Abstract
The cell cycle-dependent, ordered assembly of protein prereplicative complexes suggests that eukaryotic replication origins determine when genomic replication initiates. By comparison, the factors that determine where replication initiates relative to the sites of prereplicative complex formation are not known. In the human globin gene locus previous work showed that replication initiates at a single site 5′ to the β-globin gene when protein synthesis is inhibited by emetine. The present study has examined the pattern of initiation around the genetically defined β-globin replicator in logarithmically growing HeLa cells, using two PCR-based nascent strand assays. In contrast to the pattern of initiation detected in emetine-treated cells, analysis of the short nascent strands at five positions spanning a 40 kb globin gene region shows that replication initiates at more than one site in non-drug-treated cells. Quantitation of nascent DNA chains confirmed that replication begins at several locations in this domain, including one near the initiation region (IR) identified in emetine-treated cells. However, the abundance of short nascent strands at another initiation site ∼20 kb upstream is ∼4-fold as great as that at the IR. The latter site abuts an early S phase replicating fragment previously defined at low resolution in logarithmically dividing cells.
INTRODUCTION
The human β-globin gene locus occupies >60 kb on chromosome 11 and comprises six linked genes (ɛ, γG, γA, ψβ, δ and β) arranged 5′→3′ in the order of their expression during development (1). Southern hybridization analysis of 5′-bromodeoxyuridine (BrdUrd)-labeled nascent DNA from unsynchronized cells elutriated to enrich for cells in specific intervals of S phase showed that the β-globin gene locus replicates in the first half of S phase in erythroid cells and in late S phase in non-erythroid cells (2–4). When the amounts of globin EcoRI fragments were compared in cells early in S phase two early replicating fragments, one overlapping the γA-globin gene (near map unit 41) and another ∼20 kb downstream of the β-globin gene (near map unit 80), were significantly enriched relative to the surrounding sequences, suggesting that these early replicating fragments might contain initiation sites of DNA replication (3).
The same two sites were not revealed as origins when the polarity of leading strand synthesis in the β-globin gene locus was tested in cells treated with emetine (5), a protein synthesis poison that inhibits lagging strand DNA synthesis by an unknown mechanism (6). In emetine-treated K562, HL60 and HeLa cells a single leading strand template switch was observed in the 2 kb area 5′ to the β-globin gene. From these data it was concluded that a single initiation region (IR) for bidirectional replication is present in the globin gene locus, between the δ- and β-globin genes (5). Consistent with this interpretation, in hemoglobin Lepore cells characterized by a naturally occurring 8 kb deletion between the δ- and β-globin genes, the leading strand template switch was not observed, suggesting that the Lepore deletion includes the IR (5). In contrast, in somatic cell hybrids containing one copy of human chromosome 11 in a murine genetic background, PCR using oligonucleotide primers close to the IR revealed short nascent DNA strands at two or more sites flanking the IR (7). When an 8 kb fragment containing the human β-globin IR was targeted to an ectopic chromosomal location in monkey cells, origin activity was retained and short nascent DNA strands were detected at five positions within and flanking the IR (8).
Unwinding is a critical step in preparing the DNA template to initiate replication (9–11) and it has been shown that the unwound structure of the DNA substrate can be altered dramatically when DNA polymerase activity is chemically inhibited (12). Therefore, it remains to be determined whether multiple nascent DNA start sites exist distal to the human β-globin IR in logarithmically growing human cells. In the present study PCR was used to measure the length of nascent DNA chains at five sequence tagged sites (STSs) in a 40 kb region of the β-globin gene locus. At each STS short nascent strands were detected, with sizes between 850 and 4000 nt. Two of the STSs, near map units 40.9 and 62, were closest to initiation sites as these STSs could be amplified in nascent DNA as small as 850 nt. To confirm these findings we measured the abundance of short nascent strands (1–2 kb) at five sites by competitive PCR. The abundance of the nascent strands was consistent with the presence of multiple initiation sites in the β-globin gene locus. The highest abundance of short nascent strands was seen at STS 40.9, where nascent strands were approximately four times as abundant as at the downstream STS 72. These data suggest that there are multiple sites for replication initiation in the human β-globin gene locus and that these sites may be used with different frequencies.
MATERIALS AND METHODS
Cell culture, BrdUrd labeling and DNA isolation
HeLa cells were grown in suspension in RPMI-1640 (Gibco BRL) supplemented with 10% newborn calf serum. Exponentially growing cells (105–106 cells/ml) were pulse labeled with 30 µM BrdUrd in RPMI without serum for 15 min with constant agitation at 37°C. All subsequent steps were done in the dark or under yellow light. Cells were pelleted and washed twice with ice-cold phosphate-buffered saline (PBS). Five milliliters of lysis buffer (10 mM EDTA, 50 mM Tris–HCl, pH 8.0, 0.5% SDS) was added to each pellet and mixed by gentle inversion. RNase A was added to a final concentration of 50 µg/ml and the mixture was incubated for 30 min at 37°C. Proteinase K (Gibco BRL) was added to a final concentration of 800 µg/ml and samples were incubated at 37°C overnight. The samples were dialyzed against TE (10 mM Tris–HCl, 1 mM EDTA, pH 8.0) and concentrated by extraction with sec-butanol. The DNA was extracted with diethyl ether, which was removed by heating at 65°C.
Purification of nascent DNA
DNA was denatured by adding 10 N NaOH to a final concentration of 0.2 N and incubated for 20 min at room temperature. An aliquot of 1.2–1.5 ml of denatured DNA was added to 10 ml of saturated alkaline CsCl solution in 50 mM NaOH, 3 mM EDTA [refractive index (RI) = 1.42] and the RI was adjusted to 1.4095 (density 1.81 g/ml) with the same buffer. Gradients were centrifuged in a Beckman Ti75 rotor at 35 000 r.p.m. for 72 h at 25°C. Gradients were fractionated from the bottom into 400 µl fractions. A 20 µl aliquot was taken from every other fraction and the RI was measured. Fractions of density 1.82–1.85 g/ml (RI 1.4105–1.413) were pooled and the RI was adjusted to 1.4095 with an alkaline CsCl solution of RI 1.406. Gradients were recentrifuged under the same conditions and fractionated from below. Nascent DNA was defined as DNA uniformly substituted with BrdUrd (density 1.82–1.85 g/ml). The nascent DNA was precipitated with 3.5 vol of STE (100 mM NaCl, 10 mM Tris–HCl, pH 8.0, 1 mM EDTA) and 4.5 vol of isopropanol.
The purified nascent DNA was size fractionated by alkaline agarose gel electrophoresis. A 1.2% low melting point agarose gel (SeaPlaque GTG agarose in H2O) was soaked overnight at 4°C in 50 mM NaOH, 1 mM EDTA running buffer. Nascent DNA samples were loaded in parallel with DNA size standards and run for 17–24 h at 4°C and 35 V. The gel was neutralized with 1× TBE (89 mM Tris base, 89 mM boric acid, 2 mM EDTA, pH 8.0). The marker lanes were cut out and visualized by ethidium bromide (EtBr) staining. The nascent DNA lane was cut into size fractions according to the DNA standard. The gel slices were melted and 10 µl was directly used in PCR.
Alternatively, nascent DNA was prepared according to the method of Kumar et al. (13), modified here to eliminate handling of the nascent DNA. Twenty-four hours after seeding ∼7.5 × 107 cells were washed twice in ice-cold PBS. The resulting cell pellet was resuspended in 240 µl of ice-cold PBS with 10% glycerol and kept on ice. Seventy microliters of cell suspension was loaded into one well of a 1.25% alkaline agarose gel prechilled to 4°C. Cells were lysed in the well for 10 min before the size standards were loaded and the current applied. The gel was run for 12 h at 40 V and neutralized in 1× TAE before staining in 1× TAE with 0.5 µg/ml EtBr. The DNA was size fractionated and eluted from the gel slices using Qiagen gel extraction columns. The nascent strand abundance values from nascent DNA prepared by BrdUrd substitution (two independent preparations) were within 10–20% of the values obtained using nascent DNA prepared by alkaline cell lysis (three independent preparations).
PCR mapping
Six primer pairs were designed to amplify STSs in the β-globin gene domain. The nucleotide positions and sequences of each primer pair and its internal probe are given in Table 1. The nucleotide positions and map are derived from the 73 308 bp HUMHBB sequence in GenBank (GI 455025). In some of the PCR amplifications shown in Figure 1 primers complementary to STS A, located in the human c-myc origin (14), were mixed with the globin locus primers in multiplex reactions. PCR amplifications were set up in a final volume of 50 µl containing 10 mM Tris–HCl, pH 8.3, 50 mM KCl, 1.5 mM MgCl2, 1.25 U Taq polymerase (Perkin Elmer), 0.5 µM each upper and lower primers, 0.2 mM each dATP, dTTP, dGTP, dCTP and the template DNA. Each set of reactions included a positive control with total HeLa genomic DNA and a negative control with no template. PCR amplification was carried out in a Perkin Elmer 2400 thermal cycler for 25–27 cycles with a basic cycle profile of denaturation at 94°C for 30 s, annealing at 50°C for 30 s and extension at 72°C for 30 s, followed by a final extension at 72°C for 5 min. PCR conditions for each primer set were optimized. Products were run on 2% agarose gels and visualized by EtBr staining. Capillary blotting to nylon membranes (Hybond N+; Amersham) was done under standard conditions (15). Membranes were washed in 2× SSC (300 mM NaCl, 30 mM sodium citrate, pH 7), air dried and UV cross-linked (Stratalinker; Stratagene) to immobilize the DNA. Oligonucleotide probes were 5′-end-labeled with T4 polynucleotide kinase (Gibco BRL) and [γ-32P]ATP (6000 Ci/mmol; NEN). Labeled oligonucleotides were separated from unincorporated radioactivity using G25 Sephadex columns. Membranes were pre-hybridized at 42°C for 2 h in 1 M NaCl, 50 mM Tris–HCl, pH 7.5, 1% SDS and 0.4 mg/ml denatured salmon sperm DNA. Membranes were hybridized in the same buffer with labeled probe at 42°C for 2 h (14), washed twice for 15 min in 2× SSC, 1% SDS at 42°C and once for 60 min at 50°C in the same solution and exposed to Kodak XR-5 film at –80°C with intensifying screens for 18–24 h. For each primer set PCRs were repeated twice on at least two independent nascent DNA preparations with comparable results.
Table 1. Oligonucleotide primer sequences.
| Oligo |
5′-End |
Sequence |
Product (bp) |
| 33.3 UP | 33293 | AACAAAAGCAAAACCAAA | 431 |
| 33.3 LP | 33723 | GTATGTAGGCACCCGATG | |
| 33.3 IP | 33368 | TCCTGCCATGTTAAGTG | |
| 33.3 MP | 33293 | AACAAAAGCAAAACCAAACCAAAGAAAAA | |
| 40.9 UP | 40944 | GCAAGCAATACAAATAAT | 156 |
| 40.9 LP | 41099 | ACCACAAACACAAACAGG | |
| 40.9 IP | 41016 | TATATGAGCCACAAAGGG | |
| 40.9 MP | 41099 | ACCACAAACACAAACAGGCACTTCCCTCAATATAAAC | |
| 54.8 UP | 54806 | CTGAGGAGAAGACTGCTGT | 419 |
| 54.8 LP | 55224 | CTGGACTCACCCTGAAGTT | |
| 54.8 IP | 54977 | TCTCTGTCCCTTGGGCTG | |
| 55.5 UP | 55535 | TAATCCTTTTGTCTCTCC | 195 |
| 55.5 LP | 55729 | CCCACCTCCAGTGTAACT | |
| 55.5 IP | 55568 | AGGCTCCAACTCAAAGAT | |
| 55.5 MP | 55729 | CCCACCTCCAGTGTAACTTATTCACTTTCCTTTCC | |
| 62 UP | 62005 | CAGGTACGGCTGTCATCACTT | 210 |
| 62 LP | 62214 | CAGGGCAGTAACGGCAGA | |
| 62 MP | 62214 | CAGGGCAGTAACGGCAGAATGGTGTCTGTT | |
| 72 UPa | 71918 | TTGACAAACCTGAGAGAA | 536 |
| 72 LPa | 72453 | TGATGGCTAGTGATGATG | |
| 72 IP | 72132 | GGCATGGGCAAGGACTTC | |
| 72 UPb | 72319 | CCAGAATCTACAATGAACTC | 135 |
| 72 LPb | 72453 | TGATGGCTAGTGATGATG | |
| 72 MP | 72453 | TGATGGCTAGTGATGATGTGATTACATAAATGTCTT |
The upper (UP, rightward) and lower (LP, leftward) primers used for PCR mapping and competitive PCR are shown, as well as the internal probes (IP) for PCR mapping and the mutagenic primers (MP) for competitor construction. The 5′-end position of each oligonucleotide is given along with the size of each STS PCR product in base pairs.
aUsed for PCR mapping.
bUsed for competitive PCR.
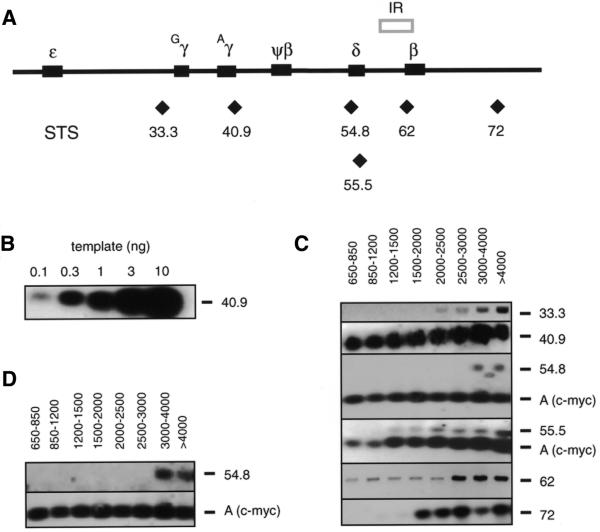
Figure 1.
PCR mapping of nascent DNA strands. (A) Map of the human globin gene locus. Solid boxes represent the position of the globin genes. The previously described IR (5) is shown above the map. Diamonds indicate the approximate position of the STSs/primer sets used in the PCR mapping of nascent strands and the nascent strand abundance assay. (B) Genomic DNA titration. Autoradiogram following PCR amplification of increasing amounts of genomic DNA with STS 40.9 primers. (C) Autoradiograms of size fractionated nascent DNA amplified with each of the STS primer pairs, hybridized to the cognate internal probes. The approximate sizes of the nascent DNA fractions used in each reaction are indicated above each lane. STSs are shown on the right. STS A, which is within the human c-myc origin, was included as an internal standard in some experiments. (D) Nascent DNA from the same preparation that had been co-amplified by primers STS A and STS 54.8 in (C) was amplified in separate reactions by the same primers. Note that the band below the STS 54.8 product in (C) is an artifact that does not reappear when the same DNA is amplified in the experiment of (D).
Competitive PCR
For each primer set a mutagenic oligonucleotide was made with a 20–30 base internal deletion compared to its wild-type counterpart (Table 1). This mutagenic primer was used with a wild-type opposite strand primer to construct a competitor product 20–30 bp shorter than the corresponding wild-type product. The competitor DNA was resolved on an 8% polyacrylamide gel and isolated from a gel slice by crushing and soaking in 200 µl of TE (10 mM Tris, 1 mM EDTA) overnight at 37°C, followed by precipitation with ethanol.
The effective concentration of each competitor was determined by titrating a fixed amount (10 ng, 3000 copies) of genomic DNA against a series of dilutions of competitor. All competitors were quantitated against the same preparation of genomic DNA. Genomic and competitor templates were co-amplified using wild-type primers. Competitive PCR was performed for 45 cycles and the products were separated on a polyacrylamide gel in 1× TBE buffer at 150 V for 4 h. Products were visualized by EtBr staining and the intensities of genomic and competitor bands were quantitated with Collage image analysis software (Fotodyne). The logarithm of the ratio of the band intensities of genomic and competitor products was plotted against the logarithm of the competitor copy number added to the reaction (16). At the interpolated value where the logarithm of the ratio of the band intensities of genomic and competitor products equals 0 the number of competitor copies equals the number of genomic copies added to each reaction. To determine the abundance of nascent strands at specific STSs, a fixed amount of 1–2 kb nascent DNA was amplified in the presence of increasing amounts of competitor and the products were resolved by polyacrylamide gel electrophoresis. Image analysis and calculations of STS abundance were done as for competitor quantitation.
RESULTS
PCR mapping of nascent strands
A modified version of the nascent strand mapping assay developed by Vassilev and Johnson (17,18) and Virta-Pearlman et al. (19) was used to improve the resolution of mapping replication initiation sites in the β-globin gene locus. Asynchronously growing HeLa cells were pulse labeled with BrdUrd to label nascent DNA strands and the population of uniformly BrdUrd-substituted nascent DNA of various lengths was purified from parental DNA chains by two rounds of alkaline CsCl density gradient centrifugation. The nascent DNA was resolved into 0.6–20 kb size fractions by alkaline agarose gel electrophoresis, for use as templates in PCR amplification by primer sets that define STSs across the human β-globin gene locus. Alternatively, nascent DNA was isolated from denatured total cellular DNA (13,20–22), with comparable results. As demonstrated previously, the proximity of an STS to an initiation site is reflected in the length of the shortest nascent DNA containing that STS (13,14,18,20,22–25).
Six PCR primer sets (Table 1) were designed to amplify selected STSs spanning a 40 kb region in the globin gene locus as shown in Figure 1A. Each primer set generated a single band corresponding to the size of the selected STS. Under the reaction conditions employed (7,8,14,20,26,27) the PCR product signals were approximately proportional to the initial amount of input DNA over the range used in these experiments (Fig. 1B). Figure 1C shows representative results of PCR amplification of size fractionated nascent DNA. Primer set 40.9 generated a strong signal with all size fractions of nascent DNA, beginning with the smallest (650–850 nt) and including nascent DNA >4–20 kb, suggesting that an initiation site is located close to STS 40.9. Primer set 33.3 amplified its genomic segment primarily in fractions containing nascent DNA of 2–2.5 kb and larger. Assuming bidirectional replication, this result indicates that there is an initiation point within 1250 bases of STS 33.3. If replication is unidirectional, the maximum distance from this segment to the nearest initiation site is ∼2500 bases.
Two primer sets were designed close to one another to check the consistency of the PCR mapping procedure. PCR at STS 54.8 yielded product most prominently in nascent DNA fractions of 3–4 kb and larger. Therefore, an initiation site can be mapped within 1.5–2 kb of STS 54.8. Slightly further downstream STS 55.5 was amplified in nascent DNAs as small as 1.2–2.0 kb, suggesting that an initiation site exists within 1 kb of this STS. Considering the locations of their respective PCR primers, the simplest interpretation of these results is that the pattern of amplification of STSs 54.8 and 55.5 derives from an initiation event near map unit 56.4, although more complex explanations are also possible.
STS 62 is located within the previously described globin IR. Amplification of STS 62 sequences was evident in all size fractions of nascent DNA, consistent with the presence of an initiation event near this site. STS 72, which is at the 3′-end of the region examined, was amplified in nascent DNAs of 1.5–2 kb and larger. If these nascent strands are generated by bidirectional replication fork movement, STS 72 is situated within ∼1000 bases of an initiation site.
The distinct patterns of amplification at different globin STSs argue that these results are not due to random degradation of the nascent DNA preparations. To test this further, oligonucleotides complementary to STS A, located in the human c-myc origin of replication (14), were used in multiplex PCRs to amplify the same nascent DNAs amplified by the STS 54.8 and 55.5 primers. STS A amplification was observed in all size fractions, implying that amplification of STS 54.8 or 55.5 would have been detectable in all size fractions had the nascent DNA been fragmented by random degradation. Under the limited amplification conditions used, the pattern of PCR products was not affected by competition for reaction components during co-amplification of these STSs, as independent amplification of STS A and STS 55.5 from the same nascent DNA preparation yielded results comparable to those obtained by co-amplification (Fig. 1D).
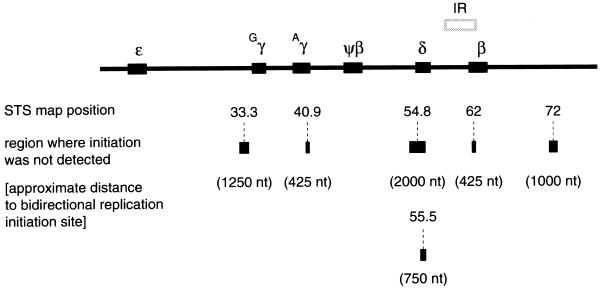
Figure 2 summarizes the results of PCR mapping of nascent strands. The initiation free zone(s) demarcates the largest zone(s) in which replication initiation does not occur and was calculated assuming bidirectional replication and equal fork progression rates. The maximum distance from a given STS to the nearest initiation event is taken to be half the length of the largest nascent DNA strands in the smallest size fraction that can be amplified. Accordingly, an initiation zone can be mapped within 425 bases of STS 40.9 or STS 62, as these sequences were detected in all size fractions beginning with the 650–850 nt fraction. Likewise, STS 33.3 is situated within 1250 bases of an initiation site and another initiation event can be found within 1000 bases of STS 72. An initiation zone can also be deduced within 750 nt of STS 55.5 and within 2 kb of STS 54.8. Taken together these data imply that there are multiple start sites for DNA replication in the β-globin gene locus.
Figure 2.
Summary of the PCR mapping of nascent DNA strands. STSs used in the PCR amplification are shown below the globin locus map. The bar centered below each STS indicates the maximum distance to an initiation zone. The deduced distance (in nucleotides) to the nearest initiation site assuming equivalent rates of bidirectional fork movement is also shown.
Quantitation of nascent DNA
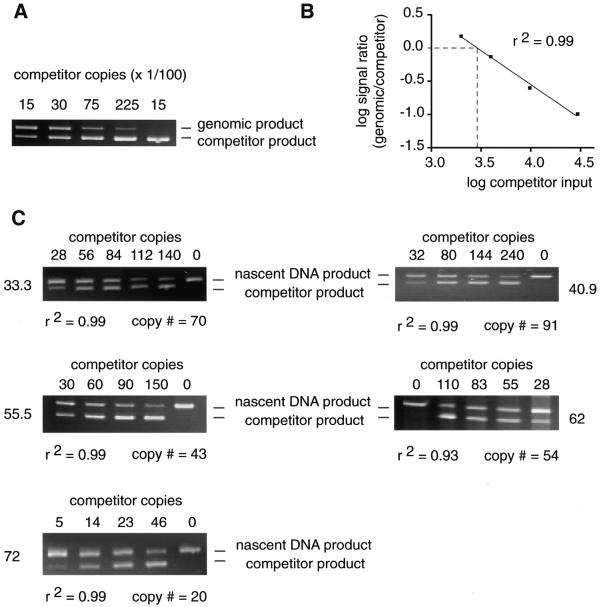
PCR mapping of nascent DNA strands can indicate the maximum distance from an STS to the nearest initiation site, but does not readily allow comparison of the relative abundance of nascent strands between STSs. To confirm that replication initiates at multiple sites in the β-globin gene locus, competitive PCR was used to measure the abundance of short nascent DNA chains (1–2 kb) at several globin STSs in nascent DNA isolated from logarithmically growing HeLa cells. The 1–2 kb size fraction was used as done previously to avoid interference by Okazaki fragments (20,22,25,28–31). Competitors specific for each STS were constructed with wild-type sequences at their termini, but containing a 20–30 bp internal deletion, so that they could be amplified by the same primers used to amplify wild-type genomic DNA. The effective concentration of each competitor template was determined by titration against a fixed amount of HeLa genomic DNA to normalize for differences in amplification activity (25; Fig. 3A and B). For quantitation of the abundance of short nascent strands at STSs 33.3, 40.9, 55.5, 62 and 72 a constant amount of nascent DNA was amplified with increasing amounts of each STS-specific competitor (Fig. 3C).
Figure 3.
Competitive PCR quantitation. (A) A constant amount of genomic DNA (3000 copies) was amplified with 1500, 3000, 7500 or 22 500 copies of the STS 40.9 competitor template. Rightmost lane, no genomic DNA. (B) The logarithm of the signal intensity ratio of genomic to competitor products is plotted against the logarithm of the number of input competitor copies. The quantity of competitor equivalent in amplification efficiency to 3000 copies of genomic DNA corresponded to the value interpolated from the experimental curve when the logarithm of the signal intensity ratio equaled 0. (C) Representative gels of competitive PCR quantitation of short nascent DNA at five STSs. The number of competitor molecules added to each reaction is shown above each lane. The positions of the nascent and competitor PCR products on the gel are indicated. The number of nascent strand copies at each STS was calculated by plotting the logarithm of the signal intensity ratio of the products against the logarithm of the amount of competitor added and is shown below each gel. The coefficient of determination for each data set, r2, is also indicated.
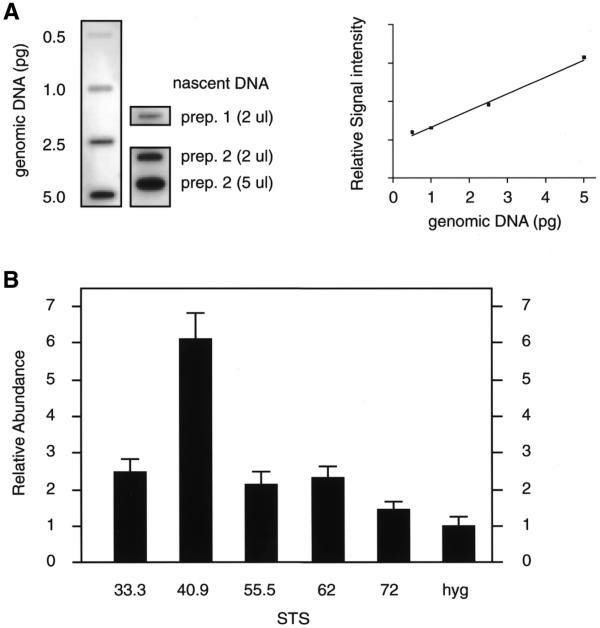
To relate the quantitation of nascent strand abundance from independent nascent DNA preparations, the total amount of nascent DNA per preparation was determined by quantitating the slot blot hybridization of a total genomic DNA probe to an aliquot of each nascent DNA against genomic DNA standards (Fig. 4A; 20). A summary of the results of the nascent strand abundance assays is shown in Figure 4B. The abundance of each STS in the 1–2 kb nascent DNA is expressed relative to the background at a site of low initiation activity, STS hyg. STS hyg is in a randomly integrated recombination target site used previously for the analysis of human c-myc origin activity at an ectopic location (20; see Discussion). The data of Figure 4 show that the abundance of nascent strands at STS 33.3, 55.5 or 62 is approximately twice the STS hyg background. However, the highest amount of nascent DNA was detected at STS 40.9, ∼6-fold above the STS hyg background. Thus, the results of both the nascent strand size mapping and competitive PCR abundance assays indicate that there are multiple initiation events within the β-globin gene locus.
Figure 4.
Summary of the nascent strand abundance assay. (A) Autoradiogram of genomic and nascent DNA (prep. 1, BrdUrd labeled; prep. 2, alkaline gel isolate) slot blotted and hybridized to total genomic DNA (left). The amount of nascent DNA in different preparations was normalized by comparison with the genomic DNA standard curve generated by PhosphorImager analysis (right) (20). (B) The nascent DNA abundance at each STS is plotted relative to the abundance at STS hyg. STS hyg is a randomly integrated target site present once per genome, used previously for the analysis of the human c-myc origin (20). The nascent strand abundance was normalized for the copy numbers of the globin and STS hyg loci per genome. Error bars, standard deviation. The results are the average of four to six separate experiments obtained from three different nascent DNA preparations.
DISCUSSION
The size distribution and abundance of nascent DNAs at selected sites across the human β-globin gene locus were examined to determine whether replication initiates at one or more sites in this region in exponentially growing HeLa cells. Based on the proximity of initiation sites to the STSs analyzed over a region of ∼40 kb, the data suggest that replication initiates at multiple locations in the β-globin gene domain. For example, short nascent strands appear to initiate within 650–850 nt of STS 40.9 and within 2000–2500 nt of STS 33.3. Since these STSs are more than 7 kb apart, these results suggest that independent initiation events are detected by the cognate primers at each site. Similarly, short nascent strands initiate within 650–850 nt of STS 62 and within 2000 nt of STS 55.5. Since these sites are more than 6 kb apart the cognate primers at each site detect nascent DNA from independent initiation events. Because we have not assayed a sufficient number of STSs to saturate the β-globin gene domain, it is possible that additional replication initiation sites exist which were not detected in this study.
The PCR mapping analysis and the nascent strand abundance assay, as well as other assays of replication origin activity (32), depend on the steady-state level of replicative intermediates. Thus, rapid fork movement or nascent strand maturation in a region of chromatin could decrease the steady-state level of short-lived intermediates at an STS relative to that in a region of slower fork movement. In the nascent strand abundance assay this would lead to an apparent decrease in the frequency of initiation near an STS. Therefore, while the presence of 1–2 kb nascent strands is suggestive of initiation near an STS, a low level of short nascent strands is not proof of the absence of initiation. In the mapping assay underestimation of nascent strand abundance could be interpreted as a greater distance from the STS to an initiation site. Hence, the PCR mapping analysis gives estimates of the maximum distances from an STS to an initiation event.
The competitive PCR nascent strand abundance assay confirmed that short nascent strands were synthesized at multiple sites in the region of the β-globin genes, including STS 33.3, STS 55.5 and the previously identified IR near STS 62 (5). However, the greatest abundance of 1–2 kb nascent strands was detected near STS 40.9, approximately six times the background level detected at STS hyg. STS hyg is within a randomly integrated FLP recombinase target previously used as an acceptor site for insertion of the human c-myc origin (20). The nascent strand abundance used as background here is of the unoccupied acceptor site. Integration of the c-myc origin DNA at this acceptor site, ∼4 kb from STS hyg, increased the nascent strand abundance at STS hyg ∼2-fold (20).
Our data agree well with the results of Dhar et al. (3) who used centrifugal elutriation and blot hybridization to identify the earliest replicating fragments in the β-globin gene domain. Despite the limitations of centrifugal elutriation to resolve cells that have differentially replicated neighboring restriction fragments, Dhar et al. (3) found the earliest replicating fragment of the β-globin gene locus in K562 or HeLa cells to abut STS 40.9. Using a modified approach to elutriate cells at higher discrimination, Epner et al. (33) found the earliest replicating fragments associated with the γG- (STS 33.3) and γA- (STS 40.9), δ- (STS 55.5) and β-globin genes (STS 62).
Subsequent PCR mapping of replication start sites in the human β-globin locus in human–mouse somatic cell hybrids indicated the presence of initiation sites flanking the globin IR (7), although the absence of a PCR signal with primer sets STS 33.3, STS 45.8 and STS 72 led to the conclusion that initiation did not occur distal to STS 62. However, when an 8 kb DNA fragment containing the globin IR was transposed to an ectopic site in the simian genome independent initiations were detected inside and from 1–3 kb outside the IR (8). In contrast, the overall polarity of leading strand synthesis in K562 and HeLa cells treated with emetine (and fluorodeoxyuridine) suggests that opposite template strands are used for leading strand synthesis beginning at a single IR between the δ- and β-globin genes (5). Because the observed pattern of template biases was not that expected for the extension of DNA strands from a single initiation site, it is possible that additional initiations occur in the globin domain. Alternatively, drug treatment to inhibit lagging strand DNA synthesis may selectively suppress some initiations.
Depending on the origin, replication initiation in metazoan cells appears to occur in a narrow region or over a dispersed zone (14,21,25,26,29,31,34–44). In this regard, the human β-globin gene locus resembles the human c-myc locus in that multiple, non-random initiation sites may be used at different frequencies over a large domain (14,20,21,26). Models that accommodate these observations involve nucleation from a replicator element of a chromatin structure permissive for initiation at underlying preferred sites (26,45). Further investigation using chromosomally integrated origin constructs (8,20) may reveal the role of DNA sequence (8), replication protein binding (46) and other epigenetic factors (47–51) in determining the breadth of metazoan replication initiation domains.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by grants from the USPHS (GM53819), the Kettering Foundation through the WSU School of Medicine and the WSU CoSM Research Challenge Program to M.L.
References
- 1.Fritsch E.F., Lawn,R.M. and Maniatis,T. (1980) Molecular cloning and characterization of the human beta-like globin gene cluster. Cell, 19, 959–972. [DOI] [PubMed] [Google Scholar]
- 2.Epner E., Rifkind,R.A. and Marks,P.A. (1981) Replication of alpha and beta globin DNA sequences occurs during early S phase in murine erythroleukemia cells. Proc. Natl Acad. Sci. USA, 78, 3058–3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dhar V., Mager,D., Iqbal,A. and Schildkraut,C.L. (1988) The coordinate replication of the human beta-globin gene domain reflects its transcriptional activity and nuclease hypersensitivity. Mol. Cell. Biol., 8, 4958–4965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dhar V., Skoultchi,A.I. and Schildkraut,C.L. (1989) Activation and repression of a beta-globin gene in cell hybrids is accompanied by a shift in its temporal replication. Mol. Cell. Biol., 9, 3524–3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kitsberg D., Selig,S., Keshet,I. and Cedar,H. (1993) Replication structure of the human beta-globin gene domain. Nature, 366, 588–590. [DOI] [PubMed] [Google Scholar]
- 6.Burhans W.C., Vassilev,L.T., Wu,J., Sogo,J.M., Nallaseth,F.S. and DePamphilis,M.L. (1991) Emetine allows identification of origins of mammalian DNA replication by imbalanced DNA synthesis, not through conservative nucleosome segregation. EMBO J., 10, 4351–4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aladjem M.I., Groudine,M., Brody,L.L., Dieken,E.S., Fournier,R.E., Wahl,G.M. and Epner,E.M. (1995) Participation of the human beta-globin locus control region in initiation of DNA replication. Science, 270, 815–819. [DOI] [PubMed] [Google Scholar]
- 8.Aladjem M.I., Rodewald,L.W., Kolman,J.L. and Wahl,G.M. (1998) Genetic dissection of a mammalian replicator in the human beta-globin locus. Science, 281, 1005–1009. [DOI] [PubMed] [Google Scholar]
- 9.Borowiec J.A. and Hurwitz,J. (1988) Localized melting and structural changes in the SV40 origin of replication induced by T-antigen. EMBO J., 7, 3149–3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Umek R.M. and Kowalski,D. (1990) Thermal energy suppresses mutational defects in DNA unwinding at a yeast replication origin. Proc. Natl Acad. Sci. USA, 87, 2486–2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang R.Y. and Kowalski,D. (1993) A DNA unwinding element and an ARS consensus comprise a replication origin within a yeast chromosome. EMBO J., 12, 4521–4531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walter J. and Newport,J. (2000) Initiation of eukaryotic DNA replication: origin unwinding and sequential chromatin association of Cdc45, RPA and DNA polymerase alpha. Mol. Cell, 5, 617–627. [DOI] [PubMed] [Google Scholar]
- 13.Kumar S., Giacca,M., Norio,P., Biamonti,G., Riva,S. and Falaschi,A. (1996) Utilization of the same DNA replication origin by human cells of different derivation. Nucleic Acids Res., 24, 3289–3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trivedi A., Waltz,S.E., Kamath,S. and Leffak,M. (1998) Multiple initiations in the c-myc replication origin independent of chromosomal location. DNA Cell Biol., 17, 885–896. [DOI] [PubMed] [Google Scholar]
- 15.Sambrook J., Fritsch,E. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 16.Connolly A.R., Cleland,L.G. and Kirkham,B.W. (1995) Mathematical considerations of competitive polymerase chain reaction. J. Immunol. Methods, 187, 201–211. [DOI] [PubMed] [Google Scholar]
- 17.Vassilev L. and Johnson,E.M. (1989) Mapping initiation sites of DNA replication in vivo using polymerase chain reaction amplification of nascent strand segments. Nucleic Acids Res., 17, 7693–7705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vassilev L. and Johnson,E.M. (1990) An initiation zone of chromosomal DNA replication located upstream of the c-myc gene in proliferating HeLa cells. Mol. Cell. Biol., 10, 4899–4904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Virta-Pearlman V.J., Gunaratne,P.H. and Chinault,A.C. (1993) Analysis of a replication initiation sequence from the adenosine deaminase region of the mouse genome. Mol. Cell. Biol., 13, 5931–5942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Malott M. and Leffak,M. (1999) Activity of the c-myc replicator at an ectopic chromosomal location. Mol. Cell. Biol., 19, 5685–5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tao L., Dong,Z., Leffak,M., Zannis-Hadjopoulos,M. and Price,G. (2000) Major DNA replication initiation sites in the c-myc locus in human cells. J. Cell. Biochem., 78, 442–457. [DOI] [PubMed] [Google Scholar]
- 22.Tao L., Nielsen,T., Friedlander,P., Zannis-Hadjopoulos,M. and Price,G. (1997) Differential DNA replication origin activities in human normal skin fibroblast and HeLa cell lines. J. Mol. Biol., 273, 509–518. [DOI] [PubMed] [Google Scholar]
- 23.Vassilev L.T., Burhans,W.C. and DePamphilis,M.L. (1990) Mapping an origin of DNA replication at a single-copy locus in exponentially proliferating mammalian cells. Mol. Cell. Biol., 10, 4685–4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shinomiya T. and Ina,S. (1994) Mapping an initiation region of DNA replication at a single-copy chromosomal locus in Drosophila melanogaster cells by two-dimensional gel methods and PCR-mediated nascent-strand analysis: multiple replication origins in a broad zone. Mol. Cell. Biol., 14, 7394–7403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kobayashi T., Rein,T. and DePamphilis,M.L. (1998) Identification of primary initiation sites for DNA replication in the hamster dihydrofolate reductase gene initiation zone. Mol. Cell. Biol., 18, 3266–3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Waltz S.E., Trivedi,A.A. and Leffak,M. (1996) DNA replication initiates non-randomly at multiple sites near the c-myc gene in HeLa cells. Nucleic Acids Res., 24, 1887–1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cimbora D.M., Schubeler,D., Reik,A., Hamilton,J., Francastel,C., Epner,E.M. and Groudine,M. (2000) Long-distance control of origin choice and replication timing in the human beta-globin locus are independent of the locus control region. Mol. Cell. Biol., 20, 5581–5591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Diviacco S., Norio,P., Zentilin,L., Menzo,S., Clementi,M., Biamonti,G., Riva,S., Falaschi,A. and Giacca,M. (1992) A novel procedure for quantitative polymerase chain reaction by coamplification of competitive templates. Gene, 122, 313–320. [DOI] [PubMed] [Google Scholar]
- 29.Pelizon C., Diviacco,S., Falaschi,A. and Giacca,M. (1996) High-resolution mapping of the origin of DNA replication in the hamster dihydrofolate reductase gene domain by competitive PCR. Mol. Cell. Biol., 16, 5358–5364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Giacca M., Zentilin,L., Norio,P., Diviacco,S., Dimitrova,D., Contreas,G., Biamonti,G., Perini,G., Weighardt,F., Riva,S. et al. (1994) Fine mapping of a replication origin of human DNA. Proc. Natl Acad. Sci. USA, 91, 7119–7123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Phi-van L. and Stratling,W.H. (1999) An origin of bidirectional DNA replication is located within a CpG island at the 3′ end of the chicken lysozyme gene. Nucleic Acids Res., 27, 3009–3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vassilev L.T. and DePamphilis,M.L. (1992) Guide to identification of origins of DNA replication in eukaryotic cell chromosomes. Crit. Rev. Biochem. Mol. Biol., 27, 445–472. [DOI] [PubMed] [Google Scholar]
- 33.Epner E., Forrester,W.C. and Groudine,M. (1988) Asynchronous DNA replication within the human beta-globin gene locus. Proc. Natl Acad. Sci. USA, 85, 8081–8085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heck M.M. and Spradling,A.C. (1990) Multiple replication origins are used during Drosophila chorion gene amplification. J. Cell Biol., 110, 903–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Delidakis C. and Kafatos,F.C. (1989) Amplification enhancers and replication origins in the autosomal chorion gene cluster of Drosophila. EMBO J., 8, 891–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Little R.D., Platt,T.H. and Schildkraut,C.L. (1993) Initiation and termination of DNA replication in human rRNA genes. Mol. Cell. Biol., 13, 6600–6613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Phi-van L., Sellke,C., von Bodenhausen,A. and Stratling,W.H. (1998) An initiation zone of chromosomal DNA replication at the chicken lysozyme gene locus. J. Biol. Chem., 273, 18300–18307. [DOI] [PubMed] [Google Scholar]
- 38.Shinomiya T. and Ina,S. (1993) DNA replication of histone gene repeats in Drosophila melanogaster tissue culture cells: multiple initiation sites and replication pause sites. Mol. Cell. Biol., 13, 4098–4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tao L., Zannis-Hadjopoulos,M., Leffak,M. and Price,G. (1998) In Zannis-Hadjopoulos,M. and Price,G. (eds), Regulation of Eucaryotic DNA Replication. St Saveur des Monts, Quebec, Canada.
- 40.Abdurashidova G., Deganuto,M., Klima,R., Riva,S., Biamonti,G., Giacca,M. and Falaschi,A. (2000) Start sites of bidirectional DNA synthesis at the human lamin B2 origin. Science, 287, 2023–2026. [DOI] [PubMed] [Google Scholar]
- 41.Araujo F.D., Knox,J.D., Ramchandani,S., Pelletier,R., Bigey,P., Price,G., Szyf,M. and Zannis-Hadjopoulos,M. (1999) Identification of initiation sites for DNA replication in the human dnmt1 (DNA-methyltransferase) locus. J. Biol. Chem., 274, 9335–9341. [DOI] [PubMed] [Google Scholar]
- 42.Ariizumi K., Wang,Z. and Tucker,P.W. (1993) Immunoglobulin heavy chain enhancer is located near or in an initiation zone of chromosomal DNA replication. Proc. Natl Acad. Sci. USA, 90, 3695–3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dijkwel P.A. and Hamlin,J.L. (1995) The Chinese hamster dihydrofolate reductase origin consists of multiple potential nascent-strand start sites. Mol. Cell. Biol., 15, 3023–3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gale J.M., Tobey,R.A. and D’Anna,J.A. (1992) Localization and DNA sequence of a replication origin in the rhodopsin gene locus of Chinese hamster cells. J. Mol. Biol., 224, 343–358. [DOI] [PubMed] [Google Scholar]
- 45.Spradling A.C. (1999) ORC binding, gene amplification and the nature of metazoan replication origins. Genes Dev., 13, 2619–2623. [DOI] [PubMed] [Google Scholar]
- 46.Austin R.J., Orr-Weaver,T.L. and Bell,S.P. (1999) Drosophila ORC specifically binds to ACE3, an origin of DNA replication control element. Genes Dev., 13, 2639–2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dimitrova D.S. and Gilbert,D.M. (1998) Regulation of mammalian replication origin usage in Xenopus egg extract. J. Cell Sci., 111, 2989–2998. [DOI] [PubMed] [Google Scholar]
- 48.Rein T., Kobayashi,T., Malott,M., Leffak,M. and DePamphilis,M.L. (1999) DNA methylation at mammalian replication origins. J. Biol. Chem., 274, 25792–25800. [DOI] [PubMed] [Google Scholar]
- 49.Lawlis S.J., Keezer,S.M., Wu,J.R. and Gilbert,D.M. (1996) Chromosome architecture can dictate site-specific initiation of DNA replication in Xenopus egg extracts. J. Cell Biol., 135, 1207–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gilbert D.M., Miyazawa,H. and DePamphilis,M.L. (1995) Site-specific initiation of DNA replication in Xenopus egg extract requires nuclear structure. Mol. Cell. Biol., 15, 2942–2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Delgado S., Gomez,M., Bird,A. and Antequera,F. (1998) Initiation of DNA replication at CpG islands in mammalian chromosomes. EMBO J., 17, 2426–2435. [DOI] [PMC free article] [PubMed] [Google Scholar]