Abstract
The Josephin domain is a conserved cysteine protease domain found in four human deubiquitinating enzymes: ataxin-3, the ataxin-3-like protein (ATXN3L), Josephin-1, and Josephin-2. Josephin domains from these four proteins were purified and assayed for their ability to cleave ubiquitin substrates. Reaction rates differed markedly both among the different proteins and for different substrates with a given protein. The ATXN3L Josephin domain is a significantly more efficient enzyme than the ataxin-3 domain despite their sharing 85% sequence identity. To understand the structural basis of this difference, the 2.6 Å x-ray crystal structure of the ATXN3L Josephin domain in complex with ubiquitin was determined. Although ataxin-3 and ATXN3L adopt similar folds, they bind ubiquitin in different, overlapping sites. Mutations were made in ataxin-3 at selected positions, introducing the corresponding ATXN3L residue. Only three such mutations are sufficient to increase the catalytic activity of the ataxin-3 domain to levels comparable with that of ATXN3L, suggesting that ataxin-3 has been subject to evolutionary restraints that keep its deubiquitinating activity in check.
Keywords: Cysteine Protease, Deubiquitination, Deubiquitylation, Protein Structure, Ubiquitin, X-ray Crystallography
Introduction
Covalent attachment of ubiquitin is a reversible signal that can alter a protein's function, control its trafficking, or mark it for degradation. Once attached, ubiquitin can be removed by deubiquitinating (DUB)3 enzymes, proteases that specifically remove adducts from the C termini of ubiquitin molecules (1). DUB activities oppose those of the ubiquitin ligases that attach ubiquitin molecules to proteins and thereby generate a dynamic balance in ubiquitin signaling pathways. DUB enzymes also maintain the pool of free ubiquitin molecules within cells by liberating ubiquitin monomers from precursor fusion proteins and by disassembling and recycling poly-ubiquitin chains. DUB enzymes make important contributions to both normal and pathogenic cellular processes, including protein degradation, DNA repair, cell-cycle regulation, transcriptional regulation, and bacterial and viral infections (2–5). DUB enzymes are typically tightly regulated so as to limit their actions to specific cellular processes and prevent the nonspecific or inappropriate proteolysis of cellular targets (6). These enzymes frequently exhibit cryptic activities, with the enzyme being only weakly active (or inactive) in the absence of appropriate substrates and/or activating partners. In many cases, however, the molecular details of DUB regulation are not fully known.
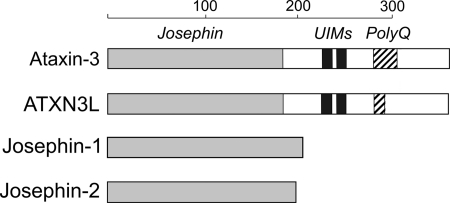
DUB enzymes are divided into five different families: the UCH, UBP, OTU, JAMM/MNN+, and Josephin proteins (7). The Josephin family is the smallest, comprising only four enzymes in humans. However, Josephin proteins are conserved throughout eukaryotes, with homologs being widely distributed among metazoans and found in plants and protozoans as well (8). Although this broad degree of conservation suggests important roles for the Josephin proteins, their biological functions remain in most cases unclear. All Josephin proteins share a common cysteine protease domain of ∼180 amino acids, known as the Josephin domain. Two of the human Josephin proteins, ataxin-3 and the ataxin-3-like protein (ATXN3L), each contain a single Josephin domain at their N terminus plus a flexible C-terminal domain of comparable length (9, 10). Josephin-1 and Josephin-2, on the other hand, are each composed solely of a single Josephin domain (Fig. 1).
FIGURE 1.
Schematic representation of the four human Josephin-domain-containing proteins. Josephin domains are shown in light gray, the ubiquitin-interacting motifs (UIM) are shown in dark gray, and the polyglutamine (polyQ) repeat regions are shown as cross-hatched. The sizes of the ubiquitin-interacting motifs and polyQ regions are not shown to scale. The scale at the top shows length in amino acids.
Ataxin-3 is the best characterized of the Josephin domain-containing proteins. It first attracted study as a polyglutamine repeat protein (11); in its expanded glutamine repeat form it causes the most common inherited ataxia, spinal cerebellar ataxia type 3 (SCA3, also known as Machado-Joseph disease). Subsequently, it became clear that ataxin-3 possesses DUB activity (12, 13) and that this activity maps to the conserved N-terminal region, which was denoted the Josephin domain (after Machado-Joseph disease). Ataxin-3 is now known to participate in numerous ubiquitin-related cellular pathways, and in many instances this participation appears to be mediated by the protein's DUB activity. For example, knocking out ataxin-3 increases levels of poly-ubiquitinated proteins in mice (14) and overexpression of catalytically inactive ataxin-3 leads to a similar accumulation of poly-ubiquitinated proteins in cell culture (15). Ataxin-3 is essential for aggresome formation in models of cellular stress triggered by high levels of misfolded proteins, and catalytically inactive mutants are unable to support aggresome formation (16). Catalytically active forms of ataxin-3 are able to suppress neurodegeneration in a fly model of polyglutamine disease but not inactive mutants (17) (interestingly, no such suppression is seen in rodent models of Machado-Joseph disease (18, 19)). Ataxin-3 regulates its own cellular turnover in a ubiquitin-dependent manner, and abolition of catalytic activity disrupts this regulation (20). Hence, it is clear that ataxin-3 DUB activity is physiologically relevant.
Initial characterization of ataxin-3 DUB activity has revealed that the enzyme prefers long poly-ubiquitin chains over short substrates and that the full-length protein preferentially cleaves Lys-63-linked and mixed-linkage ubiquitin chains (12, 21). This specificity is imparted by ubiquitin-interacting motifs found in the enzyme C-terminal region, as the isolated Josephin domain cleaves both Lys-48- and K63-linked chains with equal efficiency (21). Ataxin-3 can also cleave adducts at the C terminus of NEDD8, a protein that is closely related to ubiquitin in both structure and sequence (22). In all cases examined, however, ataxin-3 appears to cleave its substrates quite slowly. This may be due to some (as yet unknown) physiological reason that favors slow rates of cleavage, perhaps because the enzyme acts as a timer for a particular process. Alternatively, regulatory mechanisms may suppress the enzyme activity, and the in vitro analyses performed to date have not succeeded in relieving this repression (and/or in supplying appropriate activating signals). One candidate for an activating signal is mono-ubiquitination, which has been shown to increase the enzyme's rate of cleavage of Lys-63-linked substrates (23). However, the molecular mechanism by which ubiquitination increases enzyme activity is not yet known, nor is it known whether other cellular signals may also contribute to boosting the ataxin-3 DUB activity.
In contrast to ataxin-3, much less is known about the other three human Josephin domain-containing proteins. For example, solution structures are available for ataxin-3 (24, 25), but no structures have been published for any other Josephin domain proteins, and whereas both Josephin-1 and -2 have been shown to possess DUB activity (8), details of substrate specificity have been lacking. Here, we confirm that all four of the human Josephin domain-containing proteins exhibit DUB activity but that their activities vary significantly depending upon the precise substrate used. In particular, we show that the Josephin domain of the ataxin-3-like protein (ATXN3L) demonstrates substantially higher DUB activity than the ataxin-3 Josephin domain despite the high degree of sequence identity relating these two proteins. We covalently labeled ATXN3L with an activity-based ubiquitin derivative and crystallized this complex, allowing us to determine the first x-ray crystal structure of any Josephin domain as well as the first crystal structure for a Josephin-substrate complex. This structure provides clues about the structural determinants contributing to the high activity of ATXN3L relative to ataxin-3. We have verified the importance of these features by demonstrating that incorporating as few as three single-site mutations into the ataxin-3 Josephin domain can increase its DUB activity 4–5-fold, to near-ATXN3L levels. Although the crystal structure of the ATXN3L Josephin domain is highly similar to solution structures of the ataxin-3 Josephin domain, the ubiquitin substrate appears to bind at different positions in the two proteins, suggesting a possible explanation for the two enzymes' activity differences.
EXPERIMENTAL PROCEDURES
Reagents
Enzymes required for the cloning steps were purchased from New England Biolabs (Ipswich, MA). Media components were obtained from Fisher. All chemicals were purchased from Sigma unless otherwise stated. PCR primers were purchased from MWG-biotech Inc. (High Point, NC). Sequencing of constructs was performed by Genewiz (South Plainfield, NJ). All chromatography columns and resins were purchased from GE-Healthcare.
Protein Expression
The various plasmids used in this work are outlined in supplemental Table S-IV. Standard procedures were used for subcloning and mutagenesis (supplemental Table S-V). The Escherichia coli DH5α strain was used for initial vector construction and plasmid amplification (Invitrogen). All protein expression was carried out using the E. coli Rosetta (DE3) strain. For all constructs, transformed cells were grown in the auto-inducing media ZYP-5052 (26). After growth, cells were harvested by centrifugation at 5000 × g. The cell pellets were stored at −80 °C until ready for downstream processing. For selenomethionine labeling of ATXN3L, the expression plasmid was transformed into the methionine auxotroph B834(DE3), and cells grown in the auto-inducing minimal media PASM-5052 (26).
Protein Purification
Detailed information about the purification of the various protein reagents is given in the supplemental information. Briefly, the ataxin-3 and ATXN3L Josephin domains were expressed as hexahistidine-tagged SUMO fusions, using a subtractive immobilized-metal affinity chromatography purification protocol (27) followed by DEAE-Sepharose ion-exchange chromatography and gel filtration. Untagged monomeric ubiquitin constructs were isolated by treatment of cell lysates with 0.5% perchloric acid followed by dialysis versus sodium acetate, pH 5.2, and cation-exchange chromatography on HiTrap SP HP columns. The Ub-His6 protein was isolated by immobilized-metal affinity chromatography using a HiTrap Chelating HP column followed by cation-exchange chromatography. Preparation of di- and poly-ubiquitin chains was carried out using published procedures (28).
ATXN3L Josephin-Ubiquitin Complex
The ubiquitin1–75-chloroethylamine active-site-directed reagent was produced following a strategy outlined by Ploegh and co-workers (29). Residues 1–75 of ubiquitin were expressed as a fusion with a C-terminal His6-tagged Mycobacterium xenopi intein. After immobilized-metal affinity chromatography purification, the fusion was induced to self-cleave by the addition of MESNA, and the ubiquitin1–75 thioester was isolated in the flow-through from a second immobilized-metal affinity chromatography column. 2-Chloroethylamine was conjugated to the C terminus under basic conditions; the desired adduct was isolated using cation-exchange chromatography on a HiTrap SP column and used immediately for complex formation with the purified ATXN3L Josephin domain. A slight molar excess of ubiquitin1–75-chloroethylamine was added to a concentrated solution of the Josephin domain and incubated at pH 7.5 and room temperature for 72 h; the yield of the covalent complex was estimated to be ∼95% by SDS-PAGE analysis. The complex was isolated on a HiTrap Q HP column followed by gel filtration. Fractions were pooled, concentrated to ∼40 mg/ml, flash-frozen, and stored at −80 °C until needed. The preparation of the selenomethionine-labeled ATXN3L-ubiquitin conjugate was carried out in exactly the same manner. Further details are given in the supplemental information.
DUB Assays
For cleavage of poly-ubiquitin chains, a 125 μm stock solution of each DUB (Josephin domain or YUH1) was prepared in 50 mm sodium phosphate, 150 mm NaCl, 25% (w/v) glycerol, 5 mm DTT. The cleavage reactions were carried out at 30 °C in 50 mm Tris, 150 mm NaCl, 5 mm DTT. For Lys-48-linked chains, 12.5 μm enzyme was added to the equivalent of 75 μg of starting mono-ubiquitin. For the Lys-63-linked chains, the same amount of enzyme was added to the equivalent of 240 μg of starting ubiquitin. Samples were taken at 4 and 20 h and stopped by the addition of SDS-PAGE buffer.
Fluorometric Ub-AMC assays were carried out at 30 °C in 50 mm Tris-Cl, pH 7.6, 0.1 mg/ml ovalbumin, and 5 mm DTT using 50 nm substrate and enzyme concentrations of 1 μm (Josephin domains) or 0.1 nm (Yuh1). Cleavage of Ub-His6 was carried out at room temperature in 100 mm Tris-Cl, pH 7.5, 150 mm NaCl, and 5 mm β-mercaptoethanol. Di-ubiquitin cleavage was carried out at 30 °C in 10 mm Tris-Cl, pH 7.5, 30 mm NaCl, and 1 mm DTT. Trends observed in the relative activities of the different Josephin domains were consistent between enzyme preparations and were also consistent when using substrates from different sources (e.g. home-made poly-ubiquitin chains versus commercially purchased chains).
Crystallization and Diffraction Data Collection
An aliquot of the purified Josephin-ubiquitin complex was rapidly thawed under cold water, dialyzed against 10 mm Tris-Cl, 5 mm DTT, pH 7, and then diluted to 8 mg/ml. Crystals were grown by the microbatch-under-oil method (30) using paraffin oil (Hampton Research, Aliso Viejo, CA). 2-ml drops were set up manually by the addition of 0.9 μl of protein stock solution to 1.1 μl of precipitant (1.6 m sodium citrate pH 6.5, pH adjusted with HCl) in hydrophobic 96-well vapor-batch plates (Douglas Instruments, Berkshire, UK). Trays were incubated at 18 °C; crystals appeared within 24 h. Crystals were harvested in nylon loops and were cryoprotected by brief passage through 1.6 m sodium citrate, pH 6.5, before flash-freezing in liquid nitrogen. Protein containing selenomethionine crystallized isomorphously with the native protein under identical conditions. Diffraction datasets were collected at National Synchrotron Light Source beamline X6A and integrated and scaled using XDS (31). Data collection statistics are given in Table 2.
TABLE 2.
Data collection and refinement statistics
| Data collection | |
| Wavelength (Å) | 0.9780 |
| Space group | P 3 2 1 |
| Cell dimensions (Å) | a = b = 159.15, c = 146.29 |
| Resolution range (Å)a | 19.9–2.7 (2.85–2.7) |
| No. observations | 957,586 (62,188) |
| No. unique reflections | 58,889 (8,535) |
| Completeness (%) | 99.7 (99.9) |
| Multiplicity | 16.3 (7.3) |
| Mean I/σ(I) | 19.6 (2.9) |
| Rmerge | 0.104 (0.566) |
| Rmeasb | 0.107 (0.609) |
| Refinement | |
| Resolution range (Å) | 19.9–2.7 |
| Number of reflections used | 54,832 |
| Number of protein atoms | 8343 |
| Number of solvent atoms | |
| Water | 152 |
| Na+ ion | 1 |
| Mean B values (Å2) | |
| Protein (aniso)c | 79.61 |
| Water (iso) | 42.22 |
| Na+ ion (iso) | 73.95 |
| Root mean square deviations from ideal geometry | |
| Bond distances (Å) | 0.009 |
| Bond angles (degree) | 1.14 |
| Rcryst/Rfree | 0.176/0.224 |
Structure Determination and Refinement
Using a SAD dataset collected from the selenomethionine-substituted protein, the positions of the anomalous scatterers were determined and used to generate an initial map for automated model building using the AutoSol and AutoBuild wizards of PHENIX (32). Subsequent model building and correction was carried out using Coot (33) interspersed with rounds of refinement in PHENIX. After a number of rounds of ADP refinement, TLS groups and parameters were determined using the TLSMD server (34) and applied to subsequent rounds of model building and refinement. The asymmetric unit was found to contain four Josephin-ubiquitin complexes. Density for one of the complexes (chains G:H) was significantly poorer than for the other three complexes; however, omission of the fourth complex led to significantly higher R and Rfree values. Refinement statistics are given in Table 2. Structure factors and refined coordinates have been deposited in the Protein Data Bank, accession number 3O65.
RESULTS
Expression and Purification of Human Josephin Domains
Four human proteins contain the conserved Josephin domain. In two of these proteins (ataxin-3 and ATXN3L), this domain represents only the N-terminal half of the protein, whereas in Josephins-1 and -2 the domain makes up the entire protein (Fig. 1). We successfully expressed all four of the isolated human Josephin domains in E. coli and purified the recombinant proteins to homogeneity (supplemental information and Fig. S1). The Josephin domains from ataxin-3 and ATXN3L were expressed as SUMO fusions (27); both were produced at high levels and were well behaved during purification. The Josephin-1 and -2 proteins appeared to be less stable, and so we expressed them as ubiquitin-SUMO fusions, reasoning that the presence of the ubiquitin substrate might help to stabilize the enzymes. In fact, these constructs did improve stability, and for both Josephin-1 and -2 the ubiquitin fusion partner was cleaved during expression, presumably by the Josephin DUB activity.
DUB Activity of the Josephin Domain-containing Proteins
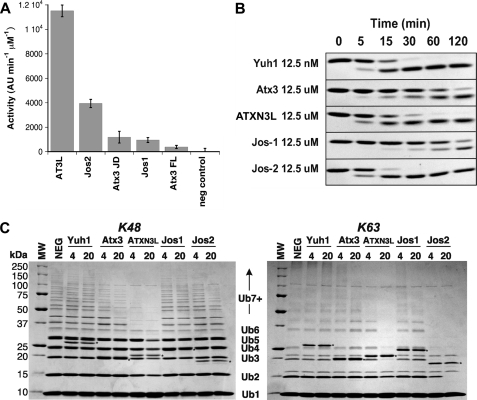
The DUB activity of ataxin-3 is well established, and the DUB activity of Josephins-1 and -2 has recently been described using the artificial substrate ubiquitin-7-amino-4-methoxycoumarin (Ub-AMC (8)). This substrate is not suitable for rigorous kinetic analysis of the Josephin domains, as its Km value appears to exceed its solubility (data not shown). However, it and other small substrates can be used to make simple comparisons of the activities of different Josephin domains. Consistent with what has been described previously, all four human Josephin domains cleave Ub-AMC but display large differences in catalytic efficiency (Fig. 2A). None of the four Josephin domains shows very high activity against the Ub-AMC substrate, requiring orders of magnitude greater enzyme concentrations to achieve comparable rates of product release to the small UCH enzyme Yuh1. For example, under the assay conditions used in Fig. 2A, 0.1 nm Yuh1 gives roughly similar initial rates to those obtained with 1 μm ATX3L Josephin domain (data not shown). The rank order of the activities of the Josephin domains is shown in Table 1; ATXN3L emerges as the most active of the four Josephin domains examined.
FIGURE 2.
DUB activities of the human Josephin domains: ataxin-3 (Atx3), ATXN3L, Josephin-1 (Jos1), and Josephin-2 (Jos2). A, shown is cleavage of the fluorogenic substrate Ub-AMC by the four different Josephin domains under identical conditions, as described under “Experimental Procedures.” AU, arbitrary units. B, shown is the time course of Ub-His6 cleavage catalyzed by Josephin proteins. The substrate concentration is 125 μm for all experiments; the enzyme concentration used is indicated on the left. C, cleavage of unanchored Lys-48- and Lys-63-linked ubiquitin chains is shown. Molecular weight markers are shown in the left-most lane of each gel, with corresponding molecular weights being indicated at the left. The positions of ubiquitin monomers (Ub1), dimers (Ub2), and larger oligomers are indicated. The negative control (NEG) contains no DUB enzyme. Time points were taken after 4 and 20 h of cleavage. Asterisks mark the bands corresponding to the DUB enzymes.
TABLE 1.
Rank order of Josephin domain activities versus various ubiquitin substrates
The activity of the small UCH domain enzyme Yuh1 is included for comparative purposes (35).
| Substrate | Rank order |
|---|---|
| Ub-AMC | Yuh1 ≫ ATXN3L > Josephin-2 ≫ ataxin-3 ≈ ataxin-3 (full-length) ≈ Josephin-1 |
| Ub-His6 | Yuh1 ≫ Josephin-2 > ATXN3L ≫ ataxin-3 ≈ ataxin-3 (full-length) > Josephin-1 |
| Lys-48-linked poly-Ub | ATXN3L > ataxin-3 ≫ Josephin-2 > Josephin-1 > Yuh1 |
| Lys-63-linked poly-Ub | Josephin-2 > ATXN3L ≫ ataxin-3 > Josephin-1 > Yuh1 |
However, Ub-AMC is a poor structural mimic for the substrates that a DUB is likely to encounter in cells, and we therefore extended our analysis to another model substrate, Ub-His6. Ub-His6 is a recombinant variant of ubiquitin having the C-terminal sequence …GG/GGPGHHHHHH, where the residues in boldface correspond to the ubiquitin C-terminal residues Gly-75 and Gly-76, and the position of the scissile bond is indicated by the backslash. The scissile bond in this molecule, although not identical to the naturally occurring isopeptide linkage, is more similar than the Ub-AMC scissile bond, and it is therefore interesting that the activity profiles for the four Josephin domain proteins differ for Ub-AMC and Ub-His6 (Table 1, Fig. 2B), with Josephin-2 showing the highest activity for the latter substrate.
We then extended this analysis to poly-ubiquitin chains, which are presumably more physiologically relevant substrates (Fig. 2C). When unanchored Lys-48-linked poly-ubiquitin chains are used as substrates, ATXN3L again emerges as the most active of the Josephin domains; however, second place is now occupied by ataxin-3, with Josephin-2 showing only very modest activity, and Josephin-1 being essentially inactive. In contrast, when Lys-63-linked poly-ubiquitin chains are used, Josephin-2 is now the most effective enzyme, with ATXN3L also showing robust activity. Ataxin-3 is modestly active against Lys-63-linked chains, and Josephin-1 again shows almost no activity. Not surprisingly, Yuh1 is unable to cleave large poly-ubiquitin chains, as the enzyme contains a loop that passes over the active site and effectively blocks ingress of any large substrate (35).
The C-terminal portion of ataxin-3 is known to affect the enzyme ubiquitin chain selectivity (21), but it does not appear to contribute to cleavage of small substrates such as Ub-AMC or Ub-His6, as no statistically significant difference was seen for the cleavage of these substrates by full-length ataxin-3 and the ataxin-3 Josephin domain (Fig. 2 and supplemental Fig. S5).
Structural Analysis of Substrate Recognition by the ATXN3L Josephin Domain
The large difference in DUB activities for ataxin-3 and ATXN3L is surprising given the 85% sequence identity shared by the Josephin domains of these two proteins. To understand the structural basis for the functional differences between ataxin-3 and ATXN3L, we used x-ray crystallography to determine the structure of the ATXN3L Josephin domain (residues 1–190). In our hands isolated Josephin domains have been refractory toward crystallization, and so we formed the covalent complex of the Josephin domain with ubiquitin using the active-site-directed reagent ubiquitin1–75-chloroethylamine (29). The resulting Josephin-ubiquitin complex provides an excellent structural mimic of the covalent enzyme-substrate intermediate formed during cleavage of ubiquitin substrates (Fig. 3). The complex yielded well ordered crystals, and we determined the structure at a resolution of 2.6 Å using single-wavelength anomalous dispersion analysis of selenomethionine-substituted protein. Statistics relating to the structure determination and refinement are given in Table 2.
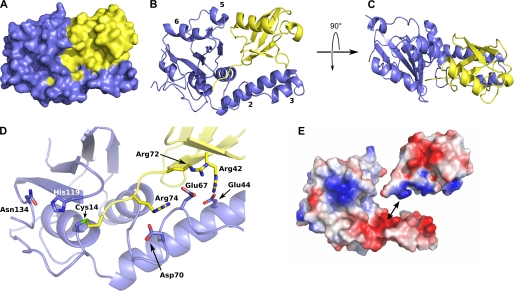
FIGURE 3.
Structure of the complex between ubiquitin and the ATXN3L Josephin domain. A, a space-filling model of the complex is shown; the Josephin domain is shown in blue, and ubiquitin in yellow. B, shown is a ribbon representation of the complex, shown in the same orientation as panel A. Helices 2, 3, 5, and 6 are labeled. C, an orthogonal view of the complex is shown. D, shown is a close-up showing the C terminus of the ubiquitin substrate threading into the ATXN3L active site and forming a covalent linkage with the cysteine nucleophile. Side chains are shown for the catalytic triad (Cys-14, His-119, and Asn-134) as well as for the three arginine-acid salt bridges that help to position the ubiquitin substrate. E, an exploded view of the complex shows the electrostatic potential surfaces of the two proteins and illustrates the charge complementarity between the helix 2/3 hairpin and the ubiquitin substrate. Figs. 3 and 5 were made using MacPyMOL (DeLano, W. L. (2008) The PyMOL Molecular Graphics System DeLano Scientific LLC, Palo Alto, CA).
There are four independent copies of the Josephin-ubiquitin complex in the crystal asymmetric unit. For three of the four complexes (chains A:B, C:D, and E:F), the N termini of the Josephin domains pack together so that the complexes form a roughly 3-fold symmetric tri-lobed arrangement; the fourth complex lies loosely packed into the space between two of the arms (supplemental information and Fig. S2). Each of the first three complexes encounter crystal contacts on multiple sides and as a result are well ordered in the crystal, as judged by the quality of the electron density and by refined atomic displacement values (B-values). However, the fourth complex (chains G:H) experiences far fewer lattice contacts, and the electron density for these chains is substantially poorer than for the other three complexes, consistent with some degree of disorder for the G:H complex within the crystals.
The ATXN3L Josephin domain adopts the same overall fold as the ataxin-3 Josephin domain, not surprising given the 85% sequence identity of these two domains (Fig. 3). The shape of the domain is similar to that of a hitchhiker's hand, which contains a closed fist with a protruding thumb. The “fist” is composed of a five-stranded antiparallel β sheet, flanked by pairs of helices on either side (helices 1 and 4 on one side and 5 and 6 on the other). The “thumb” consists of a helical hairpin formed by helices 2 and 3. The transition from helix 3 to helix 4, which lies at the base of the thumb, is formed by a serpentine structure containing two successive β turns. The active site lies at the point where the thumb joins the palm, just above this serpentine structure.
The ubiquitin molecule is held in the gap between the thumb and the fist, with its C terminus threaded into the Josephin active site. Approximately 28% of the total solvent-accessible area of the ubiquitin molecule is buried in the complex (supplemental Table S-I). Two regions on the ubiquitin surface are buried, as defined by an 80% or larger reduction in accessible surface area upon complexation; these two regions are the Ile-44 patch (36) and the extended C terminus. The Ile-44 patch is a cluster of surface-exposed hydrophobic side chains that includes Leu-8, Ile-44, and Val-70 and which appears to be a common site for recognition by all known ubiquitin-interacting domains (37, 38). The Ile-44 patch packs against the helical hairpin (thumb) formed by helices 2 and 3 (Fig. 3). However, although the patch is rendered solvent-inaccessible by complex formation, the surfaces of the enzyme and ubiquitin do not make intimate contacts with each other in this region; in the three side chains that compose the Ile-44 patch, only a single atom approaches to within 3.4 Å of any atom of the Josephin domain (a methyl carbon of ubiquitin Val-70 lies 3.4 Å from the CD carbon of the ATXN3L Glu-44).
In addition to burying the Ile-44 patch, the helical hairpin of the Josephin domain also makes a series of seven polar contacts with the bound ubiquitin molecule, including a series of three salt bridges between acidic side chains of ATXN3L and arginine side chains at or near ubiquitin C terminus (supplemental Table S-II and Fig. 3D). The electrostatic complementarity represented by these interactions is easily seen when the electrostatic potential is mapped onto the surfaces of the two interacting proteins (Fig. 3E).
At first glance the ubiquitin molecule appears to be gripped tightly between the helical hairpin (thumb) and body (fist) of the Josephin molecule. However, the interface between ubiquitin and the body of Josephin is actually quite open and fails to completely bury even a single residue of ubiquitin. Furthermore, very few specific polar interactions are seen in this interface. Weak (>3 Å) hydrogen bonds appear to form between ATXN3L His-93 and ubiquitin Gln-40, between ATXN3L Asn-95 and ubiquitin Gly-35 (carbonyl oxygen), and between ATXN3L Asn-96 and ubiquitin Gln-34. Along with a potential helix-helix dipole interaction occurring between the ubiquitin principle helix (residues 23–34) and helix 5 in ATXN3L (residues 98–103), this represents the extent of the interactions between the main body of the Josephin domain and ubiquitin.
The C terminus of ubiquitin adopts an extended conformation that leads into the partially buried active site of the Josephin domain. The ubiquitin backbone threads between the side chains of Trp-120, Phe-12, and Phe-74, which form an aromatic enclosure around the enzyme reactive center. No hydrogen bonds occur between the ubiquitin backbone atoms of Gly-75 or Gly-76 and ATXN3L. Upstream of the C terminus, the side chain of ATXN3L Ile-77 neatly interdigitates between the side chains of Leu-71 and Leu-73 of ubiquitin, but again, no polar interactions or tight van der Waals interactions are seen.
Our structure of the ATXN3L substrate-enzyme complex confirms the active site architecture that has been proposed from the solution structure of the Josephin domain of ataxin-3 (24, 25) with residues Cys-14, His-119, and Asn-134 forming a classic cysteine protease catalytic triad. Good electron density is seen for the linkage between Cys-14 and the ubiquitin C terminus (supplemental information and Fig. S3), confirming the covalent linkage arising from nucleophilic attack of the cysteine side chain upon the chloroethylamine carbon that is the positional equivalent of the carbonyl carbon in Gly-76 (see supplemental Fig. S3). The oxyanion hole is formed by the backbone amide of Cys-14 and side chain of Gln-9, which is found in a similar position to that occupied by Gln-19 in papain (39, 40).
Comparison of the Four Copies of the Josephin-Ubiquitin Complex in the Asymmetric Unit
Overall, the conformations of the four ATXN3L molecules are quite similar, superposing with root mean square differences in Cα positions ranging from 1.0 to 2.2 Å. The one aspect of the structure where the various chains differ significantly is the orientation of helix 6 (see “Discussion”). In addition, in the poorly ordered chain G, helix 3 is translated slightly relative to its position in the other three molecules in the asymmetric unit (and in the ataxin-3 solution structure). Helix 3 of chain G is involved in a lattice packing contact, which may explain the altered conformation.
Ataxin-3 Mutants with Enhanced DUB Activity
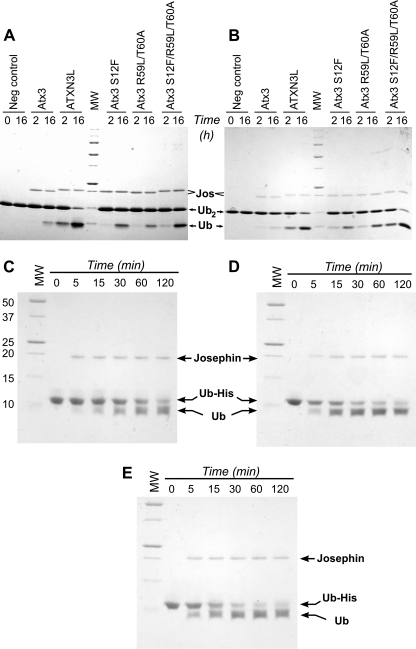
The structures of the ataxin-3 and ATXN3L Josephin domains are essentially similar (see “Discussion”), yet the DUB activities of these two proteins differ considerably for a variety of substrates. To explore this apparent paradox, we made targeted mutations in the ataxin-3 Josephin domain, replacing ataxin-3 residues with the corresponding amino acids from the ATXN3L sequence. The positions to mutate were chosen based on proximity to the active site, the potential for interacting with the ubiquitin substrate, and degree of conservation. Six ataxin-3-to-ATXN3L mutations were chosen for analysis: S12F, R59L, T60A, G67E, L93H, and E118Q. The activities of the purified mutants were tested with Lys-48- and Lys-63-linked diubiquitin, substrates with physiologically relevant linkages that nonetheless allow for easy comparison of enzyme kinetics. At three of the sites examined (residues 67, 93, and 118), conversion of the ataxin-3 residue to its ATXN3L counterpart had little effect on DUB activity. However, changes at the other three positions provided marked increases in the ataxin-3 DUB activity (Fig. 4). Furthermore, these effects were additive; the ataxin-3 double mutant R59L/T60A is more active than either of the corresponding single-site mutants (supplemental information and Fig. S4), and the S12F/R59L/T60A triple mutant shows significantly higher activity than either the S12F or the R59L/T60A mutant. Indeed, the activity of the triple mutant approaches that of the ATXN3L Josephin domain. The combined mutations produced an equivalent increase in the ataxin-3 activity toward the Ub-His6 substrate as well, enhancing the cleavage rate by ∼4–5-fold over that seen with the wild-type protein (Fig. 4, C and E).
FIGURE 4.
Increasing ataxin-3 DUB activity by mutagenesis. A, cleavage of Lys-48-linked ubiquitin dimers by the Josephin domains of wild-type and mutant ataxin-3 (Atx3) and ATXN3L is shown. Ubiquitin dimers were incubated with the Josephin domain for the times indicated. B, cleavage of Lys-63-linked ubiquitin dimers is shown. C–E, time course of cleavage of the ubiquitin-His6 construct by various Josephin domains is shown. Molecular weight markers (MW) are the same in panels C, D, and E. C, wild-type ataxin-3 is shown. D, ATXN3L is shown. E, the ataxin-3 triple mutant S12F/R59L/T60A is shown.
DISCUSSION
Many DUB enzymes are catalytically inactive in the absence of substrate but undergo some conformational change in the presence of ubiquitin that renders them active. Such regulation limits the potential for nonspecific proteolysis. It has been suggested that such a conformational switch occurs in ataxin-3, with the helical hairpin folding against the body of the protein in the absence of substrate and then opening in the presence of substrate (1, 24). However, the solution studies of Nicastro et al. (41) together with our crystal structure of the Josephin-ubiquitin complex argue strongly against this model and show that although the helical hairpin does undergo conformational changes between the free and ligand-bound states, these changes are subtle and do not involve gross obstruction of the active site cleft. Therefore, other structural regulatory mechanisms might control the proteolytic activity of ataxin-3. To probe the relationship between structure and DUB activity in ataxin-3, we reasoned it would be useful to examine the closely related protein ATXN3L.
ATXN3L is encoded by an intronless gene on the X chromosome and occurs only in primates. To date, it has only been found in Catarrhina, suggesting it appeared in the genome relatively recently, most likely between 40 million years ago, when this parvorder separated from the Platayrrhines, and 23 million years ago, when the first major division between Hominoidea and Cercopithecoidae occurred. Although the high sequence similarity (at the nucleotide level) to ataxin-3 and its existence as an intronless gene might suggest that ATXN3L is a non-transcribed pseudogene, a search of the EST data base (42) with the ATXN3L sequence finds a limited number of hits, including a full-length clone from the IMAGE consortium. Thus, the expression status of ATXN3L is not completely clear. However, it is clear that the two genes are conserved to substantially different extents, suggesting they have been shaped by very different evolutionary pressures.
Over the core sequence of the Josephin domain there are eight differences between the human and rhesus monkey ATXN3L sequences, whereas the comparable ataxin-3 sequences are 100% conserved (supplemental Table S-III). Indeed, human ATXN3L is less similar to human ataxin-3 than the latter is to orthologs from non-primate vertebrates (supplemental Table S-III). As the Josephin domain of ataxin-3 shows consistently lower DUB activity for a range of substrates than the same domain of ATXN3L, it appears that evolution has selectively maintained the ataxin-3 sequence in a less active state (and possibly maintained ATXN3L in a more active state). This may represent a regulatory mechanism that keeps ataxin-3 activity dormant until required and raises the question of how the enzyme is maintained in this less active state. Comparing the ATXN3L and ataxin-3 structure is likely to prove informative on this point, as ATXN3L appears to represent a more active form of ataxin-3.
Comparison of the ATXN3L and Ataxin-3 Josephin Domain Structures
Two solution structures have been published for the uncomplexed ataxin-3 Josephin domain (PDB entries 1YZB and 2AGA) and a third (PDB entry 2JRI) for the noncovalent complex of the ataxin-3 Josephin domain and monomeric ubiquitin (24, 25, 43). The overall conformations of PDB entries 1YZB and 2JRI are similar (root mean square difference in Cα positions <2.5 Å), but PDB 2AGA differs significantly, particularly in the helix 2/3 hairpin. PDB entries 1YZB and 2JRI both show an open structure, with the hairpin extending far from the body of the protein, whereas PDB entry 2AGA features a more closed structure, with the hairpin packed against the body of the protein. Bayesian validation calculations and x-ray solution scattering measurements both indicate that the more open structure is the correct one (41), and we therefore use PDB entries 1YZB and 2JRI for the comparisons detailed here.
Ataxin-3 and ATXN3L share 85% sequence identity in their Josephin domains, suggesting that the structures of the two proteins should be highly similar, and indeed this is true (Fig. 5). In the ATXN3L crystal structure, the helix 2/3 helical hairpin extends outward from the body of the protein, as is seen in both the complexed and uncomplexed ataxin-3 solution structures (25, 43). The hairpin conformation in the ATXN3L crystal structure is more similar to that found in the ataxin-3 complex structure (PDB entry 2JRI) than to that in the uncomplexed structure (PDB entry 1YZB); this is consistent with a proposed induced fit model (43), wherein ubiquitin binding produces a twist in the hairpin structure.
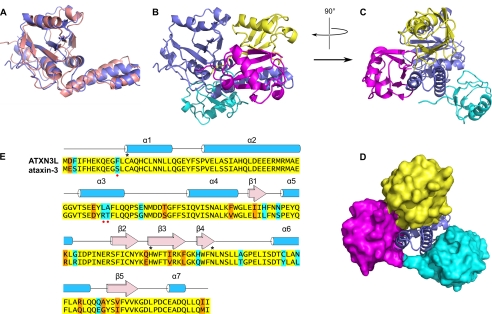
FIGURE 5.
Comparison of the ataxin-3 and ATXN3L Josephin domain structures. A, superposition of the ATXN3L crystal structure (blue) and free ataxin-3 solution structure (pink; PDB entry 1YZB). Panels A and B are oriented essentially similarly to Fig. 3, A and B. B, comparison of the ubiquitin binding modes of ATXN3L and ataxin-3. The ataxin-3 complex with ubiquitin (PDB entry 2JRI) was superposed on the ATXN3L-ubiquitin complex, aligning the Josephin domains; for simplicity's sake, only the ATXN3L Josephin domain is shown (blue). The ubiquitin molecule seen in the ATXN3L crystal structure is shown in yellow, and the two ubiquitin molecules found in the ataxin-3 solution structure are shown in magenta and cyan. The magenta molecule binds near the Josephin active site, and the cyan molecule binds in the HHR23 site. C, orthogonal view of the same superposition seen in Panel B is shown. D, shown is the same view as in panel C, but with the ubiquitin molecules shown in surface representation. E, sequence alignment of the ataxin-3 and ATXN3L Josephin domains is shown. Identical residues are highlighted in yellow, conservative differences are highlighted in orange, and non-conservative differences are highlighted in cyan. Residues of the active site triad are marked with asterisks. Red dots mark the three residues that, when mutated, increase ataxin-3 activity.
Small differences are seen between the ATXN3L and ataxin-3 structures at helix 5 (residues 105–112). In the different structures, helix 5 is displaced laterally (i.e. perpendicular to the helix axis) by as much as 5 Å. This helix lies in the middle of an extended loop that stretches across the surface of the Josephin molecule. This region features higher than average refined atomic displacement parameters (B-values), so it is likely that the differences in conformation between the solution and crystal structures may reflect some intrinsic mobility for this region. The helix formed by ubiquitin residues 23–34 makes a favorable helix dipole interaction with helix 5 in the ATXN3L Josephin domain, which may play a role in determining the position of the latter in the crystal structure.
Another region in which the ATXN3L and ataxin-3 structures differ is helix 6 (residues 146–158). This helix sits atop a β sheet, near the active site. The orientation of the helix varies considerably, not merely between the ataxin-3 solution structures and the ATXN3L crystal structure but also among the four crystallographically independent copies of ATXN3L, again suggesting intrinsic flexibility in this region.
Differences in Ubiquitin Recognition between ATXN3L and Ataxin-3
Even though the ataxin-3 and ATXN3L Josephin domains adopt similar conformations, the positions of the ubiquitin substrate are very different in the ATXN3L crystal structure and the ataxin-3 solution structure. Both structures place the C terminus of the ubiquitin molecule in or close to the Josephin's active site cleft. In the ATXN3L crystal structure, the ubiquitin C terminus is covalently bound to the active site nucleophile Cys-14. However, in the ataxin-3 solution structure the ubiquitin C terminus is displaced about 7 Å farther along the active site cleft than in the crystal structure, which moves the C-terminal carboxylate well past Cys-14 and positions the side chain of ubiquitin Arg-74 where it can interact with Thr-122 and Asn-134 of the Josephin. Moreover, the ubiquitin molecule in the ataxin-3 solution structure is pivoted around the helical hairpin, relative to the ubiquitin position in the ATXN3L crystal structure (Fig. 5). The difference in ubiquitin positions is large (∼23 Å centroid-to-centroid), which means that although similar regions of the ubiquitin molecule contact the helical hairpin in both the ataxin-3 and ATXN3L complex structures, these regions of ubiquitin pack against different parts of the hairpin surface.
It is certainly possible that the observed differences in the substrate recognition modes of ataxin-3 and ATXN3L are physiologically relevant. For example, the misalignment between the ubiquitin C terminus and the Josephin's catalytic residue Cys-14 could explain the lower catalytic efficiency of ataxin-3. However, it is important to consider whether these differences in substrate recognition might derive from the different experimental approaches used. The most significant experimental difference lies in the nature of the ubiquitin substrate used in the solution and crystal structures; the former used super-stoichiometric quantities of free ubiquitin, whereas the latter used 1 eq of a covalently bound, active-site-targeted ubiquitin derivative. The excess of ubiquitin used in the solution structure leads to two distinct ubiquitin binding sites being occupied, the first lying close to the enzyme's active center, as described above, whereas the other is found on the opposite face of the Josephin domain, overlapping a site known to bind the ubiquitin-like domain of the human Rad23 homolog HHR23B (25, 44). However, it is not obvious how the presence of this second ubiquitin molecule could alter the position of the ubiquitin bound in the catalytic site; after superposition of the ATXN3L and ataxin-3 structures, the ubiquitin bound in the ATXN3L catalytic site is separated from the ubiquitin at the HHR23 site by 8 Å at their point of closest approach. Hence, this second ubiquitin molecule does not appear to contribute any steric clashes that would preclude both sites being occupied simultaneously. Support for this comes from recent solution studies showing that the two protomers comprising Lys-48-linked di-ubiquitin can simultaneously occupy the catalytic site and the HHR23 site (45). Thus, occupancy of the second ubiquitin binding site fails to explain the differences in the position of the active site ubiquitin.
The ATXN3L crystal structure shows a covalent Josephin-ubiquitin complex, whereas the ataxin-3 solution structure captures a noncovalent complex between the Josephin domain and free ubiquitin. It is, therefore, possible that the complex seen in the ataxin-3 solution structure represents an initial encounter complex, which might rearrange to the conformation seen in the ATXN3L crystal structure at a later step in the catalytic cycle once the active site nucleophile reacts with the substrate. However, one must also consider that the presence of a charged free carboxylate group at the ubiquitin C terminus might distort the geometry of the Josephin-ubiquitin complex seen in the solution structure, causing it to adopt a conformation different from that formed during actual chain cleavage. Further experimental evidence will be required to conclusively distinguish between these possibilities and the more straightforward possibility that the sequence differences between ataxin-3 and ATXN3L simply cause the two proteins to recognize their substrates differently.
Increasing Ataxin-3 Activity via Incorporation of ATXN3L Mutations
Differences in how the ataxin-3 and ATXN3L Josephin domains recognize ubiquitin substrates could obviously affect their catalytic efficiencies either by favoring different catalytic mechanisms or by altering equilibria between conformers that occur at different points along the catalytic trajectory. To understand more precisely how differences between the two proteins affect catalysis, site-directed mutagenesis was used by altering residues at selected positions of ataxin-3 to the corresponding residue found in ATXN3L. Three of these mutations were found to significantly increase the ataxin-3 catalytic rate.
One of these mutants, S12F, lies near the enzyme active site. Together with Phe-74, Phe-12 of ATXN3L helps to form a hydrophobic lid that shields the active center from solvent (supplemental Fig. S3); it also appears to be well positioned to interact with the alkyl portion of the lysine side chain forming the isopeptide bond. Hence, it is likely that the S12F mutation in ataxin-3 exerts its effect on DUB activity by increasing the efficiency of the enzyme, perhaps by enhancing the nucleophilicity of the active site cysteine and/or enhancing affinity for substrate.
The other two mutations that markedly enhance ataxin-3 activity are R59L and T60A, which are located in the middle of helix 3, at the end of the helical hairpin and far from residue 12. The spatial separation suggests that the R59L and T60A mutations act separately from the S12F mutation, which is consistent with the additive effects of these mutations. Interestingly, the side chains of residues 59 and 60 do not contact the bound ubiquitin substrate in the ATXN3L-ubiquitin complex structure and indeed face away from the substrate toward bulk solvent. However, in the complex of the ataxin-3 Josephin domain with monomeric ubiquitin, Thr-60 is seen to be part of the molecular interface (43, 46), in agreement with our mutational data. This is consistent with the notion that substrate recognition is fundamentally different in ataxin-3 and ATXN3L; altering positions 59 and 60 in ataxin-3 might shift the substrate recognition mode to one more similar to that of ATXN3L. It is also possible that for ataxin-3, interactions between residues 59 and 60 and the substrate occur early in the catalytic process, contributing to formation of an initial encounter complex that might reflect an unproductive conformation in which the substrate is trapped, before a conformational rearrangement occurs to a more productive conformation. Mutating residues to the corresponding ATXN3L sequence would eliminate this transient unproductive interaction. Either of these scenarios could explain the enhanced activity of the mutant.
Evolutionary Restraints on Ataxin-3 Activity
We have shown that a small number of mutations in the ataxin-3 Josephin domain suffice to significantly increase its DUB activity. The failure of any such mutations to arise within this highly conserved domain supports the notion that evolutionary pressure has kept the activity of the free enzyme low, presumably as a regulatory mechanism. Such slow catalytic activity makes for a highly inefficient enzyme, however, suggesting either that the slow activity serves some useful purpose (e.g. a timer) or that the enzyme hydrolytic activity can be increased under appropriate conditions. The nature of such an activating trigger remains unclear, but an obvious candidate would be interaction with a cellular binding partner (47). One known binding partner is the ubiquitin-like domain of the human Rad23/HHR23B (25, 44), but in our hands incubation of the ataxin-3 Josephin domain with an excess of purified HHR23B ubiquitin-like domain failed to increase DUB activity4; a similar result has recently been reported (45). Hence, it is possible that binding of additional or different partners is required; alternatively, a different activation mechanism may be operating. Ataxin-3 has recently been reported to be mono-ubiquitinated in cells, with ubiquitination enhancing enzyme activity (23). The site of mono-ubiquitination, Lys-117, lies directly above the active site, and covalent attachment of a ubiquitin molecule at this site would appear uniquely qualified to modulate the enzyme activity (48). Lys-117 lies directly next to helix 6, which our structure reveals to be conformationally heterogeneous. Hence, one speculative explanation for how ubiquitination enhances activity is that it shifts the conformational equilibrium in this region toward a form that supports more efficient cleavage of substrates.
In summary, the crystal structure of the ATXN3L Josephin domain in complex with ubiquitin shows that, despite possessing high levels of sequence and structural similarity, ataxin-3 and ATXN3L adopt different binding modes for their ubiquitin substrate. These may reflect distinct means of substrate recognition or, alternatively, snapshots of how ubiquitin binds at different points along the catalytic pathway. ATXN3L is substantially more active that ataxin-3, but as few as three mutations in ataxin-3 are sufficient to nearly equalize their activities, suggesting that the DUB activity of free ataxin-3 has been evolutionarily restrained to remain low. The activating mutations might function by shifting preferences between different ubiquitin binding modes.
Supplementary Material
Acknowledgments
We gratefully acknowledge crystallization advice from Dr. Virginie Nahoum and thank Drs. Roger Greenberg, Randy Pittman, Shahri Raasi, and Wai-Kwan Tang for sharing reagents. Diffraction data were collected at the National Synchrotron Light Source, X6A beam line, funded by the NIGMS, National Institutes of Health under agreement GM-0080. The National Synchrotron Light Source at Brookhaven National Laboratory is supported by the United States Department of Energy under contract DE-AC02-98CH10886.
This work was supported, in whole or in part, by National Institutes of Health Grant R01NS065140 (NINDS).

The on-line version of this article (available at http://www.jbc.org) contains supplemental information, Tables S-I–S-V, Figs. S1–S5, and additional references.
The atomic coordinates and structure factors (code 3O65) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
K. C. Grasty and P. J. Loll, unpublished information.
- DUB
- deubiquitinating
- ATXN3L
- ataxin-3-like protein
- Ub-AMC
- ubiquitin-7-amino-4-methoxycoumarin.
REFERENCES
- 1. Komander D., Clague M. J., Urbé S. (2009) Nat. Rev. Mol. Cell Biol. 10, 550–563 [DOI] [PubMed] [Google Scholar]
- 2. Hershko A., Ciechanover A. (1998) Annu. Rev. Biochem. 67, 425–479 [DOI] [PubMed] [Google Scholar]
- 3. Mukhopadhyay D., Riezman H. (2007) Science 315, 201–205 [DOI] [PubMed] [Google Scholar]
- 4. Pickart C. M., Fushman D. (2004) Curr. Opin. Chem. Biol. 8, 610–616 [DOI] [PubMed] [Google Scholar]
- 5. Lindner H. A. (2007) Virology 362, 245–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Reyes-Turcu F. E., Ventii K. H., Wilkinson K. D. (2009) Annu. Rev. Biochem. 78, 363–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Amerik A. Y., Hochstrasser M. (2004) Biochim. Biophys. Acta 1695, 189–207 [DOI] [PubMed] [Google Scholar]
- 8. Tzvetkov N., Breuer P. (2007) Biol. Chem. 388, 973–978 [DOI] [PubMed] [Google Scholar]
- 9. Chow M. K., Mackay J. P., Whisstock J. C., Scanlon M. J., Bottomley S. P. (2004) Biochem. Biophys. Res. Commun. 322, 387–394 [DOI] [PubMed] [Google Scholar]
- 10. Masino L., Musi V., Menon R. P., Fusi P., Kelly G., Frenkiel T. A., Trottier Y., Pastore A. (2003) FEBS Lett. 549, 21–25 [DOI] [PubMed] [Google Scholar]
- 11. Kawaguchi Y., Okamoto T., Taniwaki M., Aizawa M., Inoue M., Katayama S., Kawakami H., Nakamura S., Nishimura M., Akiguchi I. (1994) Nat. Genet. 8, 221–228 [DOI] [PubMed] [Google Scholar]
- 12. Burnett B., Li F., Pittman R. N. (2003) Hum. Mol. Genet. 12, 3195–3205 [DOI] [PubMed] [Google Scholar]
- 13. Scheel H., Tomiuk S., Hofmann K. (2003) Hum. Mol. Genet. 12, 2845–2852 [DOI] [PubMed] [Google Scholar]
- 14. Schmitt I., Linden M., Khazneh H., Evert B. O., Breuer P., Klockgether T., Wuellner U. (2007) Biochem. Biophys. Res. Commun. 362, 734–739 [DOI] [PubMed] [Google Scholar]
- 15. Berke S. J., Chai Y., Marrs G. L., Wen H., Paulson H. L. (2005) J. Biol. Chem. 280, 32026–32034 [DOI] [PubMed] [Google Scholar]
- 16. Burnett B. G., Pittman R. N. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 4330–4335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Warrick J. M., Morabito L. M., Bilen J., Gordesky-Gold B., Faust L. Z., Paulson H. L., Bonini N. M. (2005) Mol. Cell 18, 37–48 [DOI] [PubMed] [Google Scholar]
- 18. Alves S., Nascimento-Ferreira I., Dufour N., Hassig R., Auregan G., Nóbrega C., Brouillet E., Hantraye P., Pedroso de Lima M. C., Déglon N., de Almeida L. P. (2010) Hum. Mol. Genet. 19, 2380–2394 [DOI] [PubMed] [Google Scholar]
- 19. Hübener J., Riess O. (2010) Neurobiol. Dis. 38, 116–124 [DOI] [PubMed] [Google Scholar]
- 20. Todi S. V., Laco M. N., Winborn B. J., Travis S. M., Wen H. M., Paulson H. L. (2007) J. Biol. Chem. 282, 29348–29358 [DOI] [PubMed] [Google Scholar]
- 21. Winborn B. J., Travis S. M., Todi S. V., Scaglione K. M., Xu P., Williams A. J., Cohen R. E., Peng J., Paulson H. L. (2008) J. Biol. Chem. 283, 26436–26443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ferro A., Carvalho A. L., Teixeira-Castro A., Almeida C., Tomé R. J., Cortes L., Rodrigues A. J., Logarinho E., Sequeiros J., Macedo-Ribeiro S., Maciel P. (2007) Biochim. Biophys. Acta 1773, 1619–1627 [DOI] [PubMed] [Google Scholar]
- 23. Todi S. V., Winborn B. J., Scaglione K. M., Blount J. R., Travis S. M., Paulson H. L. (2009) EMBO J. 28, 372–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mao Y., Senic-Matuglia F., Di Fiore P. P., Polo S., Hodsdon M. E., De Camilli P. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 12700–12705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nicastro G., Menon R. P., Masino L., Knowles P. P., McDonald N. Q., Pastore A. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 10493–10498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Studier F. W. (2005) Protein Expr. Purif. 41, 207–234 [DOI] [PubMed] [Google Scholar]
- 27. Weeks S. D., Drinker M., Loll P. J. (2007) Protein Expr. Purif. 53, 40–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pickart C. M., Raasi S. (2005) Methods Enzymol. 399, 21–36 [DOI] [PubMed] [Google Scholar]
- 29. Ovaa H., Galardy P. J., Ploegh H. L. (2005) Methods Enzymol. 399, 468–478 [DOI] [PubMed] [Google Scholar]
- 30. Chayen N. E., Shaw Stewart P. D., Blow D. M. (1992) J. Cryst. Growth 122, 176–180 [Google Scholar]
- 31. Kabsch W. (1993) J. Appl. Crystallogr. 26, 795–800 [Google Scholar]
- 32. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Painter J., Merritt E. A. (2006) J. Appl. Crystallogr. 39, 109–111 [Google Scholar]
- 35. Johnston S. C., Riddle S. M., Cohen R. E., Hill C. P. (1999) EMBO J. 18, 3877–3887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Beal R. E., Toscano-Cantaffa D., Young P., Rechsteiner M., Pickart C. M. (1998) Biochemistry 37, 2925–2934 [DOI] [PubMed] [Google Scholar]
- 37. Hicke L., Schubert H. L., Hill C. P. (2005) Nat. Rev. Mol. Cell Biol. 6, 610–621 [DOI] [PubMed] [Google Scholar]
- 38. Hurley J. H., Lee S., Prag G. (2006) Biochem. J. 399, 361–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ménard R., Carrière J., Laflamme P., Plouffe C., Khouri H. E., Vernet T., Tessier D. C., Thomas D. Y., Storer A. C. (1991) Biochemistry 30, 8924–8928 [DOI] [PubMed] [Google Scholar]
- 40. Ménard R., Plouffe C., Laflamme P., Vernet T., Tessier D. C., Thomas D. Y., Storer A. C. (1995) Biochemistry 34, 464–471 [DOI] [PubMed] [Google Scholar]
- 41. Nicastro G., Habeck M., Masino L., Svergun D. I., Pastore A. (2006) J. Biomol. NMR 36, 267–277 [DOI] [PubMed] [Google Scholar]
- 42. Boguski M. S., Lowe T. M., Tolstoshev C. M. (1993) Nat. Genet. 4, 332–333 [DOI] [PubMed] [Google Scholar]
- 43. Nicastro G., Masino L., Esposito V., Menon R. P., De Simone A., Fraternali F., Pastore A. (2009) Biopolymers 91, 1203–1214 [DOI] [PubMed] [Google Scholar]
- 44. Doss-Pepe E. W., Stenroos E. S., Johnson W. G., Madura K. (2003) Mol. Cell Biol. 23, 6469–6483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nicastro G., Todi S. V., Karaca E., Bonvin A. M., Paulson H. L., Pastore A. (2010) PLoS One 5, e12430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Takahashi H., Nakanishi T., Kami K., Arata Y., Shimada I. (2000) Nat. Struct. Biol. 7, 220–223 [DOI] [PubMed] [Google Scholar]
- 47. Ventii K. H., Wilkinson K. D. (2008) Biochem. J. 414, 161–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Todi S. V., Scaglione K. M., Blount J. R., Basrur V., Conlon K. P., Pastore A., Elenitoba-Johnson K., Paulson H. L. (2010) J. Biol. Chem. 285, 39303–39313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Diederichs K., Karplus P. A. (1997) Nat. Struct. Biol. 4, 269–275 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.