Abstract
Human acute promyelocytic leukemia is causally linked to chromosomal translocations that generate chimeric retinoic acid receptor-α proteins (x-RARα fusions). Wild-type RARα is a transcription factor that binds to the SMRT/NCoR family of corepressors in the absence of hormone but releases from corepressor and binds coactivators in response to retinoic acid. In contrast, the x-RARα fusions are impaired for corepressor release and operate in acute promyelocytic leukemia as dominant-negative inhibitors of wild-type RARα. We report that the two most common x-RARα fusions, PML-RARα and PLZF-RARα, have gained the ability to recognize specific splice variants of SMRT and NCoR that are poorly recognized by RARα. These differences in corepressor specificity between the normal and oncogenic receptors are further magnified in the presence of a retinoid X receptor heteromeric partner. The ability of retinoids to fully release corepressor from PML-RARα differs for the different splice variants, a phenomenon relevant to the requirement for supraphysiological levels of this hormone in differentiation therapy of leukemic cells. We propose that this shift in the specificity of the x-RARα fusions to a novel repertoire of corepressors contributes to the dominant-negative and oncogenic properties of these oncoproteins and helps explain previously paradoxical aspects of their behavior.
Keywords: Coregulator Transcription, Corepressor Transcription, Fusion Protein, Leukemia, Nuclear Receptors, Oncogene, Transcription Regulation
Introduction
Retinoic acid receptors (RARs)3 are members of the nuclear receptor family of ligand-regulated transcription factors. RARs bind to specific target genes and activate transcription in response to cognate agonists, such as all-trans-retinoic acid (ATRA) (1, 2). Conversely, in the absence of ATRA, RARs can repress transcription of their target genes below basal levels (1, 2). This bimodal transcriptional regulation is possible through the differential recruitment of coactivator and corepressor proteins to the RAR, which in turn creates the biochemical milieu to support or repress transcription (1, 2). Two corepressor paralogs, SMRT and NCoR, play important roles in nuclear receptor-mediated repression. Both SMRT and NCoR contain CoRNR box motifs ((I/L)XX(I/V)I) near their C termini that bind a hydrophobic groove on the surface of the unliganded nuclear receptors. The SMRT and NCoR N-terminal domains, in turn, recruit additional proteins that help confer repression, including histone deacetylases, TBL-1, TBLR-1, and GPS-2 (3–5). RARs can bind to their DNA-binding sites (retinoic acid response elements, or RAREs) as homodimers or as heterodimers with retinoic X receptors (RXRs) (1, 2).
The RARα locus, on chromosome 17, undergoes reciprocal chromosomal translocations at high frequency in human acute promyelocytic leukemia (APL), generating x-RARα fusion proteins that play a causal role in this malignancy (6, 7). In >98% of APL, the x-sequences originate from the PML (promyelocytic leukemia) open reading frame on chromosome 15. Although more rare, yet other x-RARα fusions have been identified, including PLZF (promyelocytic leukemia zinc finger)-RARα, STAT5b-RARα, NPM (nucleophosmin)-RARα, and NuMA (nuclear mitotic apparatus protein)-RARα (6, 7). Despite the diversity of these x-fusions, they inevitably include a protein dimerization motif contributed by the x-partner, fused to the DNA-binding and hormone-binding domains of RARα (8–10). The ectopic dimerization domain has been proposed to unmask the oncogenic properties of RARα by enhancing the ability of the x-RARα fusion to bind to DNA as homodimers and as heterotetramers with RXR (11, 12). These x-RARα dimers and oligomers either fail to release corepressor in response to ATRA or require higher than normal levels of agonist to do so (8–10); as a result, x-RARα fusions can function as dominant-negative inhibitors of RARα, a property that is closely linked to their oncogenic properties (11, 12). Consistent with this concept, forced release of corepressor in response to supraphysiological levels of ATRA (or other manipulations) can cause differentiation of APL cells in vitro and remission of the disease in vivo (6). However, the molecular mechanism by which x-RARα homodimerization or oligomerization impairs corepressor release is poorly understood, and paradoxically, x-RARα fusions can display a wide range of transcriptional properties, including transcriptional activation, in different cells and on different response elements (13, 14).
Both SMRT and NCoR are expressed by alternative mRNA splicing as a series of corepressor variants that possess different configurations of CoRNR box motifs and have different affinities for different nuclear receptors (15–18). As a result, the repertoire of corepressor variants available in a particular cell type can dictate the repression capability of the nuclear receptors within that cell. We report here that the PML-RARα and PLZF-RARα fusions do not simply alter corepressor binding per se, as proposed previously, but produce a striking change in their affinities for specific SMRT and NCoR variants. In general, both PML-RARα and PLZF-RARα have gained the ability to recognize corepressor variants that are poorly recognized by RARα. Heteromer formation with RXRα resulted in still greater differences in the corepressor specificities of RARα and the x-RARα oncoproteins. Further, the ability of retinoids to fully release corepressor from PML-RARα differs for the different variants. These observations help explain several previously confusing aspects of x-RARα function and may suggest better methods for clinical management of APL.
EXPERIMENTAL PROCEDURES
Cell Culture
CV-1 cells were grown at 37 °C in DMEM with high glucose plus l-glutamine (Invitrogen) and 9% heat-inactivated FBS (Hyclone, Logan, UT). NB4, HL-60, and U937/PR9 (19, 20) cells were grown at 37 °C in RPMI 1640 (plus l-glutamine) and 10% heat-inactivated FBS. Sf9 insect cells were grown in Insect-Xpress with l-glutamine (Lonza, Walkersville, MD) in suspension at 28 °C.
Protein Expression
RARα, PML-RARα, and PLZF-RARα were subcloned into the pF3A WG (BYDV) vector (Promega, Madison, WI) and were expressed using the TnT SP6 high yield protein expression system following the manufacturer's instructions (Promega, Madison, WI). Three micrograms of plasmid were used per translation. The relative yields of protein were quantified by SDS-PAGE and by immunoblotting with anti-RARα antibody (sc-551; Santa Cruz Biotechnology, Santa Cruz, CA); this antibody recognizes a C-terminal epitope shared by all the RARα constructs. RXRα was expressed using a recombinant baculovirus/Sf9 cell system.
DNAs representing the receptor-interaction domain of the corepressors (amino acid 1851 to C terminus of SMRT or amino acid 1817 to C terminus of NCoR) were inserted in-frame into Fastbac1 expressing an N-terminal GST affinity tag and were reconstituted into infectious baculovirus using the Bac-to-Bac Expression system (Invitrogen). After two or three rounds of viral amplification in Sf9 cells, 2 ml of baculovirus stock was used to infect 5 × 107 Sf9 cells grown in suspension (2 × 106 cells/ml). After 48 h at 28 °C, the cells were harvested by centrifugation, washed with TBS (10 mm Tris, pH 7.5, 150 mm NaCl), and snap frozen in liquid nitrogen. Cell pellets were thawed on ice, resuspended in 3× pellet volume in Sf9 whole cell extract buffer (20 mm HEPES, pH 7.9, 200 mm KCl, 1 mm EDTA, 0.5% Igepal CA-630, 10% glycerol, 1 mm DTT, and Complete protease inhibitor mixture; Roche Applied Science). The cell suspensions were again frozen and thawed, and clarified at 20,000 × g for 10 min at 4 °C. The supernatants were incubated with glutathione-agarose (Sigma) at 4 °C for 90 min with rocking. The immobilized GST proteins were pelleted, washed three times with PBS, 0.1% Tween 20, and were eluted with 20 mm glutathione in 100 mm TBS, pH 8.0, for 30 min at 4 °C. The proteins were analyzed by SDS-PAGE and Coomassie Brilliant Blue staining to quantify yield and quality, using a Fluorchem 8900 photography system and AlphaEase FC analysis software (Alpha Innotech, San Leandro, CA).
Plasmids expressing the individual CoRNR box motifs were described previously (21) and were expressed in Escherichia coli. Bacterial cell lysates were prepared as described (22), and the GST fusions were purified as above.
Electrophoretic Mobility Supershift Assays
Consensus DR5 oligonucleotides (top strand: 5′-agcta aAGGT CAgat aaAGG TCAgc agga-3′ bottom strand: 5′-tcgat cctgc TGACC Tttat cTGAC CTtt-3′) (IDT, Coralville, IA) were annealed and radiolabeled by Klenow polymerase fill-in using [α-32P]dGTP (3000 Ci/mmol; PerkinElmer Life Science) plus the three remaining unlabeled dNTPs. EMSAs contained the RARα or x-RARα of interest, with or without RXRα, 1 pmol of the radiolabeled DR5 probe, and the specified amount of GST-corepressor, in 20 μl of binding buffer/reaction, minus or plus ATRA. Protein-DNA complexes were resolved by native electrophoresis at 180 V for 105 min through a 4% polyacrylamide gel polymerized over an 8% polyacrylamide gel cushion, both buffered in 0.5× TBE (45 mm Tris borate, 1 mm EDTA). The gels were visualized by Storm phosphorimaging analysis (GE Healthcare). The percentage of supershift by the corepressor was calculated by quantifying the loss of shifted homodimer or heteromer band and subtracting the loss from 100. Curve fitting was performed using Prism 5.0 for Macintosh (GraphPad Software Inc., San Diego, CA).
Transfections, Coimmunoprecipitations, and Reporter Assays
For coimmunoprecipitations, CV-1 cells (two 10-cm plates/corepressor) were transfected with 0.5 μg of pSG5.1-RARα, 0.5 μg of pSG5-PML-RARα, 0.5 μg of pUC18, and 1.0 μg of either pSG5HA-mSMRTϵ or pSG5HA-mNCoRω using Effectene per the manufacturer's instructions. The cells were maintained in DMEM containing 9% heat-inactivated FBS (hormone-stripped) for 48 h, washed twice with ice-cold PBS, and harvested by scraping and centrifugation. The cell pellets were resuspended in 0.5 ml of immunoprecipitation buffer (1× PBS, 1 mm EDTA, 0.5% Triton X-100, 1× Complete protease inhibitor) and sonicated for 30 s using a Branson Sonifer 250 microtip at the number 3 setting and 50% duty cycle (Branson, Danbury, CT). After a 5-min centrifugation at 500 × g, 5% of the clarified cell lysate was set aside as an input control. The remainder was incubated with either 1 μl of anti-SMRT (Affinity Bioreagents, Rockford, IL) or 1 μl of anti-NCoR (raised in rabbits against amino acids 1953–1966 of mouse NCoRω) for 2 h at 4 °C with rocking. Forty microliters of a 50% slurry of protein A-Sepharose (Sigma) equilibrated in immunoprecipitation buffer were added, and the incubations were continued 2 h at 4 °C. Immunoprecipitates were collected by centrifugation for 1 min at 1000 × g at 4 °C, were washed four times with 1 ml of immunoprecipitation buffer, and were eluted with SDS-PAGE sample buffer at 95 °C for 5 min. Each sample was resolved by SDS (4–15%) PAGE at 100 V for 20 min, followed by 180 V for 50 min. The proteins were detected by immunoblotting using goat anti-RARα (1/500 dilution, ab28767; Abcam, Cambridge, MA) and rabbit anti-goat IgG-HRP (1/1000, ab6741-1; Abcam). The blots were developed with ECL Plus (GE Healthcare) and visualized/quantified with the Fluorchem 8900 photography system and AlphaEase FC analysis software. Luciferase reporter assays were performed as described previously (23) using 10 ng of pSG5-xRARα plasmid and 50 ng of pGL4-tk-3X-βRARE or pGL4-tk-3X-conRARE.
Quantification of Corepressor mRNAs
Total RNA was isolated from NB4, HL-60, and U937/PR9 cells using TRIzol reagent (Invitrogen) and quantified by optical absorbance. Complementary DNA was synthesized using 1 μg of RNA and the QuantiTect kit (Qiagen); semi-quantitative RT-PCR were performed for 35 cycles (94 °C for 1 min, 55 °C for 1 min, and 72 °C for 1 min) using GoTaq DNA polymerase (Promega, Madison, WI). Primers for evaluating splice variants at the SMRT S1 CoRNR box were Forward (5′-CAAGA AGCTG AACAC CCACA-3′) and Reverse (5′-CGAGT GCACT GAGGA GACAG-3′); primers for SMRT S3 CoRNR box splice variants were Forward (5′-CGGAC CCGCA CCGGG AAAAG AC-3′) and Reverse (5′-AGGTG CTTGG GGAGC CCCTT G-3′); and primers for NCoR S3 CoRNR box splice variants were Forward (5′-ACAGA GACCC AGTGT TTTCC AAG-3′) and Reverse (5′-GCAGG ACTTA TCACC TCAAT AGCA-3′). Each primer pair spans an intron to permit detection of genomic DNA contamination; none was observed. GAPDH RT-PCR was performed for 20 cycles with primers GAPDH-forward (5′GCTGA ACGGG AAGCT CACTG G-3′) and GAPDH-reverse (5′-GCCTG CTTCA CCACC TTCTT GATG-3′). PCR products were resolved by agarose gel electrophoresis, visualized by ethidium bromide staining, and quantified using AlphaEaseFC software and Fluorchem 8900 photography system.
Frozen mouse APL cells (24) were rapidly thawed and pelleted, and total RNA was extracted using the Qiagen RNeasy mini kit. cDNA synthesis and PCR were performed as before but using murine primers. Primers for evaluating splicing were: at the SMRT S1 CoRNR box: Forward (5′-AACAA GAAAC TCAAC ACCCA CAAC-3′) and Reverse (5′-CACAC GGTTG GTGAG TGGTG-3′); at the SMRT S3 CoRNR box: Forward (5′-CAGAT CTGCA CCGAG AAAAG AC-3′) and Reverse (5′-TCACC GGGCT GATGG GCTC-3′); and at the NCoR S3 CoRNR box: Forward (5′-CTGGC TGCTC TTGTG GATGCT-3′) and Reverse (5′-CTGTC CCATT CCCTC TGACTG-3′).
For quantification of total corepressor expression, gene-specific primers were designed for regions of NCoR or SMRT not subject to alternative mRNA splicing, denoted SMRT-forward (5′-ATGGC TAATG AGGCC AACAG-3′) and SMRT-reverse (5′-ACACT GCGAC ACAGT CTTGG-3′) and NCoR-forward (5′-CTGAC AGGCC TCAAG AAAGG-3′) and NCoR-reverse (5′-AACCT GTTCC AGACG TGGTC-3′). DNA standard curves were generated by PCR using the above primers and NB4 cDNA; the resulting PCR products were quantified by optical absorbance and were amplified in parallel with the NB4 cell cDNAs using a DyNamo SYBR Green qPCR kit (New England Biolabs, Ipswich, MA) for 40 cycles (10 s at 95 °C, 15 s at 60 °C, and 30 s at 72 °C), and the relative expression levels of pan-SMRT versus pan-NCoR expression were calculated as described above.
RESULTS
Acute Promyelocytic Leukemic Cells Express a Specific Subset of the Known Corepressor Variants
Most prior studies of the interaction of corepressors with RARα and with the x-RARα fusion proteins were restricted to studies of SMRTα or NCoRω (which were among the first corepressor variants discovered). Subsequent work, however, has shown that the ability of nuclear receptors to interact with corepressor depends on the specific corepressor splice variant employed (15–18, 25). We therefore investigated whether the x-RARα fusions in APL, which are known to display an altered corepressor release by ATRA, were also altered in their affinities for the different corepressor variants.
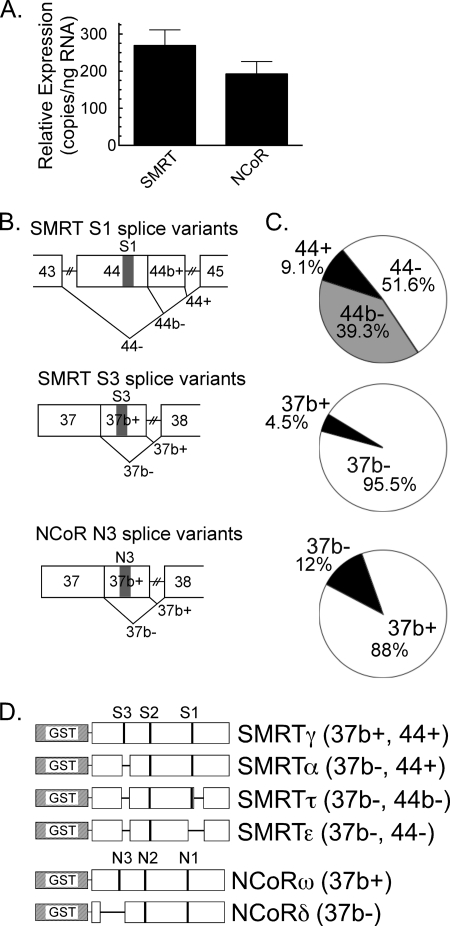
To establish the corepressor variants most relevant in the APL context, we began by analyzing the pattern of corepressor expression in NB4 cells (26). NB4 cells are a standard ex vivo model for human APL, bear a t(15:17) translocation, express detectable levels of the resulting PML-RARα fusion, and differentiate into granulocytes when treated with supraphysiological concentrations of ATRA. We first compared total SMRT expression to total NCoR expression in NB4s by RT-PCR, employing primers directed at conserved exons not influenced by alternative mRNA splicing. Slightly greater total mRNA expression was found for the SMRT locus versus the NCoR locus (Fig. 1A).
FIGURE 1.
SMRT and NCoR splice variants expressed in APL cells. A, comparable expression of SMRT versus NCoR mRNAs. Quantitative RT-PCR and splice-independent primer pairs were used to measure total SMRT and total NCoR mRNA levels in NB4 cells. The mean and S.E. of three experiments are shown. B, alternative splicing events within the SMRT and NCoR receptor interaction domains. Exons (numbered from the 5′ start site), alternative splicing events (V-shaped lines), and CoRNR motifs (S1, S3, or N3) are indicated. C, relative expression of the different SMRT and NCoR mRNA splice variants in NB4 cells. Messenger RNA isolated from NB4 cells was subjected to RT-PCR using primers spanning the splice sites in B; the products were resolved by gel electrophoresis to determine the percentage of each alternatively spliced mRNA produced at each splice site. The means of six experiments are presented. D, schematic of the GST-SMRT and GST-NCoR protein constructs. Exons deleted by alternative mRNA splicing are depicted as horizontal lines. The nomenclature is described in the text. SMRTα and SMRTτ were described previously (17, 18). SMRTγ, SMRTϵ, NCoRω, and NCoRδ were previously referred to as SMRTsp18, SMRTsp2, full-length NCoR, and RIP13Δ1, respectively (25, 46).
We next used RT-PCR primer pairs spanning the alternative splice sites to quantify the different splice variants generated at each corepressor locus. Alternative mRNA splicing at exon 44 gives rise to three distinct versions of SMRT: a full-length S1 CoRNR box (exon 44+), a deletion of S1 flanking sequences (exon 44b−), or an entire excision of the S1 CoRNR box (exon 44−) (Fig. 1B). The 44− SMRT form predominated in the NB4 cells, with the 44b− next most abundant and the full-length, 44+ variant representing a minority of the total population (Fig. 1C, top pie graph). Similarly, alternative splicing either incorporates the S3 CoRNR box into SMRT (exon 37b+) or deletes this motif (exon 37b−); the latter was the dominant version expressed in the NB4 cells (Fig. 1C, middle pie graph). Unlike SMRT, exon 44 of NCoR is not subject to alternative RNA splicing. However, alternative splicing does occur at NCoR exon 37b, either including the N3 CoRNR box (37b+) or eliminating it (37b−) (Fig. 1B); the former version predominated in the NB4 cells (Fig. 1C, bottom pie graph).
Assuming alternative mRNA splicing occurs independently at each corepressor splice site, as has been shown for other cells and tissues, the corepressor variants expressed in these NB4 cells are, in decreasing order of abundance, NCoR (37b+), SMRT (37b−, 44−), SMRT (37b−, 44b−), NCoR (37b−), SMRT (37b+, 44−), SMRT (37b+,44b−), and SMRT (37b+,44+). We assigned Greek symbols to each of these combinatorial splice variants to simplify discussion and analyzed appropriate representatives in our in vitro experiments, below (Fig. 1D).
Homodimers of PML-RARα and of PLZF-RARα Interact with Different Repertoires of Corepressor Variants Compared with Homodimers of RARα
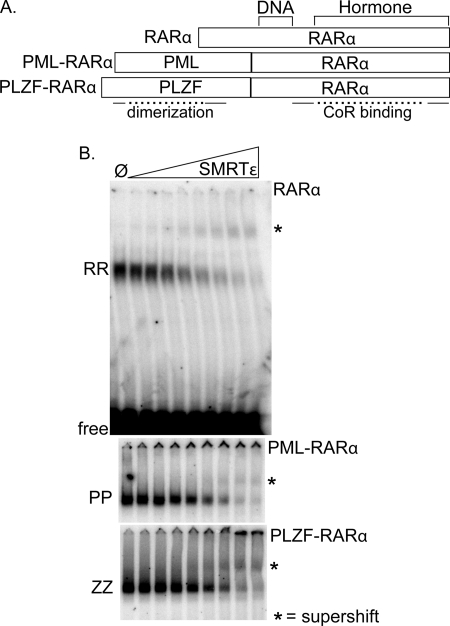
We next employed an EMSA-supershift assay to determine the interaction of wild-type and x-fusion forms of RARα with the different corepressor variants. In these assays, a constant amount of each RARα derivative (shown schematically in Fig. 2A) and 32P-labeled DR5 probe were incubated with increasing concentrations of each of the corepressor constructs depicted in Fig. 1D. A representative electrophoretogram for the different RARα derivatives interacting with the SMRTϵ construct is shown (Fig. 2B). Consistent with prior studies, all three forms of RARα bound to the DR5 DNA probe as homodimers under these conditions (e.g. in the absence of RXR) (27, 28) and were supershifted by SMRTϵ near equally (Fig. 2B). No shifted or supershifted complexes were detected using control extracts (e.g. in the absence of RARα or an x-RARα), and the identity of the various complexes was further confirmed by their interaction with anti-RARα antisera (Ref. 28 and data not shown).
FIGURE 2.
Comparable recruitment of SMRTϵ by RARα, PML-RARα, and PLZF-RARα homodimers. A, schematic representation of RARα, PML-RARα, and PLZF-RARα. DNA-binding, hormone-binding, and corepressor-binding domains in RARα and protein dimerization domains in PML and PLZF are noted. B, comparison of SMRTϵ binding by RARα, PML-RARα, and PLZF-RARα homodimers utilizing EMSA. A fixed amount of 32P-DNA probe and each RARα derivative were mixed with control extract ( ) or with a 2-fold increasing series of GST-SMRTϵ (Fig. 1D). The resulting DNA-protein complexes were resolved by native gel electrophoresis and visualized by phosphorimaging analysis. The positions of the RARα (RR), PML-RARα (PP), or PLZF-RARα (ZZ) homodimer-DNA complexes in the absence of corepressor, and the corresponding corepressor-homodimer-DNA complexes (*) are indicated. The entire electrophoretogram, including the unbound DNA probe (free), is shown for RARα, whereas only the regions containing protein-DNA complexes are shown for PML-RARα and PLZF-RARα.
) or with a 2-fold increasing series of GST-SMRTϵ (Fig. 1D). The resulting DNA-protein complexes were resolved by native gel electrophoresis and visualized by phosphorimaging analysis. The positions of the RARα (RR), PML-RARα (PP), or PLZF-RARα (ZZ) homodimer-DNA complexes in the absence of corepressor, and the corresponding corepressor-homodimer-DNA complexes (*) are indicated. The entire electrophoretogram, including the unbound DNA probe (free), is shown for RARα, whereas only the regions containing protein-DNA complexes are shown for PML-RARα and PLZF-RARα.
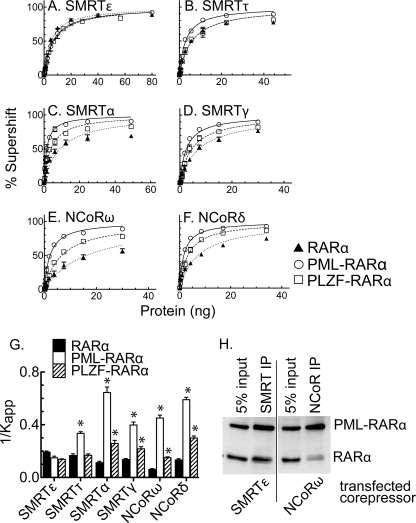
We repeated this same protocol with additional NCoR and SMRT splice variants and quantified the results (Fig. 3). All three forms of RARα and all six different corepressor splice variants displayed hyperbolic binding kinetics, consistent with theoretical expectation for these protein/DNA interactions (Fig. 3, A–F). Apparent Kd values were extracted from these curves and plotted as their reciprocal (1/Kapp) (Fig. 3G); we adopted this convention so that the stronger the affinity, the higher the bar in the plot. In agreement with prior studies (29), wild-type RARα interacted more strongly with SMRTτ than with NCoRω in these assays (Fig. 3, B versus E, dotted curves, and Fig. 3G, filled bars). However, alternative splicing clearly further influenced the apparent affinity of RARα for SMRT-derived versus NCoR-derived corepressors, with RARα binding better to NCoRδ than to NCoRω and better to SMRTϵ than to SMRTα. The difference in affinity for RARα homodimers between the strongest (SMRTϵ) and weakest corepressor (NCoRω) was ∼3-fold. These data indicate that RARα does not preferentially partner with SMRT versus NCoR corepressor per se, but rather both SMRT and NCoR produce splice variants that bind RARα with distinct and partially overlapping affinities.
FIGURE 3.
Differential recruitment of different corepressor variants by RARα, PML-RARα, and PLZF-RARα homodimers. A–F, corepressor binding assayed by EMSA supershift. The experiment in Fig. 2B was repeated using different corepressor variants (Fig. 1D), the results were quantified, and the means and S.E. were calculated (n = 3). G, relative affinities of RARα, PML-RARα, and PLZF-RARα homodimers for the different corepressor variants. The 1/Kapp values were calculated from A–F; the means and S.E. are presented. *, values that differ from that of the corresponding RARα value at a 95% CI. H, corepressor binding by coimmunoprecipitation analysis. Lysates of CV-1 cells expressing RARα, PML-RARα, and the full-length corepressor variant indicated below each panel were immunoprecipitated with antibodies to either SMRT or NCoR, as indicated above each panel. The immunoprecipitates were resolved by SDS-PAGE and visualized by immunoblotting; the positions of the proteins are indicated on the right. Comparable results were observed in three independent experiments. IP, immunoprecipitation.
Notably, both PML-RARα and PLZF-RARα possessed corepressor specificities that were substantially altered compared with that of RARα. Although displaying the same apparent affinity for SMRTϵ as did RARα, PML-RARα exhibited significantly higher affinities for most other splice versions of SMRT and for both splice versions of NCoR (Fig. 3, A–G). Specifically, PML-RARα exhibited a ∼6-fold stronger interaction with SMRTα than did RARα under these conditions and a 7.5-fold stronger interaction with NCoRω (Fig. 3, C, E, and G). The relative affinity of PML-RARα for SMRTτ and for NCoRδ were 2.0- and 4.4-fold stronger, respectively, compared with RARα (Fig. 3, B and F); although less dramatic, significant increases were also observed for the interaction of PLZF-RARα with SMRTα, SMRTγ, NCoRω, and NCoRδ (Fig. 3, C–G). Our results indicate that the x-RARα fusions, rather than exhibiting a generic increase in their affinity for corepressor as proposed previously, have instead undergone a selective shift in their ability to recognize specific corepressor variants.
To determine whether the changes in specificity of RARα and PML-RARα for the different corepressor variants were also observable in a cellular context, we explored the ability of full-length corepressor splice variants to interact with RARα and PML-RARα in CV-1 cells using a coimmunoprecipitation protocol. We simultaneously expressed RARα and PML-RARα together with full-length SMRTϵ or full-length NCoRω. We then immunoprecipitated the corepressor and visualized the amount of associated RARα or PML-RARα by SDS-PAGE and immunoblot (Fig. 3H). Under these conditions, RARα efficiently copurified with SMRTϵ, the corepressor variant with the highest apparent affinity for RARα in our EMSA supershift experiments, but much less efficiently with NCoRω, the corepressor variant with the lowest affinity for RARα in our EMSA supershifts (Fig. 3H). In contrast, PML-RARα efficiently copurified with both corepressor variants (Fig. 3H). These results are consistent with our conclusion that the PML-RARα fusion has gained the ability to recognize corepressor variants that are poorly recognized by RARα.
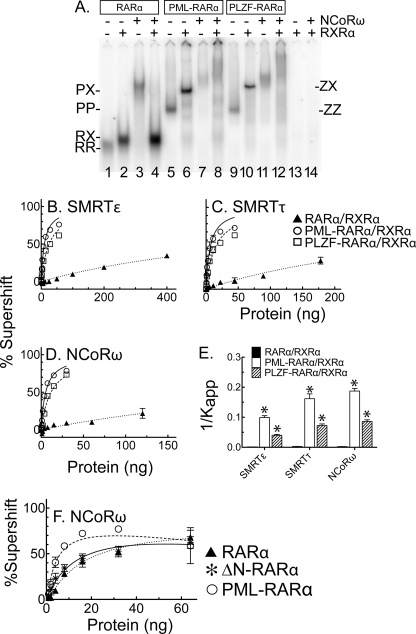
Heteromer Formation with RXRα Further Distinguishes the Different Corepressor Specificities of RARα, PML-RARα, and PLZF-RARα
RARα and the x-RARα fusions bind DNA not only as homodimers but also as heteromers with RXRs. RARα forms a RARα/RXRα heterodimer, whereas PML-RARα and PLZF-RARα generate heterotetramers representing a dimer of dimers (RXRα/x-RARα:x-RARα/RXRα) (28, 30). We next investigated the effect of RXRα on corepressor recruitment by RARα, PML-RARα, and PLZF-RARα using our EMSA/supershift assay. A representative electrophoretogram using a single concentration of the NCoRω construct is presented in Fig. 4A. RXRα, tested alone, resulted in no detectable binding to the DNA probe in either the absence or the presence of corepressor (Fig. 4A, lanes 13 and 14). In the absence of corepressor, RARα/RXRα heterodimers migrated more slowly and displayed higher affinity for the DNA probe than did RARα homodimers (Fig. 4A, compare lanes 1 and 2). Interestingly, these RARα/RXRα heterodimers lost virtually all ability to interact with NCoRω (Fig. 4, compare lanes 3 and 4). The addition of RXRα to either PML-RARα or PLZF-RARα also produced a more slowly migrating complex (Fig. 4A, lane 5 versus lane 6 and lane 9 versus lane 10, respectively) characteristic of a heterotetramer and with a more modest gain in affinity for the DNA probe. In clear contrast to the RARα/RXRα heterodimers, these x-RARα/RXRα heterotetramers retained a significant ability to interact with NCoRω (Fig. 4A, compare lanes 6 and 8 and lanes 10 and 12).
FIGURE 4.
Distinct effects of RXRα on corepressor recruitment by RARα, PML-RARα, and PLZF-RARα. A, representative EMSA supershift using the NCoRω construct. All of the reactions contained a fixed amount of 32P-labeled probe and each RARα or x-RARα protein; RXRα and/or NCoRω was included or omitted as indicated. The positions of the RARα, PML-RARα, or PLZF-RARα homodimers (labeled RR, PP, or ZZ, respectively) and RXRα heteromers (labeled RX, PX, and ZX, respectively) in the absence of corepressor are indicated. B–D, binding of RXRα heteromers of RARα, PML-RARα, or PLZF-RARα to three different corepressor variants. The experiments in Fig. 3 (A, B, and E) were repeated in the presence of RXRα; the means and S.E. are shown (n = 3). E, relative affinities of RXRα heteromers of RARα, PML-RARα, and PLZF-RARα heteromers for the different corepressor variants. The 1/Kapp values were calculated from B–D. The low affinity of RARα/RXRα for corepressor produced a poorer fit to theoretical kinetics than did the x-RARα/RXRα fusions, with R2 values of <0.9. F, corepressor binding by ΔN-RARα homodimers. EMSA supershifts were as in Fig. 3; the means and S.E. are presented (n = 2).
We repeated and quantified these heteromer experiments using a range of corepressor concentrations and focusing on SMRTϵ (recognized near equally by RARα, PML-RARα, and PLZF-RARα homodimers), NCoRω (preferentially recognized by PML-RARα and PLZF-RARα homodimers compared with RARα homodimers), and SMRTτ (preferentially recognized by PML-RARα homodimers but not by PLZF-RARα or RARα homodimers) (Fig. 4, B–E). RARα/RXRα heterodimers displayed a very weak interaction with all three corepressor variants (Fig. 4, B–D, dotted lines); SMRTϵ, the only corepressor with which we could obtain sufficient binding to RARα/RXRα heterodimers to allow determination of an accurate Kapp, displayed a near 160-fold decrease in affinity compared with RARα homodimers (Fig. 4E). In contrast, heterotetramer formation of oncogenic x-RARs with RXRα only slightly diminished their affinity for these corepressor variants (a 2.5–3-fold reduction) compared with the corresponding homodimers (Fig. 4, B–E). As a result, interaction with RXRα further magnified the differences in corepressor recruitment observed when these proteins were assayed as homodimers (compare Figs. 4 and 3).
The Altered Corepressor Specificity of the x-RARα Fusions Is Due to Addition of the x-Sequences Rather than Loss of RARα Sequences
Although the identity of the x-sequence in the oncogenic x-RARα fusions differs with the chromosomal translocation, the site of fusion within the RARα moiety always occurs between codons 60 and 61, deleting the RARα N-terminal domain. Conceivably, the altered corepressor specificity observed for the x-RARα fusions in our studies could arise either from the loss of these N-terminal RARα sequences or from the gain of PML or PLZF sequences. To differentiate these possibilities, we created an ΔN-terminal deletion of RARα identical in location to the fusion site in PML-RARα (deleting amino acids 1–60) and tested it against NCoRω, which showed the greatest difference between RARα homodimers and PML-RARα homodimers. The ΔN-RARα construct behaved like full-length RARα, exhibiting low affinity for NCoRω (Fig. 4F). We conclude that loss of the N-terminal domain does not account for the altered corepressor affinity of PML-RARα and PLZF-RARα; rather the presence of the PML and PLZF sequences is responsible for the increased affinity for NCoRω.
The Altered Corepressor Specificity of the x-RARα Proteins Corresponds with a Gain in Recognition of CoRNR Box 1, a Motif That Is Poorly Recognized by Wild-type RARα
Depending on splice variant, the SMRT and NCoR corepressors contain from 1 to 3 CoRNR boxes; these motifs represent key sites of contact by which nuclear receptors recruit corepressor, and different nuclear receptors preferentially contact different CoRNR boxes (15–18). To better understand the molecular basis behind the alterations in corepressor variant specificity observed for PML-RARα and PLZF-RARα versus RARα, we next investigated the ability of these receptor derivatives to bind to the different CoRNR box motifs.
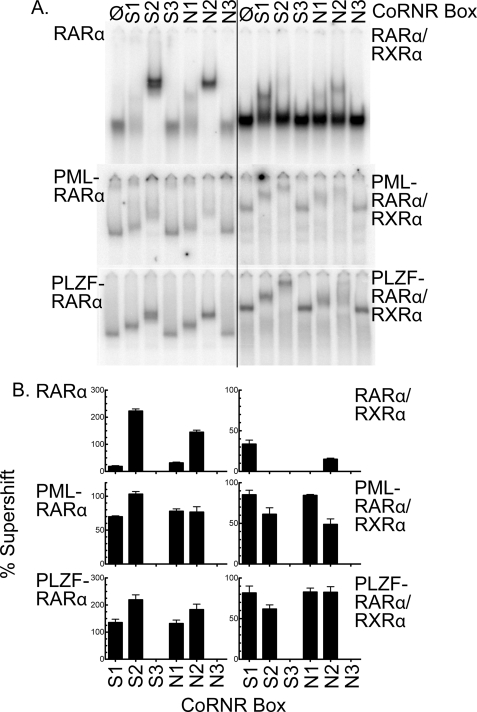
Using our EMSA/supershift protocol, we determined that RARα homodimers were efficiently supershifted by the S2 or N2 CoRNR box constructs, displayed a much weaker interaction with S1 or N1, and exhibited no detectable interaction with S3 or N3 (Fig. 5A, top left panel; and quantified in Fig. 5B). Heterodimerization of RARα with RXRα strongly inhibited binding to either S2 or N2, had no effect on the already very weak interaction with N1, but slightly increased the interaction with S1 (Fig. 5, A and B, top right panels); the latter may reflect the ability of RXRα to interact with the S1 CoRNR box (29). Intriguingly, both PML-RARα homodimers and PLZF-RARα homodimers retained the ability of RARα to bind to S2/N2 and also gained the ability to strongly interact with the S1/N1 CoRNR boxes (Fig. 5, A and B, middle and bottom left panels). This ability to efficiently recognize both S1/N1 and S2/N2 motifs was also observed for the corresponding x-RARα/RXRα heterotetramers (Fig. 5, A and B, middle and bottom right panels). Little or no binding to the S3 or N3 CoRNR box was observed for any RARα derivative. We conclude that at least one basis for the altered specificity of these x-RARα fusions for the different corepressor splice variants reflects an enhanced ability to recognize the most C-terminal CoRNR box motifs present in both SMRT and NCoR.
FIGURE 5.
Enhanced recognition of the S1/N1 CoRNR box motif by the x-RARα fusion proteins. A, binding of individual CoRNR box motifs to RARα, PML-RARα, or PLZF-RARα. EMSA supershifts were performed as in Fig. 4A but using corepressor subdomains limited to the individual S1, S2, S3, N1, N2, or N3 CoRNR motifs (as indicated above the panels). Both receptor homodimers (left panels) and receptor heteromers (right panels) were tested. B, quantification. The experiment in A was repeated (n = 3) and quantified; the means and S.E. are presented.
The Ability of PML-RARα to Fully Release Corepressor in Response to ATRA, a Property Closely Associated with Oncogenesis, Depends on the Splice Variant
PML-RARα has been reported to require significantly more ligand (ATRA) to release SMRT than does RARα, a property that is closely associated with the ability of these high doses of ATRA to induce granulocyte differentiation and disease remission in APL (8–10). However, the effects of alternative corepressor splicing were not determined in these prior experiments. Given the change in corepressor specificity of PML-RARα versus RARα in the absence of agonist, we next sought to determine whether there were also differences in the release of the different variants in response to different retinoids. We tested RARα and PML-RARα using SMRTϵ (representing a corepressor recognized strongly by both RARα and PML-RARα) and NCoRω (representing a corepressor preferentially recognized by PML-RARα compared with RARα). Both receptor homodimers and heteromers with RXRα were examined, as were a series of different retinoids.
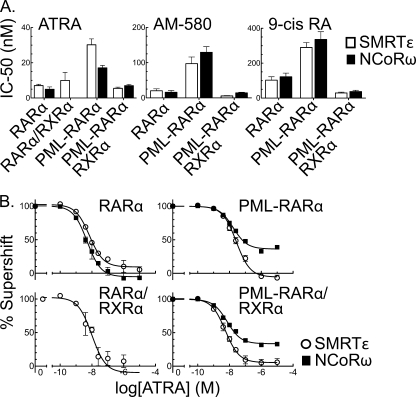
ATRA released both SMRTϵ and NCoRω from RARα homodimers at an IC50 of 5–6 nm (Fig. 6A, left panel); this value is consistent with published Kd values (31, 32). In contrast, release of either SMRTϵ or NCoRω from PML-RARα homodimers required 3–4-fold more ATRA than that required for the RARα homodimers (significant at the 95% CI) (Fig. 6A, left panel). Although RARα/RXRα heterodimers bound SMRTϵ corepressor very poorly, they released SMRTϵ with approximately the same IC50 value as did RARα homodimers (Fig. 6A, left panel; the binding of RARα/RXRα to NCoRω in the absence of hormone was too weak to permit determination of an accurate IC50 for the ATRA-driven release of this second corepressor variant). Intriguingly, heterotetramer formation by PML-RARα reduced the IC50 for corepressor release by ATRA back to values approximating those observed for RARα/RXRα heterodimers (Fig. 6A, left panel). Similar changes in the IC50 values required for corepressor release by PML-RARα compared with RARα were observed using two alternative RAR agonists: AM-580 and 9-cis-retinoic acid (Fig. 6A, middle and right panels).
FIGURE 6.
Differences in the ability of agonist to release different corepressor variants from RARα and PML-RARα. A, ATRA-mediated release of corepressor: calculated IC50 values. The EMSA supershifts in Fig. 4A were repeated using SMRTϵ or NCoRω (Fig. 1D) and increasing concentrations of the retinoid agonists ATRA, AM-50, or 9-cis-retinoic acid (9-cis RA). The agonist concentration required to release 50% of each corepressor (IC50) from the receptor-DNA complex was calculated; the means and S.E. are presented. B, ATRA-mediated release of corepressor: plotted results. The data that generated the IC50 values in A are plotted as the percentage of receptor/corepressor complex remaining at each ATRA concentration (corepressor complex formed minus agonist is defined as 100%); the means and S.E. are presented (n = 7 for PML-RARα and RARα; n = 3 for PLZF-RARα). The values slightly below 0% reflect minor changes in receptor-DNA affinity caused by binding of agonist.
A more detailed examination of the ATRA-induced corepressor release curves yielded an additional, unexpected observation. Although SMRTϵ was virtually completely released from PML-RARα at high ATRA concentrations, NCoRω was not fully displaced at even the highest ATRA concentration (10 μm) tested (Fig. 6B). This same phenomenon also observed for PML-RARα/RXRα heterotetramers and for AM-580 and 9-cis-retinoic acid but not for RARα (Fig. 6B and data not shown). Our results indicate that PML-RARα not only requires higher levels of agonist to release corepressor than does RARα but also, through its acquired ability to recognize additional corepressor variants such as NCoRω, has gained an additional, hormone-refractory component in its interaction with corepressor. It is plausible that this additional, agonist-resistant corepressor interaction may contribute to the aberrant repression properties of PML-RARα that are believed to underlie its function in the leukemic cell.
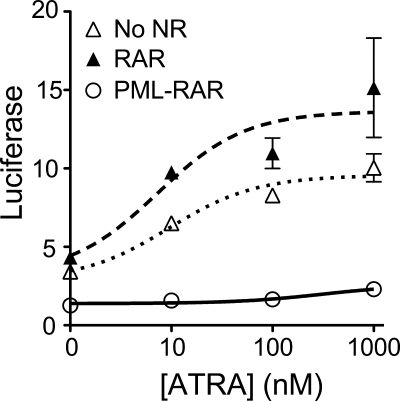
PML-RARα Displays a Hormone-refractile Repression in Transfected Cells
To determine whether the hormone-refractile release of NCoRω in vitro had parallels in vivo, we assayed the transcriptional properties of RARα and PML-RARα in transfected cells. As reported previously (23), CV1 cells express moderate levels of endogenous RARs and can activate expression of a DR5-luciferase reporter in response to ATRA (Fig. 7). Cointroduction of an ectopic RARα expression vector into these cells enhanced this ATRA-responsive reporter gene expression. In contrast, ectopic expression of PML-RARα repressed reporter gene expression at all hormone levels tested, including ATRA concentrations that, in vitro, were sufficient to release SMRTϵ but that allowed retention of NCoRω (compare Figs. 6B and 7). We suggest that this agonist-resistant transcriptional repression by PML-RARα observed in cells reflects the agonist-resistant NCoRω interaction seen in vitro.
FIGURE 7.
ATRA-refractory repression of reporter gene expression by PML-RARα. A DR5-tk-luciferase reporter was transiently transfected into CV1 cells together with expression vectors for RARα, PML-RARα, or an empty vector control, as indicated. The cells were subsequently incubated with the ATRA concentrations indicated, harvested 24 h later, and assayed for luciferase activity. The means and standard errors for three independent experiments are shown.
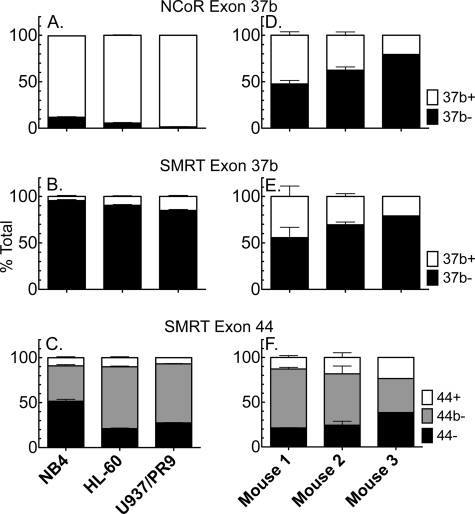
The Different Corepressor Variants Are Expressed at Different Levels in Different Leukemias
Given the altered corepressor specificity of PML-RARα, we investigated whether the abundance of the relevant corepressors might differ in different APLs and/or in different forms of leukemia. The exon 37b+ splicing event that produces NCoRω was by far the dominant form of this corepressor in all three human leukemic lines examined: NB4, HL60 (a promyelocytic leukemia-derived line lacking an x-RARα fusion), and U937/PR89 (a monocytic leukemia cell line engineered to express PML-RARα) (19, 20) (Fig. 8A). Reciprocally, the analogous exon 37b+ splicing event that produces SMRTγ was expressed at very low levels in the same three human lines, whereas the ratios of the different exon 44 splice forms (responsible for SMRTα, τ, and ϵ) varied, with the exon 44− splice form (e.g. SMRTϵ) lowest in HL60s (Fig. 8, B and C). We also compared expression of the different corepressor variants in three individual APLs obtained from a mouse PML-RARα transgenic model (24). Despite originating in genetically identical mice, there was moderate variation in the relative ratios of the different splice forms among the three samples, and in contrast to the human-derived leukemias, the murine APLs expressed the 37b− and 37b+ forms of both SMRT and NCoR at more equal ratios than did the human-derived leukemias (Fig. 8, D–F). In summary, all of the leukemias examined expressed substantial quantities of the corepressor variants that represent de novo high affinity partners for the PML-RARα fusions but in relative abundance that varied among the different leukemias examined.
FIGURE 8.
Expression of different corepressor splice variants in different leukemias. RNAs were isolated from three different human leukemia-derived cell lines (NB4, HL60, and U937/PR9, A–C) and from three different individual mouse leukemias arising from transgenic expression of human PML-RARα (D–F). These RNAs were then analyzed by reverse transcription-PCR as in Fig. 1B to determine the relative levels of the NCoR and SMRT splice variants indicated. The means and standard deviations of three separate RT reactions are presented, except for mouse 3, which yielded sufficient RNA for only one determination.
DISCUSSION
Acute Promyelocytic Leukemia and x-RAR Fusions
Virtually all human APLs characterized carry a chromosomal translocation that creates an x-RARα fusion (6, 7). The translocation breakpoint in RARα inevitably occurs at a recombinational hot spot that deletes the first 60 codons of the receptor, leaving the RARα DNA-binding and hormone-binding domains intact. The identity and nature of the N-terminal x-sequence, however, can vary considerably. Significantly, all of the x-fusion sequences encode self-dimerization domains (8, 9, 11). It has been proposed that the dominant-negative properties of the x-RARα fusions result, at least in part, from this enhanced ability to homodimerize or oligomerize, which, through an unknown mechanism, is thought to stabilize corepressor recruitment under conditions that favor corepressor release by wild-type RARα (8–12). This aberrant corepressor retention by the x-RARα fusions is thought to be a crucial mechanism by which x-RARα fusions mediate leukemogenesis; forcing corepressor release from PML-RARα with supraphysiological levels of ATRA, for example, drives APL cells into differentiation and produces disease remission in patients. Proteolysis of the fusion protein and induction of apoptosis also contribute to clinical resolution of APL (33).
The results reported here support this overall model of x-RARα function but with several significant modifications. Foremost, we observed that the ability of RARα and x-RARα fusions to recruit and release corepressors is highly dependent on the specific corepressor variant employed. For example, RARα homodimers interact well with SMRTϵ but relatively weakly with NCoRω. PML-RARα and PLZF-RARα homodimers, in contrast, interact strongly with all of the corepressor variants tested and display notably elevated interactions with SMRTα, SMRTγ, NCoRω, and NCoRδ. Further, whereas heterodimer formation with RXRα virtually abolishes the ability of RARα to recruit corepressors, the heterotetramers formed between RXRα and PML-RARα or PLZF-RARα retain substantial corepressor binding; this phenomenon helps explain how x-RARα proteins can recruit corepressors and exert dominant-negative effects in cells that express RXRs. Finally, our studies have revealed that the gain of NCoR recognition by PML-RARα results in a novel, ATRA-refractory corepressor release phenotype that may contribute to the impaired differentiation observed in APL cells. Taken as a whole, our results suggest that it is a change in corepressor specificity, not a higher affinity for all corepressors per se, that helps confer the repressive and oncogenic properties observed for these x-RARα fusions.
The x-RARα Fusions Result in Alterations in Receptor Affinity for Specific Corepressor Splice Variants
The SMRT and NCoR gene products share 50% identity at the amino acid level and undergo additional diversification through alternative mRNA splicing that modifies the CoRNR box motifs responsible for interaction with nuclear receptors (15–18). Previous studies demonstrated that RARα preferentially binds SMRTτ compared with NCoRω (34, 35). Our new results confirm this but further reveal that RARα homodimers bind to several other SMRT and NCoR splice variants, including SMRTϵ. Interestingly, SMRTϵ contains only a single S2 CoRNR box, and studies in our lab suggest that SMRTϵ may in fact represent an RARα-specific corepressor variant.4
We determined that although PML-RARα and PLZF-RARα share the ability of RARα to bind SMRTϵ, they recruit most other splice variants of SMRT and NCoR more strongly than does RARα; many of these newly recognized splice variants are highly expressed in APL cells and in other leukemia-derived cell lines. This alteration in the corepressor specificity of RARα versus x-RARα is not due to the loss of the RARα N terminus but rather due to the addition of the PML and PLZF sequences. Corepressors bind to nuclear receptors primarily through contacts between α-helical CoRNR box motifs in SMRT and NCoR and an agonist-labile docking site within the hormone binding domain of the receptor (36, 37). Notably RARα interacted primarily with CoRNR box 2 in SMRT and NCoR, whereas the x-RARα fusions also efficiently bound to CoRNR box 1; this gain in CoRNR box 1 recognition accounts, at least in part, for the broadened corepressor splice variant repertoire of these x-RARα proteins. Although the precise mechanism behind this enhanced recognition of CoRNR box 1 is unresolved, it is likely that the x-RARα homodimers display their corepressor docking surfaces in a different conformation or with different accessibility than do RARα homodimers, resulting in a more promiscuous CoRNR box recognition.
Heteromer Formation with RXRα Further Distinguishes the Corepressor Repertoire of PML-RARα and PLZF-RARα from That of RARα
Heterodimer formation with RXR inhibits corepressor binding by many nuclear receptors; this is likely to reflect, in part, an intrinsic occlusion of the corepressor docking site in RXR by helix 12 (38, 39). In our hands the RARα/RXRα heterodimers exhibited a greater than 150-fold loss in affinity for certain corepressors compared with RARα homodimers. Consistent with this observation, DNA response elements unable to bind RARα homodimers, such as the β-RARE, favor transcriptional activation over repression.5 The addition of RXRα to PML-RARα or to PLZF-RARα results in the formation of heterotetramers: two x-RARα/RXRα heterodimers held together through the homodimerization contacts within the x-moieties (40–42). Significantly, these x-RARα/RXRα heterotetramers did not display the severe inhibitory effects on corepressor affinity observed for RARα/RXRα heterodimers. As a consequence, the presence of RXRα exacerbated the divergences in corepressor recruitment observed for RARα versus the x-RARαs. For example, the difference in affinity for NCoRω between RARα and PML-RARα was 7.5-fold; the difference between RARα/RXRα and PML-RARα/RXRα was more than 100-fold.
The unusual dimer-of-dimers topology of the x-RARα/RXRα heterotetramers is likely to account for the retention of corepressor binding compared with RARα/RXRα heterodimers. Trimeric and tetrameric complexes of other nuclear receptors, for example, also display alterations in their coregulator interaction properties compared with the corresponding dimers (22, 43). This phenomenon may reflect the location or multiplicity of the coregulator binding surfaces on these higher order complexes, or it may represent an allosteric phenomenon arising from the different protein-DNA or protein-protein contacts made within these higher order complexes compared with the contacts that stabilize receptor dimers. These results help explain how x-RARα proteins can be recruited to many of their target genes as RXRα heteromers, as has been inferred from genome-wide chromosome immunoprecipitation studies (19, 44), yet retain the ability to recruit corepressors and to mediate repression. It should be noted, however, that x-RARα homodimers also strongly recruit the corepressor variants studied here, and these homodimers may also contribute to certain aspects of the APL phenotype. As a result, the precise transcriptional properties of PML-RARα may change depending upon the target gene and whether PML-RARα heteromers or homodimers are the main species recruited.
ATRA, Corepressor Variants, and Corepressor Release
PML-RARα has been reported to require significantly higher levels of ATRA to release SMRTα compared with RARα (9), a phenomenon that parallels the requirement for these supraphysiological ATRA levels to relieve dominant-negative inhibition by PML-RARα and to drive APL cells into granulocyte differentiation and disease remission. Here, we have shown that this same phenomenon applies to SMRTϵ, which requires 3–4-fold more ATRA to release from PML-RARα than from RARα when these receptors are assayed as homodimers. Interestingly, heterotetramer formation with RXRα restored ATRA sensitivity to PML-RARα to approximately that observed for RARα homodimers and RARα/RXRα heterodimers. However, PML-RARα, either as a homodimer or as a heterotetramer with RXRα, displayed an incomplete ability to release NCoRω even at saturating ATRA concentrations. PML-RARα/RXRα heteromers are thought to be the dominant receptor species operative in APL cells (40, 41); given that SMRTϵ release by these heterodimers is near normal, this inefficient release of NCoRω may be help explain the impaired ability of the APL cells to respond to physiological levels of ATRA. These results also suggest that the ability of hormone to release a given corepressor variant may differ on different target genes (depending on whether PML-RARα heteromers or homodimers are the main species recruited) and in the presence of different levels of RXR expression. In support of this model, we observed that ATRA concentrations able to fully release SMRTϵ in vitro failed to fully reverse PML-RARα-mediated repression in transfected cells, a consequence, we suggest, of the hormone-refractory association of PML-RARα with NCoRω.
More globally, many of the changes observed for the x-RARα fusions compared with their wild-type RARα progenitors are gains-of-function that result in a widening of their recognition properties. PML-RARα recognizes a wider variety of DNA elements than does RARα (45) and with less discrimination between homomeric and RXR-heteromeric binding affinity. As reported here, both PML-RARα and PLZF-RARα bind a wider variety of corepressor variants than does RARα, also with less discrimination between homomeric and RXR heteromeric binding affinities. The widening of corepressor recognition by these x-RARα, fusions has potential repercussions not only in regard to release by ATRA but also in the response of these functions to extracellular signaling cascades. Many, but not all, corepressor variants are functionally inactivated by phosphorylation through the MAPK signaling cascade, which causes release of these corepressors from the nuclear receptor (46, 47). Arsenite is used clinically in the treatment of APL and has been shown to induce partial differentiation of APL cells, in part through activation of this MAPK cascade and the release of SMRT from PML-RARα (48). The specific SMRT variants operating in this context and the possible contributions of NCoR family members were not determined in this prior study. The work presented here reveals that x-RARα fusion proteins actually recruit a mix of corepressors that includes variants both sensitive and resistant to inactivation by this MAPK cascade. It will be valuable to determine how this broadening of the corepressor repertoire of PML-RARα and PLZF-RARα impacts the response of leukemic cells to both ATRA and arsenite, whether changes in the repertoire of corepressor variants expressed in different patients or at different stages of the disease can alter presentation or prognosis, and whether targeting therapies to specific corepressor variants can improve future clinical interventions in APL.
Acknowledgments
We thank Scott Kogan for the generous gift of mouse PML-RARα APL cells, Joost Martens and Henk Stunnenberg for the generous gift of PR9 cells, and Liming Liu for superb technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grant RO1DK53528.
B. J. Mengeling, M. L. Goodson, and M. L. Privalsky, unpublished observations.
B. J. Mengeling and M. L. Privalsky, unpublished observations.
- RAR
- retinoic acid receptor
- APL
- acute promyelocytic leukemia
- RXR
- retinoid X receptor
- ATRA
- all-trans-retinoic acid
- RARE
- retinoic acid response element.
REFERENCES
- 1. Privalsky M. L. (2008) in NR Coregulators and Human Diseases (Kumar R., O'Malley B. W. eds) pp. 243–280, World Scientific, Hackensack, NJ [Google Scholar]
- 2. Germain P., Chambon P., Eichele G., Evans R. M., Lazar M. A., Leid M., De Lera A. R., Lotan R., Mangelsdorf D. J., Gronemeyer H. (2006) Pharmacol. Rev. 58, 712–725 [DOI] [PubMed] [Google Scholar]
- 3. Perissi V., Jepsen K., Glass C. K., Rosenfeld M. G. (2010) Nat. Rev. Genet. 11, 109–123 [DOI] [PubMed] [Google Scholar]
- 4. Lazar M. A. (2003) Nucl. Recept. Signal 1, e001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ordentlich P., Downes M., Evans R. M. (2001) Curr. Top. Microbiol. Immunol. 254, 101–116 [DOI] [PubMed] [Google Scholar]
- 6. Collins S. J. (2008) Curr. Opin. Hematol. 15, 346–351 [DOI] [PubMed] [Google Scholar]
- 7. Redner R. L. (2002) Leukemia 16, 1927–1932 [DOI] [PubMed] [Google Scholar]
- 8. Minucci S., Maccarana M., Cioce M., De Luca P., Gelmetti V., Segalla S., Di Croce L., Giavara S., Matteucci C., Gobbi A., Bianchini A., Colombo E., Schiavoni I., Badaracco G., Hu X., Lazar M. A., Landsberger N., Nervi C., Pelicci P. G. (2000) Mol. Cell 5, 811–820 [DOI] [PubMed] [Google Scholar]
- 9. Lin R. J., Evans R. M. (2000) Mol. Cell 5, 821–830 [DOI] [PubMed] [Google Scholar]
- 10. Guidez F., Ivins S., Zhu J., Söderström M., Waxman S., Zelent A. (1998) Blood 91, 2634–2642 [PubMed] [Google Scholar]
- 11. Sternsdorf T., Phan V. T., Maunakea M. L., Ocampo C. B., Sohal J., Silletto A., Galimi F., Le Beau M. M., Evans R. M., Kogan S. C. (2006) Cancer Cell 9, 81–94 [DOI] [PubMed] [Google Scholar]
- 12. Kwok C., Zeisig B. B., Dong S., So C. W. (2006) Cancer Cell 9, 95–108 [DOI] [PubMed] [Google Scholar]
- 13. Kakizuka A., Miller W. H., Jr., Umesono K., Warrell R. P., Jr., Frankel S. R., Murty V. V., Dmitrovsky E., Evans R. M. (1991) Cell 66, 663–674 [DOI] [PubMed] [Google Scholar]
- 14. de Thé H., Lavau C., Marchio A., Chomienne C., Degos L., Dejean A. (1991) Cell 66, 675–684 [DOI] [PubMed] [Google Scholar]
- 15. Sutanto M. M., Symons M. S., Cohen R. N. (2006) Mol. Cell Endocrinol. 259, 43–49 [DOI] [PubMed] [Google Scholar]
- 16. Short S., Malartre M., Sharpe C. (2005) Biochem. Biophys. Res. Commun. 334, 845–852 [DOI] [PubMed] [Google Scholar]
- 17. Goodson M. L., Jonas B. A., Privalsky M. L. (2005) J. Biol. Chem. 280, 7493–7503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Goodson M., Jonas B. A., Privalsky M. A. (2005) Nucl. Recept. Signal 3, e003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Martens J. H., Brinkman A. B., Simmer F., Francoijs K. J., Nebbioso A., Ferrara F., Altucci L., Stunnenberg H. G. (2010) Cancer Cell 17, 173–185 [DOI] [PubMed] [Google Scholar]
- 20. Grignani F., Ferrucci P. F., Testa U., Talamo G., Fagioli M., Alcalay M., Mencarelli A., Peschle C., Nicoletti I. (1993) Cell 74, 423–431 [DOI] [PubMed] [Google Scholar]
- 21. Rosen M. D., Privalsky M. L. (2009) Mol. Endocrinol. 23, 1183–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mengeling B. J., Lee S., Privalsky M. L. (2008) Mol. Cell Endocrinol. 280, 47–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Farboud B., Hauksdottir H., Wu Y., Privalsky M. L. (2003) Mol. Cell Biol. 23, 2844–2858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kogan S. C., Hong S. H., Shultz D. B., Privalsky M. L., Bishop J. M. (2000) Blood 95, 1541–1550 [PubMed] [Google Scholar]
- 25. Malartre M., Short S., Sharpe C. (2004) Nucleic Acids Res. 32, 4676–4686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lanotte M., Martin-Thouvenin V., Najman S., Balerini P., Valensi F., Berger R. (1991) Blood 77, 1080–1086 [PubMed] [Google Scholar]
- 27. Phan T. Q., Jow M. M., Privalsky M. L. (2010) Mol. Cell Endocrinol. 319, 88–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hauksdóttir H., Privalsky M. L. (2001) Cell Growth Differ. 12, 85–98 [PMC free article] [PubMed] [Google Scholar]
- 29. Wong C. W., Privalsky M. L. (1998) Mol. Cell Biol. 18, 5724–5733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Perez A., Kastner P., Sethi S., Lutz Y., Reibel C., Chambon P. (1993) EMBO J. 12, 3171–3182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Xiao J. H., Durand B., Chambon P., Voorhees J. J. (1995) J. Biol. Chem. 270, 3001–3011 [DOI] [PubMed] [Google Scholar]
- 32. Allenby G., Janocha R., Kazmer S., Speck J., Grippo J. F., Levin A. A. (1994) J. Biol. Chem. 269, 16689–16695 [PubMed] [Google Scholar]
- 33. Nasr R., Lallemand-Breitenbach V., Zhu J., Guillemin M. C., de Thé H. (2009) Clin. Cancer Res. 15, 6321–6326 [DOI] [PubMed] [Google Scholar]
- 34. Cohen R. N., Brzostek S., Kim B., Chorev M., Wondisford F. E., Hollenberg A. N. (2001) Mol. Endocrinol. 15, 1049–1061 [DOI] [PubMed] [Google Scholar]
- 35. Cohen R. N., Putney A., Wondisford F. E., Hollenberg A. N. (2000) Mol. Endocrinol. 14, 900–914 [DOI] [PubMed] [Google Scholar]
- 36. Perissi V., Staszewski L. M., McInerney E. M., Kurokawa R., Krones A., Rose D. W., Lambert M. H., Milburn M. V., Glass C. K., Rosenfeld M. G. (1999) Genes Dev. 13, 3198–3208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hu X., Lazar M. A. (1999) Nature 402, 93–96 [DOI] [PubMed] [Google Scholar]
- 38. Zhang J., Hu X., Lazar M. A. (1999) Mol. Cell Biol. 19, 6448–6457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zamir I., Zhang J., Lazar M. A. (1997) Genes Dev. 11, 835–846 [DOI] [PubMed] [Google Scholar]
- 40. Zhu J., Nasr R., Pérès L., Riaucoux-Lormière F., Honoré N., Berthier C., Kamashev D., Zhou J., Vitoux D., Lavau C., de Thé H. (2007) Cancer Cell 12, 23–35 [DOI] [PubMed] [Google Scholar]
- 41. Zeisig B. B., Kwok C., Zelent A., Shankaranarayanan P., Gronemeyer H., Dong S., So C. W. (2007) Cancer Cell 12, 36–51 [DOI] [PubMed] [Google Scholar]
- 42. Kamashev D., Vitoux D., De Thé H. (2004) J. Exp. Med. 199, 1163–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lin B. C., Wong C. W., Chen H. W., Privalsky M. L. (1997) J. Biol. Chem. 272, 9860–9987 [DOI] [PubMed] [Google Scholar]
- 44. Wang K., Wang P., Shi J., Zhu X., He M., Jia X., Yang X., Qiu F., Jin W., Qian M., Fang H., Mi J., Yang X., Xiao H., Minden M., Du Y., Chen Z., Zhang J. (2010) Cancer Cell 17, 186–197 [DOI] [PubMed] [Google Scholar]
- 45. Zhou J., Pérès L., Honoré N., Nasr R., Zhu J., de Thé H. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 9238–9243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jonas B. A., Varlakhanova N., Hayakawa F., Goodson M., Privalsky M. L. (2007) Mol. Endocrinol. 21, 1924–1939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jonas B. A., Privalsky M. L. (2004) J. Biol. Chem. 279, 54676–54686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hong S. H., Yang Z., Privalsky M. L. (2001) Mol. Cell Biol. 21, 7172–7182 [DOI] [PMC free article] [PubMed] [Google Scholar]