Abstract
Mitochondrial dynamics have been extensively studied in the context of classical cell death models involving Bax-mediated cytochrome c release. Excitotoxic neuronal loss is a non-classical death signaling pathway that occurs following overactivation of glutamate receptors independent of Bax activation. Presently, the role of mitochondrial dynamics in the regulation of excitotoxicity remains largely unknown. Here, we report that NMDA-induced excitotoxicity results in defects in mitochondrial morphology as evident by the presence of excessive fragmented mitochondria, cessation of mitochondrial fusion, and cristae dilation. Up-regulation of the mitochondrial inner membrane GTPase, Opa1, is able to restore mitochondrial morphology and protect neurons against excitotoxic injury. Opa1 functions downstream of the calcium-dependent protease, calpain. Inhibition of calpain activity by calpastatin, an endogenous calpain inhibitor, significantly rescued mitochondrial defects and maintained neuronal survival. Opa1 was required for calpastatin-mediated neuroprotection because the enhanced survival found following NMDA-induced toxicity was significantly reduced upon loss of Opa1. Our results define a mechanism whereby breakdown of the mitochondrial network mediated through loss of Opa1 function contributes to neuronal death following excitotoxic neuronal injury. These studies suggest Opa1 as a potential therapeutic target to promote neuronal survival following acute brain damage and neurodegenerative diseases.
Keywords: Cell Death; Glutamate Receptors Ionotropic (AMPA, NMDA); Mitochondria; Neurodegeneration; Neuroscience; Calpain; Excitotoxicity; NMDA; Opa1
Introduction
Excitotoxicity is a distinct mode of neuronal death that is attributed to increased presynaptic glutamate release. Disturbance of extracellular glutamate levels acting on NMDA3 receptors (NMDAR) results in enhanced calcium influx and death of the cells. Overactivation of NMDAR is a major cause of cell death following acute neuronal injury, such as stroke and trauma, and is also implicated in neurodegenerative diseases, such as Parkinson, Huntington, and Alzheimer diseases (reviewed in Refs. 1 and 2). Mitochondria play a central role in directing cell death signaling generated at the NMDAR. The influx of cytoplasmic calcium resulting from excitotoxicity leads to the accumulation of calcium in the mitochondria. Blocking the mitochondrial calcium overload through manipulation of mitochondrial membrane potential has been shown to block neuronal death following NMDAR overactivation (3).
Recent studies suggest a link between components of mitochondrial dynamics and calcium signaling. In mammals, optic atrophy 1 (Opa1) and mitofusins (Mfn1/2) regulate mitochondrial fusion, and dynamin-related protein 1 (Drp1) regulates mitochondrial fission (reviewed in Ref. 4). Calcium-regulated activity of Drp1 is mediated, at least in part, by Ca2+/calmodulin-dependent protein kinase Iα and calcineurin (5–7). An increase in mitochondrial calcium influx results in mitochondrial fragmentation (5, 8–10), and this fragmentation can be rescued by down-regulation of Drp1 activity (8, 10). Our knowledge of how mitochondrial fusion components respond to calcium transients is limited. Mfn2 was recently reported to bridge endoplasmic reticulum and mitochondria in cell lines and to facilitate mitochondrial calcium influx from endoplasmic reticulum stores (11). Components of the mitochondrial fusion machinery play important roles in the nervous system. For example, mutations in Mfn2 result in Charcot-Marie-Tooth neuropathy type II, and mutations in Opa1 result in autosomal dominant optic atrophy. Drp1 and Mfn2 have been shown to impact mitochondrial morphology and neuronal survival in the Bax-dependent mechanisms of neuronal injury (12, 13). Excitotoxicity exhibits a partial apoptotic-necrotic-like cell death, which does not require Bax/Bak-mediated signaling (14–16). Because much of the function of mitochondria in cell death has focused on Bax-mediated model systems (reviewed in Ref. 17), the role of mitochondrial dynamics in nonclassical death signaling pathways, such as excitotoxicity, remains largely unknown. In the present study, we have taken a combination of loss and gain of function approaches and real-time imaging to investigate the mechanisms by which excitotoxicity results in loss of mitochondrial integrity and neuronal survival. We demonstrate that excitotoxic injury results in a fragmented mitochondrial phenotype and an impairment of mitochondrial fusion. Importantly, the inner mitochondrial membrane GTPase, Opa1, can promote neuronal survival and rescue mitochondrial morphology defects following NMDA-induced toxicity. We identify Opa1 as a key regulator of mitochondrial integrity that is modulated by calpain activation. Inhibition of calpains by calpastatin can preserve mitochondrial morphology and protect neurons against excitotoxic cell death. Opa1 is required for calpastatin-mediated protection because protection was significantly reduced in the absence of Opa1. Our study provides a new mechanistic link between components of inner mitochondrial membrane fusion and death signaling following excitotoxic neuronal injury. These dynamin GTPases may serve as key molecular targets by which to preserve neuronal survival after acute brain damage.
MATERIALS AND METHODS
Cell Culture and Virus Construction
Cerebellar granule neurons (CGNs) were cultured from CD1 mice at postnatal day 7 or 8, as described previously (18). For 4-well plates and 60-mm dishes, 0.5 and 6 million neurons were plated, respectively, in the DMEM (Sigma) that contained 10% dialyzed FBS (Sigma), 25 mm KCl, 2 mm glutamine (Invitrogen), 25 mm glucose, and 0.1 mg/ml gentamycin (Sigma). Recombinant adenoviral vectors carrying the Opa1 expression cassette were prepared using AdEasy system, as described previously (19). Lentivirus vectors carrying photoactivable GFP-ornithine carbamyltransferase (OCT:PAGFP) and MitoDsRed were prepared using the ViraPower lentiviral expression system (catalog no. K4990-0, Invitrogen). For studies with lentiviruses, neurons were transduced with a multiplicity of infection of 2–3 at 1 day in vitro (DIV), and for studies with adenoviruses, neurons were infected with a multiplicity of infection of 100 at 5 DIV.
NMDA Treatment, Mitochondrial Length Measurements, and Cell Viability Assays
Neurons were infected with adenoviruses for either calpastatin (green fluorescent protein (GFP)-tagged), Mfn2 (cyan fluorescent protein (CFP)-tagged), human Opa1 (yellow fluorescent protein (YFP)-tagged), ShOpa1 (YFP-tagged), or GFP at 5 DIV. The efficiency of infection was about 10%. To model excitotoxicity, neurons were treated with 100 μm NMDA and 10 μm glycine at 7 DIV (1 h, 100 μm in the 4-well plates), after which they were switched to conditioned media from parallel cultures. Because most dead neurons round up and are released from coverslips following treatment with NMDA, the total number of live infected neurons remaining in the entire well was counted by an individual blinded to the treatment conditions as described previously (20). To count all of the infected live neurons, the cells were first stained with an antibody against MAP2 (to ensure that only neurons were counted), an antibody against the fluorescent tag (to identify all adenoviruses tagged with GFP, CFP, or YFP), and Hoechst as described previously (21). The number of live infected neurons in the entire well was counted by identifying the cells that were (a) positive for MAP2, (b) contained the fluorescence tag, and (c) had intact nuclei in the corresponding wells. The ratio of live infected neurons was taken as a percentage of the total number of neurons plated multiplied by infection efficiency. Mitochondrial imaging and quantification were performed as described previously (12). Briefly, images were acquired by exciting at 549 nm with the CY3 filter (Chroma Technology Corp., Rockingham, VT). Mitochondrial length was measured by tracing the mitochondria using Northern Eclipse software. For comparison purposes, mitochondria were classified into different categories with a length ranging from less than 0.5 μm to 0.5–1 μm, 1–2 μm, 2–3 μm, and greater than 3 μm.
Time Lapse Imaging and Mitochondrial Fusion Assay
CGNs were seeded on 4-well plates (Nalgene Nunc International, Rochester, NY) with attached glass coverslips coated with poly-d-lysine (Fisher) and transduced with the photoactivable GFP tagged to ornithine carbamyltransferase (OCT:PAGFP) and/or MitoDsRed lentiviruses at 1 DIV. The coverslip was mounted in a temperature-controlled chamber (37 °C) in regular growth medium supplemented with 20 mm HEPES (pH 7.4) and visualized with an LSM-510 confocal laser-scanning microscope (Axiovert 200), with a ×63 oil immersion objective, numerical aperture 1.4. For time lapse studies, the MitoDsRed was excited with the 594-nm line of a multiple line argon laser. Mitochondrial fusion assays were performed as described previously with minor modifications (22). Briefly, the OCT:PAGFP construct was photoactivated with a 405-nm laser (three scans, 50–60% intensity), and the spreading of the signal was imaged every 5 min using a 488-nm line for a total of 25 min. The fusion rate was expressed as a percentage of the area with pixel intensity at 25 min over that of 0 min (where 0 min represents the signal detected right after photoactivation).
Immunofluorescence
At each time point, neurons were fixed for 30 min with ice-cold 4% paraformaldehyde in phosphate-buffered saline (1× PBS) and then rinsed twice with 1× PBS. Cells were permeabilized with 300 μl of ice-cold 0.4% Triton X-100 in 1× PBS for 10 min. Cells were stained with the primary antibodies in 10% normal goat serum, 0.4% Triton X-100, PBS for 1 h. The cells were washed three times for 5 min each with ice-cold 1× PBS. Cells were incubated with the secondary antibodies in 10% normal goat serum, 0.4% Triton X-100, PBS for 1 h. The cells were washed for 5 min and stained with Hoechst for 5 min. Following Hoechst staining, neurons were washed with 1× PBS three times for 5 min each and mounted. Representative samples were photographed using a Zeiss 510 meta confocal microscope (Oberkochen, Germany). The mitofluor red was excited with the 543-nm line of a helium/neon laser, the Alexa 647 was excited with the 633-nm line of a helium/neon red laser, and the Hoechst was excited with the 405-nm line and the GFP was excited with the 488-nm line of a helium/neon green laser. All images shown demonstrate cells that are representative of moderate infection efficiencies and that have been obtained from at least three independent experiments.
Electron Microscopy (EM) Analysis
Neurons were seeded on glass coverslips and treated with NMDA at different time points. Neurons were fixed with 4% glutaraldehyde and subjected to EM analysis as described previously (23). All counts were quantified by an individual blinded to the treatment conditions.
Cellular Fractionation and Western Analysis
Mitochondria were isolated from CGNs as described previously for cortical neurons (16). Protein was extracted and subjected to Western blot analysis. Opa1 expression was visualized in mitochondrial and cytoplasmic fractions using an Opa1 antibody (1:1000; BD Biosciences).
Quantification and Statistical Analysis
The data represent the mean and S.D. from three independent experiments (n = 3). n represents independently prepared neuronal cultures obtained from different animals and on different days. p values were obtained using two-way analysis of variance and Student's t tests. A p value of <0.05 was considered significant and is indicated on the graphs by an asterisk. p values of <0.01 and <0.005 are indicated by two and three asterisks, respectively.
RESULTS
Mitochondrial Morphology Defects following Excitotoxic Injury
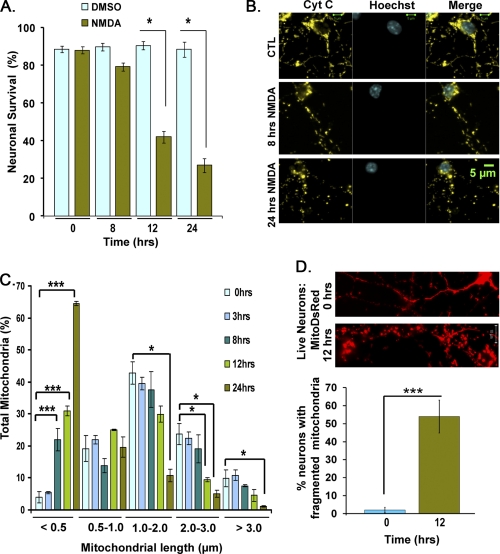
Excitotoxicity is a key mechanism of cell death following acute neuronal injury that involves deregulation of intracellular calcium. To model excitotoxicity, we used the agonist, NMDA, to activate NMDA receptors in primary CGNs. NMDA receptors are ionotropic receptors for glutamate, and their overactivation is implicated as a main cause of brain damage following stroke and trauma. CGNs at 7 DIV were treated with 100 μm NMDA and 10 μm glycine for 1 h, followed by return to conditioned media. This concentration of NMDA results in 60–70% neuronal death at 24 h (Fig. 1A). To analyze mitochondrial morphology changes relative to cell death, mitochondria and nuclei were analyzed over a time course at 0, 3, 8, 12, and 24 h. Representative images of mitochondrial morphology under different treatment conditions are shown in Fig. 1B. A corresponding quantitative analysis of mitochondrial length over time after NMDA treatment is shown in Fig. 1C. In order to perform a comparative analysis at different time points, mitochondrial length was binned into different categories of <0.5, 0.5–1, 1–2, 2–3, and >3 μm as described previously (12). Mitochondria with a length of >3 μm were considered elongated. Mitochondria with a length of 1–3 μm were considered tubular, and those with a length of 0.5–1 and <0.5 were considered short and fragmented, respectively. In control neurons at 7 DIV, 42.82 ± 3.52% of mitochondria exhibited a tubular shape with a length of 1–2 μm (Fig. 1C, n = 3). In contrast, NMDA-treated neurons exhibited fragmented mitochondria, with 30.9 ± 1.47 and 64.5 ± 0.68% having a length of less than 0.5 μm at 12 and 24 h, respectively (compared with 3.9 ± 1.79% at 0 h) (Fig. 1C, n = 3, p < 0.005). Mitochondrial fragmentation is first detected at 8 h following NMDA treatment with 21.9 ± 3.52% of mitochondria exhibiting a length of less than 0.5 μm (Fig. 1C, n = 3, p < 0.005).
FIGURE 1.
Mitochondrial morphology defects following NMDA-induced neuronal death. Primary neurons were treated with 100 μm NMDA and 10 μm glycine for 1 h followed by switching to conditioned media. A, neuronal survival was analyzed at different time points. B, mitochondrial and nuclear morphology was assessed by staining with an antibody against cytochrome c and Hoechst. Representative panels show mitochondrial morphology switches at 0, 8, and 24 h. C, mitochondrial length was binned into different length categories of <0.5, 0.5–1, 1–2, 2–3, and >3.0 μm. Panels demonstrate quantification of mitochondrial length (expressed as a percentage) following NMDA-induced neuronal death. D, mitochondrial morphology in the absence and presence of NMDA was assessed in live neurons targeted with MitoDsRed. The percentage of live neurons exhibiting fragmented mitochondria at 0 and 12 h is plotted. Three independent experiments were performed (n = 3). *, p < 0.05; ***, p < 0.005. Error bars, S.D.; CTL, control.
Because cytochrome c release is a very late event in excitotoxic cell death (16, 24), mitochondrial morphology was assessed by immunostaining with cytochrome c. To confirm results with immunostaining, mitochondrial fragmentation was also evaluated by live imaging of neurons expressing mitochondrial targeted Ds-red. In live imaging studies, 54 ± 9.1% of neurons exhibited fragmented morphology following NMDA treatment at 12 h compared with 2.0 ± 1.5% at 0 h (p < 0.004, n = 3; Fig. 1D). Our results with live imaging were entirely consistent with those obtained with immunostaining with cytochrome c. Together, these results demonstrate that mitochondria become significantly fragmented following NMDA-induced neuronal injury.
Excitotoxicity Results in a Defect in Mitochondrial Fusion
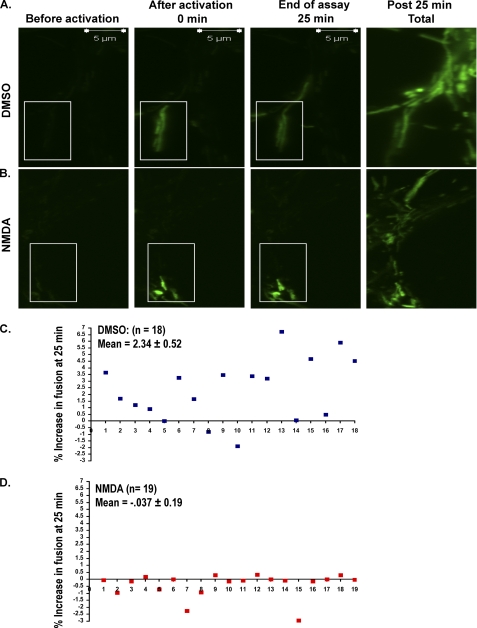
We next asked if an inhibition of mitochondrial fusion contributed to the fragmented mitochondrial phenotype observed following NMDA receptor overactivation. To measure the rate of fusion in a quantitative manner we employed a mitochondrial fusion assay involving photoactivable GFP, as previously described (22), with minor modification. A viral vector was constructed carrying the photoactivable GFP (PAGFP) fused to the 32-amino acid-matrix-targeting signal of ornithine carbamyltransferase (OCT:PAGFP). Neurons were infected with lentiviral vectors expressing OCT:PAGFP at 1 DIV. At 7 DIV, neurons were treated with NMDA or DMSO control. 7–8 h following treatment, the GFP signal was activated in a small region aimed at 2–4 mitochondria (Fig. 2, A and B). A small region was selected in order to avoid illuminating mitochondria of the neighboring neuronal processes. The spread of the signal was examined every 5 min over a 25-min period. The assay of 25 min with scanning intervals of every 5 min was chosen to reassure that there was no photobleaching of the signal. At the end of the 25-min period, all of the PAGFP within the field was activated using the laser at 405 nm (Total in Fig. 2). By creating a mask that scored one binary signal per voxel, we calculated the percentage of the voxels containing a PAGFP signal at each time point within the field relative to the total signal upon complete illumination of the mitochondria. For mitochondria to share their signal, a fusion event must occur. Thus, increased fusion will enhance the spread of the PAGFP signal. The increase in the area containing the PAGFP signal over the 25-min period was plotted for the DMSO-treated (Fig. 2C) and NMDA-treated (Fig. 2D) neurons. A minimum of 18 assays were performed in the absence (Fig. 2C) and presence (Fig. 2D) of NMDA. Our data show that mitochondria of control neurons shared their signal with 2.34 ± 0.52% of inactivated mitochondria (18 assays; Fig. 2C). In contrast, this value decreased dramatically in NMDA-treated neurons (19 assays; Fig. 2D). Our results show that overactivation of NMDA receptors results in an arrest in mitochondrial fusion.
FIGURE 2.
Mitochondrial fusion rates are attenuated following NMDA-induced cell death. A, neurons were transduced with a lentivirus for OCT:PAGFP at 1 DIV. At 7 DIV, the GFP signal was activated in a small region (aimed at 2–4 mitochondria) using a laser at 405 nm (boxed area). The spread of the signal throughout the axonal processes was captured by obtaining images of the neurons every 5 min for a total of 25 min. At the end of the assay, all of the PAGFP within the targeted neuron was activated at 405 nm (Total). B, the assays were repeated as explained in A on the NMDA-treated neurons. C and D, the increase in the area containing mitochondria pool over the 25-min period was plotted. 18 assays (n = 18) (C) and 19 assays (n = 19) (D) were performed in the absence and presence of NMDA, respectively.
EM Links Mitochondrial Morphology Defects to Cristae Remodeling
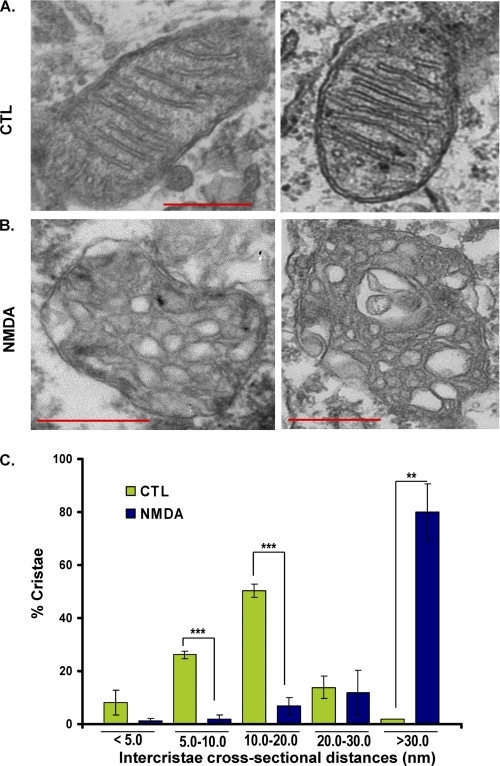
To investigate whether these mitochondrial defects following excitotoxicity are associated with changes in inner mitochondrial membrane structure, an ultrastructural analysis was performed on neurons exposed to NMDA using EM. Our results indicate that NMDAR overactivation results in defects in the inner mitochondrial membrane structure and opening of intercristal distances and the cristae junctions (Fig. 3). In the control neurons, the majority of mitochondrial cristae appeared tubular with regular intercristal cross-sectional distances (9, 25) of 10–20 nm across (Fig. 3A). Following 8 h of treatment with NMDA, the majority of cristae (80.11 ± 10.59%) exhibited an intercristal distance greater than 30 nm compared with control at 1.82 ± 0.02% (p < 0.0089). In conclusion, NMDA resulted in aberrant mitochondrial morphology in which the cristae were dilated (Fig. 3). Together, our studies indicate that mitochondria cease fusion (Fig. 2) and undergo dramatic membrane remodeling (Fig. 3) following overactivation of NMDA receptors.
FIGURE 3.
Mitochondrial morphology defects following NMDAR overactivation. Neurons were plated on glass coverslips and treated with NMDA for 8 h. Neurons were fixed with 4% glutaraldehyde and examined by EM (A and B). Mitochondrial images are shown prior to (A) and following treatment with NMDA (B). C, the intercristal distance of mitochondria was measured and binned into different length categories of <5.0, 5.0–10.0, 10.0–20.0, 20.0–30.0, and >30.0 nm. The difference between NMDA and control group is plotted for each binned category. **, p < 0.01; ***, p < 0.005. Error bars, S.D.; CTL, control.
The Inner Membrane Fusion GTPase, Opa1, Preserves Mitochondrial Structure and Protects Neurons against NMDA-induced Excitotoxicity
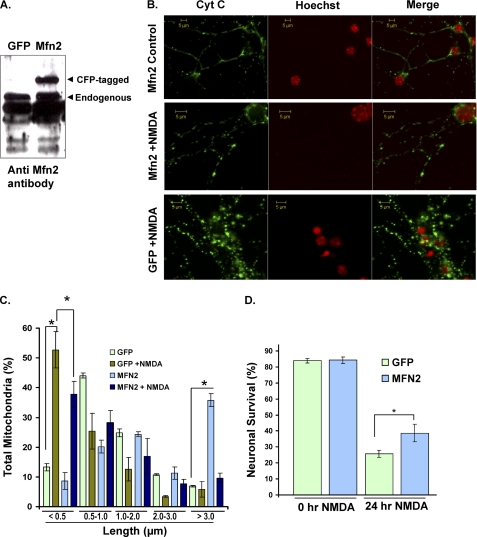
Given that we have found an arrest in mitochondrial fusion, we first asked whether up-regulation of the outer membrane fusion protein, Mfn2, could protect neurons against excitotoxic cell death. Neurons were infected with viral vectors expressing Mfn2 or GFP control and after 7 days were subjected to NMDA treatment as described above. Mfn2 expression levels were assessed by an antibody against Mfn2 (Fig. 4A). Our results revealed that, although Mfn2 results in robust mitochondrial elongation in control neurons (Fig. 4B), up-regulation of Mfn2 had little effect in restoring mitochondrial length following exicitotoxicity (Fig. 4, B and C). In addition, the Mfn2-expressing group exhibited a slight improvement in neuronal survival, where 38.8 ± 5.44% of neurons survived compared with 25.9 ± 2.26% in GFP-expressing controls, p < 0.05, n = 3) (Fig. 4D). Our results clearly indicate that other mechanisms must contribute to mitochondrial morphology defects and neuronal loss following excitotoxicity. We therefore examined the involvement of other mitochondrial proteins.
FIGURE 4.
Effect of Mfn2 on mitochondrial morphology defects and cell death following excitotoxicity. A, primary neurons were infected with adenoviruses for Mfn2 or GFP control at 5 DIV. Western blot shows Mfn2 protein expression. B, neurons were treated with NMDA at 7 DIV and stained with an antibody against cytochrome c (Cyt C) and Hoechst. Mitochondrial morphology was analyzed in the infected neurons expressing either Mfn2 or GFP control. C, mitochondrial length was binned into different categories and quantified as described in the legend to Fig. 1. D, live neurons were scored in each group by scoring the live infected neurons remaining in the entire well following 24 h of treatment with NMDA. *, p < 0.05; three independent experiments (n = 3). Error bars, S.D.
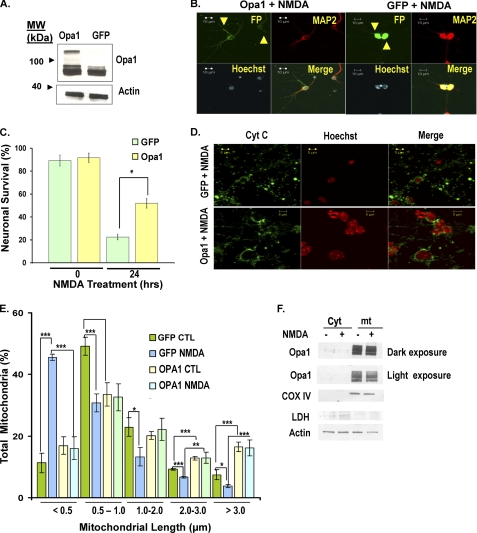
Given that NMDA-treated neurons exhibited severe cristae deformation, we asked whether the function of the inner membrane GTPase, Opa1, might be disrupted, contributing to the loss of mitochondrial integrity. Not only is Opa1 an important component of the mitochondrial fusion machinery (26); Opa1 is also essential for the assembly of cristae junctions (25, 27). In the yeast model system, the function of the Opa1 orthologue has been differentially shown, where the GTPase domain is required for fusion activity, and the coiled-coil domains play a role in maintaining the complex cristae architecture (28). To ask if increasing the levels of Opa1 could preserve mitochondrial integrity and neuronal survival, primary neurons were plated at equal numbers, infected with adenovirus vectors carrying Opa1 or GFP at 5 DIV, and then treated with NMDA at 7 DIV. Overexpression of Opa1 was confirmed by Western blot (Fig. 5A). Neurons were fixed following 24 h of treatment and stained with an antibody against MAP2 (to ensure that only neurons were counted) and Hoechst (Fig. 5B). The number of live infected neurons (indicated with an arrow in Fig. 5B) in the entire well was counted as described under “Materials and Methods.” Following treatment with NMDA, 22.5 ± 2.49% of GFP-treated cells survived, whereas more than twice as many cells survived in the Opa1 group (51.98 ± 4.17%). These results show that up-regulation of Opa1 provides significant protection against NMDA-induced excitotoxic cell death (p < 0.02, n = 3; Fig. 5C). We then asked whether up-regulation of Opa1 could preserve mitochondrial morphology following excitotoxicity. Following NMDA exposure, cultures expressing GFP alone exhibited 45.5 ± 0.99% of neurons with mitochondrial lengths less than 0.5 μm (Fig. 5, D and E). In contrast, neurons expressing elevated levels of Opa1 had only 15.4 ± 3.75% of mitochondria with lengths less than 0.5 μm. This indicates that Opa1 significantly rescues the mitochondrial morphology defect caused by overactivation of NMDA receptors (p < 0.005, n = 3; Fig. 5, D and E).
FIGURE 5.
Opa1 promotes neuronal survival and rescues mitochondrial morphology defects following NMDA-induced neuronal death. A, neurons were infected with an adenovirus for Opa1 or GFP control at 5 DIV. The lysates were collected at 7 DIV, and the Opa1 expression levels were assessed using an antibody for Opa1. B, neurons were infected with Opa1 or GFP adenoviruses at 5 DIV and treated with NMDA at 7 DIV. Neurons were stained with Hoechst and antibodies against MAP2 and fluorescence tag (FP). The yellow arrows represent live infected neurons in GFP and Opa1 groups. C, neuronal survival was plotted as described under “Materials and Methods,” whereby the totals of live infected neurons were compared between the Opa1 group and GFP control. D, images of mitochondria in the GFP and Opa1 overexpressing neurons are shown following treatment with NMDA. Mitochondrial morphology was evaluated by staining for cytochrome c (Cyt C). E, mitochondrial length was binned into different categories and quantified as described in the legend to Fig. 1. F, neurons were subjected to subcellular fractionation to compare Opa1 levels between NMDA-treated (+) and nontreated control (−) in the mitochondrial (mt) and cytoplasmic (Cyt) fractions. Cytochrome c oxidase IV (COX IV) and lactate dehydrogenase (LDH) were used as mitochondrial and cytoplasmic controls, respectively. Our results show that there is no major release of Opa1 from mitochondria following NMDA treatment. *, p < 0.05; **, p < 0.01; ***, p < 0.005. Error bars, S.D.; CTL, control.
Opa1 Functions Downstream of Calpain Activation
Our results suggest that disruption of Opa1 function may be a key defect underlying aberrant mitochondrial architecture following exposure to excitotoxic injury. One possibility is that Opa1 is lost from the mitochondria following NMDA treatment. Opa1 expression was measured in mitochondrial and cytoplasmic fractions of neurons subjected to NMDA treatment (Fig. 5F). Western blot analysis revealed no major change in Opa1 isoforms following exposure to NMDA. Our results furthermore suggest that NMDA treatment does not result in loss of Opa1 from mitochondria within 24 h of NMDA treatment (Fig. 5F). We therefore examined other potential mechanisms by which Opa1 function may be affected.
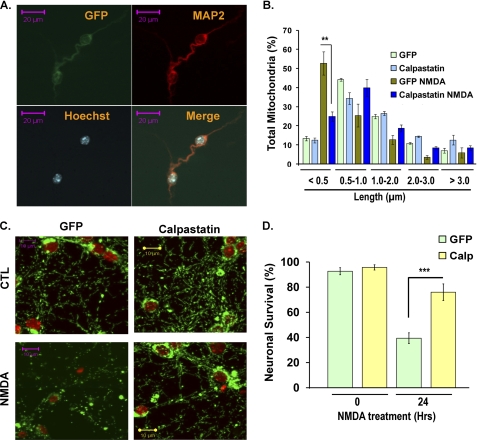
One of the essential players contributing to excitotoxic cell death is activation of multiple calpain family members upon calcium influx (reviewed in Ref. 29). In the nervous system, the μ-calpain is among the calcium-activated cysteine proteases that cleave structural proteins and function at the mitochondria. μ-Calpain contains an N-terminal mitochondrial targeting sequence (30), and it is found in the inner membrane space fractions (31). We therefore asked if calpain activation might be involved in disrupting Opa1 function. We first investigated whether calpain inhibition could rescue the mitochondrial morphology defect. Primary neurons were infected with a recombinant adenoviral vector carrying either calpastatin (an endogenous inhibitor of calpain) or GFP control at 5 DIV. Expression of calpastatin was confirmed by immunocytochemistry (Fig. 6A) and Western blot (Fig. 7A). Neurons were treated with NMDA at 7 DIV and analyzed for mitochondrial morphology at 8 DIV. In NMDA-treated neurons expressing GFP, 52.69 ± 6.19% of mitochondria exhibited a highly fragmented phenotype with a length of less than 0.5 μm. In contrast, neurons expressing calpastatin exhibited a less severe phenotype, with only 24.77 ± 2.50% having a length of less than 0.5 μm (Fig. 6B). Images of mitochondrial morphology are shown in Fig. 6C. Calpastatin significantly rescued mitochondrial morphology defects induced by NMDA (p < 0.006, n = 3). Primary neurons expressing calpastatin also exhibited a significant rescue of neuronal death such that 76.02 ± 6.61% of neurons survived compared with 39.41 ± 4.31% in the GFP-treated group (p < 0.001, n = 3; Fig. 6D). In conclusion, these results suggest that calpains play an important role in the disruption of mitochondrial architecture and function.
FIGURE 6.
Effect of calpastatin on mitochondrial morphology defects and cell death following excitotoxicity. A, neurons were infected with adenoviruses carrying GFP-tagged calpastatin or GFP control at 5 DIV and treated with NMDA at 7 DIV. Cells were stained with an antibody against fluorescence tag (FP) and Map2 to identify live infected neurons in the calpastatin-overexpressing neurons and GFP control. B, mitochondrial morphology was assessed as described, and quantification of mitochondrial length in the absence and presence of NMDA is plotted. C, representative images of mitochondrial morphology in the GFP- and calpastatin-overexpressing neurons in the absence and presence of NMDA. D, the graph demonstrates percentage of neuronal survival in calpastatin (Calp)- and GFP-overexpressing neurons in the absence and presence of NMDA (n = 3). Error bars, S.D.; CTL, control.
FIGURE 7.
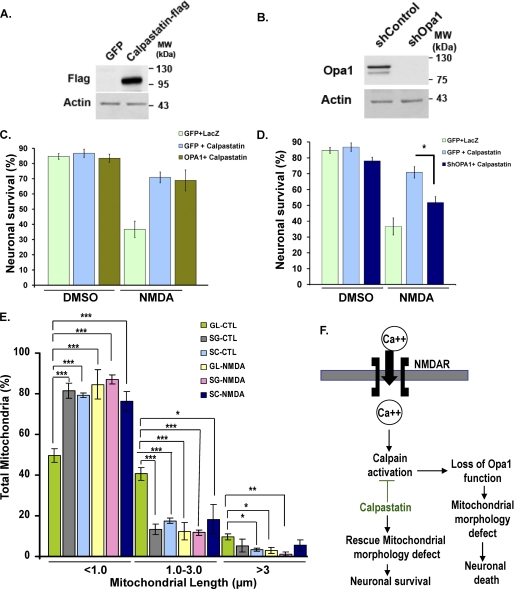
Opa1 functions downstream of calpain activation. A, neurons were infected with adenoviruses carrying FLAG-tagged calpastatin or GFP control at 5 DIV. Cell lysates were collected at 8 DIV, and the expression levels were analyzed by an antibody against FLAG. Actin was used as a loading control. B, neurons were infected with adenoviruses carrying ShOpa1 or control (shControl). Cell lysates were collected at 8 DIV, and the expression levels were analyzed by an antibody against Opa1. Actin was used as a loading control. C and D, neuronal survival was plotted in neurons co-expressing control vectors alone (GFP + LacZ), calpastatin alone (GFP + Calpastatin), calpastatin and Opa1 (Opa1 + Calpastatin) or calpastatin and ShOpa1 (ShOpa1 + Calpastatin) at 0 and 24 h following treatment with NMDA. Infectivity (MOI) was equalized using control vectors GFP and LacZ. The results were compared with control neurons expressing LacZ + GFP. E, mitochondrial length was quantified in Opa1-depleted neurons (ShOpa1) and compared with the GFP control group in the absence (LacZ) and presence of calpastatin without and with NMDA. SG, ShOpa1 + GFP; SC, ShOpa1 + calpastatin; GL, GFP + LacZ (control). Three independent experiments (n = 3) are shown. *, p < 0.05; **, p < 0.01; ***, p < 0.005. F, our present model proposes that Opa1 functions downstream of calpain to regulate mitochondrial morphology and neuronal survival following calcium influx associated with excitoxicity. Error bars, S.D.; CTL, control.
To determine whether Opa1 indeed functions on the same pathway as calpain activation, we asked if simultaneous activation of both Opa1 and calpastatin in primary neurons could provide synergistic protection against cell death. If Opa1 and calpastatin function by the same mechanism, then no further protection would be expected. Alternatively, if they function by distinct pathways, one would predict that protection would be enhanced by adding both Opa1 and calpastatin. Neurons were infected with calpastatin and either Opa1 or GFP and compared with control neurons co-infected with LacZ and GFP. Our results indicate that there was no significant increase in protection against NMDA-induced toxicity between neurons expressing both calpastatin and Opa1 (68.87 ± 6.87%) versus neurons expressing calpastatin and GFP (70.9 ± 3.58%; Fig. 7C). This suggests that calpastatin and Opa1 may be acting on a common pathway to modulate mitochondrial inner membrane structure in response to calcium influx following excitotoxic injury. To ask if Opa1 is required for protection conferred by calpastatin-mediated inhibition of calpain, neuronal survival was compared in neurons expressing calpastatin in the absence and presence of Opa1 by using an adenovirus for ShOpa1 to knock down Opa1 expression. Efficient knockdown of Opa1 with ShOpa1 was confirmed by Western blot (Fig. 7B). Neurons expressing calpastatin and GFP exhibited a 70.9 ± 3.58% survival rate, whereas those expressing both calpastatin and ShOpa1 exhibited a significant reduction in survival at 51.8 ± 3.64% (Fig. 7D).
Finally, we questioned if loss of Opa1 in the absence and presence of calpastatin induces mitochondrial fragmentation and whether these defects are rescued by calpastatin. To test this, mitochondrial morphology was analyzed in primary neurons co-expressing ShOpa1 and calpastatin (SC in Fig. 7) or ShOpa1 and GFP (SG) and compared with control neurons co-infected with GFP + LacZ (GL) in the absence and presence of NMDA. Mitochondria of the ShOpa1-expressing neurons were significantly short and fragmented, with 81.36 ± 3.7% exhibiting a length of less than 1 μm compared with control at 49.79 ± 3.32% even in the absence of NMDA (Fig. 7E). Interestingly, mitochondria fragmentation induced by ShOpa1 could not be rescued by expression of calpastatin, and 79.30 ± 1.0% of mitochondria remained fragmented in the ShOpa1 + calpastatin group. These data support the hypothesis that Opa1 regulates mitochondrial morphology downstream of calpain activation (Fig. 7E). Together, our results support a model (Fig. 7F) whereby the loss of Opa1 function is a key defect underlying excitotoxic cell death.
DISCUSSION
The results of these studies support a number of conclusions. First, mitochondrial morphology defects are key events in the progression of excitotoxic neuronal injury. Second, defects in mitochondrial fusion and aberrant inner mitochondrial membrane structure are identified as major contributors to the disrupted mitochondrial morphology following excitotoxic injury. Third, increased levels of Opa1 promote neuronal survival and rescue mitochondrial morphology defects. Importantly, inhibition of calpain proteases restores mitochondrial morphology and neuronal survival. Neuroprotection conferred by calpastatin requires Opa1 because protection was significantly reduced following knockdown of Opa1. These findings are the first to identify Opa1 as a key regulator of neuronal fate during excitotoxic cell death and show that Opa1 function can be modulated by calpain activation.
These studies suggest extensive evidence supporting a role for mitochondrial dynamics as a modulator of excitotoxic signaling. Proteins that control mitochondrial fission and fusion have been previously shown to be involved in apoptotic cell death regulation. Most studies described so far implicate these mitochondrial GTPases in classical apoptotic cell death that require the proapoptotic Bcl family proteins. For example, Drp1, which is essential for mitochondrial fission, is recruited to the mitochondria at puncta with Bax (32). Drp1 activity is associated with the opening of cristae and the release of cytochrome c (9, 33). The importance of Drp1 in apoptotic cell death is demonstrated by the expression of dominant negative mutants of Drp1, which protect cells against multiple apoptotic insults (9, 13). The fusion protein Mfn2 has also been found in puncta with Bax and increased expression of Mfn2 protects against exposure to staurosporine, reactive oxygen species, and DNA damage (12, 32, 34). Thus, it has become increasingly clear that the fission/fusion machinery is involved in the regulation of classical apoptotic cell death.
Because much of the function of mitochondria in cell death has focused on Bax-mediated model systems, the role of mitochondrial dynamics in nonclassical death signaling pathways remains unknown. Excitotoxic cell death is a Bax-independent mechanism of cell death that results from the release of excessive levels of the excitatory neurotransmitter, glutamate (15, 16). There are several receptors for glutamate; however, the major receptors implicated in excitotoxic cell death are the NMDA receptors (35, 36). Glutamate overstimulation results in an excessive influx of calcium, which triggers calcium-mediated activation of second messengers. Unlike classical apoptotic cell death, the absence of Bax does not affect the rate of neuronal cell death induced by excitotoxicity (16). In addition, Mfn2, which provides robust protection against Bax-mediated pathways of neuronal injury (13), could only provide modest protection against excitotoxic injury (Fig. 4D). This clearly implicates distinct signaling mechanisms, causing neuronal loss in each pathway. In the present study, we questioned the role of Opa1 in death signaling involving these Bax-independent pathways.
Following NMDA receptor activation, mitochondria transition from tubular morphology into fragmented structures. In addition, results using the photoactivable fusion assay indicate that mitochondrial fusion is significantly blocked following 7–8 h of treatment with NMDA. These data show for the first time that defects in the rate of mitochondrial fusion are major contributors of mitochondrial morphology defects following excitotoxicity. Although decreased mitochondrial fusion plays a significant role in mitochondrial dysfunction, other possibilities cannot be excluded. For example, Drp1-mediated fission may be in part accountable for the dramatic fragmented phenotype observed following excitotoxicity. This hypothesis is supported by studies in cell lines where an increase in mitochondrial calcium influx results in mitochondrial fragmentation (5, 8, 10), in part rescued by inhibition of Drp1 (9). In the present study, defects in mitochondrial fusion were clearly evident following exposure to NMDA, which may be attributable to loss of function of the inner membrane fusion GTPase, Opa1.
Our EM studies revealed that excitoxicity resulted in a dramatic deformation of cristae structure. In addition to a role in fusion, Opa1 has been shown to play a key role in the regulation of cristae structure by forming oligomers at cristae junctions (25). Thus, we asked whether Opa1 promotes neuronal survival following excitotoxicity. Opa1 up-regulation confers significant protection against NMDA-induced neuronal death and associated mitochondrial defects. The rescue with Opa1 overexpression, although very clear, is not 100%, probably due to the complexity of this mode of injury, involving multiple signaling pathways.
We investigated whether loss of Opa1 from mitochondria and its release into cytoplasm contributes to mitochondrial dysfunction. Opa1 expression levels were therefore evaluated in the cytoplasmic and mitochondrial fractions in the absence and presence of NMDA (Fig. 5F). No major release of Opa1 from mitochondria was found. In addition, Opa1 isoforms and expression levels were similar following NMDA treatment. These findings suggest that NMDA and increased calcium levels may affect Opa1 function by either disrupting interactions with other proteins or modifying Opa1 oligomerization (25). Future studies are required to explore these different possibilities.
Due to the excessive intracellular calcium levels induced by NMDA, we asked whether calpains might have a role in this process. The present studies identify a novel mechanism that place Opa1 downstream of calpain activation following excitotoxic injury. Inhibition of calpains with calpastatin maintained mitochondrial morphology and protected neurons against excitotoxic cell death. Furthermore, this protective effect is significantly attenuated upon down-regulation of Opa1. Whether calpains directly target Opa1 or whether they modulate Opa1 activity through an indirect mechanism is a subject for future studies.
In conclusion, our studies identify a link between Opa1, a key regulator of mitochondrial dynamics, and NMDA-induced toxicity. We propose a model in which overactivation of NMDAR followed by calcium influx results in loss of Opa1 oligomers and deformation of cristae junctions downstream of calpain activation. This may explain the mitochondrial morphology defects, disruption of respiratory chain complex, and ATP depletion associated with excitotoxic neuronal loss. Because Opa1 belongs to the family of large GTPases and GTPases are highly accessible to therapeutic modulation (37–43), Opa1 is a promising target for the study of novel pharmaceutical agents to rescue neuronal death following acute neuronal injury.
Acknowledgments
We thank Peter Ripstein for technical assistance and Marc Germain and Carmen Hamze for critical review of the manuscript.
This work was supported by Heart and Stroke Foundation of Canada Grant NA6030 and a grant from the Canadian Institutes of Health Research (CIHR) (to R. S. S.).
- NMDA
- N-methyl-d-aspartic acid
- NMDAR
- NMDA receptor(s)
- CGN
- cerebellar granule neuron
- DIV
- days in vitro
- EM
- electron microscopy
- OCT
- orinthine carbamyltransferase
- PAGFP
- photoactivable GFP
- ShOpa1
- short hairpin Opa1.
REFERENCES
- 1. Arundine M., Tymianski M. (2003) Cell Calcium 34, 325–337 [DOI] [PubMed] [Google Scholar]
- 2. Hardingham G. E., Bading H. (2003) Trends Neurosci. 26, 81–89 [DOI] [PubMed] [Google Scholar]
- 3. Seo S. Y., Kim E. Y., Kim H., Gwag B. J. (1999) J. Neurosci. 19, 8849–8855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Knott A. B., Perkins G., Schwarzenbacher R., Bossy-Wetzel E. (2008) Nat. Rev. Neurosci. 9, 505–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cereghetti G. M., Stangherlin A., Martins de Brito O., Chang C. R., Blackstone C., Bernardi P., Scorrano L. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 15803–15808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cribbs J. T., Strack S. (2007) EMBO Rep. 8, 939–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Han X. J., Lu Y. F., Li S. A., Kaitsuka T., Sato Y., Tomizawa K., Nairn A. C., Takei K., Matsui H., Matsushita M. (2008) J. Cell Biol. 182, 573–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Breckenridge D. G., Stojanovic M., Marcellus R. C., Shore G. C. (2003) J. Cell Biol. 160, 1115–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Germain M., Mathai J. P., McBride H. M., Shore G. C. (2005) EMBO J. 24, 1546–1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hom J. R., Gewandter J. S., Michael L., Sheu S. S., Yoon Y. (2007) J. Cell Physiol. 212, 498–508 [DOI] [PubMed] [Google Scholar]
- 11. de Brito O. M., Scorrano L. (2008) Nature 456, 605–610 [DOI] [PubMed] [Google Scholar]
- 12. Jahani-Asl A., Cheung E. C., Neuspiel M., MacLaurin J. G., Fortin A., Park D. S., McBride H. M., Slack R. S. (2007) J. Biol. Chem. 282, 23788–23798 [DOI] [PubMed] [Google Scholar]
- 13. Barsoum M. J., Yuan H., Gerencser A. A., Liot G., Kushnareva Y., Gräber S., Kovacs I., Lee W. D., Waggoner J., Cui J., White A. D., Bossy B., Martinou J. C., Youle R. J., Lipton S. A., Ellisman M. H., Perkins G. A., Bossy-Wetzel E. (2006) EMBO J. 25, 3900–3911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Stout A. K., Raphael H. M., Kanterewicz B. I., Klann E., Reynolds I. J. (1998) Nat. Neurosci. 1, 366–373 [DOI] [PubMed] [Google Scholar]
- 15. Dargusch R., Piasecki D., Tan S., Liu Y., Schubert D. (2001) J. Neurochem. 76, 295–301 [DOI] [PubMed] [Google Scholar]
- 16. Cheung E. C., Melanson-Drapeau L., Cregan S. P., Vanderluit J. L., Ferguson K. L., McIntosh W. C., Park D. S., Bennett S. A., Slack R. S. (2005) J. Neurosci. 25, 1324–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jahani-Asl A., Germain M., Slack R. S. (2010) Biochim. Biophys. Acta 1802, 162–166 [DOI] [PubMed] [Google Scholar]
- 18. Cregan S. P., MacLaurin J. G., Craig C. G., Robertson G. S., Nicholson D. W., Park D. S., Slack R. S. (1999) J. Neurosci. 19, 7860–7869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. He T. C., Zhou S., da Costa L. T., Yu J., Kinzler K. W., Vogelstein B. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 2509–2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rashidian J., Iyirhiaro G., Aleyasin H., Rios M., Vincent I., Callaghan S., Bland R. J., Slack R. S., During M. J., Park D. S. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 14080–14085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. O'Hare M. J., Kushwaha N., Zhang Y., Aleyasin H., Callaghan S. M., Slack R. S., Albert P. R., Vincent I., Park D. S. (2005) J. Neurosci. 25, 8954–8966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zunino R., Schauss A., Rippstein P., Andrade-Navarro M., McBride H. M. (2007) J. Cell Sci. 120, 1178–1188 [DOI] [PubMed] [Google Scholar]
- 23. Cheung E. C., Joza N., Steenaart N. A., McClellan K. A., Neuspiel M., McNamara S., MacLaurin J. G., Rippstein P., Park D. S., Shore G. C., McBride H. M., Penninger J. M., Slack R. S. (2006) EMBO J. 25, 4061–4073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cregan S. P., Fortin A., MacLaurin J. G., Callaghan S. M., Cecconi F., Yu S. W., Dawson T. M., Dawson V. L., Park D. S., Kroemer G., Slack R. S. (2002) J. Cell Biol. 158, 507–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Frezza C., Cipolat S., Martins de Brito O., Micaroni M., Beznoussenko G. V., Rudka T., Bartoli D., Polishuck R. S., Danial N. N., De Strooper B., Scorrano L. (2006) Cell 126, 177–189 [DOI] [PubMed] [Google Scholar]
- 26. Chan D. C. (2006) Dev. Cell 11, 592–594 [DOI] [PubMed] [Google Scholar]
- 27. Herlan M., Vogel F., Bornhovd C., Neupert W., Reichert A. S. (2003) J. Biol. Chem. 278, 27781–27788 [DOI] [PubMed] [Google Scholar]
- 28. Meeusen S., DeVay R., Block J., Cassidy-Stone A., Wayson S., McCaffery J. M., Nunnari J. (2006) Cell 127, 383–395 [DOI] [PubMed] [Google Scholar]
- 29. Won S. J., Kim D. Y., Gwag B. J. (2002) J. Biochem. Mol. Biol. 35, 67–86 [DOI] [PubMed] [Google Scholar]
- 30. Badugu R., Garcia M., Bondada V., Joshi A., Geddes J. W. (2008) J. Biol. Chem. 283, 3409–3417 [DOI] [PubMed] [Google Scholar]
- 31. Polster B. M., Basañez G., Etxebarria A., Hardwick J. M., Nicholls D. G. (2005) J. Biol. Chem. 280, 6447–6454 [DOI] [PubMed] [Google Scholar]
- 32. Karbowski M., Lee Y. J., Gaume B., Jeong S. Y., Frank S., Nechushtan A., Santel A., Fuller M., Smith C. L., Youle R. J. (2002) J. Cell Biol. 159, 931–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Scorrano L., Ashiya M., Buttle K., Weiler S., Oakes S. A., Mannella C. A., Korsmeyer S. J. (2002) Dev. Cell 2, 55–67 [DOI] [PubMed] [Google Scholar]
- 34. Neuspiel M., Zunino R., Gangaraju S., Rippstein P., McBride H. (2005) J. Biol. Chem. 280, 25060–25070 [DOI] [PubMed] [Google Scholar]
- 35. Sattler R., Tymianski M. (2001) Mol. Neurobiol. 24, 107–129 [DOI] [PubMed] [Google Scholar]
- 36. Forder J. P., Tymianski M. (2009) Neuroscience 158, 293–300 [DOI] [PubMed] [Google Scholar]
- 37. Aznar S., Fernández-Valerón P., Espina C., Lacal J. C. (2004) Cancer Lett 206, 181–191 [DOI] [PubMed] [Google Scholar]
- 38. Kloog Y., Cox A. D. (2000) Mol. Med. Today 6, 398–402 [DOI] [PubMed] [Google Scholar]
- 39. Kume H. (2008) Curr. Med. Chem. 15, 2876–2885 [DOI] [PubMed] [Google Scholar]
- 40. Marchioni F., Zheng Y. (2009) Curr. Pharm. Des. 15, 2481–2487 [DOI] [PubMed] [Google Scholar]
- 41. Wakino S., Kanda T., Hayashi K. (2005) Drug News Perspect. 18, 639–643 [DOI] [PubMed] [Google Scholar]
- 42. Xing X. Q., Gan Y., Wu S. J., Chen P., Zhou R., Xiang X. D. (2006) Drug News Perspect. 19, 517–522 [DOI] [PubMed] [Google Scholar]
- 43. Zhao D., Pothoulakis C. (2003) Expert Opin. Ther. Targets 7, 583–592 [DOI] [PubMed] [Google Scholar]