiak-Baldys J., Bielawski J., Taha T. A., Jenkins R. W., Senkal C. E., Ogretmen B., Obeid L. M., Hannun Y. A. (2006) J. Biol. Chem. 281, 36793–36802 [DOI] [PubMed] [Google Scholar]
- 74. Noureddine L., Azzam R., Nemer G., Bielawski J., Nasser M., Bitar F., Dbaibo G. S. (2008) Prostaglandins Other Lipid Mediat. 86, 49–55 [DOI] [PubMed] [Google Scholar]
- 75. Sot J., Aranda F. J., Collado M. I., Goñi F. M., Alonso A. (2005) Biophys. J. 88, 3368–3380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Wang X., Carlsson Y., Basso E., Zhu C., Rousset C. I., Rasola A., Johansson B. R., Blomgren K., Mallard C., Bernardi P., Forte M. A., Hagberg H. (2009) J. Neurosci. 29, 2588–2596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Potter D. A., Tirnauer J. S., Janssen R., Croall D. E., Hughes C. N., Fiacco K. A., Mier J. W., Maki M., Herman I. M. (1998) J. Cell Biol. 141, 647–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Ishihara I., Minami Y., Nishizaki T., Matsuoka T., Yamamura H. (2000) Neurosci. Lett. 279, 97–100 [DOI] [PubMed] [Google Scholar]
- 79. Joshi A., Bondada V., Geddes J. W. (2009) Exp. Neurol. 218, 221–227 [DOI] [PMC free article] [PubMed] [Google Scholar]