Abstract
The diverse roles of TopBP1 in DNA replication and checkpoint signaling are associated with the scaffolding ability of TopBP1 to initiate various protein-protein interactions. The recognition of the BACH1/FANCJ helicase by TopBP1 is critical for the activation of the DNA replication checkpoint at stalled replication forks and is facilitated by the C-terminal tandem BRCT7/8 domains of TopBP1 and a phosphorylated Thr1133 binding motif in BACH1. Here we provide the structural basis for this interaction through analysis of the x-ray crystal structures of TopBP1 BRCT7/8 both free and in complex with a BACH1 phospho-peptide. In contrast to canonical BRCT-phospho-peptide recognition, TopBP1 BRCT7/8 undergoes a dramatic conformational change upon BACH1 binding such that the two BRCT repeats pivot about the central BRCT-BRCT interface to provide an extensive and deep peptide-binding cleft. Additionally, we provide the first structural mechanism for Thr(P) recognition among BRCT domains. Together with systematic mutagenesis studies, we highlight the role of key contacts in governing the unique specificity of the TopBP1-BACH1 interaction.
Keywords: DNA Damage, DNA Replication, Peptide Interactions, Protein Domains, X-ray Crystallography
Introduction
DNA damage checkpoints coordinate the cellular events necessary to ensure that DNA is repaired and faithfully replicated before cell cycle progression. A critical checkpoint triggered by DNA damage encountered during DNA replication (1, 2) involves ATR (Ataxia telangiectasia and Rad3 related), a Ser/Thr kinase that phosphorylates an array of proteins including CHK1 to regulate checkpoint control (3). The ability of ATR to function at the replication fork is dependent on the assembly of a growing list of proteins, including replication protein A, the ATR-ATRIP heterodimer, the trimeric Rad9-Hus1-Rad1 (9-1-1) clamp complex, BACH1/FANCJ (BRCA1-associated C-terminal helicase/Fanconi anemia group J protein), and topoisomerase IIβ-binding protein 1 (TopBP1)2 (4–6). In particular, the emergence of TopBP1 as a key regulator in the DNA replication checkpoint pathway is underscored by its multiple roles contributing to the activation of ATR.
TopBP1 possesses nine BRCT domains, the most of any BRCT domain-containing protein (7, 8). Originally identified with eight BRCT domains using sequence analysis, recent structural studies have confirmed an additional cryptic BRCT domain (BRCT0) at the extreme N terminus of TopBP1 (9, 10). The roles of BRCT domains as phosphorylated protein-binding modules have been demonstrated in studies of the BRCT domains in BRCA1 (breast cancer-associated protein 1) and other BRCT-containing proteins (11, 12). Although the phospho-peptide binding ability of single BRCTs is still poorly defined, the role of tandem BRCT domains in recognizing phospho-peptide motifs is well established. For example, the tandem BRCT domains of BRCA1 form a functional unit to recognize the Ser(P)-Xaa-Xaa-Phe binding motif in a number of proteins involved in the DNA damage response (11–16), and the structural principles governing these interactions have been elucidated through a number of structural studies (17–22). Because of the abundance of BRCT repeats present in TopBP1, it is speculated that the diverse roles of TopBP1 in DNA replication and checkpoint signaling are associated with the ability of TopBP1 to act as a scaffolding protein and facilitate various protein-protein interactions. TopBP1 BRCT5 is responsible for the localization of TopBP1 to DNA damage foci (23), although the interactions mediated by this domain are not well understood. Recent evidence suggests that TopBP1 BRCT4/5 interacts with 53BP1 to mediate the checkpoint function of 53BP1 in G1 (24). BRCT6 is a target for poly(ADP) ribosylation (25), but unlike other BRCT repeats, it lacks a functional phosphate-binding pocket (26). Studies of the TopBP1 yeast homolog show that the N-terminal BRCT1/2 domains of Dpb11 interact with phosphorylated Sld3, which is required for replication initiation (27, 28). A phosphorylation-dependent interaction involving Treslin and TopBP1 BRCT1/2 was also shown to be pivotal for replication initiation (29). There is also evidence that TopBP1 C-terminal BRCT7/8 domains bind to an internal Ser(P) binding motif in TopBP1, inducing TopBP1 oligomerization that is needed for E2F1-mediated apoptosis (30).
It is now apparent that regulation of the DNA replication checkpoint by TopBP1 is governed by two distinct BRCT-mediated interactions at the N- and C-terminal ends of TopBP1. Activation of ATR through the ATR-activating domain of TopBP1 depends on the interaction between the TopBP1 BRCT1/2 domains and the phosphorylated tail of the Rad9 component of the 9-1-1 complex (31, 32). Recently, we identified an interaction between TopBP1 and BACH1/FANCJ required for replication protein A chromatin loading, which is a prerequisite for the loading of the ATR-ATRIP complex to stalled replication forks and subsequent replication checkpoint activation (5). This interaction is mediated by the S phase-specific phosphorylation of BACH1 at Thr1133 and the TopBP1 BRCT7/8 domains. The discovery of a BACH1-binding motif for TopBP1 is particularly intriguing, because BACH1 was originally identified as a BRCA1 BRCT-interacting partner (12, 33).
Previous work suggests that BRCT domains specifically bind Ser(P)-containing peptide motifs (34). For example, in vitro peptide library studies show that BRCA1, MDC1 (mediator of DNA damage checkpoint protein 1), BARD1, and DNA ligase IV BRCT repeats preferentially bind Ser(P) peptides (11, 35). However, given that checkpoint Ser/Thr kinases such as ATM, ATR, and cyclin-dependent kinases can phosphorylate both Ser and Thr sites in target proteins, it is reasonable to suspect that a subset of BRCT domains could have Thr(P) peptide binding ability. Indeed, other conserved Ser(P)/Thr(P)-binding modules such as 14-3-3 and WW domains can recognize Ser(P)- and Thr(P)-binding motifs. On the other hand, the FHA-binding domain has a unique selectivity for Thr(P)-binding motifs only (36).
Crystal structures of complexes involving tandem BRCT repeats with their cognate phospho-peptides have provided insight into the molecular basis of BRCT domain interactions. Studies of BRCA1, MDC1, Schizosaccharomyces pombe Brc1, and S. pombe Crb2 BRCT domain-peptide complexes reveal a conserved mode of recognition that can be divided into two key regions: a Ser(P)-binding pocket in the N-terminal BRCT and a +3 specificity pocket at the BRCT-BRCT interface (18–22, 37–40). Comparison of the bound and unbound forms of the tandem BRCT domains reveal only subtle changes in structure, suggesting that the binding pocket is largely preformed for peptide binding. Although the current structures provide mechanistic detail of Ser(P) peptide recognition, how BRCT domains can recognize Thr(P) peptide motifs remains elusive.
Here we present the molecular basis of the TopBP1 BRCT7/8-BACH1 interaction involved in DNA replication checkpoint control. In combination with systematic mutagenesis studies in vitro and in vivo, we illustrate the role of key contact residues in the specificity of TopBP1-BACH1 interactions. Comparison of the apo and bound structures reveal a dramatic rearrangement of the BRCT domains that is required for specific phospho-peptide recognition. The structure of the TopBP1 BRCT7/8-BACH1 complex also establishes the basis for Thr(P) recognition of BRCT domains. Taken together, our studies provide insight into novel roles of BRCT-phospho-peptide recognition.
EXPERIMENTAL PROCEDURES
Cloning, Expression, and Purification
TopBP1 BRCT7/8 (1264–1493) was cloned into the pGEX-6P-1 vector (GE Healthcare) encoding an N-terminal GST tag. The GST fusion protein was expressed in Escherichia coli BL21-Gold cells and purified using glutathione affinity chromatography. TopBP1 BRCT7/8 was then cleaved from GST with PreScission protease at 4 °C overnight, and the TopBP1 BRCT7/8 polypeptide was purified from GST by cation exchange chromatography. Further purification was achieved using gel filtration chromatography on a Superdex 75 column (Amersham Biosciences) in storage buffer (400 mm NaCl, 1 mm Tris(2-carboxyethyl)phosphine, and 10 mm Tris-HCl, pH 7.5). TopBP1 BRCT7/8 missense variants were engineered using mutagenesis by PCR-directed overlap extension (41) and cloned into pGEX-6P-1 vector. Selenomethionine-incorporated TopBP1 BRCT7/8 was expressed in E. coli BL21-Gold pLys S cells and purified in the same manner as native TopBP1 BRCT7/8.
Crystallization
Purified Selenomethionine TopBP1 BRCT7/8 was concentrated to 18 mg/ml for crystallization. Selenomethionine-derivative crystals were grown at room temperature using hanging drop vapor diffusion by mixing 2 μl of protein with 1 μl of reservoir containing 1.35 m Li2SO4 and 0.1 m Tris-HCl, pH 8. The crystals were flash-cooled in a cryo-protectant consisting of mother liquor supplemented with 23% glycerol. Native TopBP1 BRCT7/8 concentrated to 12 mg/ml was incubated in a 1:2 molar ratio of BACH1 phospho-peptide (Ac-ESIYFpTPELYDPEDTKK-NH2, Biomatik) for co-crystallization. Co-crystals were grown at room temperature by mixing 2 μl of protein with 1 μl of reservoir solution (3.5 m sodium formate, pH 8) and flash-cooled in mother liquor supplemented with 15% glycerol.
Data Collection and Structure Determination
The data were collected at the CMCF-1 beamline at the Canadian Light Source (Saskatoon, Canada). Data for a single-wavelength anomalous dispersion experiment was collected at the selenium peak from a selenomethionine crystal, and intensity data were processed using the HKL-2000 package (42). Two selenium atom positions were found using SHELXD (43) and refined using SOLVE (44). The phases were improved by density modification with RESOLVE (44), resulting in a figure of merit of 0.64. Automated model building was carried out in ARP/wARP (45) using experimental phases and phase restraints to produce 214 of 235 built residues with side chains. Further model building was carried out in COOT (46) and refinement using TLS and restrained refinement in REFMAC5 (47, 48). The final model lacks the N-terminal residues 1264–1265 and loop residues 1442–1449, which are presumed to be disordered in the crystals. The Ramachandran plot contained 94.8% of all residues in the core and 5.2% in allowed regions.
Intensity data from a TopBP1 BRCT7/8-BACH1 peptide complex crystal was reduced and scaled using the HKL-2000 package. Phases for the TopBP1 BRCT7/8-BACH1 peptide complex were solved by molecular replacement using the apo TopBP1 BRCT7/8 structure as a model in PHASER (49). TopBP1 BRCT7/8 N-terminal residues 1264–1265, side chain of residue 1265, loop residues 1442–1449, and C-terminal residues 1492–1493 are disordered and missing from the model. We were also unable to model BACH1 phospho-peptide residues −5 and +6 to +9. The Ramachandran plot contained 92.0% of all residues in the core and 8.0% in allowed regions.
The data collection and refinement statistics are listed in Table 1. Secondary structure prediction of the models was performed with DSSP (50) and converted using DSSP2PDB. Hydrogen bonding was verified using HBPLUS (51). All of the figures were made with PyMOL. The morph movie was generated with the program CNS (52) using the input file morph_dist.inp (53, 54). The coordinates for TopBP1 BRCT7/8 and TopBP1 BRCT7/8-BACH1 peptide complexes are in the Protein Data Bank (accession codes 3AL2 and 3AL3).
TABLE 1.
Data collection, phasing, and refinement statistics
| TopBP1 BRCT7/8 (selenomethionine) | TopBP1 BRCT7/8-peptide complex | |
|---|---|---|
| Data collection | ||
| Space group | C2 | P6222 |
| Cell dimensions | ||
| a, b, c (Å) | 102.58, 32.75, 81.76 | 78.07, 78.07, 136.77 |
| α, β, γ (°) | 90.0, 109.3, 90.0 | 90.0, 90.0, 120.0 |
| Wavelength | 0.97879 | 0.97949 |
| Resolution (Å) | 50.0-2.0 | 50.0-2.15 |
| Rsyma | 6.2 (31.8) | 4.6 (46.5) |
| I/σI | 14.1 (2.4) | 29.0 (2.6) |
| Completeness (%) | 99.1 (92.9) | 99.9 (100.0) |
| Redundancy | 3.6 (2.7) | 6.8 (6.2) |
| Refinement | ||
| Resolution (Å) | 31.0-2.00 | 33.9-2.15 |
| No. reflections | 17624 (902) | 14021 (699) |
| Rwork/Rfreeb | 17.0/20.4 | 19.6/23.6 |
| No. atoms | ||
| Protein | 1755 | 1731 |
| Peptide | 92 | |
| Ligand | 10 | 3 |
| Water | 182 | 96 |
| B-factors | ||
| Protein | 21.3 | 47.1 |
| Peptide | 39.8 | |
| Ligand | 34.8 | 55.7 |
| Water | 32.9 | 57.1 |
| Root mean square deviations | ||
| Bond lengths (Å) | 0.009 | 0.009 |
| Bond angles (°) | 1.346 | 1.258 |
a Rsym = Σ|I − <I>|2/Σ|I2|.
b R = Σ||Fo| − |Fc||/Σ|Fo|. Rfree was calculated from 5% of the data excluded from refinement.
Fluorescence Polarization
FP measurements were carried out using an Envision multilabel plate reader (PerkinElmer Life Sciences). For TopBP1 BRCT7/8 FP assays, each well consisted of 10 nm FITC-labeled BACH1 phospho-peptide (FITC-ESIYFpTPELYDPEDT-NH2; Biomatik) and increasing protein concentrations in FP assay buffer (10 mm Tris-HCl, pH 7.5, 400 mm NaCl, 1 mm Tris(2-carboxyethyl)phosphine, 0.5% Tween 20). Competition assays were done by titrating increasing peptide concentrations with a saturated concentration of protein and 10 nm FITC-phospho-peptide. Competition assays for MDC1 and BRCA1 were done as described previously (17). The competition peptides used for the assays are: Thr(P) (Ac-ESIYFpTPELYDPEDTKK-NH2 for TopBP1, Ac-pTPTF-OH for BRCA1, and Ac-pTQEY-OH for MDC1) and Ser(P) (Ac-ESIYFpSPELYDPEDTKK-NH2 for TopBP1, Ac-pSPTF-OH for BRCA1, and Ac-pSQEY-OH for MDC1). FP assays were incubated for 15 min at room temperature prior to taking FP measurements using an excitation wavelength of 485 nm and an emission wavelength of 538 nm. Triplicate data points are represented in graphs as the means ± S.E. Curve fitting, Kd, and IC50 measurements were obtained using PRISM software (GraphPad). Ki calculations were calculated using the IC50-to-Ki converter (55).
Immunoprecipitation
For transient transfection and co-immunoprecipitation experiments, 293T cells were transfected with indicated plasmids for 24 h. The cells were collected and lysed with NTEN buffer (20 mm Tris-HCl, pH 8.0, 100 mm NaCl, 1 mm EDTA, 0.5% Nonidet P-40) containing protease inhibitors on ice for 20 min. After removal of cell debris by centrifugation, the soluble fractions were collected and incubated with S protein-agarose (Novagen) for 3 h at 4 °C. The precipitates were then washed four times with NTEN buffer and boiled in SDS loading buffer. The samples were resolved on SDS-PAGE, transferred to PVDF membrane, and immunoblotted with antibodies as indicated.
RESULTS
Structures of TopBP1 BRCT7/8 and BACH1 Phospho-peptide Complex
The crystal structures of TopBP1 BRCT7/8 and its complex with a BACH1 phospho-peptide were determined to 2.0 and 2.15 Å, respectively. Like other established tandem BRCT domain structures (19), TopBP1 BRCT7/8 packs in a head to tail manner via a hydrophobic interface between the two BRCT domains (Fig. 1A). Each BRCT domain consists of a central four-stranded parallel β-sheet packed on opposite sides by helical elements. Separating BRCT7 and 8 is a prominent linker helix (αL), which is part of an unusually long linker region compared with other phospho-peptide binding tandem BRCT domains (supplemental Fig. S1). The conserved phospho-peptide recognition of tandem BRCT repeats is evident in the TopBP1 BRCT7/8-BACH1 phospho-peptide complex, where the Thr(P) sits in a conserved phosphate-binding pocket in the N-terminal BRCT7 and the +3 residue is complemented by a hydrophobic cavity formed at the interface of BRCT7 and 8 (Fig. 1B). Contacts between the BRCT7/8 domains and BACH1 phospho-peptide span the −2 to +5 positions of the peptide, burying a solvent-accessible surface area of 1208 Å2 at the interface.
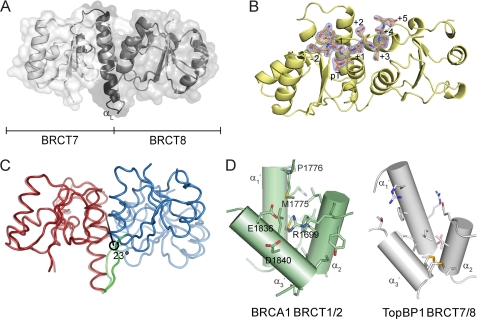
FIGURE 1.
Structure of TopBP1 BRCT7/8 and TopBP1 BRCT7/8-BACH1 peptide complex. A, cartoon representation of the apo structure of TopBP1 BRCT7/8. The linker region is colored black, and the αL helix is labeled. B, the BACH1 phospho-peptide (orange) binds in a region spanning the TopBP1 BRCT7/8 domains (yellow). The (2Fo − Fc) electron density map at 2σ for the phospho-peptide is shown. The residue positions in the phospho-peptide are labeled. C, representation of the structural rearrangement of TopBP1 BRCT7/8 around the central rotation axis. The initial apo state is represented at 50% transparency. The fixed domain (red), moving domain (blue), and interdomain bending residues (green) are colored. D, comparison of the hydrophobic packing interface of BRCA1 BRCT1/2 and TopBP1 BRCT7/8. The helices are represented as cylinders and labeled. Residues involved in interface packing are shown as sticks. Residues involved in BRCA1 and not in TopBP1 BRCT packing are labeled.
Surprisingly, the apo TopBP1 BRCT7/8 structure adopts a conformation that is significantly more open than the TopBP1 BRCT7/8-BACH1 phospho-peptide complex. This is unusual for tandem BRCT repeats, where comparisons of the apo and peptide-bound crystal structures in BRCA1, MDC1, Brc1, and Crb2 BRCT domains only yield subtle structural changes (18–21, 37–39, 56). This can be further illustrated, for example, in structural alignments of both the apo and bound crystal structures of MDC1 BRCT1/2 with the TopBP1 BRCT7/8-BACH1 peptide complex, which clearly show a better agreement compared with the apo TopBP1 BRCT7/8 structure (supplemental Fig. S2). The open structure of TopBP1 BRCT7/8 also does not appear to be a consequence of the crystal packing, because the structure of apo TopBP1 BRCT7/8 crystallized under different conditions in a different space group yielded the same conformation (data not shown). To quantify the degree of conformational change between the apo and peptide-bound structure, we used the program DYNDOM (57) to define and measure protein domain motion. Both a fixed domain (BRCT7 and the αL helix (residues 1268–1392)) and moving domain (BRCT8 (residues 1393–1489)) were identified, with the moving domain rotating 23° around a central axis (Fig. 1C). The residues defined in interdomain bending (residues 1388–1393) also conveniently flank the C-terminal end of the αL helix.
The plasticity of TopBP1 BRCT7/8 may be explained by the difference in the packing interface compared with canonical tandem BRCT repeats. Because of the extensive hydrophobic interface contributed by the α2-α1′-α3′ helices, such as in BRCA1 BRCT1/2 (56), tandem BRCT repeats are typically rigid (Fig. 1D). Consequently, their peptide-binding surfaces are considered preformed, aside from minor changes to the backbone or side chain conformation, and the +3 binding cavity is relatively shallow. In contrast, the smaller hydrophobic packing surface at the interface between BRCT7 and 8 of TopBP1 results in a larger cavity (Fig. 1D). In particular, the TopBP1 equivalent residues of BRCA1 Arg1699, Met1775, Pro1776, Glu1836, and Asp1840, all of which contribute to the interface packing at the top of the pocket in BRCA1, do not do so in TopBP1. This allows for a more dynamic interaction of the BRCT domains in TopBP1 BRCT7/8 and explains its conformational change upon peptide binding.
Conserved Thr(P)-binding Pocket of TopBP1 BRCT7/8
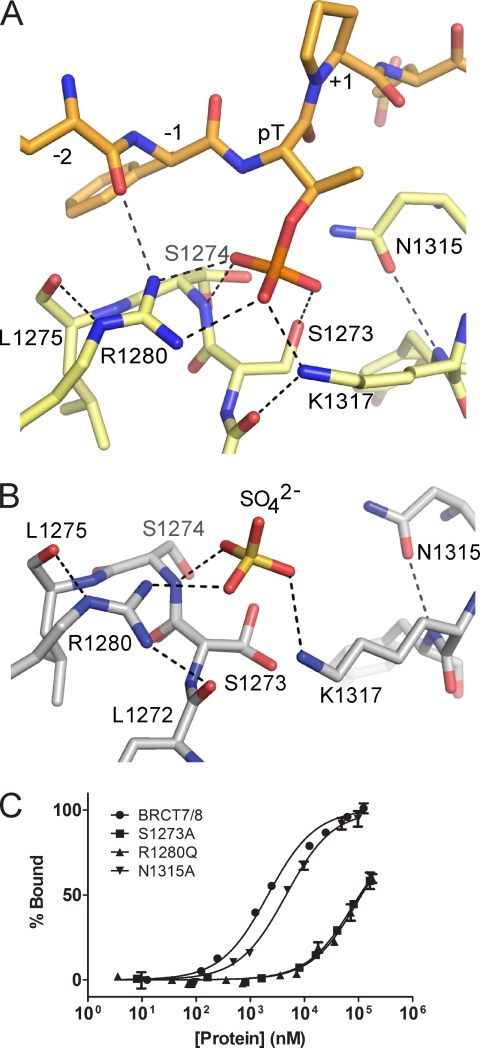
The Thr(P)-binding pocket of TopBP1 BRCT7/8 is made up of phosphate-binding residues that are conserved in other phosphate-binding BRCT repeats. The phosphate moiety makes conserved interactions with the side chains of Ser1273 and Lys1317, as well as with the main chain amide of Ser1274 (Fig. 2A and supplemental Fig. S1). In addition, the guanidinium group of Arg1280 makes a novel bidentate interaction with two phosphate oxygen atoms and is supported by a secondary hydrogen bond with the main chain of the peptide at the −2 position. In comparison with the bound state, the phosphate-binding pocket of TopBP1 BRCT7/8 in the apo form does not appear to be in a favorable conformation for binding. A sulfate ion is bound in the apo crystal structure and mimics the phosphate group in the binding pocket (Fig. 2B). Because of the open conformation in the apo state, Lys1317 is pulled away from the phosphate pocket and does not interact with the correct oxygen on the sulfate. Additionally, loss of the peptide backbone contact with Arg1280 shifts the guanidinium group, which instead contacts the sulfate and the main chain of Leu1272. In the phosphate-binding pocket of canonical BRCT repeats, the conserved Ser1273 side chain (Ser1655/Thr1898 in BRCA1/MDC1, respectively) is normally held in the optimal rotamer by hydrogen bonding with a conserved threonine residue (Thr1700/Thr1934 in BRCA1/MDC1, respectively) across the pocket. Because the residue at this position in TopBP1, Asn1315, is incapable of making such an interaction, Ser1273 is free to adopt multiple side chain conformations and is not held in the proper hydrogen bonding distance or orientation in the apo structure (Fig. 2B).
FIGURE 2.
Phosphate-binding pocket of TopBP1 BRCT7/8. A, stick representation of the TopBP1 BRCT7/8 (yellow) in complex with the BACH1 phospho-peptide (orange). Hydrogen bonding and electrostatic interactions are indicated by dotted lines. B, phosphate-binding pocket of apo TopBP1 BRCT7/8. C, FP binding results for TopBP1 BRCT7/8 phosphate-binding pocket mutants. GST fusion BRCT7/8 variants were purified and used in the assay. Triplicate data points are represented as the means ± S.E.
To assess the role of Arg1280 and other residues within the conserved phosphate-binding pocket in recognition of the Thr(P) BACH1 peptide in vitro, we used a FP assay. Mutation of the conserved Ser1273 or Arg1280 residues in GST-tagged TopBP1 BRCT7/8 markedly reduced binding to the peptide compared with wild type (Kd = 92 ± 8 μm for S1273A, Kd = 104 ± 11 μm for R1280Q, and Kd = 2.1 ± 0.1 μm for wild type), providing further evidence that Arg1280 is required for phosphate binding (Fig. 2C). In contrast, the N1315A mutation (Kd = 4.6 ± 0.3 μm) showed similar levels of binding as wild type, suggesting that Asn1315 does not have a role in the phosphate-binding pocket.
Thr(P)/Ser(P) Specificity of Tandem BRCT Domains
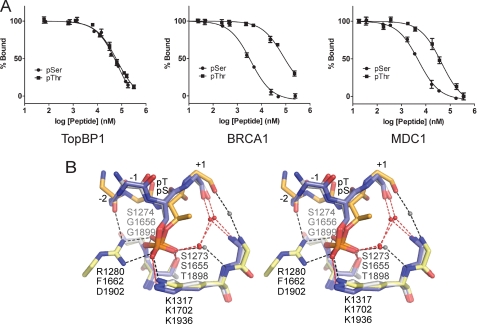
Given that BACH1 Thr(P)1133 is the first validated Thr(P) target for any BRCT domain, we were interested in whether TopBP1 BRCT7/8 also has the ability to bind Ser(P) phospho-peptides. We compared the ability of a Ser(P)-1133 derivative of the BACH1-binding motif to bind TopBP1 BRCT7/8 in a FP competition assay. Surprisingly, both Thr(P) and Ser(P) BACH1 phospho-peptides competed with the FITC-BACH1 phospho-peptide similarly (Ki = 7.7 ± 0.2 μm for Thr(P) and Ki = 8.6 ± 2.4 μm for Ser(P)), demonstrating that TopBP1 BRCT7/8 has equal specificity for both Thr(P) and Ser(P) (Fig. 3A). In contrast, previous studies of tandem BRCT repeats indicate a preference for Ser(P) phospho-peptide targets over Thr(P) (11, 35). Using the FP competition assay, we confirm that both the BRCA1 and MDC1 BRCT repeats prefer Ser(P) in their respective minimal tetra-peptide targets (Ki = 1.5 ± 0.1 μm for Ser(P) and Ki = 39.7 ± 8.0 μm for Thr(P) in BRCA1, Ki = 0.9 ± 0.1 μm for Ser(P), and Ki = 9.4 ± 3.2 μm for Thr(P) in MDC1) (Fig. 3A). The degree of preference for Ser(P), however, is more dramatic for BRCA1.
FIGURE 3.
TopBP1 BRCT7/8 has specificity for Thr(P)- and Ser(P)-binding motifs. A, FP competition assays in which cognate FITC-labeled phospho-peptide complexes of TopBP1 BRCT7/8, BRCA1 BRCT1/2, and MDC1 BRCT1/2 are challenged with their respective Thr(P) and Ser(P) peptides. B, stereo view of the superimposed Ser(P) peptide-specific coordination of BRCA1 (blue, Protein Data Bank code 1T15) and MDC1 (gray, Protein Data Bank code 2AZM) with Thr(P) peptide-specific coordination of TopBP1 (orange-yellow). The residues are labeled for TopBP1 (top), BRCA1 (middle), and MDC1 (bottom). Hydrogen bonding and electrostatic interactions in the complex are represented as dotted lines for TopBP1 (black) and BRCA1/MDC1 (red). Conserved waters mediating peptide-BRCT domain interactions are shown as spheres for TopBP1 (gray) and BRCA1/MDC1 (red).
A comparison of the structure of the TopBP1 BRCT7/8-BACH1 complex with those of the BRCA1 and MDC1 BRCT repeats bound to their respective targets suggest why these proteins exhibit different Thr(P) versus Ser(P) binding specificities. Superposition of the phosphate-binding residues in BRCA1, MDC1, and TopBP1 reveals that although the conserved phosphate contacts (S1273/S1655/T1898, K1317/K1702/K1936, and S1274/G1656/G1899 in TopBP1/BRCA1/MDC1, respectively) are maintained, the position of the phosphate group and orientation of the peptide backbone differ between the TopBP1 complex and either the BRCA1 or MDC1 complexes (Fig. 3B). The specific backbone and Thr(P)1133 position in the BACH1 phospho-peptide bound to TopBP1 is supported by the additional contacts TopBP1 Arg1280 makes with the −2 main chain and the phosphate group. The difference in phosphate position of Ser(P) compared with Thr(P) is a result of the large discrepancy in χ2 angles. The Thr(P) residue in the TopBP1 complex is gauche+, but the Ser(P) residues in the BRCA1 and MDC1 complexes are trans (supplemental Table S1), which may account for the selectivity of BRCA1/MDC1 for Ser(P) phospho-peptides. Modeling of the Thr(P) derivative in the BRCA1 and MDC1 structures clearly illustrates how the addition of the γ-methyl group in a χ2 trans orientation causes a steric clash with the phosphate oxygen atom (supplemental Fig. S3). Introduction of this methyl group also clashes with two conserved waters that mediate interactions between the phospho-peptide and phosphate-binding pocket, which may also impact phospho-peptide binding. The more dramatic preference for Ser(P) observed by BRCA1 over MDC1 may arise from the differences in the +1 residue of their cognate peptides (Pro/Gln for BRCA1/MDC1, respectively). The presence of a +1 Pro in the BRCA1 target peptide restricts the backbone geometry, perhaps limiting conformational changes that might otherwise facilitate binding of the Thr(P) peptide. In contrast to the Ser(P)-specific rotamer in the BRCA1 and MDC1 complexes, the gauche+ χ2 orientation of Thr(P)1133 in the TopBP1 complex permits the co-existence of the γ-methyl group and phosphate oxygens without steric hindrance. This specific orientation of Thr(P) also resembles that seen in structures of Thr(P) in complex with FHA domains, which are known to be selective for Thr(P)-binding motifs (supplemental Table S1).
+3/+4 Binding Pocket of TopBP1 BRCT7/8
To gain a better understanding of the specificity of the BACH1-binding motif for TopBP1 BRCT7/8, we performed alanine scanning mutagenesis to identify the residues in BACH1 important for the TopBP1 interaction in vitro. Using a FP competition assay, we show that alanine mutations of BACH1 + 5 (D1138A) and +1 (P1134A) result in little or no change in competition compared with wild type (Fig. 4A). In contrast, mutations at +3 (L1136A) and +4 (Y1137A) almost completely abolish any competition with the FITC-labeled cognate peptide, providing evidence that the +3 and +4 residues of BACH1 are the most critical specificity determinants. The fact that the +3 residue is absolutely required for specific binding is characteristic of the common mode of BRCT repeat recognition, although the additional importance of the +4 residue is surprisingly different. Mutation at +2 (E1135A) also yields a smaller but significant reduction in competition.
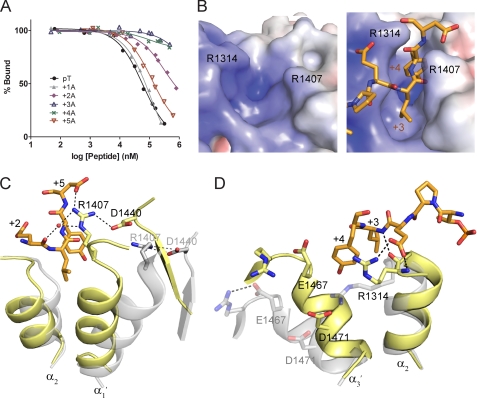
FIGURE 4.
TopBP1-BACH1 interaction at the +3/+4 binding pocket. A, FP competition analysis of the BACH1-binding motif using alanine scanning mutagenesis. BACH1 phospho-peptides mutated to alanine at +1 to +5 positions were used to compete with the FITC-labeled phospho-peptide bound to TopBP1 BRCT7/8. B, electrostatic potential surface of the TopBP1 BRCT7/8 + 3/+4 binding pocket in the apo (left) and peptide-bound (right) structures. TopBP1 Arg1314 and Arg1407 residues are mapped on the surface. C, role of Arg1407 in the TopBP1-BACH1 complex. TopBP1 BRCT7/8 in the apo (gray) and complex (yellow) structures are superimposed. Residues involved in interacting with Arg1407 are labeled. D, role of Arg1314 in +2/+3 binding of the BACH1 peptide. TopBP1 BRCT7/8 in the apo (gray) and complex (yellow) structures are superimposed.
The observed peptide binding specificity can be explained by the TopBP1 BRCT7/8-BACH1 phospho-peptide complex crystal structure. Both +3 and +4 BACH1 residues are nestled in a deep hydrophobic cavity in TopBP1 BRCT7/8 that is sculpted upon peptide binding (Fig. 4B). In the apo structure, the +3/+4 pocket is held open by the open conformation of TopBP1 BRCT7/8. Binding of the peptide initiates closure of the walls of the +3/+4 pocket, creating a tight, narrow cleft that accommodates the hydrophobic side chains of the peptide. The pocket in TopBP1 BRCT7/8 is both larger and deeper than the +3 binding pockets of other established phospho-peptide binding BRCT repeats. For example, although the same leucine +3 peptide residue exists in the S. pombe Brc1-γH2A complex (39), the leucine side chain packs horizontally across the shallow +3 pocket created by Brc1 BRCT repeats. The base of the TopBP1 BRCT7/8 + 3/+4 binding pocket is made up of a number of residues at the hydrophobic BRCT interface: Leu1319, Phe1411, Leu1414, Ile1469, and Ala1470. Additional residues (Arg1314, Glu1316, Arg1407, Gly1410, Thr1466, and Glu1467) constitute the sides of the +3/+4 pocket (supplemental Fig. S1). The pocket complements the charge and shape of the +3 leucine and +4 tyrosine residues perfectly. In addition to the hydrophobic interactions made between the hydrophobic residues, the +4 tyrosine side chain also hydrogen bonds with the main chain of Thr1466 and stacks against the guanidinium group of Arg1314 of TopBP1.
Two essential arginine residues (Arg1314 and Arg1407), neatly placed on opposite sides of the +3/+4 binding pocket, effectively shape the sides of the pocket by making a number interactions with the phospho-peptide. Mutations of Arg1314 (R1314Q) and Arg1407 (R1407A) markedly reduce binding to the FITC-BACH1 phospho-peptide in the FP assay (Kd = 56.4 ± 3.8 μm for R1314Q and Kd = 63.4 ± 4.5 μm for R1407A), highlighting their importance in phospho-peptide binding (supplemental Fig. S4). As a function of the rotational movement of BRCT8, Arg1407 makes a dramatic switch by breaking an existing salt bridge with Asp1440 to form a new salt bridge with the +5 side chain and make a water-mediated interaction with the +2 main chain (Fig. 4C). Arg1314 is conserved in other tandem BRCT domains and has a major role in recognition of the +3 main chain or carboxyl tail of the cognate peptide (17). In canonical tandem BRCT repeats, the conserved arginine (Arg1699 in BRCA1 and Arg1933 in MDC1) contributes to the BRCT interface and is held in place by salt bridge interaction(s) with conserved acidic residues (Glu1836 and Asp1840 in BRCA1 and Glu2063 in MDC1) across the interface. Although the role of the Arg1314 main chain in binding the +3 main chain is preserved, the Arg1314 side chain has a number of additional roles that appears to be unique to TopBP1 BRCT7/8. In the apo state, Arg1314 is too far from the conserved Glu1467 and Asp1471 residues across the BRCT interface to make contact (Fig. 4D). The inherent mobility of Arg1314 is further supported by the relatively poor electron density and higher B-factors associated with the Arg1314 side chain. Consequently in the peptide-bound state, Arg1314 is free to adopt a different rotamer to interact with the +2 side chain and form a cation-π interaction with +4 Tyr side chain. Taken together, the structural plasticity of the +3/+4 pocket, which is imparted by the rearrangement of TopBP1 BRCT7/8, is required for the specific TopBP1-BACH1 interaction.
In Vivo Binding Specificity of TopBP1 BRCT7/8-BACH1
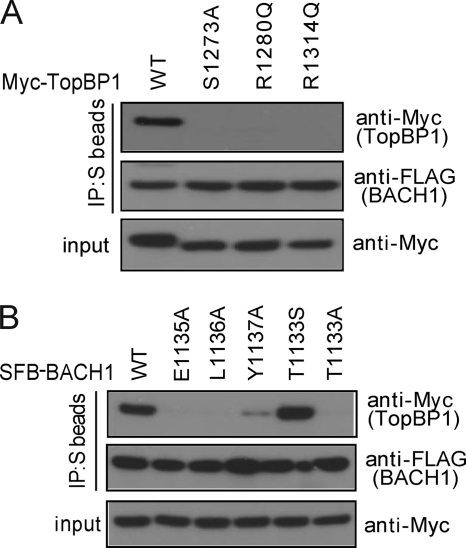
To further characterize the TopBP1 BRCT7/8-BACH1 binding specificities, we assessed the effects of mutations on the TopBP1 BRCT7/8-BACH1 interaction in vivo. From our in vitro FP binding results, we showed that mutations in the conserved phosphate-binding pocket residues (S1273A and R1280Q) and in the +3/+4 binding pocket (R1314Q) significantly reduce binding to the FITC-labeled BACH1 phospho-peptide. To determine the effects of these mutations in human cells, Myc-tagged full-length TopBP1 harboring these mutations were co-transfected with SFB-tagged BACH1. None of the mutants formed a complex with BACH1, as indicated by the absence of Myc-TopBP1 in BACH1 immunoprecipitates (Fig. 5A). Using alanine scanning mutagenesis, we also concluded that BACH1 + 2 to +4 residues are critical for TopBP1 binding specificity. In support of this, mutants of SFB-tagged BACH1 + 2 (E1135A), +3 (L1136A), and +4 (Y1137A) failed to bind Myc-TopBP1 in BACH1 immunoprecipitates (Fig. 5B). However, mutation at +2 seemed to have a more detrimental effect on binding than at +4 in our in vivo co-immunoprecipitation. Although the TopBP1 BRCT7/8 mutants failed to bind BACH1 in vivo, we note that this is not a consequence of a defect in overall TopBP1 function because these mutants still co-localize with γH2AX to IR-induced foci like wild-type TopBP1 (supplemental Fig. S5). This is consistent with our previous reports suggesting that TopBP1 BRCT7/8 does not have a role in TopBP1 localization following DNA damage (5, 23).
FIGURE 5.
In vivo binding specificities of the TopBP1-BACH1 interaction. A, effects of TopBP1 BRCT7/8 mutations in binding BACH1. Constructs encoding Myc-tagged wild type and mutants of TopBP1 were co-transfected with plasmids encoding SFB-BACH1. Immunoprecipitation (IP) reactions were performed using S protein beads and then subjected to Western blot analyses using antibodies as indicated. B, effects of BACH1 mutations in binding TopBP1. Lysates for immunoprecipitation were prepared from cells expressing Myc-tagged TopBP1 along with SFB-tagged wild type or mutants of BACH1.
To address the Thr(P)/Ser(P) specificity of TopBP1 BRCT7/8 in vivo, we tested the ability of BACH1 T1133A and BACH1 T1133S mutants to bind TopBP1. As expected, TopBP1 failed to co-immunoprecipitate with SFB-tagged BACH1 T1133A (Fig. 5B); however, BACH1 T1133S restored binding to TopBP1, suggesting that BACH1 T1133S is similarly phosphorylated in cells and interacts with TopBP1 BRCT7/8 in the same manner as wild-type BACH1. Consistent with our in vitro results, this indicates that the TopBP1 BRCT7/8, unlike the BRCA1 and MDC1 BRCT repeats, is competent to bind both Thr(P) and Ser(P) peptide motifs with similar affinities.
DISCUSSION
The role of TopBP1 as an integrator of diverse signals that control the replication stress response critically depends on its nine BRCT domains that dictate diverse molecular interactions. Here we present the first structural information providing insight into how TopBP1 binds one of its critical partners in the DNA damage response, BACH1. BACH1 was initially identified through its interactions with BRCA1 (33), which involve the recognition of a pSPTF motif (residues 990–993 of BACH1) by the tandem BRCT repeats of BRCA1 (12). This interaction is critical for regulation of cell cycle checkpoint function in response to ionizing radiation. Additionally, mutations in BACH1 are found in Fanconi anemia patients and are associated with defects in DNA cross-link repair, and indeed BACH1 is recognized as a member of the family of Fanconi-associated genes, designated FANCJ (58–60). The BRCT7/8 tandem pair of TopBP1 functionally interacts with another phosphorylated region of BACH1 that is distinct from that recognized by the BRCA1 BRCT repeats (5). This interaction is required for the proper loading of replication protein A onto single-stranded DNA near stalled replication forks in a manner that is also dependent upon the helicase activity of BACH1 and ultimately leads to ATR-dependent phosphorylation signaling. Our structural and functional analysis reveals intriguing differences in the way that TopBP1 recognizes its BACH1 target compared with BRCA1 recognition of BACH1 that govern how these two tandem BRCT repeats bind their respective phospho-targets with a high degree of specificity.
A major difference is the fact that phospho-peptide binding induces a large scale rearrangement in the packing of the TopBP1 BRCT7/8 repeats, whereas the conventional BRCT repeats are much more rigid and fixed in a conformation very similar to the bound form. The conformational change in TopBP1 BRCT7/8 upon peptide binding corresponds to a ∼20° rotation of one BRCT with respect to the other about the extended linker helix (αL) (supplemental Movie S1). The bound form of TopBP1 BRCT7/8 closely resembles the standard packing of BRCT domains found in the other tandem BRCT structures, whereas the unbound form represents a more relaxed structure, with opened binding pockets for both the phosphate and peptide +3/+4 residues. The plasticity of TopBP1 BRCT7/8 compared with the other BRCT repeats is likely due to reduced packing of the BRCT repeats. This increased flexibility could allow TopBP1 BRCT7/8 to recognize a more divergent array of peptide targets. In addition to the interaction with BACH1, BRCT7/8 has also been shown to interact with an AKT-phosphorylated region of TopBP1 between the sixth and seventh BRCT to regulate the oligomerization of TopBP1 (30). The +3/+4 positions of this target (1159pSNLQWPS) are not conserved with the BACH1 target sequence, and it may be that recognition of this peptide involves a further rearrangement of the TopBP1 BRCT7/8 specificity pocket. Although TopBP1 is more flexible than the other tandem BRCT proteins, a certain degree of flexibility in packing of the tandem repeats of BRCA1 likely also exists in solution, as suggested by both NMR (61) and thermodynamic stability studies (62, 63). A BRCT interface rotation is also observed in the Nbs1 BRCT domains, although it appears to be initiated by a mechanism dependent on the neighboring FHA domain. Binding of the peptide with the FHA domain initiates a helical rearrangement of the FHA-BRCT1 interface and ∼20° rotation at the BRCT1-BRCT2 interface (64). In contrast to the rearrangement in TopBP1 BRCT7/8, however, this rotation is in an orthogonal direction, and its significance is still unknown.
The other intriguing difference between TopBP1 BRCT7/8 and other tandem BRCT proteins is its recognition of a Thr(P) target peptide. We demonstrate that both BRCA1 and MDC1 BRCTs are specific for Ser(P)-containing targets and not Thr(P)-containing targets, whereas TopBP1 has a relaxed specificity, binding both Ser(P) and Thr(P) peptides. The sequence conservation of the different BACH1-binding motifs also complements this notion, because BACH1 Thr1133 exists as a Ser residue in some mammalian species, whereas BACH1 Ser990 is absolutely conserved throughout. The difference in specificity between BRCA1/MDC1 and TopBP1 relies on subtle differences in the geometry of recognition of the phosphorylated amino acid, likely driven by the presence of the additional Arg1280 residue in the phosphate-binding pocket of TopBP1 BRCT7/8 (Fig. 3B). Arg1280 is also conserved in a number of BRCT domains, suggesting a possible conserved mechanism of phosphate recognition (supplemental Fig. S6). For example, the equivalent arginine side chain in S. pombe Crb2 (Arg558) makes a single water-bridged interaction with the phosphate of γ-H2A in the Crb2 BRCT1/2-γH2A crystal structure (37). The bidentate mode of recognition by Arg1280 may also be conserved, as suggested by the interaction of TopBP1 Arg121 with a sulfate ion in the TopBP1 BRCT0/1/2 crystal structure (10).
Interestingly, this recognition of the Thr(P) side chain is similar to that employed by FHA domains, which specifically bind Thr(P) phospho-peptides through conserved arginine residues (65, 66). For example, Arg61 of the RNF8 FHA domain makes a bidentate interaction with the phosphate of its target Thr(P), whereas Arg42 mediates an interaction between the phosphate group and −2 of the peptide backbone. These interactions anchor the Thr(P) side chain and the backbone configuration of the peptide in a similar conformation to that observed in the TopBP1 BRCT7/8 complex (supplemental Fig. S7). TopBP1 Arg1280 fulfills the roles of the conserved RNF8 Arg61 and Arg42 residues by concurrently contacting both the phosphate and the −2 of the peptide backbone. It has been suggested that FHA domains prefer Thr(P) because binding of the Thr(P)-binding motif places the Thr(P) in an orientation that allows the γ-methyl group to pack in a small cavity in the FHA domain. Substitution to a Ser(P) would result in loss of these particular van der Waal's interactions (65). In the case of TopBP1 BRCT7/8, the Ser(P) binding ability can be explained by the absence of the FHA γ-methyl cavity, which is substituted by the water-mediated network of interactions (Fig. 3B).
TopBP1 BRCT7/8 also has in vitro DNA binding activity, and similar activities have been found for several other BRCT domains (67, 68). Perhaps the best characterized interaction is that between the single BRCT of the RFC p140 subunit and double-stranded DNA ends containing a 5′-phosphate. This BRCT domain harbors an arginine, Arg423, which is analogous to TopBP1 Arg1280, and directly participates in the recognition of the 5′-phosphate of dsDNA (69, 70). Binding of the single RFC BRCT to dsDNA also requires an additional N-terminal helix, which binds to the major groove of the dsDNA.
The interaction between TopBP1 and BACH1 is crucial for the response to DNA replication stress. We have previously shown that disruption of the TopBP1-BACH1 interaction impairs replication protein A chromatin loading, which is a prerequisite for ATR activation and DNA replication checkpoint control. This study provides the molecular mechanism that underlies this critical interaction, as well as new insights into the surprising versatility of BRCT domain function in the DNA damage response.
Supplementary Material
Acknowledgments
We thank Dr. Pawel Grochulski and the Canadian Light Source staff for assistance with synchrotron data collection. We also thank Dr. Ross Edwards and Stephen Campbell for assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants CA92584 (to J. N. M. G.) and CA089239 (to J. C.). This work was also supported by grants from the Canadian Cancer Society and the Howard Hughes Medical Institute International Scholar program (to J. N. M. G.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Movie S1, Table S1, and Figs. S1–S7.
- TopBP1
- topoisomerase IIβ-binding protein 1
- BRCT
- BRCA1 C-terminal
- FHA
- forkhead-associated
- FP
- fluorescence polarization
- SFB
- S protein, FLAG, and streptavidin binding peptide tag.
REFERENCES
- 1. Nyberg K. A., Michelson R. J., Putnam C. W., Weinert T. A. (2002) Annu. Rev. Genet. 36, 617–656 [DOI] [PubMed] [Google Scholar]
- 2. Osborn A. J., Elledge S. J., Zou L. (2002) Trends Cell Biol. 12, 509–516 [DOI] [PubMed] [Google Scholar]
- 3. Cimprich K. A., Cortez D. (2008) Nat. Rev. Mol. Cell Biol. 9, 616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Burrows A. E., Elledge S. J. (2008) Genes Dev. 22, 1416–1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gong Z., Kim J. E., Leung C. C., Glover J. N., Chen J. (2010) Mol. Cell 37, 438–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Paulsen R. D., Cimprich K. A. (2007) DNA Repair 6, 953–966 [DOI] [PubMed] [Google Scholar]
- 7. Yamane K., Kawabata M., Tsuruo T. (1997) Eur. J. Biochem. 250, 794–799 [DOI] [PubMed] [Google Scholar]
- 8. Garcia V., Furuya K., Carr A. M. (2005) DNA Repair 4, 1227–1239 [DOI] [PubMed] [Google Scholar]
- 9. Huo Y. G., Bai L., Xu M., Jiang T. (2010) Biochem. Biophys. Res. Commun. 401, 401–405 [DOI] [PubMed] [Google Scholar]
- 10. Rappas M., Oliver A. W., Pearl L. H. (2010) Nucleic Acids Res., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Manke I. A., Lowery D. M., Nguyen A., Yaffe M. B. (2003) Science 302, 636–639 [DOI] [PubMed] [Google Scholar]
- 12. Yu X., Chini C. C., He M., Mer G., Chen J. (2003) Science 302, 639–642 [DOI] [PubMed] [Google Scholar]
- 13. Wang B., Matsuoka S., Ballif B. A., Zhang D., Smogorzewska A., Gygi S. P., Elledge S. J. (2007) Science 316, 1194–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yu X., Chen J. (2004) Mol. Cell. Biol. 24, 9478–9486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liu Z., Wu J., Yu X. (2007) Nat. Struct. Mol. Biol. 14, 716–720 [DOI] [PubMed] [Google Scholar]
- 16. Kim H., Huang J., Chen J. (2007) Nat. Struct. Mol. Biol. 14, 710–715 [DOI] [PubMed] [Google Scholar]
- 17. Campbell S. J., Edwards R. A., Glover J. N. (2010) Structure 18, 167–176 [DOI] [PubMed] [Google Scholar]
- 18. Clapperton J. A., Manke I. A., Lowery D. M., Ho T., Haire L. F., Yaffe M. B., Smerdon S. J. (2004) Nat. Struct. Mol. Biol. 11, 512–518 [DOI] [PubMed] [Google Scholar]
- 19. Glover J. N., Williams R. S., Lee M. S. (2004) Trends Biochem. Sci. 29, 579–585 [DOI] [PubMed] [Google Scholar]
- 20. Williams R. S., Lee M. S., Hau D. D., Glover J. N. (2004) Nat. Struct. Mol. Biol. 11, 519–525 [DOI] [PubMed] [Google Scholar]
- 21. Shiozaki E. N., Gu L., Yan N., Shi Y. (2004) Mol. Cell 14, 405–412 [DOI] [PubMed] [Google Scholar]
- 22. Varma A. K., Brown R. S., Birrane G., Ladias J. A. (2005) Biochemistry 44, 10941–10946 [DOI] [PubMed] [Google Scholar]
- 23. Yamane K., Wu X., Chen J. (2002) Mol. Cell. Biol. 22, 555–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cescutti R., Negrini S., Kohzaki M., Halazonetis T. D. (2010) EMBO J. 29, 3723–3732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wollmann Y., Schmidt U., Wieland G. D., Zipfel P. F., Saluz H. P., Hänel F. (2007) J. Cell. Biochem. 102, 171–182 [DOI] [PubMed] [Google Scholar]
- 26. Leung C. C., Kellogg E., Kuhnert A., Hänel F., Baker D., Glover J. N. (2010) Protein Sci. 19, 162–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tanaka S., Umemori T., Hirai K., Muramatsu S., Kamimura Y., Araki H. (2007) Nature 445, 328–332 [DOI] [PubMed] [Google Scholar]
- 28. Zegerman P., Diffley J. F. (2007) Nature 445, 281–285 [DOI] [PubMed] [Google Scholar]
- 29. Kumagai A., Shevchenko A., Shevchenko A., Dunphy W. G. (2010) Cell 140, 349–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu K., Paik J. C., Wang B., Lin F. T., Lin W. C. (2006) EMBO J. 25, 4795–4807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Delacroix S., Wagner J. M., Kobayashi M., Yamamoto K., Karnitz L. M. (2007) Genes Dev. 21, 1472–1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lee J., Kumagai A., Dunphy W. G. (2007) J. Biol. Chem. 282, 28036–28044 [DOI] [PubMed] [Google Scholar]
- 33. Cantor S. B., Bell D. W., Ganesan S., Kass E. M., Drapkin R., Grossman S., Wahrer D. C., Sgroi D. C., Lane W. S., Haber D. A., Livingston D. M. (2001) Cell 105, 149–160 [DOI] [PubMed] [Google Scholar]
- 34. Mohammad D. H., Yaffe M. B. (2009) DNA Repair 8, 1009–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rodriguez M., Yu X., Chen J., Songyang Z. (2003) J. Biol. Chem. 278, 52914–52918 [DOI] [PubMed] [Google Scholar]
- 36. Yaffe M. B., Elia A. E. (2001) Curr. Opin. Cell Biol. 13, 131–138 [DOI] [PubMed] [Google Scholar]
- 37. Kilkenny M. L., Doré A. S., Roe S. M., Nestoras K., Ho J. C., Watts F. Z., Pearl L. H. (2008) Genes Dev. 22, 2034–2047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Stucki M., Clapperton J. A., Mohammad D., Yaffe M. B., Smerdon S. J., Jackson S. P. (2005) Cell 123, 1213–1226 [DOI] [PubMed] [Google Scholar]
- 39. Williams J. S., Williams R. S., Dovey C. L., Guenther G., Tainer J. A., Russell P. (2010) EMBO J. 29, 1136–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shen Y., Tong L. (2008) Biochemistry 47, 5767–5773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Heckman K. L., Pease L. R. (2007) Nat. Prot. 2, 924–932 [DOI] [PubMed] [Google Scholar]
- 42. Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 43. Sheldrick G. M. (2008) Acta Crystallogr. A 64, 112–122 [DOI] [PubMed] [Google Scholar]
- 44. Terwilliger T. C., Berendzen J. (1999) Acta Crystallogr. D Biol. Crystallogr. 55, 849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cohen S. X., Ben Jelloul M., Long F., Vagin A., Knipscheer P., Lebbink J., Sixma T. K., Lamzin V. S., Murshudov G. N., Perrakis A. (2008) Acta Crystallogr. D Biol. Crystallogr. 64, 49–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 47. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 48. Winn M. D., Isupov M. N., Murshudov G. N. (2001) Acta Crystallogr. D Biol. Crystallogr. 57, 122–133 [DOI] [PubMed] [Google Scholar]
- 49. McCoy A. J. (2007) Acta Crystallogr. D Biol. Crystallogr. 63, 32–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kabsch W., Sander C. (1983) Biopolymers 22, 2577–2637 [DOI] [PubMed] [Google Scholar]
- 51. McDonald I. K., Thornton J. M. (1994) J. Mol. Biol. 238, 777–793 [DOI] [PubMed] [Google Scholar]
- 52. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 53. Echols N., Milburn D., Gerstein M. (2003) Nucleic Acids Res. 31, 478–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Krebs W. G., Gerstein M. (2000) Nucleic Acids Res. 28, 1665–1675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Cer R. Z., Mudunuri U., Stephens R., Lebeda F. J. (2009) Nucleic Acids Res. 37, W441–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Williams R. S., Green R., Glover J. N. (2001) Nat. Struct. Biol. 8, 838–842 [DOI] [PubMed] [Google Scholar]
- 57. Hayward S., Berendsen H. J. (1998) Proteins 30, 144–154 [PubMed] [Google Scholar]
- 58. Bridge W. L., Vandenberg C. J., Franklin R. J., Hiom K. (2005) Nat. Genet. 37, 953–957 [DOI] [PubMed] [Google Scholar]
- 59. Levran O., Attwooll C., Henry R. T., Milton K. L., Neveling K., Rio P., Batish S. D., Kalb R., Velleuer E., Barral S., Ott J., Petrini J., Schindler D., Hanenberg H., Auerbach A. D. (2005) Nat. Genet. 37, 931–933 [DOI] [PubMed] [Google Scholar]
- 60. Litman R., Peng M., Jin Z., Zhang F., Zhang J., Powell S., Andreassen P. R., Cantor S. B. (2005) Cancer Cell 8, 255–265 [DOI] [PubMed] [Google Scholar]
- 61. Botuyan M. V., Nominé Y., Yu X., Juranic N., Macura S., Chen J., Mer G. (2004) Structure 12, 1137–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ekblad C. M., Wilkinson H. R., Schymkowitz J. W., Rousseau F., Freund S. M., Itzhaki L. S. (2002) J. Mol. Biol. 320, 431–442 [DOI] [PubMed] [Google Scholar]
- 63. Nominé Y., Botuyan M. V., Bajzer Z., Owen W. G., Caride A. J., Wasielewski E., Mer G. (2008) Biochemistry 47, 9866–9879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Williams R. S., Dodson G. E., Limbo O., Yamada Y., Williams J. S., Guenther G., Classen S., Glover J. N., Iwasaki H., Russell P., Tainer J. A. (2009) Cell 139, 87–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Yaffe M. B., Smerdon S. J. (2001) Structure 9, R33–38 [DOI] [PubMed] [Google Scholar]
- 66. Liang X., Van Doren S. R. (2008) Acc. Chem. Res. 41, 991–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Choi J. H., Lindsey-Boltz L. A., Sancar A. (2009) Nucleic Acids Res. 37, 1501–1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Yamane K., Tsuruo T. (1999) Oncogene 18, 5194–5203 [DOI] [PubMed] [Google Scholar]
- 69. Kobayashi M., Ab E., Bonvin A. M., Siegal G. (2010) J. Biol. Chem. 285, 10087–10097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kobayashi M., Figaroa F., Meeuwenoord N., Jansen L. E., Siegal G. (2006) J. Biol. Chem. 281, 4308–4317 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.