Abstract
Toll-like receptors (TLRs) are the key molecular sensors used by the mammalian innate immune system to detect various types of pathogens. Tlr13 is a novel and uncharacterized member of the mammalian TLR family. Here we report the cloning and characterization of tlr13. Tlr13 is predominantly expressed in the spleen, particularly in dendritic cells and macrophages. Tlr13 appears to activate a MyD88- and TAK1-dependent TLR signaling pathway, inducing the activation of NF-κB. This receptor can also activate type 1 interferon through IRF7. Furthermore, Tlr13 seems to be another intracellular TLR. Remarkably, cells expressing tlr13 fail to respond to known TLR ligands but instead respond specifically to vesicular stomatitis virus. Cells with the knockdown of tlr13 are highly susceptible to vesicular stomatitis virus infection. Thus, these results provide an important insight into the potential role of the novel Toll-like receptor tlr13 in the recognition of viral infection.
Keywords: Innate Immunity, Interferon, NF-κB, Toll-like Receptors (TLR), Viral Immunology
Introduction
The best characterized molecular sensors used by the mammalian innate immune system to detect invading pathogens are the Toll-like receptors (TLRs)3 (1–3). TLRs are type I transmembrane proteins that contain an amino-terminal leucine-rich repeat (LRR) domain and a carboxyl-terminal Toll-interleukin-1 receptor (TIR) domain (4). The leucine-rich repeat domain is responsible for the recognition of pathogen-associated molecular patterns, whereas the TIR domain is required for initiating intracellular signaling (3, 4). Signal transduction by TLRs after ligand engagement is initiated with the recruitment of the cytosolic TIR-containing adaptor proteins such as MyD88 and TRIF (also known as TICAM1) (5–7). MyD88 is utilized by all TLRs except for TLR3 (5). For the MyD88-dependent pathway, MyD88 subsequently recruits the serine/threonine interleukin-1 receptor-associated kinase (IRAK) to the receptor complex through a homophilic interaction of the death domains (8). The recruited IRAK is then auto-phosphorylated and, after associating with the cytosolic adaptor protein TRAF6, dissociates from the receptor and is degraded (5). Finally, TRAF6 activates the IκB kinase complex through the adaptor protein TAK1 (5–7). The MyD88-independent pathway is the TRIF pathway. TLR3 and TLR4 recognize double-stranded RNA (dsRNA) and LPS, respectively, to activate this pathway. This results in the activation of IRF3 and the subsequent induction of type I interferons and IFN-inducible genes (9–11). IRF7 is a key transcription factor for the induction of type I interferons, and its activation occurs via both the MyD88-dependent pathway and the TRIF-dependent pathway (12, 13).
The subcellular localization of different TLRs correlates with the nature of their ligands. In the TLR family, TLR1, -2, -4, -5, -6, and -11 are present on the cell surface membrane, whereas TLR3, -7, -8, and -9 are expressed in the intracellular endosomal compartments. Intracellular TLRs are sensors of nucleic acids that have been well studied in the recognition of viral infection. After viruses are internalized and delivered to the endosome, the released nucleic acids might be recognized by these TLRs.
Although Toll-like receptors share common sequence features including an amino-terminal leucine-rich repeat domain and a carboxyl-terminal TIR domain, there is as little as ∼30% homology in the full-length amino acid sequence among different TLR family members. However, the TIR domain among TLRs is highly conserved. We used the TIR domain of our recently identified tlr11 (1) to search NCBI databases for related sequences. We identified a novel TIR domain-containing sequence (GenBankTM accession number AY510706), which represents a potential novel TLR, dubbed tlr13, based on the sequence of its discovery. Although a partial sequence was reported (2), the function of this novel TLR is completely unknown. Here we report the cloning and characterization of tlr13. This TLR displays a pattern of expression in dendritic cells and macrophages. Tlr13 is an intracellular TLR and activates the MyD88-dependent pathway. Moreover, consistent with its subcellular localization, tlr13 is able to recognize the vesicular stomatitis virus to activate innate immune antiviral responses.
EXPERIMENTAL PROCEDURES
Cell Lines and Reagents
RAW 264.7, NIH3T3, and HEK293 cells were purchased from ATCC. Mouse embryonic fibroblasts (MEFs) of MyD88−/− were kindly provided by R. Medzhitov (Yale University). TAK1-deficient MEFs were obtained from M. Schneider (Baylor College of Medicine). Cells were cultured at 37 °C in 5% CO2 incubator in DMEM (Invitrogen) and supplemented with 10% (v/v) heat-inactivated fetal bovine serum (HyClone), 100 units/ml penicillin, and 100 g/ml streptomycin. TLR ligands were purchased from Invivogen, including polyinosine-polycytidylic acid (poly(I:C)), a synthetic analog of viral dsRNA that is recognized by TLR3; single-stranded RNA40 is a U-rich single-strand RNA derived from the HIV-1 long terminal repeat that is recognized by TLR8; R848, a low molecular weight synthetic imidazoquinoline compound, activates immune cells via TLR7/8 MyD88-dependent signaling pathway.
Cloning of tlr13 and Construction of CD4-tlr13
Mouse tlr13 open reading frame was amplified from cDNA made from mRNA isolated from RAW 264.7 cells using the following primers 5′-CACCATGAGTGGGCTCTACAGGATC-3′ and 5′-AGCCGCCTCAACAACAATTAGATGTG-3′ and was cloned into pcDNA3.1/V5/His-TOPO (Invitrogen). Constitutively active CD4/tlr13 was constructed by fusing cDNAs encoding the extracellular domain of murine CD4 (amino acids 1–391) to the transmembrane and cytoplasmic domains of murine tlr13 (amino acids 769–991). CD4/TLR4 plasmid was kindly provided by R. Medzhitov (Yale University).
Cell Sorting
Various cell types were sorted from single-cell suspensions of C57BL/6 mouse spleens. Cells were stained with combinations of fluorescence-conjugated monoclonal antibodies (BD Pharmingen) and were sorted by a FACSAria (BD Biosciences) with the following sorting criteria for each cell type: B cell, CD19+; CD4 T cell, CD3+CD4+; macrophage, CD11b+F4/80+; myeloid dendritic cell, CD11c+CD11b+. Dendritic cells were further sorted with CD11c+CD45RAhighCD11blow for plasmatocytoid dendritic cells (pDCs) and CD11c+CD45RAnegCD11bhigh for cDCs. The purity of all the sorted cell types was greater than 96%, as determined by post-sorting flow cytometry with a FACSCalibur (BD Biosciences).
RNA Isolation, RT-PCR, and Real-time PCR
These procedures were performed as previously described (3).
Transfection and Luciferase Assays
HEK293, MEFs, or NIH3T3 cells were seeded into 24-well plates at a density of 1 × 105 cells/well in antibiotic-free media. The next day cells were transfected with Lipofectamine 2000 (Invitrogen). Briefly, 0.8 μg of DNA including TLRs, CD4-TIRs, and reporter plasmids were diluted with Opti-MEM and then mixed with diluted Lipofectamine 2000. After 20 min of incubation at room temperature, the mixtures were added to each well. Dual-luciferase assays (Promega) were performed 24 h after transfection according to the manufacturer's protocol.
Immunoprecipitation and Western Blot
After a PBS wash, cells were lysed with lysis buffer (50 mm Tris-HCl (pH 7.4), 150 mm NaCl, 1 mm EDTA, 1% Nonidet P-40, 0.25% sodium deoxycholate, 0.5 mm phenylmethylsulfonyl fluoride, phosphatase inhibitor mixture (Sigma). The lysate was centrifuged at 14,000 rpm for 15 min at 4 °C, and then the supernatant was incubated with 0.5 μg of antibody and rotated for 3 h at 4 °C. After adding a protein G-agarose bead suspension (Santa Cruz), the mixture was further incubated with rotation for 3 h at 4 °C. After washing with the washing buffer (50 mm Tris-HCl (pH 7.4), 150 mm NaCl, 1 mm EDTA, 1% Nonidet P-40, 0.25% sodium deoxycholate) 3 times, the beads were resuspended in Laemmli sample buffer and boiled for 5 min. Immunoprecipitates or whole cell lysates were resolved by SDS-PAGE and transferred to a PVDF transfer membrane (Thermo Scientific). The membranes were probed with appropriate antibodies. IgG horseradish peroxidase-conjugated antibodies followed. The proteins on the membrane were detected using the ECL-Plus Western blotting detection system (Amersham Biosciences).
Confocal Microscopy
HEK293 cells and NIH3T3 cells seeded on glass coverslips were transiently transfected with tlr13-GFP and UNC93B1-RFP using Lipofectamine 2000. After 24 h, coverslips were washed with PBS and fixed with 4% paraformaldehyde in PBS. Subcellular localizations of tlr13-GFP and UNC93B1 were visualized by an LSM 510 laser confocal system (Zeiss) with an oil immersion lens (magnification, ×63).
For retroviral gene transduction, tlr13-GFP and UNC93B1-RFP were cloned into pBabe-puro plasmids. To produce the replication incompetent virions, HEK293T cells were transfected with respective pBabe-puro-tlr13-GFP or pBabe-puro-UNC93B1-RFP and helper plasmids (Oligoengine) using Lipofectamine 2000. Twenty-four and forty-eight hours after transfection, the viral supernatant was collected, filtered, and frozen. The viruses were added to RAW264.7 macrophages or bone marrow dendritic cells seeded onto glass coverslips in the presence of Polybrene (8 μg/ml) (Sigma). Twenty-four hours after viral infection, cells were washed and fixed, then visualized by laser confocal microscope as described above.
Viruses
VSV-AV1, a VSV harboring a point mutation in the M gene, was a gift from Glen Barber (University of Miami). Vesicular stomatitis virus (VSV) was generated and titered on baby hamster kidney (BHK21) cells. In brief, BHK21 cells were infected with VSV-AV1 at a multiplicity of infection of 0.01, incubated until more than 70% cells exhibit clear cytopathic effects. Cell supernatant was harvested and separated into aliquots at −80 °C. To titer VSV-AV1, BHK21 cells were seeded on a 6-well plate at 8 × 105 cells per well. The next day serial dilutions of the stocked virus were made in DMEM without serum. 300 μl of the diluted virus suspension was then added to PBS-washed BHK21 cells. After 1 h of incubation at 37 °C, viruses were removed. Subsequently, wells were overlaid with 2 ml of growth medium with 1% low-melting agarose and incubated at 37 °C for 36 h. After staining with crystal violet/methanol mix, the viral titer was estimated by counting the plaque number.
Plasmid and Short Hairpin RNAs (shRNA) Constructions
Mouse cDNA encoding full-length TLR7, UNC93B1, IRF3, and IRF7 were amplified from the mouse spleen cDNA. Tlr13 cDNA was cloned into pCMV-Tag2 (Stratagene) and pEGFP-N3 (Clontech) vectors. UNC93B1 was inserted into both pCMV-Tag2 and pDsRed1-N1 (Clontech) vectors. Others were cloned into pCMV-Tag2 vector. IFN-β promoter-driven luciferase reporter construct was kindly provided by Dr. S. Akira (Japan).
A pSuper-retro vector (Ambion) was used to generate shRNA plasmids for tlr13 knockdown. The following target sequences have been selected: 5′-TGAAAAAACTCCAGTCTTT-3′ (shTLR13–1), 5′-CAATCTGTCTGCTCTGGTG-3′ (shTLR13–2). The authenticity of these plasmids was confirmed by sequencing. The silencing vector expressing siRNA targeting TRIF was obtained from InvivoGen.
RESULTS
Tlr13 Is Strongly Expressed by Dendritic Cells and Macrophages
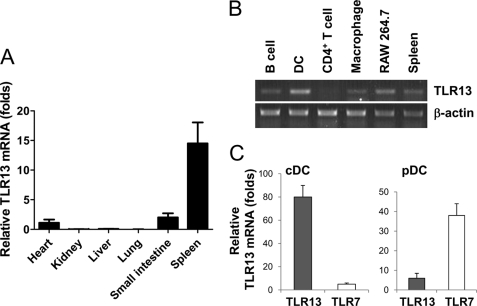
We used quantitative real-time RT-PCR to examine the profile of expression of tlr13 in different mouse organs and found that tlr13 was expressed strongly only in the spleen (Fig. 1A). tlr13 expression in different cell types of the spleen was further analyzed by RT-PCR after cell sorting of splenocytes by FACS. We isolated CD4+T cells, B cells, macrophages, and DCs cells. RNA from these cells was isolated and used for tlr13 expression analysis by RT-PCR. β-Actin was used as a control to normalize the level of tlr13 expression. tlt13 was expressed strongly in DCs and macrophages (Fig. 1B), indicating that tlr13 is involved in innate immune responses. The dendritic cells were further sorted to isolate pDCs and cDCs. Unlike TLR7, which was expressed in pDCs, tlr13 was expressed predominately in cDCs (Fig. 1C).
FIGURE 1.
Characterization of TLR13 expression profile. A, real-time RT-PCR analysis of TLR13 mRNA expression in various mouse organs is shown. The graph shows the mean ± S.D. of three independent experiments. B, shown is RT-PCR of TLR13 expression in various cell types in spleen, including B and CD4+ T lymphocytes, DCs, macrophages. Single-cell suspensions of splenocytes were sorted by FACS. β-Actin was used as a control. C, shown is quantitative RT-PCR analysis of TLR13 and TLR7 mRNA expression in pDCs and cDCs.
Tlr13 Is a Novel Functional TLR That Activates Both NF-κB and IFN-β
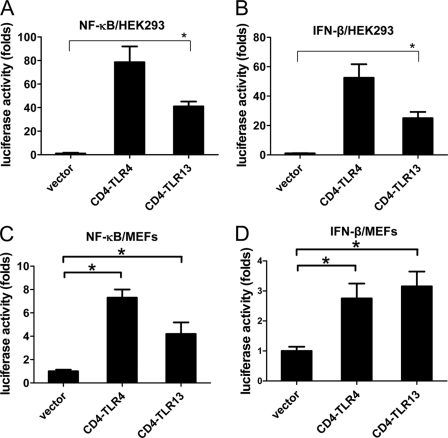
We then determined whether tlr13 is capable of activating signal transduction pathways that lead to the activation of NF-κB and interferon (4, 5). A constitutive activated TLR can be generated by using the CD4 extracellular domain and the TLR intracellular TIR domain chimera (6). Because CD4 autodimerizes, the functional TLR-TIR domain will activate TLR signaling pathways. We generated a CD4-tlr13 fusion construct and overexpressed it along with an NF-κB or IFN-β luciferase reporter into 293 cells and MEF cells. Like the expression of CD4-TLR4 (6), the expression of CD4-tlr13 led to the activation of both NF-κB and IFN-β (Fig. 2). This result suggests that tlr13 is a functional TLR that activates the TLR signal transduction pathway leading to the activation of both NF-κB and IFN-β.
FIGURE 2.
Constitutively active TLR13 (CD4-TLR13) activates both NF-κB and IFN-β. HEK293 cells (A and B) and MEFs (C and D) transiently transfected with expression vectors for CD4-TLR13, CD4-TLR4, or empty expression vector (control) together with an NF-κB or IFN-β luciferase reporter are shown. Luciferase activity was measured with a luminometer after 24 h transfection. Asterisk, p < 0.05. The data are representative of three similar experiments.
Signal Transduction by Tlr13 Uses the MyD88- and TAK1-dependent Pathway
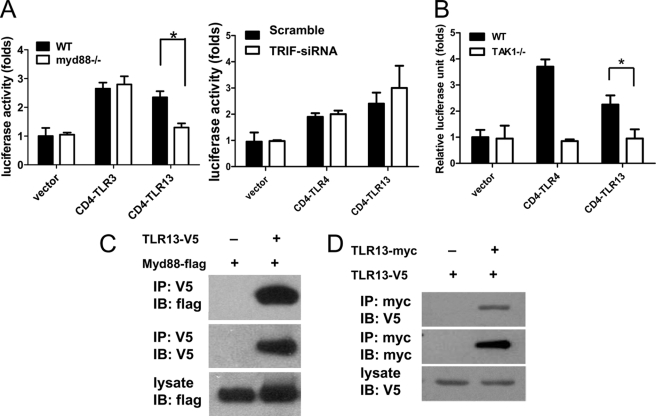
Signal transduction by TLRs after ligand engagement is initiated with the recruitment of cytosolic TIR-containing adaptor proteins, including MyD88, TIRAP (MAL), TRIF (TICAM1), and TRAM (TICAM2). MyD88 is utilized by all TLRs except TLR3. To determine the tlr13 signaling pathway, we cotransfected constitutive active CD4/tlr13 alone with IFN-β luciferase reporter into MyD88 knock-out, TRIF knockdown, or TAK1 knock-out MEFs. The IFN-β activation was abolished in both MyD88- and TAK1-deficient cells but not in TRIF knockdown cells (Fig. 3, A and B), suggesting that overexpression of tlr13 activates the MyD88- and TAK1-dependent pathway. Then we determined whether tlr13 was able to interact directly with TRIF or MyD88. Indeed, we found that tlr13 was able to interact with MyD88 (Fig. 3C) but not with TRIF (data not shown).
FIGURE 3.
Signal transduction by TLR13 is dependent on MyD88 and TAK1. A and B, MyD88−/−, TRIF-siRNA knockdown, TAK1−/−, and wild-type control MEFs were transiently transfected with expression vectors for CD4-TLR13, CD4-TLR3, CD4-TLR4, or empty expression vector (control) together with an IFN-β luciferase reporter. Luciferase activity was measured 48 h after transfection. Asterisk, p < 0.05. The data are representative of three similar experiments. C, MyD88 interacts with TLR13. Immunoblot (IB) analysis of 293 cells expressing TLR13-V5 and MyD88-FLAG directly or after immunoprecipitates (IP) with anti-V5 antibody is shown. D, homodimerization of TLR13 is shown. HEK293 cells were transfected with myc and V5-tagged TLR13 plasmids, TLR13-myc was immunoprecipitated using anti-myc antibody from lysates 48 h post-transfection, and the presence of TLR13-V5 in the immunocomplex was tested.
Homodimerization of tlr13
TLR2 requires heterodimerization with either TLR1 or TLR6 to promote signal transduction (7, 8). This heterodimerization may help to significantly increase the spectrum of ligands recognized by these receptors. Does tlr13 collaborate with any of the other Toll-like receptors?
It is well known that the majority of the current commercial TLR antibodies lack specificity, whereas small epitope-tag antibodies such as anti-V5, anti-Myc, and anti-FLAG tags do work well. For this reason, we generated FLAG epitope-tagged mouse TLR constructs and a V5 and Myc epitope-tagged mouse tlr13 constructs. We co-transfected these constructs into 293 cells and immunoprecipitated the tagged tlr13 from transfected cell extracts. The immunoprecipitates were analyzed on SDS-PAGE followed by immunoblotting with anti-V5, anti-FLAG, or anti-myc antibodies to identify interactions between TLRs and tlr13. We did not identify heterodimerization between tlr13 with other TLRs (data not shown); however, tlr13 can become a homodimer (Fig. 3D).
Tlr13 Localizes Intracellularly and Interacts with UNC93B
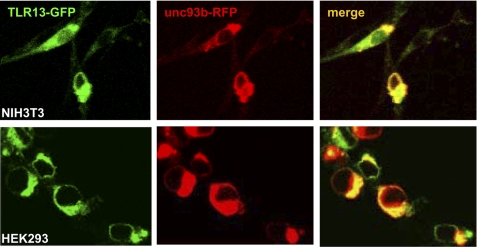
Among TLRs, TLR1, -2, -4, -5, -6, and -11 are expressed on the cell surface and specialize in the detection of bacteria; in contrast, TLR3, -7, -8, and -9 are expressed intracellularly and specialize in viral detection. The mechanisms by which their ligands, including double and single-stranded RNA and unmethylated CpG DNA, activate their respective intracellular TLRs have not been determined, although it has been shown that stimulation by CpG requires its internalization into late endosomal or lysosomal compartments. We first used immune-staining analysis with confocal microscopy to determine the subcellular localization of tlr13 in its expressed 293, NIH3T3 cells, and Raw264.7 cells (Fig. 4 and data not shown). Tlr13 appeared to be an intracellular receptor.
FIGURE 4.
TLR13 localizes intracellularly and interacts with UNC93B. NIH3T3 and HEK293 cells were cotransfected with TLR13-GFP and UNC93B-RFP plasmids. Twenty-four hours after transfection, cells were directly visualized for TLR13-GFP and UNC93B co-localization by confocal microscopy. The images are representative of three independent experiments.
It has been demonstrated that UNC93B, an endoplasmic reticulum membrane protein, associates with TLR3, TLR7, and TLR9, and it resides in the endoplasmic reticulum and endosomes (9, 10). UNC93B also plays an important role in the activation of these intracellular TLRs. We then wanted to determine whether tlr13, as a novel intracellular TLR, behaved the same way as other intracellular TLRs. Indeed, our results showed tlr13 and UNC93B displayed co-localization when we cotransfected them into both HEK293 and NIH3T3 cells (Fig. 4) (9, 10).
Tlr13 Is Involved in the Recognition of Vesicular Stomatitis Virus
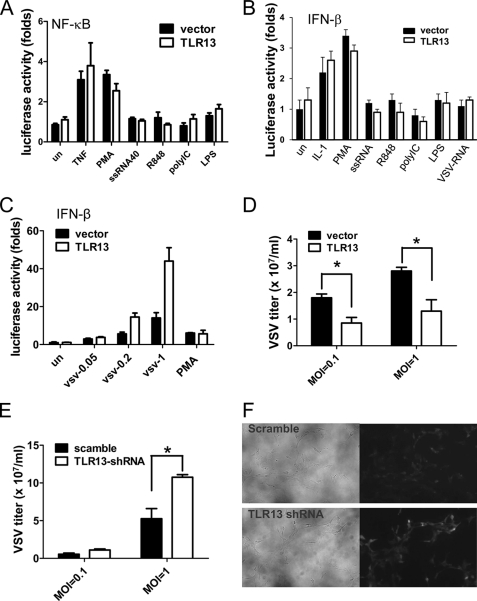
Tlr13 is a novel and poorly characterized member of the Toll-like receptor family, and elucidation of the function of this novel TLR depends mainly on the identification of its natural ligand. To begin to identify and characterize the ligand for tlr13, it was necessary to have a cell line that can demonstrate tlr13-specific activity. After testing many cell lines, we found that NIH3T3 is a good cell model as it does not respond to TLR ligands unless TLRs are overexpressed (11). We first tested whether the known ligands of other TLRs can activate the tlr13 signaling pathways by measuring NF-κB or IFN-β activity. As shown in Fig. 5, A and B, none of the tested cognate ligands for TLRs was able to activate NF-κB or IFN-β in the tlr13-expressing NIH3T3 cells, indicating that tlr13 recognizes a novel tlr13 specific ligand. To seek the pathogen that might be recognized by tlr13, we tested the Gram-positive Staphylococcus K2 strain and the Gram-negative Salmonella SR11 strain to stimulate NF-κB activity in the cells. Our results demonstrate that tlr13 recognizes neither Gram-positive nor Gram-negative bacteria (data not shown).
FIGURE 5.
VSV contains TLR13-stimulating activity. A and B, TLR13 does not recognize the known TLR ligands. NIH3T3 cells were transfected with TLR13 or empty expression vector together with NF-κB (A) or IFN-β (B) luciferase reporter. Twenty-four hours after transfection, cells were challenged with different stimuli (10 ng/ml TNF-α, 200 μg/ml phorbol 12-myristate 13-acetate (PMA), 1 μg/ml of single-stranded RNA 40, 1 μg/ml of R848, 10 μg/ml of polyIC, 100 ng/ml LPS, and 10 μg/ml VSV RNA) for 6 h. The luciferase activity was measured with a luminometer. C, the transfected cells were treated with VSV at different multiplicities of infection (0.05, 0.2, and 1) or phorbol 12-myristate 13-acetate (control) for 12 h, and luciferase activity driven by IFN-β promoter was measured. Asterisk, p < 0.05. The data are representative of three similar experiments. D, the transfected NIH3T3 cells were infected with VSV (multiplicity of infection 0.1 and 1), and the virus titer was determined in BHK21 cells 24 h post-infection. E and F, MEFs transiently transfected with pSUPER.retro.puro-scramble or pSUPER.retro.puro-TLR13 shRNA by Lipofectamine 2000 are shown. Twenty-four hours after transfection, cells were infected with VSV for 24 h. Virus titer was determined (E), and the VSV in TLR13 wild-type and knockdown cells were visualized by confocal microscopy (F).
Although we do not know the natural ligand for tlr13, we do know that tlr13 is an intracellular TLR (Fig. 4) (10, 12). We then made an assumption that the intracellular tlr13 might recognize viruses, similar to other intracellular TLRs including TLR3, -7, -8, and -9. Indeed, tlr13 is involved in the recognition of VSV, and it induces tlr13-specific INF-β activity (Fig. 5, C and D); however, tlr13 appeared not to respond to VSV RNA (Fig. 5B). To confirm tlr13 specificity we then knocked down tlr13 by using of RNA interference. We designed shRNAs targeting tlr13 and cloned them into the pSUPER Retro vector (13). An shRNA with the most efficient suppression of the expression of tlr13 was used. To test the role of tlr13 in VSV infection, we transfected the shRNA or scramble vector into MEF cells. The cells with silenced tlr13 almost lost the ability to inhibit VSV (Fig. 5, E and F), indicating that VSV contains a tlr13-specific activity.
Tlr13 Activates IFN-β via IRF-7, Not IRF-3, in VSV Infection
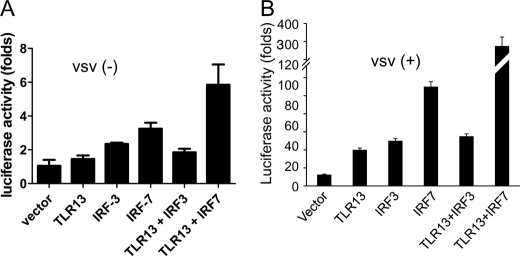
TLRs sense viruses that enter the endosome through endocytosis. Then the downstream pathway leads to the activation of the transcription factors NF-κB, IRF3, and IRF7, ultimately inducing the production of type I interferons (14, 15). IRF3 and IRF7 are the key regulators that potently induce IFN-β production during viral infection in different cell types (16, 17). To analyze which IRF is utilized by tlr13 to activate IFN-β, NIH3T3 cells were overexpressed with tlr13 and vector plasmids or IRFs followed by VSV infection for 12 h. We found that IRF7, not IRF3, is the downstream molecule of tlr13 responsible for triggering IFN-β activation (Fig. 6, A and B).
FIGURE 6.
TLR13 activates IFN-β via IRF7, not IRF3 in VSV infection. NIH3T3 cells were transiently transfected with vector, TLR13, IRF3, IRF7, TLR13 plus IRF3, and TLR13 plus IRF7. Luciferase activity was measured after 24 h (A) or challenged with VSV (multiplicity of infection 1) for another 12 h (B).
DISCUSSION
Tlr13 is a novel and uncharacterized member of the mammalian TLR family, although only its partial sequence has been reported (2). Based on phylogenetic analysis, Beutler and co-workers (2), using a multiple sequence alignment of leucine-rich repeat domains of TLRs, claimed that tlr13 belongs to the TLR3 subfamily. In contrast, Aderem and co-workers (18), by performing a multiple sequence alignment of the full-length amino acids of TLRs and by using systemic evolutionary analysis, indicated that tlr13 belongs to the tlr11 subfamily. However, our experimental results demonstrate that tlr13 function does not belong to either the TLR3 or the tlr11 subfamily. TLR3 is an intracellular TLR that activates only the MyD88-independent TRIF pathway (19), whereas tlr11 is a cell-surface TLR and activates the MyD88-dependent pathway (1, 20). As for tlr13, although it is an intracellular TLR, it activates the MyD88-dependent pathway, a characteristic similar to TLR7/8 (21).
The elucidation of the function of tlr13 depends mainly on the identification of its natural ligand. An intracellular TLR almost exclusively functions as a nucleic acid sensor. For example, TLR3 recognizes dsRNA, TLR7/8 recognizes single-stranded RNA, and TLR9 recognizes CpG DNA (21–23). Therefore tlr13, as another intracellular TLR, might recognize DNA or RNA. Tlr13 appears to be able to recognize VSV, but the exact natural ligand of tlr13 is currently unknown and needs to be further identified.
VSV is a single-stranded RNA virus member of the Rhabdoviridae family. The role of innate immunity in the recognition of this virus represents an insight of antiviral responses triggered by the host. Beyond tlr13, TLR7 has been implicated in the recognition of VSV. Lund et al. (24) demonstrated that in pDCs TLR7 was necessary for the production of IFN-α, a cytokine implicated in the induction of innate and the adaptive immunity during viral infections. Moreover, it was shown that the recognition of VSV by TLR7 was in endosomal compartments. Additionally, in vivo studies demonstrated a reduction in the response to VSV in mice deficient in TLR7 or MyD88. Another study found that although VSV intranasal infection was MyD88-dependent, VSV intravenous infection was not (25). Other studies suggested that as VSV replicated in lung epithelial cells, the viral ribonucleoprotein induced the activation of TBK1 and the up-regulation of IκB kinase ϵ, which are noncanonical IκB kinase-related kinases involved in the production of type 1 interferons (26). Furthermore, Oganesyan et al. (27) showed that TRAF3-deficient MEF cells and Flt3-ligand-derived dendritic cells from Traf3−/− mice had higher susceptibility to VSV infection. Taken together, their findings suggest that this molecule is necessary for the production of type I interferons induced both by TLR-dependent and TLR-independent pathways. Besides TLRs, the dsRNA-dependent protein kinase PKR and the retinoic acid-inducible protein I (RIG-I) have been related in the protection against VSV infection. Mice deficient in PKR showed an increased in susceptibility to this kind of infection (28). Interestingly, Akira and co-workers demonstrated that RIG-1 was important for the response against VSV in several cells but not in pDCs (29). Moreover, bone marrow-derived DCs generated by granulocyte-macrophage-stimulating factor and transfected with RNAs prepared from VSV showed a RIG-I-dependent production of IFN-α. Also, RIG-I−/− mice presented higher susceptibility to VSV infection and a diminished interferon response (30). Our data showed that tlr13 represents another molecule that recognizes VSV. The relationships between tlr13 and TLR7 as well as tlr13 with RIG-I-like receptor (RLR) family members need to be further defined.
In summary, tlr13 is a novel and functional TLR that is predominantly expressed in DCs and macrophages. Tlr13 not only activates a MyD88- and TAK1-dependent TLR signaling pathway to activate NF-κB but also induces type 1 interferon through IRF7. As an intracellular TLR, Tlr13 is involved in the recognition of VSV infection.
Acknowledgments
We thank Lida Keene for critical reading of the manuscript and S. Ghosh for advice.
This work was supported, in whole or in part, by National Institutes of Health Grant AI085164 (to D. Z.). This study was also supported by Texas A&M University (to D. Z.).
- TLR
- Toll-like receptor
- DC
- dendritic cell
- cDC
- conventional DC
- pDC
- plasmatocytoid DC
- TRIF
- TIR-domain-containing adapter-inducing interferon-β
- IRF
- interferon regulatory factor
- VSV
- vesicular stomatitis virus
- TIR
- Toll-interleukin-1 receptor
- MEF
- mouse embryonic fibroblasts
- RIG-I
- retinoic acid-inducible protein I.
REFERENCES
- 1. Zhang D., Zhang G., Hayden M. S., Greenblatt M. B., Bussey C., Flavell R. A., Ghosh S. (2004) Science 303, 1522–1526 [DOI] [PubMed] [Google Scholar]
- 2. Tabeta K., Georgel P., Janssen E., Du X., Hoebe K., Crozat K., Mudd S., Shamel L., Sovath S., Goode J., Alexopoulou L., Flavell R. A., Beutler B. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 3516–3521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Shi Z., Cai Z., Wen S., Chen C., Gendron C., Sanchez A., Patterson K., Fu S., Yang J., Wildman D., Finnell R. H., Zhang D. (2009) J. Biol. Chem 284, 20540–20547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Takeda K., Kaisho T., Akira S. (2003) Annu. Rev. Immunol. 21, 335–376 [DOI] [PubMed] [Google Scholar]
- 5. O'Neill L. A. (2008) Immunity 29, 12–20 [DOI] [PubMed] [Google Scholar]
- 6. Medzhitov R., Preston-Hurlburt P., Janeway C. A., Jr. (1997) Nature 388, 394–397 [DOI] [PubMed] [Google Scholar]
- 7. Bulut Y., Faure E., Thomas L., Equils O., Arditi M. (2001) J. Immunol. 167, 987–994 [DOI] [PubMed] [Google Scholar]
- 8. Ozinsky A., Underhill D. M., Fontenot J. D., Hajjar A. M., Smith K. D., Wilson C. B., Schroeder L., Aderem A. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 13766–13771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brinkmann M. M., Spooner E., Hoebe K., Beutler B., Ploegh H. L., Kim Y. M. (2007) J. Cell Biol. 177, 265–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kim Y. M., Brinkmann M. M., Paquet M. E., Ploegh H. L. (2008) Nature 452, 234–238 [DOI] [PubMed] [Google Scholar]
- 11. Burger-Kentischer A., Abele I. S., Finkelmeier D., Wiesmüller K. H., Rupp S. (2010) J. Immunol. Methods 358, 93–103 [DOI] [PubMed] [Google Scholar]
- 12. Tabeta K., Hoebe K., Janssen E. M., Du X., Georgel P., Crozat K., Mudd S., Mann N., Sovath S., Goode J., Shamel L., Herskovits A. A., Portnoy D. A., Cooke M., Tarantino L. M., Wiltshire T., Steinberg B. E., Grinstein S., Beutler B. (2006) Nat. Immunol. 7, 156–164 [DOI] [PubMed] [Google Scholar]
- 13. Brummelkamp T. R., Bernards R., Agami R. (2002) Science 296, 550–553 [DOI] [PubMed] [Google Scholar]
- 14. Akira S., Uematsu S., Takeuchi O. (2006) Cell 124, 783–801 [DOI] [PubMed] [Google Scholar]
- 15. Xagorari A., Chlichlia K. (2008) Open Microbiol. J. 2, 49–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Honda K., Takaoka A., Taniguchi T. (2006) Immunity 25, 349–360 [DOI] [PubMed] [Google Scholar]
- 17. Honda K., Yanai H., Negishi H., Asagiri M., Sato M., Mizutani T., Shimada N., Ohba Y., Takaoka A., Yoshida N., Taniguchi T. (2005) Nature 434, 772–777 [DOI] [PubMed] [Google Scholar]
- 18. Roach J. C., Glusman G., Rowen L., Kaur A., Purcell M. K., Smith K. D., Hood L. E., Aderem A. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 9577–9582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Alexopoulou L., Holt A. C., Medzhitov R., Flavell R. A. (2001) Nature 413, 732–738 [DOI] [PubMed] [Google Scholar]
- 20. Yarovinsky F., Zhang D., Andersen J. F., Bannenberg G. L., Serhan C. N., Hayden M. S., Hieny S., Sutterwala F. S., Flavell R. A., Ghosh S., Sher A. (2005) Science 308, 1626–1629 [DOI] [PubMed] [Google Scholar]
- 21. Takeuchi O., Akira S. (2010) Cell 140, 805–820 [DOI] [PubMed] [Google Scholar]
- 22. Iwasaki A., Medzhitov R. (2010) Science 327, 291–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Beutler B. (2009) Immunol. Rev. 227, 248–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lund J. M., Alexopoulou L., Sato A., Karow M., Adams N. C., Gale N. W., Iwasaki A., Flavell R. A. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 5598–5603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhou S., Kurt-Jones E. A., Fitzgerald K. A., Wang J. P., Cerny A. M., Chan M., Finberg R. W. (2007) J. Immunol. 178, 5173–5181 [DOI] [PubMed] [Google Scholar]
- 26. tenOever B. R., Sharma S., Zou W., Sun Q., Grandvaux N., Julkunen I., Hemmi H., Yamamoto M., Akira S., Yeh W. C., Lin R., Hiscott J. (2004) J. Virol. 78, 10636–10649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Oganesyan G., Saha S. K., Guo B., He J. Q., Shahangian A., Zarnegar B., Perry A., Cheng G. (2006) Nature 439, 208–211 [DOI] [PubMed] [Google Scholar]
- 28. Balachandran S., Roberts P. C., Brown L. E., Truong H., Pattnaik A. K., Archer D. R., Barber G. N. (2000) Immunity 13, 129–141 [DOI] [PubMed] [Google Scholar]
- 29. Kato H., Sato S., Yoneyama M., Yamamoto M., Uematsu S., Matsui K., Tsujimura T., Takeda K., Fujita T., Takeuchi O., Akira S. (2005) Immunity 23, 19–28 [DOI] [PubMed] [Google Scholar]
- 30. Kato H., Takeuchi O., Sato S., Yoneyama M., Yamamoto M., Matsui K., Uematsu S., Jung A., Kawai T., Ishii K. J., Yamaguchi O., Otsu K., Tsujimura T., Koh C. S., Reis e Sousa C., Matsuura Y., Fujita T., Akira S. (2006) Nature 441, 101–105 [DOI] [PubMed] [Google Scholar]