Abstract
Contraction and insulin promote glucose uptake in skeletal muscle through GLUT4 translocation to cell surface membranes. Although the signaling mechanisms leading to GLUT4 translocation have been extensively studied in muscle, the cellular transport machinery is poorly understood. Myo1c is an actin-based motor protein implicated in GLUT4 translocation in adipocytes; however, the expression profile and role of Myo1c in skeletal muscle have not been investigated. Myo1c protein abundance was higher in more oxidative skeletal muscles and heart. Voluntary wheel exercise (4 weeks, 8.2 ± 0.8 km/day), which increased the oxidative profile of the triceps muscle, significantly increased Myo1c protein levels by ∼2-fold versus sedentary controls. In contrast, high fat feeding (9 weeks, 60% fat) significantly reduced Myo1c by 17% in tibialis anterior muscle. To study Myo1c regulation of glucose uptake, we expressed wild-type Myo1c or Myo1c mutated at the ATPase catalytic site (K111A-Myo1c) in mouse tibialis anterior muscles in vivo and assessed glucose uptake in vivo in the basal state, in response to 15 min of in situ contraction, and 15 min following maximal insulin injection (16.6 units/kg of body weight). Expression of wild-type Myo1c or K111A-Myo1c had no effect on basal glucose uptake. However, expression of wild-type Myo1c significantly increased contraction- and insulin-stimulated glucose uptake, whereas expression of K111A-Myo1c decreased both contraction-stimulated and insulin-stimulated glucose uptake. Neither wild-type nor K111A-Myo1c expression altered GLUT4 expression, and neither affected contraction- or insulin-stimulated signaling proteins. Myo1c is a novel mediator of both insulin-stimulated and contraction-stimulated glucose uptake in skeletal muscle.
Keywords: Actin, Glucose Transport, Insulin, Insulin Resistance, Mouse, Myosin, Skeletal Muscle Metabolism, Contraction, Exercise Training, High Fat Diet
Introduction
Glucose uptake in skeletal muscle provides energy for muscle fibers and is also critical in the maintenance of whole body glucose homeostasis. In people with type 2 diabetes, defects in insulin-stimulated glucose uptake in skeletal muscle are a major factor in impaired glucose homeostasis. Therefore, elucidating the molecular mechanisms regulating glucose uptake in muscle has been the focus of considerable research and could eventually lead to the development of new treatments for type 2 diabetes.
Contraction and insulin are the major physiological stimulators of glucose uptake in muscle, and both stimuli increase uptake by increasing the number of glucose transporter proteins at the sarcolemma and transverse tubules (1–5). Glucose transporter 4 (GLUT4) is the major glucose transporter protein expressed in skeletal muscle, and in the basal state, it is contained in specialized intracellular vesicular structures. Insulin stimulates GLUT4 translocation in muscle via a distinct, well characterized intracellular signaling cascade that includes phosphorylation of the insulin receptor and insulin receptor substrate-1/2 (IRS-1/2), activation of phosphatidylinositol 3-kinase (PI3-kinase), and phosphorylation of Akt (6). Although the signaling pathways by which muscle contraction stimulates glucose uptake are not yet fully elucidated, contraction signaling is distinct from that of insulin, and LKB1 and Ca2+/calmodulin-dependent protein kinase II (CaMKII)3 have emerged as important mediators (5, 7–9). Recent findings suggest that contraction- and insulin-stimulated signals converge at the Akt substrate of 160 kDa (AS160, also known as TBC1D4) and its homologue TBC1D1 (10–12). These Rab GTPase-activating proteins (Rab-GAP) are thought to function by releasing intracellular GLUT4 vesicles, allowing movement to the sarcolemma and transverse tubules. Beyond AS160 and TBC1D1, little is known about the molecules that bring about the physical movement of GLUT4 toward the cell surface membranes in skeletal muscle.
The cytoskeleton consists of an elaborate array of distinct protein filaments that, among other functions, participates in the intracellular transport of cellular proteins. Actin and microtubule networks are the major components of the cytoskeleton and have been reported to play roles in GLUT4 translocation in cultured 3T3-L1 adipocytes, whereas only the actin network has been shown to be involved in glucose uptake in rat skeletal muscle (13, 14). Insulin causes actin filament (F-actin) formation in L6 skeletal muscle cells (15, 16), and in rat epitrochlearis muscle, F-actin has been observed as mesh-like structures beneath the inner face of the sarcolemma membrane (14). Disruption of the actin network by latrunculin B, an actin monomer-binding compound, inhibited insulin-stimulated glucose uptake and GLUT4 translocation in rat epitrochlearis muscle without affecting proximal insulin signaling (14). Furthermore, F-actin staining was decreased in epitrochlearis muscles isolated from insulin-resistant hyperinsulinemic Zucker fatty rats (17), muscles that are characterized by reduced insulin-stimulated GLUT4 translocation. In contrast, depolymerization of microtubules with colchicine, an agent that binds free tubulin monomers and acts by disrupting the microtubule lattice at the growing ends, did not inhibit insulin- or contraction-stimulated glucose uptake in multiple rat skeletal muscles despite the disappearance of the majority of the microtubule network (13). Taken together, these observations suggest that the actin cytoskeleton, but not the microtubules, plays a role in the movement or fusion of GLUT4 to the cell surface membrane in skeletal muscle.
Myo1c (previously called MM1β/Myr2) is a single-headed, actin-associated molecular motor protein of ∼120 kDa belonging to the class I myosin family. Myo1c consists of three domains: an amino-terminal motor domain, a regulatory domain (neck domain or lever arm), and a carboxyl-terminal tail domain (18). The motor domain is conserved and contains the ATPase catalytic and actin-binding sites. In 3T3-L1 adipocytes, Myo1c functions as one component of the GLUT4-vesicle carrier system that promotes GLUT4 translocation to the cell surface membrane (19–22). In 3T3-L1 adipocytes, Myo1c was detected in GLUT4 vesicles and was found to be associated with the G protein RalA, which interacted with the exocyst, an evolutionarily conserved vesicle-tethering complex (22). In addition, reduced expression of Myo1c in adipocytes using siRNA or overexpression of the Myo1c tail domain as a dominant inhibitor impaired insulin-stimulated GLUT4 translocation to the cell surface membrane in 3T3-L1 adipocytes (19–23). In 3T3-L1 adipocytes, it was also demonstrated that insulin regulates Myo1c function via CaMKII-dependent phosphorylation that increased Myo1c ATPase activity, the domain responsible for motor activity (23).
In the current study, we tested the hypothesis that Myo1c is expressed in skeletal muscle and functions in the regulation of glucose uptake. For this purpose, we determined the expression profile of Myo1c in various metabolic states in mouse skeletal muscle and used in vivo gene transfection and glucose uptake measurements to determine whether Myo1c regulates contraction- and insulin-stimulated glucose uptake. We found that Myo1c protein abundance is associated with the metabolic phenotype of the muscle and that this protein functions in the control of glucose uptake in skeletal muscle.
EXPERIMENTAL PROCEDURES
Animals
Female ICR mice (8–9 weeks, 25–30 g) were obtained from Taconic Laboratory (Hudson, NY) or Charles River Laboratories (Wilmington, MA), and female db/db mice were obtained from Charles River Laboratories. Mice were housed at 20–22 °C with a 12-h light/dark cycle. For the exercise training studies, female ICR mice were housed in wheel cages for 4 weeks, and the corresponding control mice for these experiments were housed in separate cages that did not have a wheel. These mice received LabDiet® rodent chow (Purina Mills Inc., St. Louis, MO) and water ad libitum. A separate cohort of 6-week-old C57BL/6NHsd male mice was fed a high fat diet (60% kcal fat) or normal chow diet from Harlan Labs for 9 weeks (TD 06414). All experiments were performed in accordance with the Institutional Animal Care and Use Committee of the Joslin Diabetes Center and the National Institutes of Health guidelines for the care and use of laboratory animals.
Plasmid cDNA Constructs and Transfection of Plasmid DNA
A mouse wild-type Myo1c cDNA construct was purchased from OriGene (MC201330, Rockville, MD). Myo1c DNA was excised from the original pCMV6 vector, affixed to a single hemagglutinin (HA) tag sequence at the NH2-terminal via specific primers and PCR amplification, and subcloned into pCS2+ plasmids. An ATPase-inactive mutant of Myo1c (K111A-Myo1c) was generated by mutating Lys111 to Ala111 using site-directed mutagenesis (200522; Stratagene, La Jolla, CA). This mutation has previously been shown to eliminate Myo1c ATPase activity (23). Myo1c cDNA constructs were sequenced to confirm accuracy using the high throughput DNA sequencing service at the Brigham and Women's Hospital (Boston, MA). Plasmid DNA was amplified in Escherichia coli TOP10 cells (Invitrogen), extracted using an endotoxin-free plasmid mega kit (Qiagen), and solubilized in saline at 4 μg/μl. Myo1c plasmids and empty vector (100 μg) were injected into mouse tibialis anterior muscles followed by electroporation as we have previously described (7, 24, 25).
In Vivo Muscle Contraction and Insulin Treatments and Glucose Uptake Measurements
The mice were fasted overnight (9 p.m. to 8 a.m.) prior to study and anesthetized (100 mg of pentobarbital/kg of body weight), and one leg was sham-treated and the other leg was contracted in situ by peroneal nerve stimulation for 15 min as we have previously described (25). For maximal insulin stimulation, insulin (16.6 units/kg of body weight) was administered through the retro-orbital sinus together with 20% glucose bolus to prevent hypoglycemia. For immunoblotting studies, muscles were dissected after 15 min of in situ contractions or 15 min after insulin administration, and muscles were snap-frozen in liquid nitrogen for subsequent analysis. Glucose uptake was measured in vivo using 2-[3H]deoxyglucose as described previously (25–27).
Sample Preparation and Immunoblots
Muscles were processed as described previously (25–27), and lysate protein concentrations were determined by the Bradford method (28) using a dye reagent from Bio-Rad. Lysates (40 μg of protein) were separated by SDS-PAGE before immunoblotting (29). Antibody-bound proteins were visualized with chemiluminescence detection reagents (PerkinElmer Life Sciences) and detected on a film or using the FluorChemTM 8000 (Alpha Innotech Co., San Leandro, CA). Protein bands in the film were scanned with an ImageScanner (GE Healthcare). Images were quantitated by densitometry (Fluor-Chem 2.0, Alpha Innotech). Quantification of protein expression was calculated relative to the loading control. Fold increases were expressed relative to the average of the empty vector-injected control group (Basal or Saline). Anti-Myo1c was a gift from Michael P. Czech (University of Massachusetts, School of Medicine, Worcester, MA) (19). The commercially available primary antibodies that were used were: anti-phospho-IRS-1 Tyr612 (44-816G) obtained from BIOSOURCE (Carlsbad, CA); horseradish peroxidase (HRP)-conjugated anti-goat (V805A) from Promega (Madison, WI); anti-HA-HRP (12 013 819 001) from Roche Applied Science; HRP-conjugated anti-rabbit (31460) and streptavidin HRP for acetyl-CoA carboxylase (21126) detection from Thermo Fisher Scientific; anti-CaMKII (sc-9035) and anti-hexokinase II (sc-6521) from Santa Cruz Biotechnology (Santa Cruz, CA); purified anti-TBC1D1 antibody (5929) (12), anti-Akt (9272), anti-phospho-Akt Thr308 (9275), anti-phospho-Akt Ser473 (4058), anti-PAS (9611), anti-phospho-AMPK Thr172 (2535), and anti-phospho-CaMKII Thr286 (3361) from Cell Signaling Technologies (Beverly, MA); and anti-phospho-acetyl-CoA carboxylase Ser212 (07-303), anti-AMPK α1 (07-350), anti-AMPK α2 (07-363), anti-IRS-1 (06-248), anti-AS160 (07-741), anti-GLUT4 (07-1404), anti-GLUT1 (07-1401), and HRP-conjugated anti-mouse (12-349) from Millipore (Billerica, MA).
Glycogen Measurements
Aliquots of muscle lysates were added to microcentrifuge tubes containing 30% KOH, 5% Na2SO4 and then heated at 60 °C for 10 min. Glycogen was precipitated by adding 100% ethanol and placed at −20 °C overnight. Tubes were centrifuged and decanted; the precipitates were washed with 100% ethanol and then centrifuged and decanted once more. The precipitates were hydrolyzed by heating at 95 °C for 1 h in 6 n H2SO4 (30). Neutralized supernatants were used for measurement of glucose (31), and glycogen content was expressed as glucose units in muscle lysates.
Statistics
Data are expressed as the means ± S.E. Statistical analyses were performed using unpaired t tests, one-way analysis of variance, or two-way analysis of variance (SigmaStat 3.5, Systat, San Jose, CA). When differences between means were detected by one- or two-way analysis of variance, the Student-Newman-Keuls test was used for post hoc testing. The differences between groups were considered significant when p < 0.05.
RESULTS
Myo1c Is Highly Expressed in Oxidative Mouse Skeletal Muscle
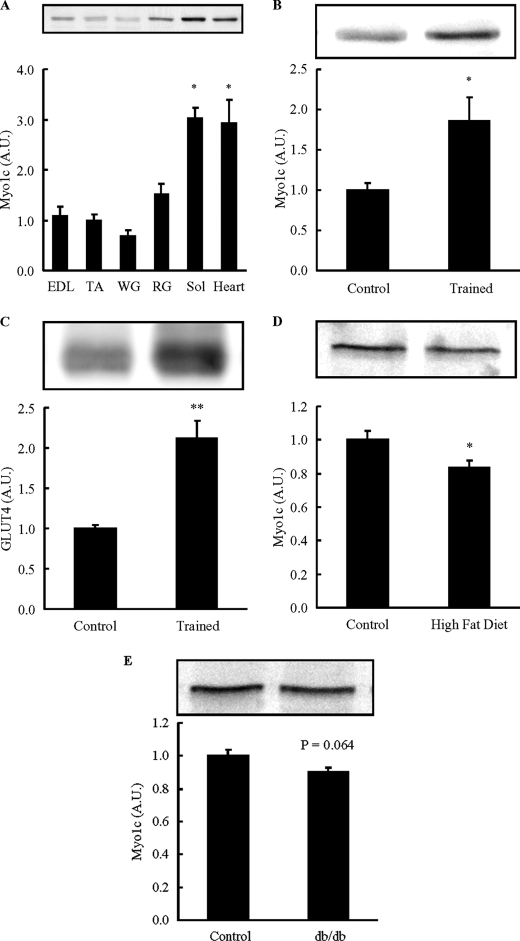
The expression profile of Myo1c in skeletal muscle has not been previously investigated. Because muscles composed of a higher percentage of oxidative fibers have greater levels of GLUT4 and glucose transport when compared with muscles containing more glycolytic fiber types (32, 33), we hypothesized that oxidative muscles would express greater levels of Myo1c. Myo1c protein abundance was greatest in soleus muscle followed by red gastrocnemius, extensor digitorum longus, tibialis anterior muscle, and white gastrocnemius (Fig. 1A). The heart, which is also a highly oxidative muscle, also had high levels of Myo1c expression (Fig. 1A). There was a significant correlation between Myo1c and GLUT4 protein abundance in red and white gastrocnemius muscle (supplemental Fig. S1). Thus, Myo1c is expressed at higher levels in more oxidative muscle types that also have a greater abundance of GLUT4.
FIGURE 1.
Myo1c protein abundance is high in oxidative muscle. A, female ICR mice (12 weeks old) were sacrificed by cervical dislocation. Extensor digitorum longus (EDL), tibialis anterior (TA), white gastrocnemius (WG), red gastrocnemius (RG), soleus (Sol) muscles, and hearts were dissected, frozen, and processed, and muscle lysates were subjected to immunoblotting. Myo1c protein abundance was expressed relative to tibialis anterior. A.U., arbitrary units. B and C, female ICR mice (8–10 weeks old) were individually housed in cages without (Control) or with a wheel cage (Trained) for 4 weeks, and triceps muscles were dissected, frozen, processed, and immunoblotted for Myo1c (B) and GLUT4 (C). D, tibialis anterior muscles were dissected from mice fed a high fat diet for 9 weeks or a control diet. E, tibialis anterior muscles were dissected from db/db mice or control misty mice. Data are means ± S.E., *, p < 0.05 versus tibialis anterior or control. n = 6 muscles/group (A), 8 muscles/group (B and C), 15 muscles/group (D), and 10 muscles/group (E).
We next tested the effects of chronic metabolic alterations on Myo1c expression. Exercise training can induce profound changes in muscle phenotype including increases in oxidative capacity and GLUT4 expression in skeletal muscle (34–36). To determine whether exercise training alters Myo1c abundance in skeletal muscle, mice were housed in exercise wheel cages for 4 weeks. The mice completed high levels of voluntary exercise during the 4-week period (8.2 ± 0.8 km/day), which we have previously shown results in significant increases in oxidative muscle fiber types and increases in citrate synthase activity (37–41). Wheel cage training significantly increased Myo1c and GLUT4 protein in triceps muscle by 1.9- and 2.1-fold, respectively, when compared with sedentary controls (Fig. 1, B and C). Taken together, these results demonstrate that muscles that have a higher level of oxidative capacity have a higher level of Myo1c expression.
Myo1c Protein Is Decreased in Mouse Models of Insulin Resistance
High fat-fed mice and db/db mice are animal models of insulin resistance that have impaired insulin-stimulated glucose uptake in skeletal muscle (42–44). To determine whether Myo1c abundance is altered with high fat feeding, 6-week-old mice were fed a high fat diet (60% kcal from fat) for 9 weeks, and tibialis anterior muscles were studied. Myo1c protein was significantly decreased in the muscles when compared with chow-fed control mice (Fig. 1D). Consistent with these findings, Myo1c protein tended to be decreased in tibialis anterior muscles from db/db mice (Fig. 1E), a model of insulin resistance by virtue of an inactivating mutation of the leptin receptor. Thus, Myo1c has a propensity to be decreased in muscles from insulin-resistant animal models, consistent with the idea that Myo1c may have a specialized function to regulate glucose metabolism in skeletal muscle.
Overexpression of Wild-type Myo1c Increases Contraction- and Insulin-stimulated Glucose Uptake in Mouse Skeletal Muscle
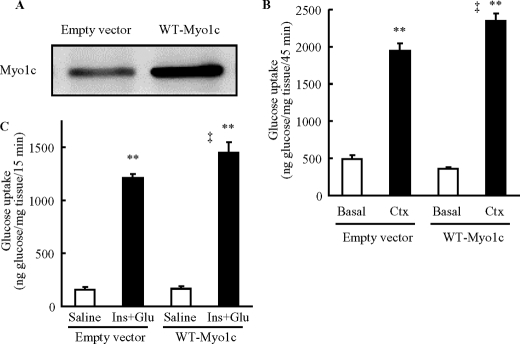
To study Myo1c function in skeletal muscle, we used in vivo injection and electroporation methods to overexpress wild-type Myo1c in tibialis anterior muscles of mice. Seven days after cDNA plasmid injection, expression of Myo1c was severalfold higher in mouse tibialis anterior muscles when compared with endogenous Myo1c obtained from empty vector-injected controls (Fig. 2A). To determine whether Myo1c regulates contraction-stimulated glucose uptake in skeletal muscle in vivo, mice were anesthetized and administered 2-[3H]deoxyglucose via the retro-orbital sinus. One leg was sham-treated, and the other leg was contracted by electrical stimulation of the peroneal nerve for 15 min, and tibialis anterior muscles were obtained 30 min later. Overexpression of wild-type Myo1c had no effect on basal rates of glucose uptake. However, contraction-stimulated glucose uptake was significantly increased when compared with the empty vector control-injected muscles (Fig. 2B).
FIGURE 2.
Overexpression of wild-type Myo1c increases contraction-stimulated glucose uptake in vivo. Mouse tibialis anterior muscles were transfected with DNA vectors containing either wild-type Myo1c (WT-Myo1c) or empty vector as control (Empty vector). One week after transfection, muscles were analyzed. A, muscles were harvested to assess Myo1c protein expression. B, mice were anesthetized and injected with a bolus of 2-[3H]deoxyglucose (333 μCi/kg via the orbital vein), muscles were stimulated to contract in situ (Ctx, 15 min) and obtained 30 min later. 2-[3H]Deoxyglucose uptake was measured for the duration encompassing the contraction and 30 min after contraction periods. C, mice were injected with a bolus of 2-[3H]deoxyglucose (333 μCi/kg), glucose (1 g/kg), and insulin (16.6 units/kg of insulin), and muscles were obtained 15 min later (Ins+Glu). Open bar = control group (Basal or Saline); black bar = stimulated group (Ctx or Ins+Glu). Data are means ± S.E., **, p < 0.01 versus Basal, ‡, p < 0.01 versus empty vector. n = 17–18 muscles/group (B), 9 muscles/group (Saline), and 8 muscles/group (Ins+Glu) (C).
We next examined whether Myo1c regulates insulin-stimulated glucose uptake in skeletal muscle in vivo. Anesthetized mice were administered 2-[3H]deoxyglucose together with both insulin (16.6 units/kg of body weight) and 20% glucose (1 g of glucose/kg of body weight) to prevent hypoglycemia via the retro-orbital sinus. Fifteen min after injection, muscles were harvested and processed for measurements of muscle glucose uptake. Overexpression of wild-type Myo1c had no effect on basal rates of glucose uptake (Fig. 2C). However, muscles overexpressing wild-type Myo1c exhibited a significant increase in maximal insulin-stimulated glucose uptake (Fig. 2C). Thus, exogenous Myo1c can enhance maximal insulin-stimulated glucose uptake.
Overexpression of Wild-type Myo1c Does Not Affect Key Proteins That Regulate Glucose Uptake
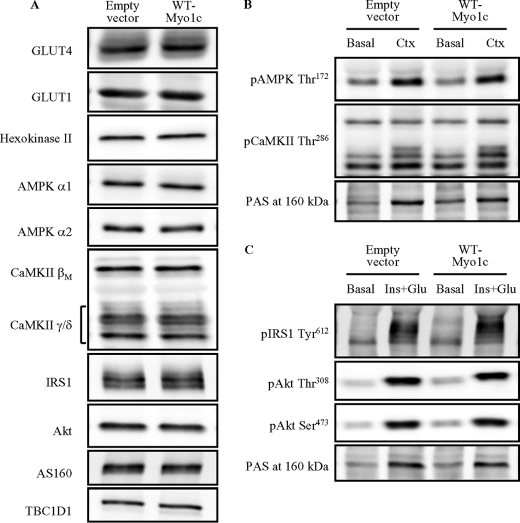
Myo1c has been reported to associate with GLUT4-containing vesicles (19, 22). Therefore, we determined whether overexpression of wild-type Myo1c alters the abundance of GLUT4 in the transfected muscles. There was no difference in GLUT4 protein expression among muscles injected with empty vector or wild-type Myo1c constructs (Fig. 3A and supplemental Table S1). In addition, overexpression of wild-type Myo1c did not alter GLUT1 protein expression or the glycolytic enzyme hexokinase II, which can be a critical regulator of glucose uptake in skeletal muscle (Fig. 3A and supplemental Table S1).
FIGURE 3.
Overexpression of wild-type Myo1c does not alter the abundance of key proteins that regulate glucose uptake. Mouse tibialis anterior muscles were transfected with DNA vectors containing either wild-type Myo1c (WT-Myo1c) or the empty vector as control (Empty vector). After 1 week, mice were anesthetized, and muscles were harvested. Muscles were without treatment (A), stimulated to contract in situ for 15 min (Ctx, B), or stimulated by injection of insulin (16.6 units/kg of insulin) and a glucose bolus to elicit maximal insulin effect (Ins+Glu). pAMPK, phosphorylated AMPK; pCaMKII, phosphorylated CaMKII. C, muscles were harvested to assess protein abundance 15 min after stimulation. The images are representative of 7–8 muscles. Quantification results are shown in the supplemental tables. pIRS1, phosphorylated IRS-1; pAkt, phosphorylated Akt.
Contraction and insulin activate numerous signaling proteins in skeletal muscle that could regulate glucose uptake. Therefore, we determined whether overexpression of wild-type Myo1c altered the protein expression of the AMPKα1 and -α2 catalytic subunits, the CaMKII βM and γ/δ isoforms, IRS-1, Akt, and the Rab-GAP proteins AS160 and TBC1D1. The immunoblots showed that there was no effect of overexpressing wild-type Myo1c on the expression of any of these key signaling proteins (Fig. 3A and supplemental Table S1). To determine whether overexpression of wild-type Myo1c was associated with increased contraction-stimulated AMPK and CaMKII signaling, we measured phosphorylation of AMPKα1/2 at Thr172 and CaMKII at Thr286, sites that regulate enzyme activity. We also measured phospho-Akt-substrate (PAS) at 160 kDa, which is a measure of AS160 and TBC1D1 phosphorylation. Contraction significantly increased phosphorylation of all proteins, but none of these contraction-stimulated increases in phosphorylation were augmented by overexpression of wild-type Myo1c in the tibialis anterior muscles (Fig. 3B and supplemental Table S2). To determine whether overexpression of wild-type Myo1c altered maximal insulin-stimulated IRS-Akt and CaMKII signaling, we measured phosphorylation of IRS-1 Tyr612, Akt Thr308, Akt Ser473, PAS at 160 kDa, and CaMKII Thr286. Maximal insulin stimulation increased phosphorylation of all proteins except for CaMKII (Fig. 3C and supplemental Table S2), but phosphorylation was not enhanced by overexpression of wild-type Myo1c in the muscles (Fig. 3C and supplemental Table S2). These results indicate that Myo1c overexpression alters glucose uptake in the absence of changes in multiple contraction- and insulin-stimulated signaling molecules.
Overexpression of Wild-type Myo1c Does Not Alter Muscle Glycogen Concentrations
High levels of intracellular glycogen have been shown to decrease rates of glucose uptake in response to contraction and insulin in skeletal muscle (45–47), and under some conditions, glycogen concentrations can be increased transiently by sustained increases in glucose uptake (48). Therefore, we determined whether alterations in Myo1c concentrations affected glycogen concentrations in the tibialis anterior muscle overexpressing wild-type Myo1c. However, our results show that overexpression of wild-type Myo1c did not alter glycogen concentrations when compared with empty vector-injected control muscles (supplemental Table S1). This suggests that overexpression of wild-type Myo1c increases glucose uptake in the absence of decreases in intracellular glycogen and that overexpression of wild-type Myo1c does not cause excess glycogen accumulation.
Expression of K111A-Myo1c Impairs Contraction- and Insulin-stimulated Glucose Uptake
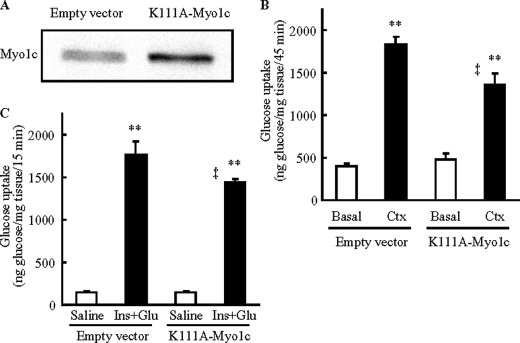
We next examined whether an active motor domain of Myo1c is necessary for contraction- and insulin-stimulated glucose uptake in skeletal muscle. We generated a point mutant of Myo1c in which the ATPase in the motor domain was inactivated by mutating Lys111 to Ala111 (K111A-Myo1c) (23), and we overexpressed K111A-Myo1c in tibialis anterior muscles (Fig. 4A). Seven days after transfection of K111A-Myo1c, we measured contraction- and insulin-stimulated glucose uptake in vivo. Expression of K111A-Myo1c inhibited both contraction-stimulated (Fig. 4B) and insulin-stimulated (Fig. 4C) glucose uptake when compared with the empty vector-injected control. These decreases in glucose uptake occurred in the absence of changes in the expression of GLUT4, GLUT1, and hexokinase II (Fig. 5A and supplemental Table S3). Similar to the data from wild-type Myo1c overexpression, K111A-Myo1c expression did not alter expression of the α1 and α2 subunits of AMPK, AS160, TBC1D1, CaMKII, IRS-1, and Akt (Fig. 5A and supplemental Table S3). In addition, K111A-Myo1c expression did not impair contraction-stimulated phosphorylation of AMPKα1/α2 and PAS at 160 kDa or insulin-stimulated phosphorylation of IRS, Akt, and PAS at 160 kDa (Fig. 5, B and C, and supplemental Table S4). Glycogen concentrations were also normal in muscles expressing the K111A-Myo1c mutant (supplemental Table S3). These results suggest that normal ATPase activity is necessary for Myo1c to regulate glucose uptake and that Myo1c functions downstream or independent of proximal contraction- and insulin-stimulated signaling proteins in skeletal muscle.
FIGURE 4.
Expression of Myo1c mutated at ATPase catalytic site (K111A-Myo1c) inhibits glucose uptake in vivo in skeletal muscle. Mouse tibialis anterior muscles were transfected with DNA vectors containing either Myo1c mutated at Lys111 to Ala (K111A-Myo1c), which has been shown to inactivate the ATPase activity of Myo1c, or the empty vector as control (Empty vector). One week after transfection, muscles were analyzed. A, muscles were harvested to assess Myo1c protein expression. B and C, mice were anesthetized, and then muscles were stimulated to contract in situ for 15 min (Ctx, B) or stimulated by injection of insulin (16.6 units/kg of insulin) and a glucose bolus to elicit maximal insulin effect (Ins+Glu, C), and tracer (2-[3H]deoxyglucose) was injected via the orbital vein. Muscles were harvested to assess 2-[3H]deoxyglucose uptake 45 min (B) or 15 min (C) after tracer injection. Open bar = basal muscle; black bar = stimulated to contract or stimulated by insulin + glucose injection. Values are means ± S.E., **, p < 0.01 versus Basal or Saline, ‡, p < 0.01 versus empty vector. The images are representative of 8 muscles (A). n = 19–20 (empty vector), 11–12 muscles/group (K111A-Myo1c) (B), 6 muscles/group (Saline), and 12 muscles/group (Ins+Glu) (C).
FIGURE 5.
Expression of K111A-Myo1c does not alter the abundance of key proteins that regulate glucose uptake. Mouse tibialis anterior muscles were transfected with DNA vectors containing either K111A-Myo1c or the empty vector as control (Empty vector). After 1 week, mice were anesthetized, and then muscles were harvested. Muscles were without treatment (A), stimulated to contract in situ for 15 min (Ctx, B), or stimulated by injection of insulin (16.6 units/kg of insulin) and a glucose bolus to elicit maximal insulin effect (Ins+Glu)(C). Muscles were harvested to assess protein abundance 15 min after stimulation. The images are representative of eight muscles. Quantification results are shown in the supplemental tables. pAMPK, phosphorylated AMPK; pCaMKII, phosphorylated CaMKII; pIRS1, phosphorylated IRS-1; pAkt, phosphorylated Akt.
DISCUSSION
Skeletal muscle has the unique ability to promote glucose uptake; it responds not only to stimuli from outside the cell such as insulin but also to intracellular environmental changes elicited by contraction. It is well established that these stimuli utilize distinct proximal signaling pathways that eventually converge, leading to an increase in the amount of GLUT4 in the sarcolemma and transverse tubule membranes and a subsequent increase in glucose uptake (49, 50). Over the past several years, these signaling pathways have been intensively studied; however, the molecules responsible for the physical movement of GLUT4 to the cell surface membranes have not been identified. Here, we provide direct evidence that the actin motor protein Myo1c regulates contraction- and insulin-stimulated glucose uptake in skeletal muscle in vivo. To our knowledge, this is the first report showing that a motor protein is necessary for the regulation of glucose uptake in adult skeletal muscle. In addition, our data reveal an association between Myo1c abundance and oxidative capacity in muscle.
Actin filaments have been found to be co-localized with GLUT4 in cultured skeletal muscle cells (15), and disruption of the actin filament network inhibits insulin-stimulated GLUT4 translocation and glucose uptake in skeletal muscle (14). Myo1c has been reported to move along actin filaments by in vitro motility assay (51, 52). In cultured 3T3-L1 adipocytes, overexpression of wild-type Myo1c increased GLUT4 translocation (19) and rescued impaired GLUT4 translocation in adipocytes with knockdown of endogenous Myo1c by siRNA (23). In the current study, there was enhanced glucose uptake in muscles expressing wild-type Myo1c and impaired glucose uptake in muscles expressing mutant Myo1c. Although the current model does not allow us to directly measure GLUT4 translocation, taking these studies together, we hypothesize that Myo1c functions as a motor protein that physically moves GLUT4 to the cell surface membrane in skeletal muscle, regulating glucose uptake.
Contraction and insulin activate distinct proximal signaling pathways leading to glucose uptake in adult skeletal muscle, and in the current study, we show that multiple contraction- and insulin-stimulated signaling proteins were not altered with overexpression of wild-type or K111A-Myo1c. Consistent with our data, disruption of the actin filament network inhibited insulin-stimulated glucose uptake in rat skeletal muscle without affecting proximal insulin signaling, including IRS-1-associated PI3-kinase activity and Akt phosphorylation (14). AS160 and TBC1D1 phosphorylation and subsequent inhibition of Rab-GAP activity have been the most distal steps identified to date in the contraction- and insulin-stimulated signaling pathways in adult skeletal muscle. Our finding that TBC1D1 and AS160 expression and phosphorylation were not changed in muscles overexpressing wild-type or K111A-Myo1c, along with the purported function of AS160 and TBC1D1 to regulate GLUT4-vesiclular movement, suggests that Myo1c mediates GLUT4 translocation at a step distal to AS160 and/or TBC1D1 signaling. Our data showing that expression of wild-type Myo1c or K111A-Myo1c did not change the expression of GLUT4, GLUT1, and hexokinase II indicates that Myo1c does not function by altering transcription of critical proteins that directly regulate glucose uptake. Moreover, muscle glycogen concentrations, which can affect rates of glucose uptake (45–47), were not altered in muscles expressing wild-type Myo1c or K111A-Myo1c. Thus, Myo1c does not regulate glucose uptake by altering glycogen concentrations, proximal signaling, or gene transcription, supporting the hypothesis that Myo1c functions as a motor protein.
Expression of the K111A-Myo1c mutant only partially inhibited glucose uptake, indicating that, in addition to Myo1c, there may also be a Myo1c-independent mechanism for glucose uptake in muscle. Alternately, it is possible that full inhibition does not occur because our in vivo electroporation system does not result in 100% transfection efficiency of the muscle fibers. Future studies with Myo1c knock-out mice will be important to elucidate the relative contribution of Myo1c in the regulation of glucose uptake in skeletal muscle.
In 3T3-L1 adipocytes, insulin has been shown to increase Myo1c phosphorylation at Ser701 in a CaMKII-dependent manner (23). These authors concluded that CaMKII-induced Myo1c Ser701 phosphorylation increases Myo1c ATPase activity and facilitates GLUT4 translocation in adipocytes (23). However, in this same report, insulin stimulation of skeletal muscle did not result in 14-3-3 binding to Myo1c, a predictor of Myo1c Ser701 phosphorylation. Consistent with these findings, in the current study, insulin did not increase CaMKII phosphorylation, and in another report, we recently demonstrated that CaMKII does not regulate insulin-stimulated glucose uptake in skeletal muscle (7). Thus, although CaMKII-dependent regulation of Myo1c may play a fundamental role in insulin-stimulated glucose uptake in adipocytes, this mechanism of Myo1c regulation is unlikely to be important in skeletal muscle. Instead, Myo1c may be regulated by other, CaMKII-independent phosphorylation sites that are insensitive to 14-3-3 binding in skeletal muscle. One possibility is that binding partners directly regulate Myo1c activity or regulate the accessibility of Myo1c to GLUT4-containing vesicles and/or the actin filament network (22).
Chronic exercise training can have significant effects on skeletal muscle metabolism, including a shift toward a more oxidative phenotype. We found that Myo1c abundance was increased in triceps muscles after 4 weeks of exercise training (Fig. 1B). The mechanism for the increase in Myo1c is not known, but this finding raises the possibility that Myo1c expression is regulated by transcriptional factors and/or co-activators that also regulate the expression of oxidative enzymes, such as nuclear respiratory factors 1 and 2 (53), peroxisome proliferator-activated receptor-γ co-activator-1 α (53, 54), and hypoxia-inducible factor-1 α (55). We also found that Myo1c expression was correlated with GLUT4 expression in gastrocnemius muscle. In addition, muscles with insulin-resistant phenotypes or lower oxidative capacity were associated with lower Myo1c expression, and muscles with more insulin-sensitive phenotypes or higher oxidative capacity were associated with higher Myo1c expression. It is unlikely that changes in Myo1c regulate GLUT4 expression because 7-fold overexpression of wild-type Myo1c did not alter GLUT4 expression in the muscle. Instead, Myo1c expression may be regulated by the amount of glucose utilization and the level of GLUT4 in the muscle, which could occur through a feedback mechanism. Most individuals with diabetes have a distinct skeletal muscle phenotype with impaired oxidative capacity, and a number of studies have shown that fiber type distribution correlates with insulin resistance (56, 57). The decrease in Myo1c may exacerbate impaired glucose uptake under certain pathological conditions in insulin-resistant muscle. In fact, not only insulin-stimulated but also contraction-stimulated glucose uptake is impaired in some types of insulin-resistant models (58). Thus, the expression profile of Myo1c supports the idea that Myo1c participates in glucose metabolism, specifically glucose uptake in skeletal muscle.
In summary, Myo1c expression in skeletal muscle directly correlates with the oxidative capacity and insulin sensitivity of this tissue. Although proximal signaling proteins regulating contraction- and insulin-stimulated glucose uptake are distinct in skeletal muscle, Myo1c functions in the regulation of glucose uptake with both stimuli, at a point distal to established signaling mechanisms. To our knowledge, this is the first study to investigate the metabolism-based expression profiles and role of an unconventional myosin in skeletal muscle.
Supplementary Material
Acknowledgment
We are grateful to Michael P. Czech (University of Massachusetts, School of Medicine, Worcester, MA) for the generous donation of the Myo1c antibody.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 AR45670 and R01 AR42238 (to L. J. G.), F32AR051663 and K99AR056298 (to C. A. W.), and the Diabetes and Endocrinology Research Center grant P30 DK36836 (to the Joslin Diabetes Center). This work was also supported by funding from the Graetz Challenge Grant Program of the Joslin Diabetes Center (to L. J. G.); the Japan Society for the Promotion of Science Kakenhi 21240063 (to N. F.); grants from the Nakatomi Foundation, the Naito Foundation, the Uehara Memorial Foundation, and the Japan Society for the Promotion of Science (to T. T.); an American Diabetes Association Mentor-based Award (to L. J. G.); a fellowship from the Canadian Institute for Health Research (MFE-83802) and a Canadian Diabetes Association Incentive Award PF-3-07-2255-DA (to D. A.); and the American Physiological Society (APS) Fellowship in Physiological Genomics (to H.-J. K.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S4 and Fig. S1.
- CaMKII
- calcium/calmodulin-dependent protein kinase II
- PAS
- phospho-Akt-substrate
- AMPK
- AMP-activated protein kinase
- Ctx
- contraction
- Ins
- insulin.
REFERENCES
- 1. Huang S., Czech M. P. (2007) Cell Metab. 5, 237–252 [DOI] [PubMed] [Google Scholar]
- 2. Marette A., Burdett E., Douen A., Vranic M., Klip A. (1992) Diabetes 41, 1562–1569 [DOI] [PubMed] [Google Scholar]
- 3. Ploug T., van Deurs B., Ai H., Cushman S. W., Ralston E. (1998) J. Cell Biol. 142, 1429–1446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wilson C. M., Cushman S. W. (1994) Biochem. J. 299, 755–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jessen N., Goodyear L. J. (2005) J. Appl. Physiol. 99, 330–337 [DOI] [PubMed] [Google Scholar]
- 6. Kido Y., Nakae J., Accili D. (2001) J. Clin. Endocrinol. Metab. 86, 972–979 [DOI] [PubMed] [Google Scholar]
- 7. Witczak C. A., Jessen N., Warro D. M., Toyoda T., Fujii N., Anderson M. E., Hirshman M. F., Goodyear L. J. (2010) Am. J. Physiol. Endocrinol. Metab. 298, E1150–E1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sakamoto K., McCarthy A., Smith D., Green K. A., Grahame Hardie D., Ashworth A., Alessi D. R. (2005) EMBO J. 24, 1810–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Koh H. J., Toyoda T., Fujii N., Jung M. M., Rathod A., Middelbeek R. J., Lessard S. J., Treebak J. T., Tsuchihara K., Esumi H., Richter E. A., Wojtaszewski J. F., Hirshman M. F., Goodyear L. J. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 15541–15546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cartee G. D., Funai K. (2009) Exerc. Sport. Sci. Rev. 37, 188–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kramer H. F., Witczak C. A., Fujii N., Jessen N., Taylor E. B., Arnolds D. E., Sakamoto K., Hirshman M. F., Goodyear L. J. (2006) Diabetes 55, 2067–2076 [DOI] [PubMed] [Google Scholar]
- 12. Taylor E. B., An D., Kramer H. F., Yu H., Fujii N. L., Roeckl K. S., Bowles N., Hirshman M. F., Xie J., Feener E. P., Goodyear L. J. (2008) J. Biol. Chem. 283, 9787–9796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ai H., Ralston E., Lauritzen H. P., Galbo H., Ploug T. (2003) Am. J. Physiol. Endocrinol. Metab. 285, E836–E844 [DOI] [PubMed] [Google Scholar]
- 14. Brozinick J. T., Jr., Hawkins E. D., Strawbridge A. B., Elmendorf J. S. (2004) J. Biol. Chem. 279, 40699–40706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tong P., Khayat Z. A., Huang C., Patel N., Ueyama A., Klip A. (2001) J. Clin. Invest. 108, 371–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kanzaki M. (2006) Endocr. J. 53, 267–293 [DOI] [PubMed] [Google Scholar]
- 17. McCarthy A. M., Spisak K. O., Brozinick J. T., Elmendorf J. S. (2006) Am. J. Physiol. Cell Physiol. 291, C860–C868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Reizes O., Barylko B., Li C., Südhof T. C., Albanesi J. P. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 6349–6353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bose A., Guilherme A., Robida S. I., Nicoloro S. M., Zhou Q. L., Jiang Z. Y., Pomerleau D. P., Czech M. P. (2002) Nature 420, 821–824 [DOI] [PubMed] [Google Scholar]
- 20. Bose A., Robida S., Furcinitti P. S., Chawla A., Fogarty K., Corvera S., Czech M. P. (2004) Mol. Cell Biol. 24, 5447–5458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Huang J., Imamura T., Babendure J. L., Lu J. C., Olefsky J. M. (2005) J. Biol. Chem. 280, 42300–42306 [DOI] [PubMed] [Google Scholar]
- 22. Chen X. W., Leto D., Chiang S. H., Wang Q., Saltiel A. R. (2007) Dev. Cell 13, 391–404 [DOI] [PubMed] [Google Scholar]
- 23. Yip M. F., Ramm G., Larance M., Hoehn K. L., Wagner M. C., Guilhaus M., James D. E. (2008) Cell Metab. 8, 384–398 [DOI] [PubMed] [Google Scholar]
- 24. Fujii N., Boppart M. D., Dufresne S. D., Crowley P. F., Jozsi A. C., Sakamoto K., Yu H., Aschenbach W. G., Kim S., Miyazaki H., Rui L., White M. F., Hirshman M. F., Goodyear L. J. (2004) Am. J. Physiol. Cell Physiol. 287, C200–C208 [DOI] [PubMed] [Google Scholar]
- 25. An D., Toyoda T., Taylor E. B., Yu H., Fujii N., Hirshman M. F., Goodyear L. J. (2010) Diabetes 59, 1358–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ho R. C., Alcazar O., Fujii N., Hirshman M. F., Goodyear L. J. (2004) Am. J. Physiol. Regul. Integr. Comp. Physiol. 286, R342–R349 [DOI] [PubMed] [Google Scholar]
- 27. Fujii N., Hirshman M. F., Kane E. M., Ho R. C., Peter L. E., Seifert M. M., Goodyear L. J. (2005) J. Biol. Chem. 280, 39033–39041 [DOI] [PubMed] [Google Scholar]
- 28. Bradford M. M. (1976) Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 29. Laemmli U. K. (1970) Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 30. Lo S., Russell J. C., Taylor A. W. (1970) J. Appl. Physiol. 28, 234–236 [DOI] [PubMed] [Google Scholar]
- 31. Bondar R. J., Mead D. C. (1974) Clin. Chem. 20, 586–590 [PubMed] [Google Scholar]
- 32. Henriksen E. J., Bourey R. E., Rodnick K. J., Koranyi L., Permutt M. A., Holloszy J. O. (1990) Am. J. Physiol. 259, E593–E598 [DOI] [PubMed] [Google Scholar]
- 33. Hayasaki H., Shimada M., Kanbara K., Watanabe M. (2001) Acta Histochem. 103, 355–363 [DOI] [PubMed] [Google Scholar]
- 34. Rodnick K. J., Holloszy J. O., Mondon C. E., James D. E. (1990) Diabetes 39, 1425–1429 [DOI] [PubMed] [Google Scholar]
- 35. Terjung R. L. (1976) Am. J. Physiol. 230, 946–950 [DOI] [PubMed] [Google Scholar]
- 36. Röckl K. S., Witczak C. A., Goodyear L. J. (2008) IUBMB Life 60, 145–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rodnick K. J., Henriksen E. J., James D. E., Holloszy J. O. (1992) Am. J. Physiol. 262, C9–C14 [DOI] [PubMed] [Google Scholar]
- 38. Vihko V., Salminen A., Rantamäki J. (1979) J. Appl. Physiol. 47, 43–50 [DOI] [PubMed] [Google Scholar]
- 39. Sexton W. L. (1995) J. Appl. Physiol. 79, 287–296 [DOI] [PubMed] [Google Scholar]
- 40. Röckl K. S., Hirshman M. F., Brandauer J., Fujii N., Witters L. A., Goodyear L. J. (2007) Diabetes 56, 2062–2069 [DOI] [PubMed] [Google Scholar]
- 41. Goodyear L. J., Hirshman M. F., Valyou P. M., Horton E. S. (1992) Diabetes 41, 1091–1099 [DOI] [PubMed] [Google Scholar]
- 42. Fujii N., Ho R. C., Manabe Y., Jessen N., Toyoda T., Holland W. L., Summers S. A., Hirshman M. F., Goodyear L. J. (2008) Diabetes 57, 2958–2966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Vicario P., Brady E. J., Slater E. E., Saperstein R. (1987) Life Sci. 41, 1233–1241 [DOI] [PubMed] [Google Scholar]
- 44. Bray G. A., York D. A. (1979) Physiol. Rev. 59, 719–809 [DOI] [PubMed] [Google Scholar]
- 45. Kawanaka K., Nolte L. A., Han D. H., Hansen P. A., Holloszy J. O. (2000) Am. J. Physiol. Endocrinol. Metab. 279, E1311–E1318 [DOI] [PubMed] [Google Scholar]
- 46. Derave W., Hansen B. F., Lund S., Kristiansen S., Richter E. A. (2000) Am. J. Physiol. Endocrinol. Metab. 279, E947–E955 [DOI] [PubMed] [Google Scholar]
- 47. Lai Y. C., Zarrinpashneh E., Jensen J. (2010) J. Appl. Physiol. 108, 1106–1115 [DOI] [PubMed] [Google Scholar]
- 48. Bergström J., Hultman E. (1966) Nature 210, 309–310 [DOI] [PubMed] [Google Scholar]
- 49. Cartee G. D., Wojtaszewski J. F. (2007) Appl. Physiol. Nutr. Metab. 32, 557–566 [DOI] [PubMed] [Google Scholar]
- 50. Lauritzen H. P., Schertzer J. D. (2010) Am. J. Physiol. Endocrinol. Metab. 299, E169–E179 [DOI] [PubMed] [Google Scholar]
- 51. Gillespie P. G., Albanesi J. P., Bahler M., Bement W. M., Berg J. S., Burgess D. R., Burnside B., Cheney R. E., Corey D. P., Coudrier E., de Lanerolle P., Hammer J. A., Hasson T., Holt J. R., Hudspeth A. J., Ikebe M., Kendrick-Jones J., Korn E. D., Li R., Mercer J. A., Milligan R. A., Mooseker M. S., Ostap E. M., Petit C., Pollard T. D., Sellers J. R., Soldati T., Titus M. A. (2001) J. Cell Biol. 155, 703–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhu T., Beckingham K., Ikebe M. (1998) J. Biol. Chem. 273, 20481–20486 [DOI] [PubMed] [Google Scholar]
- 53. Patti M. E. (2004) Curr. Opin. Clin. Nutr. Metab. Care 7, 383–390 [DOI] [PubMed] [Google Scholar]
- 54. Holloszy J. O. (2008) J. Physiol. Pharmacol. 59, Suppl. 7, 5–18 [PubMed] [Google Scholar]
- 55. Mason S., Johnson R. S. (2007) Adv. Exp. Med. Biol. 618, 229–244 [DOI] [PubMed] [Google Scholar]
- 56. Zierath J. R., Hawley J. A. (2004) PLoS Biol. 2, e348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lowell B. B., Shulman G. I. (2005) Science 307, 384–387 [DOI] [PubMed] [Google Scholar]
- 58. Tanaka S., Hayashi T., Toyoda T., Hamada T., Shimizu Y., Hirata M., Ebihara K., Masuzaki H., Hosoda K., Fushiki T., Nakao K. (2007) Metabolism 56, 1719–1728 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.