Abstract
Gelonin-based immunotoxins vary widely in their cytotoxic potency as a function of antigen density, target cell internalization and trafficking kinetics, and conjugate properties. We have synthesized novel gelonin immunotoxins using two different binding scaffold types (single-chain antibody variable fragments and fibronectin domains) targeting two different tumor antigens (carcinoembryonic antigen and EGF receptor). Constructs were characterized using an antigen-negative cell line (HT-1080), cell lines positive for each antigen (HT-1080(CEA) for carcinoembryonic antigen and A431 for EGF receptor), and a cell line positive for both antigens (HT-29). Immunotoxins exhibited Kd values between 8 and 15 nm and showed 20–2000-fold enhanced cytotoxicity compared with gelonin (IC50 ∼ 0.25–30 nm versus 500 nm). Using quantitative fluorescence flow cytometry, we measured internalization of gelonin (via pinocytosis) and gelonin-based immunotoxins (via antigen-dependent, receptor-mediated endocytosis). Results were matched with cytotoxicity measurements made at equivalent concentration and exposures. Unexpectedly, when matched internalization and cytotoxicity data were combined, a conserved internalized cytotoxicity curve was generated that was common across experimental conditions. Considerable variations in antigen expression, trafficking kinetics, extracellular immunotoxin concentration, and exposure time were all found to collapse to a single potency curve on the basis of internalized immunotoxin. Fifty percent cytotoxicity occurred when ∼5 × 106 toxin molecules were internalized regardless of the mechanism of uptake. Cytotoxicity observed at a threshold internalization was consistent with the hypothesis that endosomal escape is a common, highly inefficient, rate-limiting step following internalization by any means tested. Methods designed to enhance endosomal escape might be utilized to improve the potency of gelonin-based immunotoxins.
Keywords: Antibodies, Anticancer Drug, Drug Transport, Fusion Protein, Intracellular Trafficking, Protein Translocation, Toxins, Tumor Therapy, Immunotoxin, Gelonin
Introduction
Immunotoxins are a promising approach to the targeted delivery of highly potent, cancer-specific, cytotoxic agents. Immunotoxins are frequently composed of a targeting moiety (derived from antibodies or other cell-binding proteins) either chemically conjugated or genetically fused to highly cytotoxic plant or bacterial protein toxins. Clinical success for immunotoxins has been mostly limited to hematological malignancies due to transport limitations in solid tumors (1). Such limitations have been extensively studied experimentally (2) and with several computational models (3, 4).
The potency of a particular immunotoxin is dependent on the ability to deliver the toxin to the cytoplasm, which is commonly considered to be the rate-limiting step. For some native toxins such as ricin, intracellular delivery is achieved through lectin binding, followed by internalization and toxin release with membrane fusion or retrograde trafficking (5). Immunotoxins attempt to recreate this scenario by replacing the indiscriminate lectin binding with cancer-specific antigen binding as a means of targeting and internalization (6). Subsequent intracellular trafficking, release, and endosomal escape are often achieved using existing toxin characteristics, translocation domains, protease cleavage sites, disulfide bonds, and/or signaling peptides (7–10). However, the inclusion of toxins with domains facilitating cytoplasmic access can also lead to increased nonspecific toxicity in vivo (11, 12).
Gelonin is a plant toxin and classified as a type I ribosome-inactivating protein because it lacks any cell-binding or cytoplasmic delivery domains. Recombinant gelonin (rGel)2 is an ∼30-kDa N-glycosidase with activity similar to the ricin A chain but exhibiting better stability and lower immunogenicity (13, 14). The use of rGel in tumor-targeted cytotoxic agents has been well studied (15, 16). Furthermore, rGel has been shown to be active without cleavage from the binding domain and without negative impact on the targeting agent's pharmacokinetics (17). The necessity of internalization for activity of rGel immunotoxins and the antigen to which it is directed have been demonstrated previously (18, 19).
Carcinoembryonic antigen (CEA) is a 180-kDa membrane glycoprotein that exhibits depolarized overexpression in numerous epithelial tumor types (20). The utility of CEA as a tumor-targeting tool for both therapy and imaging has been well established (21–23). Experiments in our laboratory have shown that CEA is internalized with a half-life between 10 and 16 h and thus represents a potential target for immunotoxins (24). CEA has been used previously as a target for the early development of immunotoxins (25–27). The EGF receptor (EGFR) has a strikingly faster internalization rate (∼30 min). However, many such internalized molecules return to the cell surface by recycling (28). Like CEA, EGFR is a well established cancer-associated antigen. EGFR has also been used as a target for designed immunotoxins (29, 30). Previous studies have suggested that antigens displaying similar expression levels but different internalization rates can lead to profoundly different immunotoxin potencies (6).
MFE-23 is an antibody single-chain variable fragment (scFv) directed against CEA. Originally identified by phage library selection, this scFv was later humanized by resurfacing and engineered in yeast for greater stability and solubility (shMFE) as well as affinity (sm3E) (31–33). Both of these engineered molecules are well expressed in yeast and have Kd values of ∼7 nm and ∼30 pm, respectively. The tenth human fibronectin type III domain (Fn3) has been designed using various directed evolution approaches for specific affinity toward numerous different targets (34–36). We describe engineered fibronectin fragments binding EGFR and CEA (designated E246 and C743, respectively).
In this study, we generated several novel immunotoxins targeting CEA and EGFR, including the first published report of Fn3-based immunotoxins. Comparing the different immunotoxin constructs, we investigated the mechanisms of cellular intoxication, including the cell binding-dependent internalization of immunotoxins and the subsequent loss of cell viability. Using a novel analysis of viability versus net internalized antigen, a universal potency relationship was found that was independent of the antigen, binding affinity/scaffold, internalization/recycling rate, external immunotoxin concentration, and incubation time. This work may be useful in understanding the mechanisms of immunotoxin-based cell killing and which factors influence cellular intoxication. With a better understanding of these mechanisms and factors, we can engineer more effective agents as cell-targeted therapeutics for cancer.
EXPERIMENTAL PROCEDURES
Cell Lines
The human fibrosarcoma cell line HT-1080 was used throughout as an antigen-negative control. HT-1080 cells were transfected with a plasmid for CEA expression, and those cells, denoted HT-1080(CEA), were maintained under antibiotic selection pressure from Geneticin as described previously (24). The human epidermoid carcinoma cell line A431 was used as an EGFR-positive line, and the human colorectal carcinoma cell line HT-29 was used as a double-positive cell line for both CEA and EGFR.
Construction of Expression Plasmids
The gene encoding the recombinant form of the gelonin toxin was codon-optimized for Escherichia coli expression and ordered from DNA 2.0 (Menlo Park, CA). The gene was digested out of the synthetic vector using designed PstI and HindIII restriction sites and cloned into the pMal-c2x expression vector encoding a maltose-binding protein fusion product. Into this construct, Fn3 clones were inserted by amplification of Fn3 genes out of their own expression vectors and using the purified amplification products as primers for a QuikChangeTM insertion similar to the protocol described by Geiser et al. (37). The linker between Fn3 and rGel was modified to consist strictly of the amino acids encoded by the necessary restriction sites for binder cloning and a G4S sequence. In this setting, various Fn3 and scFv genes could be inserted by restriction digestion and cloning using NheI and BamHI. As an alternative vector for expression, we cloned the immunotoxin construct by EcoRI and HindIII digestion into pET32a (Novagen), expressing the product as a TrxA fusion. We further modified this vector by mutating the protease site designed to remove the fusion tag from enterokinase to tobacco etch virus.
Protein Expression and Purification
The pMal-c2x vector containing the rGel gene was transformed into Origami 2(DE3) (Novagen, San Diego, CA) and grown on LB-agar plates containing ampicillin and tetracycline. Colonies were picked from the plate and grown overnight at 37 °C in 5-ml aliquots of selective media, which were then used to seed 1 liter of antibiotic-free rich LB medium and allowed to grow to logarithmic phase. When the culture reached an A600 between 0.5 and 1.0, 5 ml of 0.1 m isopropyl 1-thio-β-d-galactopyranoside was added, and the induction was allowed to continue at 37 °C for 4 h. Following induction, cultures were centrifuged at 15,000 × g for 12 min, and cell pellets were frozen at −20 °C. Pellets were resuspended in amylose column buffer containing Complete EDTA-free protease inhibitor (Roche Applied Science) and then sonicated on a Branson Sonifier 450 at 50% duty cycle and power level 5 for three 1-min intervals. The resulting solution was centrifuged at 50,000 × g for 30 min to pellet cell debris, and the supernatant was applied to an amylose column as described by the manufacturer (New England Biolabs, Ipswich, MA). Purified recombinant proteins were concentrated, and the buffer was exchanged into Factor Xa digestion buffer using Amicon columns with a 30-kDa molecular mass cutoff and then incubated overnight at room temperature with 5 μl of Factor Xa (New England Biolabs). rGel was isolated from the removed maltose-binding protein and Factor Xa by ion-exchange chromatography with a HiTrap Q column (GE Healthcare).
For production of immunotoxins, the Rosetta-gami 2(DE3) bacterial host cell line was used, and chloramphenicol was added to plates and overnight growth medium as a selection agent. For Fn3-rGel immunotoxins, all other purification steps were the same as for rGel expression. In the case of scFv immunotoxins, the TrxA fusion construct was used to facilitate more efficient formation of stabilizing disulfide bonds. In this case, induction was carried out in standard LB medium using 10 ml 0.1 m isopropyl 1-thio-β-d-galactopyranoside at 20 °C for 6 h. Purification of TrxA-scFv-rGel was achieved using immobilized metal affinity chromatography with TALON resin (Clontech, Mountain View, CA). The TrxA tag was removed by digestion with tobacco etch virus protease, scFv-rGel was isolated from TrxA and tobacco etch virus by size-exclusion chromatography on Superdex 200 and 75 10/300 columns connected in series.
Yields for rGel were ∼3 mg/liter after all purification steps. We synthesized two Fn3 immunotoxins, one targeting CEA (C7rGel) and one targeting EGFR (E4rGel), based on the affinity-matured parent Fn3 fragments C743 and E246. Yields for C7rGel and E4rGel were ∼2.2 and ∼3 mg/liter, respectively. We synthesized two scFv immunotoxins (3ErGel and FErGel), both targeting CEA, based on disulfide-stabilized versions of the affinity-matured scFv fragments sm3E and shMFE (33). Yields for 3ErGel and FerGel were ∼125 and ∼750 μg/liter, respectively. Relative yields of each of the different immunotoxins and analysis by PAGE suggests that the primary reason for differences in yields may be the proper folding and solubility for each immunotoxin. The disulfide-stabilized scFv fragments appeared to disrupt folding (and thus soluble expression yields) to a greater degree than Fn3 fragments (although some of this difference may be due to alternative fusion partners TrxA versus maltose-binding protein). Similarly, the 3E scFv destabilizes to a greater extent than the FE scFv in the context of the immunotoxin.
Antigen Binding Affinity Titration
Immunotoxins were biotinylated using amine-reactive EZ-Link Sulfo-NHS-LC-biotin (Pierce). Antigen-positive cell lines HT-1080(CEA) and A431 were lifted from culture plates with trypsin and resuspended in 4% formalin for 30 min before being washed and stored in PBS with 1% (w/v) BSA. Fixed cells were incubated with varying concentrations of biotinylated immunotoxins overnight at 37 °C, washed once, and resuspended in 250 μl of PBS with 1% (w/v) BSA with 1 μl of goat anti-biotin-FITC antibody (Sigma) for 1 h at 4 °C. Cells were washed once again and resuspended in 150 μl of PBS with 1% (w/v) BSA before being analyzed for fluorescence on an Accuri C6 flow cytometer. For each titrating concentration,the median fluorescent intensity was determined, and data sets for each immunotoxin were fitted to a standard binding isotherm using least-squares regression.
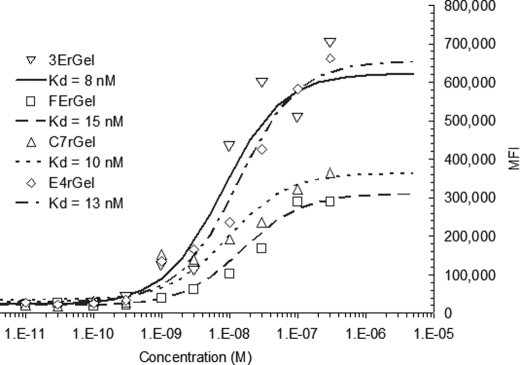
Early attempts to titrate directly labeled immunotoxins on fixed cells resulted in a high nonspecific background signal. Alternative approaches to titration were attempted before we eventually arrived at the secondary detection method described above. Titration of all immunotoxins (see Fig. 1) was performed on either fixed HT-1080(CEA) cells or A431 cells. Titration of the scFv immunotoxins 3ErGel and FErGel resulted in fitted Kd values of 8 and 15 nm, respectively, which were somewhat higher than the titrated affinity of their parent sm3E and shMFE scFv fragments (Kd ∼ 30 pm and 9 nm, respectively) (data not shown). We fitted the titration data for Fn3 immunotoxins C7rGel and E4rGel and found Kd values of 10 and 13 nm, respectively. These Kd values were also slightly higher than those found for the parent Fn3 fragments, 2 nm for C743 and 3 nm for E246.3 Differences in overall signal were attributed to differences in the degree of biotinylation of immunotoxins. Biotinylated rGel showed no significant signal when titrated over the same range of concentrations.
FIGURE 1.
Antigen affinity of binding fragments is retained in their respective immunotoxin constructs. The four immunotoxins target either CEA or EGFR via either scFv or Fn3. The 3E and FE scFv clones targeting CEA, the C7 fibronectin clone targeting CEA, and the E4 fibronectin clone targeting EGFR were each fused to rGel and titrated for binding affinity on HT-1080(CEA) or A431 cells for CEA or EGFR, respectively. Binding on fixed cells was detected with goat anti-biotin-FITC antibody by flow cytometry. Data were fitted using least-squares regression with a binding isotherm, giving Kd values of 8 nm for 3ErGel, 15 nm for FErGel, 10 nm for C7rGel, and 13 nm for E4rGel.
Cytotoxicity
Log-phase tumor cells were removed by trypsinization, counted, and seeded on 96-well plates at 2500 cells/well. Cells were allowed to adhere overnight, after which fresh growth medium containing varying concentrations of rGel or immunotoxin was added to triplicate wells. Toxins were incubated with the cells for 72 h before the toxin-containing medium was removed and replaced with medium containing the WST-1 reagent (Roche Applied Science). The assay was allowed to develop for 1–3 h under normal culture conditions, after which plates were measured for absorbance at 450 nm. Untreated cells and cells lysed with a 1% Triton X-100 solution were used as positive and negative controls, respectively. Measurements were compared with the base line and normalized to control treatments, triplicates were averaged, and standard errors were calculated. Time-dependent cytotoxicity data were obtained by treating cells as described, removing the toxin-containing medium, washing once with PBS, replacing with fresh medium for wells at each time point, and then following identical assay procedures after 72 h. rGel cytotoxicity measurements were made on concentrations between 1 × 10−9 and 3 × 10−6 m and times between 1 and 72 h. High antigen-expressing cells were incubated with 10 nm scFv immunotoxin or 30 nm Fn3 immunotoxin, whereas the low antigen-expressing line was incubated with 30 nm immunotoxin, with all incubations lasting between 12 and 72 h.
Quantitative Internalization
Cell lines were incubated with immunotoxins directly labeled with Alexa Fluor 488. At various times, cells were washed with PBS and incubated for 30 min with the quenching rabbit anti-Alexa Fluor 488 (Invitrogen). Cells were then scraped from the wells, washed again with PBS, and analyzed for an internal fluorescent signal. Quantum Simply Cellular anti-mouse IgG beads (Bangs Laboratories, Fishers, IN) with different quantified binding capacities were incubated with Alexa Fluor 488-labeled mouse IgG for 30 min, washed with PBS, and then measured for fluorescence. The number of fluorescent molecules/protein on both immunotoxins and mouse IgG was determined using absorbance measurements at 280 and 494 nm. Bead fluorescence measurements were used to generate a standard curve for fluorescent signal/fluorescent molecule. Immunotoxin internalization data were quantified by mapping the fluorescent signal to the bead fit and converting the signal to fluorescent molecules and then translated into immunotoxin molecules using the labeling ratio.
Internalized Cytotoxicity
Data obtained in time-dependent cytotoxicity experiments were combined with those from quantitative internalization experiments and plotted to suggest the former as the dependent variable and the latter as the independent variable. Accumulated results were fitted using nonlinear least-squares regression of an exposure-response curve with variable slopes using MATLAB (MathWorks, Natick, MA). From this fitting, a near-full response metric was calculated and reported.
RESULTS
Novel Gelonin-based Immunotoxins
Four new immunotoxins were constructed that target CEA or EGFR. Three different immunotoxins targeting CEA were constructed using antibody scFv fragments (3ErGel and FErGel) or a newly selected fibronectin scaffold-based binding domain with specificity for CEA (C7rGel). 3ErGel incorporates a high-affinity anti-CEA scFv (sm3E) described previously (33), whereas FErGel incorporates a lower affinity precursor scFv (shMFE). As separate scFv molecules, the affinities of these two binding modules are 30 pm (sm3E) and 9 nm (shMFE). Using a yeast-displayed library of fibronectin scaffold proteins with randomized loops, a new binding module targeting CEA was isolated (C743) with a Kd of 2 nm, and another module that binds EGFR (E246) was isolated with a Kd of 3 nm. Each of these four binding modules was expressed as N-terminal fusions to rGel (see “Experimental Procedures”), and binding of the resultant immunotoxin constructs was assessed with tumor cell lines (Fig. 1).
Cytotoxicity of rGel and Immunotoxins
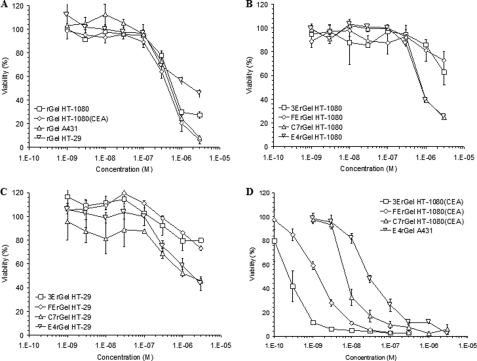
The results of concentration-dependent cytotoxicity (Fig. 2) suggest that immunotoxin potency varied over 2 orders of magnitude and was determined by a complex combination of antigen density, binding affinity, binding scaffold, and antigen internalization/recycling rate. On all cell lines tested, gelonin showed IC50 values of ∼500 nm (Fig. 2A). We found that all immunotoxins had an IC50 of ∼1 μm on the antigen-negative HT-1080 cell line (Fig. 2B). When incubated with the double-positive low antigen-expressing HT-29 cell line, all of the immunotoxins again displayed IC50 values of ≥1 μm (Fig. 2C). The exposure response curves of Fig. 2D are consistent with previous results that apparent immunotoxin potency varies widely when expressed solely as a function of the extracellular concentration of the agent and that each of the immunotoxins shows a different IC50 on the high antigen-expressing cell lines HT-1080(CEA) and A431 (3ErGel = 250 pm, FErGel = 1.5 nm, C7rGel = 8 nm, and E4rGel = 30 nm).
FIGURE 2.
Fusion of scFv and Fn3 binding domains to rGel leads to enhanced cytotoxicity specific for antigen-positive cells. A, using the WST-1 assay, the cytotoxicity of soluble rGel was tested on all four cell lines used in the study. Across all cell lines, rGel showed an IC50 of ∼500 nm. B, antigen-negative cells (HT-1080) were treated with the four different immunotoxins that displayed roughly equivalent potency to soluble toxin. C, immunotoxins were also tested for cytotoxicity on the double-positive, low-antigen density HT-29 cell line. Surprisingly, none of the immunotoxins showed enhanced cytotoxicity compared with the IC50 of rGel. D, on high antigen-expressing cells (HT-1080(CEA) and A431), significantly greater potency was observed for the immunotoxins compared with the soluble toxin. Against cells expressing their respective antigen targets, they had IC50 values of 250 pm for 3ErGel, 1.5 nm for FErGel, 8 nm for C7rGel, and 30 nm for E4rGel.
rGel Time-dependent Internalization, Cytotoxicity, and Internalized Cytotoxicity
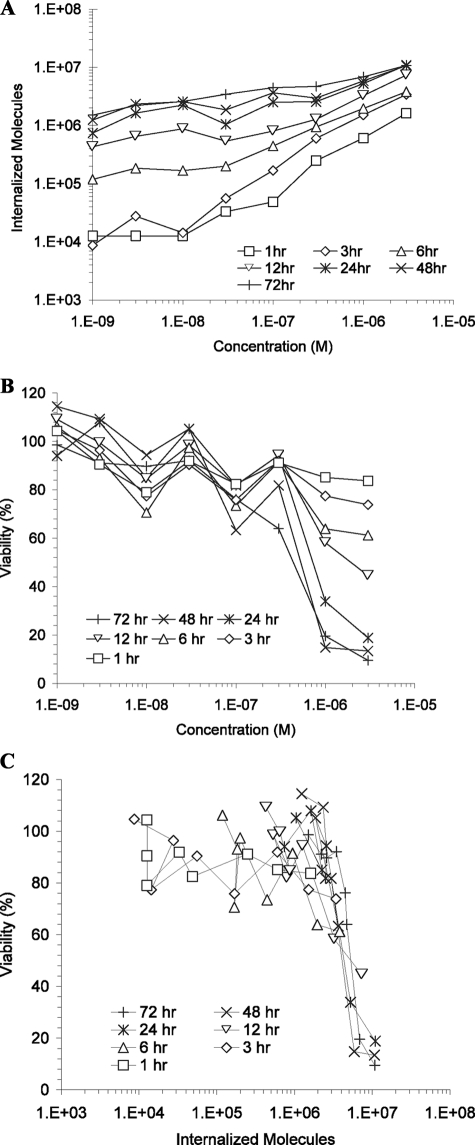
rGel was incubated with HT-1080 cells for various lengths of time up to 72 h and assessed for both internalization and cytotoxicity as described above, after which data were combined to determine the internalized cytotoxicity profile (Fig. 3). Time-dependent quantitative internalization results (Fig. 3A) indicate that, for very low concentrations and very short times, a minimum signal of ∼1 × 104 molecules could be detected, likely due to autofluorescence from the cells above the base-line bead autofluorescence. As treatment concentrations and incubation times were increased, the number of internalized rGel molecules increased, peaking at nearly 1 × 107 molecules at 3 μm for >24 h. Time-dependent cytotoxicity results (Fig. 3B) show a similarly consistent theme in which low-concentration, short-time treatments consistently resulted in viabilities between 80 and 100%. At the highest concentrations and longest times, viability dropped as low as 10%, whereas 50% viability was achieved either by 3 μm treatment for 12 h or by lower concentration treatments for somewhat longer durations. By combining time-dependent quantitative internalization and cytotoxicity data to remove time as a variable, an internalized cytotoxicity profile was obtained (Fig. 3C). For the HT-1080 cells treated with rGel, we found a wide variation in uptake and viability as a function of concentration and time (Fig. 3, A and B). These data collapsed to a fairly tight relationship between viability and the number of internalized gelonin molecules, and we noted a steep reduction in viability at ∼5 × 106 internalized molecules (Fig. 3C).
FIGURE 3.
Correlated internalization and cytotoxicity measurements indicate that a precise number of rGel molecules must be internalized by a single cell before cytotoxicity is observed. A, time and concentration dependence of rGel internalization by HT-1080 cells using the described quantitative internalization flow cytometry assay. B, concentration- and exposure-matched cytotoxicity was measured using the WST-1 assay. C, data from A and B were combined and plotted to show the dependence of cytotoxicity on the number of internalized gelonin molecules, resulting in the determination of the TN50 near 5 × 106.
Immunotoxin Time-dependent Internalization, Cytotoxicity, and Internalized Cytotoxicity
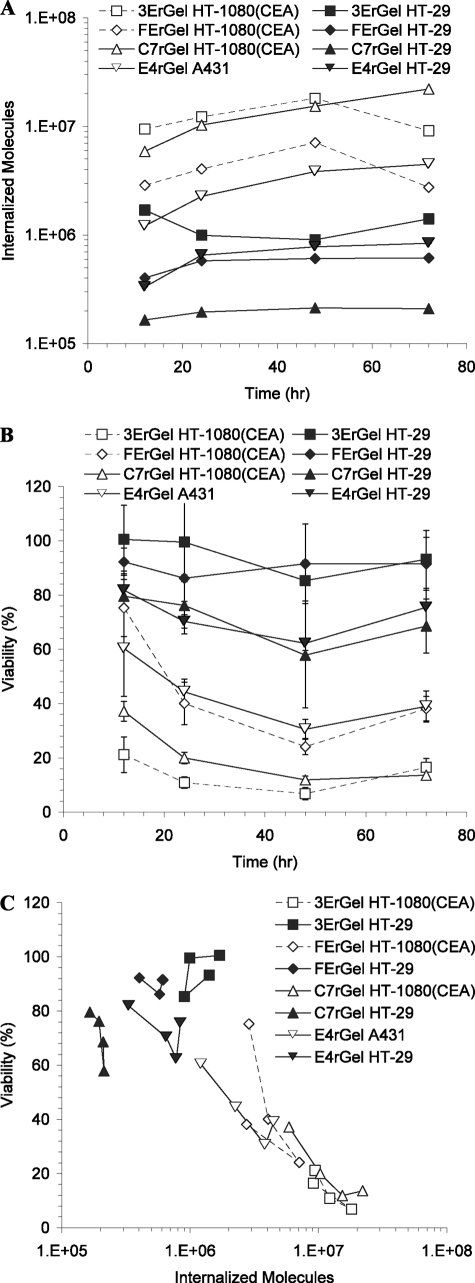
High antigen-expressing HT-1080(CEA) and A431 cell lines, as well as the double-positive low antigen-expressing HT-29 cell line, were incubated with immunotoxins targeting the appropriate antigens for various times at the concentrations selected to show a wide change in viability over the range in time. We assessed cells for immunotoxin internalization as well as viability following incubations as described above, and the data from the two measurements were combined to determine the internalized cytotoxicity of the immunotoxins (Fig. 4). In Fig. 4, A and B, a wide variation in uptake and viability with the different cell lines and immunotoxins was observed. However, the combined data for internalized cytotoxicity surprisingly produced a curve consistent with that for pinocytosed rGel, with a sharp reduction in viability once cells internalized >5 × 106 molecules.
FIGURE 4.
Rate-limiting toxicity step measured in rGel is similarly observed in scFv- and Fn3-targeted immunotoxins using antigen-dependent internalization. A, the internalization of immunotoxins by antigen-positive cells was measured by the quantitative internalization flow cytometry assay at varying times and concentrations. All HT-29 cells treatments were made at 30 nm, as were the C7rGel and E4rGel treatments on HT-1080(CEA) and A431 cells, respectively, whereas the 3ErGel and FErGel treatments on HT-1080(CEA) cells were made at 10 nm. For all treatments, strictly antigen-dependent internalization is reported by subtracting signal from cells blocked with an unlabeled competitor. B, concentration- and exposure-matched cytotoxicity was measured using the WST-1 assay. C, data from A and B were combined and plotted to show the dependence of cytotoxicity on the number of internal immunotoxins, resulting in the determination of the TN50 near 3 × 106 for all species.
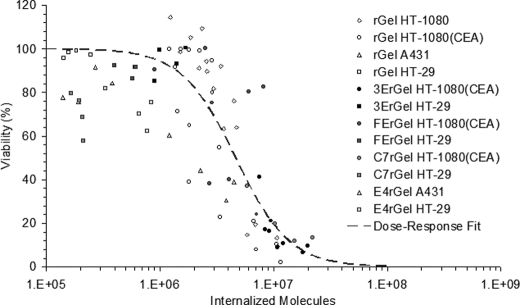
Cumulative Internalized Cytotoxicity
Data collected from multiple experiments using each of the described cell lines with various concentrations and exposures of rGel or one of the immunotoxins were matched for internalized toxin levels and cytotoxicity as described and combined into one cumulative internalized cytotoxicity plot (Fig. 5). The entire data set was accounted for irrespective of experimental conditions and used to fit an exposure-response curve with variable slopes of the form y = 100/(1 + (x/TN50)slope), where both TN50 and slope are fitted parameters. Fitting the exposure-response curve parameters resulted in a TN50 of 4.68 × 106 and a slope of 1.86. The TN50 fit describes the number of internalized immunotoxins necessary to induce a 50% loss of viability, in contrast to the traditional LC50 metric (38). Additionally, in situations such as this where we observe a strong step-function response characterized by a large slope parameter, it is useful to consider values associated with a more complete response. Here, we consider TN90 as calculated from the fit using the equation TN90 = TN50(90/(100 − 90))1/slope.
FIGURE 5.
Aggregated data from experiments using different binding scaffolds, antigen targets, affinities, and cell lines all converge to the same TN50 curve. Shown are the accumulated internalized cytotoxicity data for rGel on all cell lines and immunotoxins on high and low antigen-expressing cells. The cumulative data set was fit using an exposure-response curve with variable slopes giving a TN50 of 4.7 × 106.
The resulting TN90 indicates that 1.53 × 107 molecules of toxin must be internalized on average for 90% of the population to undergo apoptosis. In other words, when the average population uptake reaches TN90, only one-tenth of the population has failed to achieve the ∼5 × 106 molecule limit for toxicity. In Fig. 5, the plotted cumulative internalized cytotoxicity data are each a median value of a Gaussian distribution of cells internalizing different numbers of toxin molecules as measured by flow cytometry. Thus, for each data point, half of the cells in the treatment population internalized more than the recorded number, and half took up less. So at TN50, 50% of the cells died, and 50% internalized at least ∼5 × 106 molecules, indicating that this is indeed the average threshold for loss of viability in a single cell.
DISCUSSION
The importance of internalization for the efficacy of immunotoxins is well appreciated (6, 18, 39, 40). However, these studies clearly indicate that a type I toxin such as gelonin exhibits quantitatively the same low intracellular potency regardless of the particular pathway that drives internalization. Delivery of the cytotoxic domain of an immunotoxin to the cytoplasm of a cell is dependent upon a series of steps, each with a varying degree of efficiency depending on the cell type, antigen density, binding affinity, internalization/recycling, and subcellular trafficking or endosomal escape. Because of the enzymatic potency of ribosome-inactivating proteins, only a few toxin molecules successfully delivered to the cytoplasm (or ribosomal compartment) may be lethal to the cell (41). Quantitative measurement of levels this low is exceedingly difficult, and it is therefore difficult to directly determine a rate of translocation or escape of molecules into the active compartment. Alternatively, we chose to quantify the total intracellular level of immunotoxins and infer their cytoplasmic access by the resulting cytotoxicity. Strikingly, there is an apparently near-universal requirement for ∼5 × 106 gelonin molecules to be internalized to kill a tumor cell, regardless of the route of vesicular internalization.
Build-up of such a significant number of toxins intracellularly, given the efficiency of the enzyme once in the cytoplasm, suggests not only that binding and internalization of immunotoxin are quite efficient but also that endosomal escape is the rate-limiting step in the process of intoxication. If endosomal escape were more efficient, cells would become intoxicated much faster, and such high levels of internalization would not be observed. Nevertheless, it should be acknowledged that these data might also be explained by a rate-limiting degradation of toxin assuming retention of fluorescence.
Here, we have synthesized a novel set of immunotoxins based on binding domain fusions to the plant toxin gelonin. We used disulfide-stabilized scFv fragments engineered for affinity toward CEA as well as Fn3 fragments engineered for affinity toward CEA and EGFR. To our knowledge, this is one of the first reports of Fn3-based immunotoxins. Studies describing rGel fusions with fibronectin fragments targeting IGF1R were reported recently (50). We examined the ability of these binding scaffolds to retain affinity within the fusion construct and found that although affinity was retained to a considerable degree, there was some loss of affinity compared with the parent molecule, likely attributable to partial misfolding and instability in the fusion construct. This is consistent with work by others designing direct fusion immunotoxins (42, 43). We believe that this misfolding and instability also influence the synthetic yields of each of the immunotoxins. Those constructs showing the greatest loss of parent affinity also have the lowest yields. Despite losses of relative affinity, all of the immunotoxins still bound their respective antigens with low nanomolar affinities.
Development of the new class of Fn3-based immunotoxins is an attempt to overcome two of the most substantial limitations to immunotoxin therapy. Commonly, these therapies are limited by vascular toxicity and a failure to infiltrate the tumor mass. The smaller Fn3 scaffold (∼10 kDa) should allow for more rapid clearance from the vasculature, thereby minimizing exposure, and maximize the diffusion coefficient to optimize tumor penetration under fast clearance conditions. It is well established that molecules smaller than the 60–70-kDa molecular mass cutoff for renal filtration are rapidly cleared (44). Although this clearance itself limits total tumor targeting, our labortory's mathematical models suggest that the smaller molecular mass should lead to a more homogeneous immunotoxin distribution within the tumor (4). We plan to test whether these hypotheses regarding the pharmacokinetics of Fn3 immunotoxins are validated by in vivo experiments. This work and others have shown that the Fn3 scaffold is capable of mediating antigen-specific binding at affinities on par with scFv or IgG at a fraction of the molecular mass.
Antigen binding by scFv or Fn3 domains was sufficient to enhance gelonin potency toward antigen-positive cells between 20- and 2000-fold depending on scaffold, affinity, antigen internalization/recycling, and antigen density. Immunotoxins showed no increase in cytotoxicity compared with rGel on antigen-negative cells and cells expressing low levels of antigens. These results support previous examples using scFv fusions to enhance toxin potency (17) and validate Fn3 as a new targeting agent with improved stability and expression, in part due to an absence of disulfide bonds, for immunotoxin design. Fn3-based immunotoxins have the added advantage of being smaller in size, allowing them to potentially better penetrate tumors due to improved capillary permeability and diffusion.
In an attempt to better understand the subcellular barrier to cytotoxicity, we measured the molecular internalization of rGel in HT-1080 cells. Using identical treatments, we determined the cytotoxicity resulting from the measured internalization. As toxin concentrations and incubation times were increased, the number of molecules internalized per cell increased, and the population viability decreased. When viability was plotted as a function of internalized molecules, a strong nonlinear relationship was found, independent of incubation time and concentration. We hypothesize that the number of internalized molecules required to induce a 50% loss in viability, TN50, is determined primarily by the rate of endosomal escape and cytoplasmic access. Conversely, one might have expected that antigen binding might actually deter the rate of intoxication following internalization, based on the success of methods enhancing cytotoxicity by incorporating release elements between binding and toxin domains (45, 46). However, the overlap in the internalized immunotoxin cytotoxicity curve for antigen-bound and internalized immunotoxin versus pinocytosed free gelonin indicates that release of immunotoxins from antigen binding plays a negligible role in intoxication.
The described technique for determining TN50 does not address the precise fate of the immunotoxins following endocytosis. Murphy and co-workers (16, 47, 48) have modeled the intoxication process for diphtheria and gelonin immunotoxins in great subcellular detail and fitted model parameters to protein synthesis inhibition data, finding that the translocation rate constant for gelonin, ∼5 × 10−8 min−1, is several orders of magnitude slower than that for diphtheria alternatives and independent of the targeting moiety. In this work, they concluded that for every 106 internalized gelonin immunotoxin molecules, only one reaches the cytoplasm (16), a result in striking quantitative agreement with the measured TN50 of ∼5 × 106 in the present work. It should be noted that those experiments focused on protein synthesis inhibition rather than a direct measure of cell viability. Utilizing the reported technique, it is likely that immunotoxins incorporating toxins with dedicated translocation domains will show lower TN50 values depending on the efficiency of their endosomal escape machinery. However, such added functionality is likely to contribute directly to off-target toxicity in vivo.
The subcellular barrier to delivery is common to all therapeutic macromolecules requiring cytoplasmic access. Many groups interested in the delivery of siRNA have investigated various tools for overcoming this barrier, including endosome-disrupting polymers (49). Our measurements of the number of toxin molecules required to achieve a single translocation event should be a useful assay for investigating the potential of these tools to enhance intracellular delivery in a quantitative way.
In the future, assessment of novel immunotoxin constructs should be more thorough when potency is evaluated both by the traditional extracellular concentration response metric and the internalized cytotoxicity measurement presented here. Having designed novel potent immunotoxins, engineering efforts should now be properly directed toward designing either molecules with improved intracellular uptake (e.g. tumor delivery/penetration, antigen density, binding affinity, internalization kinetics, and tumor retention) or ones that have improvements in the efficiency of intracellular trafficking to the biologically active compartment. These results should have a significant impact on the rational design of immunotoxins and their combination therapies.
This work was supported, in whole or in part, by National Institutes of Health Grants CA96504 and CA101830. This work was also supported by a pilot project grant from the Massachusetts Institute of Technology Center for Cancer Nanotechnology Excellence and by National Science Foundation graduate research fellowships (to C. M. P. and B. J. H.).
B. Hackel, unpublished data.
- rGel
- recombinant gelonin
- CEA
- carcinoembryonic antigen
- EGFR
- EGF receptor
- scFv
- single-chain variable fragment
- Fn3
- fibronectin type III domain.
REFERENCES
- 1. Pastan I., Hassan R., FitzGerald D. J., Kreitman R. J. (2007) Annu. Rev. Med. 58, 221–237 [DOI] [PubMed] [Google Scholar]
- 2. Ackerman M. E., Pawlowski D., Wittrup K. D. (2008) Mol. Cancer Ther. 7, 2233–2240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wenning L. A., Murphy R. M. (1999) Biotechnol. Bioeng. 62, 562–575 [PubMed] [Google Scholar]
- 4. Thurber G. M., Schmidt M. M., Wittrup K. D. (2008) Adv. Drug Delivery Rev. 60, 1421–1434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Roberts L. M., Lord J. M. (2004) Mini-Rev. Med. Chem. 4, 505–512 [DOI] [PubMed] [Google Scholar]
- 6. Du X., Beers R., Fitzgerald D. J., Pastan I. (2008) Cancer Res. 68, 6300–6305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kondo T., FitzGerald D., Chaudhary V. K., Adhya S., Pastan I. (1988) J. Biol. Chem. 263, 9470–9475 [PubMed] [Google Scholar]
- 8. Keller J., Heisler I., Tauber R., Fuchs H. (2001) J. Controlled Release 74, 259–261 [DOI] [PubMed] [Google Scholar]
- 9. Rathore D., Batra J. K. (1996) Biochem. Biophys. Res. Commun. 222, 58–63 [DOI] [PubMed] [Google Scholar]
- 10. Seetharam S., Chaudhary V. K., FitzGerald D., Pastan I. (1991) J. Biol. Chem. 266, 17376–17381 [PubMed] [Google Scholar]
- 11. Kreitman R. J., Hassan R., Fitzgerald D. J., Pastan I. (2009) Clin. Cancer Res. 15, 5274–5279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kreitman R. J., Stetler-Stevenson M., Margulies I., Noel P., Fitzgerald D. J., Wilson W. H., Pastan I. (2009) J. Clin. Oncol. 27, 2983–2990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Stirpe F., Olsnes S., Pihl A. (1980) J. Biol. Chem. 255, 6947–6953 [PubMed] [Google Scholar]
- 14. Rosenblum M. G., Kohr W. A., Beattie K. L., Beattie W. G., Marks W., Toman P. D., Cheung L. (1995) J. Interferon Cytokine Res. 15, 547–555 [DOI] [PubMed] [Google Scholar]
- 15. Scott C. F., Jr., Lambert J. M., Goldmacher V. S., Blatter W. A., Sobel R., Schlossman S. F., Benacerraf B. (1987) Int. J. Immunopharmacol. 9, 211–225 [DOI] [PubMed] [Google Scholar]
- 16. Yazdi P. T., Murphy R. M. (1994) Cancer Res. 54, 6387–6394 [PubMed] [Google Scholar]
- 17. Rosenblum M. G., Cheung L. H., Liu Y., Marks J. W., 3rd (2003) Cancer Res. 63, 3995–4002 [PubMed] [Google Scholar]
- 18. Goldmacher V. S., Scott C. F., Lambert J. M., McIntyre G. D., Blättler W. A., Collnhson A. R., Stewart J. K., Chong L. D., Cook S., Slayter H. S. (1989) J. Cell Physiol. 141, 222–234 [DOI] [PubMed] [Google Scholar]
- 19. Lambert J. M., Senter P. D., Yau-Young A., Blättler W. A., Goldmacher V. S. (1985) J. Biol. Chem. 260, 12035–12041 [PubMed] [Google Scholar]
- 20. Hammarström S. (1999) Semin. Cancer Biol. 9, 67–81 [DOI] [PubMed] [Google Scholar]
- 21. Wegener W. A., Petrelli N., Serafini A., Goldenberg D. M. (2000) J. Nucl. Med. 41, 1016–1020 [PubMed] [Google Scholar]
- 22. Sharma S. K., Pedley R. B., Bhatia J., Boxer G. M., El-Emir E., Qureshi U., Tolner B., Lowe H., Michael N. P., Minton N., Begent R. H., Chester K. A. (2005) Clin. Cancer Res. 11, 814–825 [PubMed] [Google Scholar]
- 23. Kraeber-Bodéré F., Rousseau C., Bodet-Milin C., Ferrer L., Faivre-Chauvet A., Campion L., Vuillez J. P., Devillers A., Chang C. H., Goldenberg D. M., Chatal J. F., Barbet J. (2006) J. Nucl. Med. 47, 247–255 [PubMed] [Google Scholar]
- 24. Schmidt M. M., Thurber G. M., Wittrup K. D. (2008) Cancer Immunol. Immunother. 57, 1879–1890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Levin L. V., Griffin T. W., Childs L. R., Davis S., Haagensen D. E., Jr. (1987) Cancer Immunol. Immunother. 24, 202–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Avila A. D., Mateo de Acosta C., Lage A. (1989) Int. J. Cancer 43, 926–929 [DOI] [PubMed] [Google Scholar]
- 27. Akamatsu Y., Murphy J. C., Nolan K. F., Thomas P., Kreitman R. J., Leung S. O., Junghans R. P. (1998) Clin. Cancer Res. 4, 2825–2832 [PubMed] [Google Scholar]
- 28. Sorkin A., Goh L. K. (2009) Exp. Cell Res. 315, 683–696 [DOI] [PubMed] [Google Scholar]
- 29. Di Massimo A. M., Di Loreto M., Pacilli A., Raucci G., D'Alatri L., Mele A., Bolognesi A., Polito L., Stirpe F., De Santis R. (1997) Br. J. Cancer 75, 822–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Beers R., Chowdhury P., Bigner D., Pastan I. (2000) Clin. Cancer Res. 6, 2835–2843 [PubMed] [Google Scholar]
- 31. Chester K. A., Begent R. H., Robson L., Keep P., Pedley R. B., Boden J. A., Boxer G., Green A., Winter G., Cochet O., et al. (1994) Lancet 343, 455–456 [DOI] [PubMed] [Google Scholar]
- 32. Boehm M. K., Corper A. L., Wan T., Sohi M. K., Sutton B. J., Thornton J. D., Keep P. A., Chester K. A., Begent R. H., Perkins S. J. (2000) Biochem. J. 346, 519–528 [PMC free article] [PubMed] [Google Scholar]
- 33. Graff C. P., Chester K., Begent R., Wittrup K. D. (2004) Protein Eng. Des. Sel. 17, 293–304 [DOI] [PubMed] [Google Scholar]
- 34. Koide A., Bailey C. W., Huang X., Koide S. (1998) J. Mol. Biol. 284, 1141–1151 [DOI] [PubMed] [Google Scholar]
- 35. Hackel B. J., Kapila A., Wittrup K. D. (2008) J. Mol. Biol. 381, 1238–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Koide A., Koide S. (2007) Methods Mol. Biol. 352, 95–109 [DOI] [PubMed] [Google Scholar]
- 37. Geiser M., Cèbe R., Drewello D., Schmitz R. (2001) BioTechniques 31, 88-90, 92 [DOI] [PubMed] [Google Scholar]
- 38. Zhang M., Aguilera D., Das C., Vasquez H., Zage P., Gopalakrishnan V., Wolff J. (2007) Anticancer Res. 27, 35–38 [PubMed] [Google Scholar]
- 39. Preijers F. W., Tax W. J., De Witte T., Janssen A., vd Heijden H., Vidal H., Wessels J. M., Capel P. J. (1988) Br. J. Haematol. 70, 289–294 [DOI] [PubMed] [Google Scholar]
- 40. May R. D., Wheeler H. T., Finkelman F. D., Uhr J. W., Vitetta E. S. (1991) Cell Immunol. 135, 490–500 [DOI] [PubMed] [Google Scholar]
- 41. Yamaizumi M., Mekada E., Uchida T., Okada Y. (1978) Cell 15, 245–250 [DOI] [PubMed] [Google Scholar]
- 42. Reiter Y., Brinkmann U., Jung S. H., Lee B., Kasprzyk P. G., King C. R., Pastan I. (1994) J. Biol. Chem. 269, 18327–18331 [PubMed] [Google Scholar]
- 43. Thompson J., Stavrou S., Weetall M., Hexham J. M., Digan M. E., Wang Z., Woo J. H., Yu Y., Mathias A., Liu Y. Y., Ma S., Gordienko I., Lake P., Neville D. M., Jr. (2001) Protein Eng. 14, 1035–1041 [DOI] [PubMed] [Google Scholar]
- 44. Deen W. M., Lazzara M. J., Myers B. D. (2001) Am. J. Physiol. Renal Physiol. 281, F579–F596 [DOI] [PubMed] [Google Scholar]
- 45. Neville D. M., Jr., Srinivasachar K., Stone R., Scharff J. (1989) J. Biol. Chem. 264, 14653–14661 [PubMed] [Google Scholar]
- 46. Chiron M. F., Fryling C. M., FitzGerald D. (1997) J. Biol. Chem. 272, 31707–31711 [DOI] [PubMed] [Google Scholar]
- 47. Wenning L. A., Yazdi P. T., Murphy R. M. (1998) Biotechnol. Bioeng. 57, 484–496 [DOI] [PubMed] [Google Scholar]
- 48. Chan M. C., Murphy R. M. (1999) Cancer Immunol. Immunother. 47, 321–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Convertine A. J., Benoit D. S., Duvall C. L., Hoffman A. S., Stayton P. S. (2009) J. Controlled Release 133, 221–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Liu Z., Zhang W., Ellis L. M., Fan F., Cao Y., Cheung L. H., Marks J. W., Zhou H., Camphausen R., Rosenblum M. G. (2010) ASCR 101st Annual Meeting, Washington D.C., April 17–21, 2010, Abstr. 2576, American Society for Cancer Research, Philadelphia [Google Scholar]