Abstract
In Saccharomyces cerevisiae, the key components of the nonhomologous end joining (NHEJ) pathway that repairs DNA double-strand breaks (DSBs) are yeast Ku (yKu), Mre11-Rad50-Xrs2, Dnl4-Lif1, and Nej1. Here, we examined the role of Nej1 in NHEJ by a combination of molecular genetic and biochemical approaches. As expected, the recruitment of Nej1 to in vivo DSBs is dependent upon yKu. Surprisingly, Nej1 is required for the stable binding of yKu to in vivo DSBs, in addition to Dnl4-Lif1. Thus, Nej1 and Dnl4-Lif1 are independently recruited by yKu to in vivo DSBs, forming a stable ternary complex that channels DSBs into the NHEJ pathway. In accord with these results, purified Nej1 interacts with yKu and preferentially binds to DNA ends bound by yKu. Furthermore, the binding of a mixture of Nej1 and Dnl4-Lif1 to DNA ends bound by yKu is greater than the sum of the binding of the individual proteins, indicating that pairwise interactions among yKu, Nej1, and Dnl4-Lif1 contribute to complex assembly at DNA ends. Nej1 stimulates intermolecular ligation by Dnl4-Lif1, but, more interestingly, the addition of Nej1 results in more than one intermolecular ligation per Dnl4 molecule. Thus, Nej1 not only plays an important role in determining repair pathway choice by participating in the initial NHEJ complex formed at DSBs but also contributes to the reactivation of Dnl4-Lif1 after repair is complete, thereby increasing the capacity of the NHEJ repair pathway.
Keywords: Chromatin Immunoprecipitation (ChiP), DNA-binding Protein, DNA-Protein Interaction, DNA Repair, Yeast Genetics, DNA Double-strand Breaks, DNA Ligase, Nonhomologous End Joining
Introduction
DNA double-strand breaks (DSBs)3 are particularly difficult to repair because there is no intact template strand to guide the repair process. Consequently, these lesions are extremely cytotoxic and, if misrepaired, can give rise to mutations ranging from gross chromosomal rearrangements to small deletions and insertions. DSBs can be repaired by two fundamentally different types of repair pathways (1, 2). In homology-dependent repair, a DNA duplex that is homologous to the break site, frequently a sister chromatid, is used to guide the repair of the DSB (1). Usually, this type of repair accurately restores the DSB site, whereas the repair of DSBs by nonhomologous end joining (NHEJ) is error-prone. In NHEJ, the ends of broken DNA molecules are brought together in the absence of extensive DNA sequence homology. This end bridging or synapsis is the key reaction that defines the NHEJ pathway (1, 2).
Key players in the NHEJ pathway were initially identified by the cloning of the genes that complemented the IR sensitivity of mutant rodent cell lines (2, 3). The pronounced IR sensitivity of NHEJ-deficient mammalian cells reflects the major contribution of this repair pathway to cell survival in response to chromosomal DSBs. Although homologs or orthologs of most of the key mammalian NHEJ factors have been identified in the lower eukaryote Saccharomyces cerevisiae, genetic inactivation of the yeast NHEJ factors does not significantly increase IR sensitivity unless the predominant homologous recombination pathway is also inactivated (2–4). The yeast Ku (yKu) and Dnl4-Lif1 complexes are functional homologs of mammalian Ku70-Ku80 (Ku) and DNA ligase IV-XRCC4, indicating that the mechanism of NHEJ in eukaryotes is at least partially conserved. In mammals, the DNA end-binding factor Ku recruits DNA-PKcs, a protein kinase catalytic subunit to DNA ends, forming the DNA-dependent protein kinase (5). DNA-PKcs itself and Artemis, a DNA structure-specific endonuclease, are important in vivo targets of the kinase activity of DNA-PK, which is critical for NHEJ (6, 7). In addition, DNA-PKcs appears to be the end-bridging factor in mammalian NHEJ (8). Although yeast lacks a homolog of DNA-PKcs, there is compelling genetic and biochemical evidence that the yeast Mre11-Rad50-Xrs2 complex is an essential NHEJ component (4) and is the major end-bridging activity in yeast NHEJ (9). In vivo, the yKu, Mre11-Rad50-Xrs2, and Dnl4-Lif1 complexes are all required for the efficient rejoining of linear plasmid DNA molecules with cohesive ends (4, 10–15). Consistent with these genetic analyses, the intermolecular ligation of linear DNA molecules with cohesive ends in vitro is also dependent on these three complexes at physiological salt concentrations and appears to be mediated by functional interactions among them (9).
Studies examining the assembly of the core NHEJ factors at both in vitro and in vivo DSBs have shown that the binding of yKu to the DSB is required for the recruitment of Dnl4-Lif1 (3, 16–19). Although not absolutely dependent upon either yKu or Dnl4-Lif1, the binding of Mre11-Rad50-Xrs2 to in vitro and in vivo DSBs is altered in the presence of these factors (16, 17, 19). Because the binding of yKu to in vivo DSBs is dynamic while its binds stably to in vitro DSBs (19), it appears that yKu is actively displaced from in vivo DSBs, presumably by competing DSB repair pathways. Interestingly, Dnl4-Lif1 stabilizes the binding of yKu to in vivo DSBs, and both yKu and Dnl4-Lif1 accumulate at unrepaired DSBs in the absence of Mre11-Rad50-Xrs2, suggesting that Dnl4-Lif1 and yKu form a complex at in vivo DSBs that sequesters the ends away from the homologous recombination machinery (19).
Several groups identified Nej1 as a novel essential NHEJ factor. NEJ1 is a haploid-specific gene whose inactivation causes a defect in NHEJ that is epistatic with DNL4 (20–23). There is now compelling evidence that Nej1 is a direct participant in NHEJ, in addition to regulating the subcellular localization of Dnl4-Lif1 (24). This notion is supported by parallel studies with the mammalian ortholog of Nej1, XLF (XRCC4-like factor; Cernunnos) (25–27). Notably, XLF not only stimulates the joining of cohesive DNA ends by DNA ligase IV-XRCC4 but also the joining of mismatched DNA ends (28–34). Interestingly, XRCC4 and XLF are structurally similar, forming homodimers with a globular head domain and coiled-coil regions (28, 30). Despite this structural information, the mechanisms by which XLF and Nej1 modulate DNA joining are poorly understood.
In this study, we examined the role of Nej1 in NHEJ by a combination of in vitro and in vivo approaches. Notably, we show that Nej1 changes the mode of ligation by Dnl4-Lif1 from single to multiple turnover. More surprisingly, our studies also reveal that Nej1 acts at the initial DNA-binding step of NHEJ and inhibits Rad51-dependent repair of DSBs by acting in concert with Dnl4-Lif1 to increase the stability of yKu binding to in vivo DSBs.
EXPERIMENTAL PROCEDURES
Plasmid Constructs
The yeast plasmids expressing calmodulin-binding peptide (CBP)-Nej1 and FLAG-Nej1 from a GAL promoter were gifts from Dr. Thomas Wilson (35). Fragments encoding full-length Nej1 and derivatives lacking either the N-terminal 129 amino acids (ΔN-Nej1, amino acids 129–343) or the C-terminal 120 amino acids (Nej1-ΔC, amino acids 1–223) were amplified by PCR and, after verification by DNA sequencing, were subcloned in-frame with a V5 epitope tag into the plasmid vector pYES2.1 using the pYES2.1-TOPO-TA expression kit (Invitrogen).
Yeast Strains and Induction of HO Endonuclease Expression
All yeast strains were derived from SLY1A and have been described previously (36) except for SLY1A ΔNEJ1. This strain was constructed by one-step gene replacement by transforming a PCR fragment of ΔNEJ1::KanMX into wild-type SLY1A and selecting for G418-resistant colonies. For chromatin immunoprecipitation experiments, HO endonuclease expression was induced by the addition of galactose (2% final concentration) to logarithmic cultures (107 cells/ml) grown in Yeast Extract Peptone-glycerol medium at 30 °C. To examine the effect of HO endonuclease expression on cell survival, dilutions of cultures of SLY1A and its derivatives were spotted onto agar plates containing complete medium and galactose. To measure the in vivo repair of DSBs by NHEJ, SLY1A and its derivatives were transformed with the plasmid pBEVY-GL (37) that had been linearized by EcoRV cleavage within the LEU2 gene. The number of colonies growing on plates lacking leucine reflects the efficiency of the plasmid recircularization by NHEJ.
Yeast NHEJ Proteins and Antibodies
His-tagged versions of yKu and Dnl4-Lif1 complexes were purified as described previously (9, 38). CBP-tagged Nej1 was purified from ΔNEJ1 cells harboring the plasmid encoding CBP-Nej1. Expression of CBP-Nej1 was induced by switching from medium containing 2% glucose to medium containing 2% galactose. After growth for 14 h at 30 °C, cells were harvested, resuspended in 50 mm Tris-HCl (pH 7.5), 1 mm EDTA, 1 mm dithiothreitol, 50 mm NaCl, 10% glycerol, and 0.2% IGEPAL CA-630 containing a mixture of protease inhibitors (1 mm PMSF, 1 mm benzamidine, 1 μg/ml leupetin, 1 μg/ml aprotinin, and 1 μg/ml pepstatin), and then lysed in a French press. The lysate was clarified by centrifugation and applied to a phosphocellulose column. CBP-Nej1, which was detected in the column flow-through fractions by immunoblotting with the anti-CBP antibody (Millipore), was further purified on a calmodulin affinity resin, followed by Resource S, Superdex S-200, and Mono Q chromatography. Approximately 10 μg of CBP-Nej1 was obtained from a 10-liter culture. Antibodies against yKu and Rad51 have been described previously (19).
ChIP Assay
ChIP was carried out as described previously (19, 36). Briefly, HO endonuclease was induced in exponentially growing yeast cultures as described above. After in vivo cross-linking of nucleic acids and proteins by 1% formaldehyde (1% final volume), cells were lysed, and genomic DNA was sonicated to yield fragments with an average size of 0.5 kb. Sonicated extracts were incubated with anti-yKu, anti-Rad51, or anti-V5 antibody (for V5-tagged Nej1) at 4 °C for 2 h and then with protein G-agarose beads for 1 h. After cross-link reversal, genomic DNA was purified and amplified by real-time quantitative PCR using the ABI Prism 7900 instrument (Applied Biosystems) with multiple primers sets that anneal 0.1–0.9-kb proximal (L) or distal (R) to the DSB, as well as primers specific for the PRE1 gene situated on chromosome V. PCRs were set up by following the manufacturer's standard real-time PCR protocol (Applied Biosystems). The recruitment of protein at the HO break site is expressed as relative immunoprecipitate, which represents the ratio of a specific PCR signal at the HO break site to the nonspecific signal at a distant locus (PRE1), normalized to the value obtained from the uninduced sample (0 h).
Pulldown Assays
To detect the association between Nej1 and yKu, purified His-tagged yKu (1 μg) was immobilized on nickel beads (10 μl) and then incubated with extracts (200 μg) from yeast cells expressing V5-tagged versions of Nej1 in 50 mm Tris-HCl (pH 7.5), 1 mm EDTA, 5% mecaptoethanol, and 150 mm NaCl containing the protease inhibitor mixture. After washing, bound proteins were released from the nickel beads by heating at 95 °C for 5 min in the presence of SDS loading buffer (15 μl). Tagged derivatives of Nej1 were detected among the proteins retained on the nickel beads by immunoblotting with anti-V5 antibody.
To demonstrate that there is a direct interaction between Nej1 and yKu, purified His-tagged yKu (1 μg) was immobilized on nickel beads (10 μl) and then incubated with purified CBP-Nej1 (1 μg). The binding of CBP-Nej1 to the beads was detected by immunoblotting. In reciprocal experiments, CBP-Nej1 (1 μg) was immobilized on protein G beads liganded by anti-CBP antibody and then incubated with purified yKu (1 μg).
EMSA
A linear blunt-ended DNA fragment of 200 bp was generated by PCR. DNA concentration is expressed as DNA molecules. The DNA substrate (38 nm) was incubated with Nej1 and yKu either alone or in combination for 15 min at room temperature in 20 mm Tris-HCl (pH 7.5), 50 mm NaCl, 2 mm dithiothreitol, and 5% glycerol (final volume of 20 μl). To confirm the presence of specific NHEJ proteins in DNA-protein complexes, antibodies for the NHEJ factors were added, and incubation was continued for 10 min. After separation by agarose gel electrophoresis (0.7% (w/v) agarose), DNA was visualized by ethidium bromide staining.
Surface Plasmon Resonance
DNA binding experiments were performed on a Biacore 3000 instrument (Biacore, Inc.) as described previously (19). A 5′-biotinylated forward primer and a nonbiotinylated reverse primer were used to amplify a product of 669 bp by PCR using Pfu polymerase (Stratagene). The biotinylated duplex DNA was immobilized on the chip surface at a concentration of 10 nm. All DNA concentrations are expressed as DNA molecules. The purified NHEJ proteins yKu, Dnl4-Lif1, and Nej1 were injected and analyzed for association and dissociation individually and in various combinations. All sensorgrams were generated at a flow rate of 10 μl/min. Proteins were diluted in running buffer (10 mm HEPES/potassium hydroxide (pH 7.5), 100 mm NaCl, 3 mm EDTA, and 0.05% surfactant P-20) prior to injection for 3 min, followed by a 2.5-min dissociation phase in which running buffer was passed through the flow cell. The proteins were simultaneously injected over a cell surface lacking the oligonucleotide to detect nonspecific binding, which was subtracted from the binding observed in the DNA cell, to generate the reported sensorgram. All experiments were performed in triplicate. Kinetic evaluation was performed using the manufacturer's software (Evaluation Version 4.1, Biacore, Inc.). The chip binding surface was regenerated by incubation for 60 s with 0.05% SDS solution, followed by a 30-s incubation with running buffer containing 500 mm NaCl.
Deadenylation and Adenylation Assays
To remove the AMP moiety from preadenylated Dnl4, the His-tagged Dnl4-Lif1 complex (1 pmol) was incubated with 10 mm pyrophosphate in a final volume of 20 μl for 30 min at 25 °C in AMP buffer (60 mm Tris-HCl (pH 8.0), 10 mm MgCl2, 5 mm DTT, 50 μg/ml BSA, and 150 mm NaCl) to reverse the first step of the ligation reaction in the presence or absence of CBP-Nej1 (2 pmol). To deadenylate Dnl4 by transferring the AMP moiety to DNA, the His-tagged Dnl4-Lif1 complex (1 pmol) was incubated in a final volume of 20 μl with a linear duplex DNA substrate with ligatable cohesive ends (100 pmol) in the presence or absence of CBP-Nej1 (2 pmol) for 30 min at 25 °C in ligation buffer. Dnl4-Lif1 was immobilized on nickel beads (10 μl), washed with 50 mm Tris-HCl (pH 7.5), 1 mm DTT, 150 mm NaCl, and 10% glycerol containing the protease inhibitor mixture, and then incubated with [α-32P]ATP (3000 Ci/mmol, 10 mCi/ml, 3 μCi, 1 pmol) in the presence or absence of CBP-Nej1 (2 pmol) for 20 min at 25 °C in AMP buffer. Bound proteins were released from the nickel beads by heating at 95 °C for 5 min in the presence of SDS loading buffer (15 μl), and after separation by SDS-PAGE, labeled Dnl4 was detected and quantitated by PhosphorImager analysis. To directly quantitate the number of labeled adenylated Dnl4 molecules, labeled bands were cut out and radioactivity measured by liquid scintillation counting (Beckman Instruments).
Ligation Assays
DNA ligation assays with radioactively labeled DNA substrates were performed as described previously (39). The DNA substrate to measure nick ligation was a labeled 44-bp linear DNA duplex with a single ligatable nick. The DSB substrates for intermolecular ligation were two 25-mer oligonucleotide duplexes, each with one blunt nonligatable end and a cohesive 5′-overhang of four nucleotides. One of the 5′-overhangs was end-labeled, whereas the other 5′-ends were not phosphorylated so that alignment of the duplexes via their cohesive ends generated one ligatable nick. Dnl4-Lif1 (1 pmol) was incubated with the DNA substrate (1 pmol) in the absence or presence of CBP-Nej1 at 25 °C for 150 min in AMP buffer (final volume of 10 μl) to measure ligation under single-turnover conditions. Dnl4-Lif1 (1 pmol) was incubated with the DNA substrate (10 pmol) in the absence or presence of CBP-Nej1 at 25 °C for 150 min unless indicated in ligation buffer (final volume of 10 μl) to measure ligation under multiple-turnover conditions.
RESULTS
Nej1 Is Recruited by and Stabilizes the Binding of yKu to in Vivo DSBs
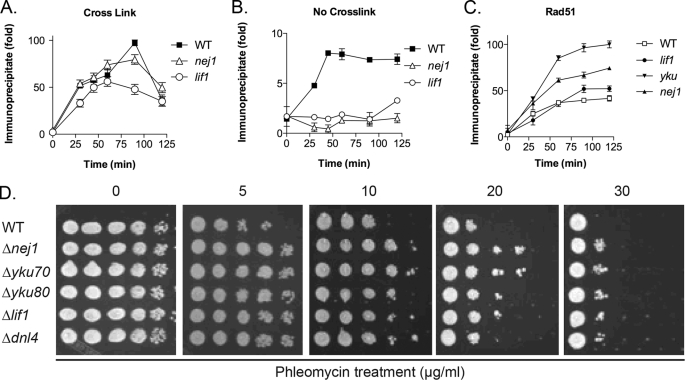
Although Nej1 is a direct participant in yeast NHEJ (24), the mechanisms by which it contributes to NHEJ are not well defined. In accord with published studies (20–23), deletion of NEJ1 in the SLY1 genetic background resulted in a defect in NHEJ, measured either by growth when HO endonuclease was constitutively expressed (Fig. 1A) or by repair of linearized plasmid DNA (Fig. 1B) that could be complemented by expression of tagged versions of Nej1 (Fig. 1, A and B). In ChIP assays to measure the steady-state levels of Nej1 at a site-specific DSB generated by HO endonuclease, genetic inactivation of yKu markedly reduced the recruitment of Nej1, whereas the absence of either the Lif1 or Dnl4 subunit of the Dnl4-Lif1 complex resulted in a relatively minor increase in Nej1 recruitment (Fig. 1C). Thus, the recruitment of Nej1 to in vivo DSBs is largely dependent upon yKu but independent of Dnl4-Lif1. Previously, ChIP assays performed without cross-linking were used to demonstrate that although the recruitment of Dnl4-Lif1 to an in vivo DSB was dependent upon yKu, Dnl4-Lif1 was critical for the stable retention of yKu at the DSB (19). This prompted us to examine the role of Nej1 in the binding of yKu to an in vivo DSB. In ChIP assays with cross-linking, the binding of yKu at the DSB site was not dependent upon either Nej1 or Lif1 (Fig. 2A). However, the binding of yKu to the DSB site in ChIP assays without cross-linking was dramatically reduced in the absence of either Nej1 or Lif1 (Fig. 2B). Thus, it appears that like Dnl4-Lif1 (19), Nej1 is required to stabilize the binding of yKu to in vivo DSBs.
FIGURE 1.
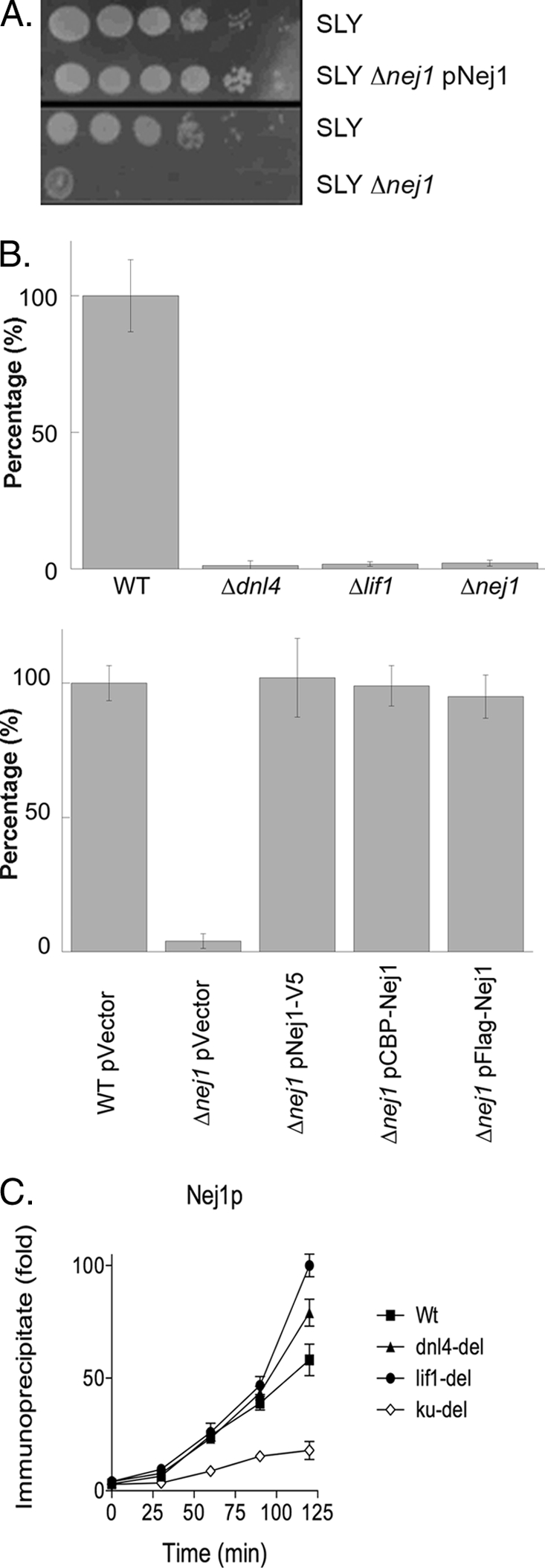
Effect of genetic inactivation of NEJ1 on NHEJ and the recruitment of Nej1 to an in vivo DSB. A, cultures of haploid yeast strains, wild-type SLY1A (SLY), SLY1A ΔNEJ1 (SLY Δnej1), and a derivative of SLY1A ΔNEJ1 harboring a plasmid expressing V5-tagged Nej1 (SLY Δnej1 pNej1) were diluted and spotted onto Yeast Extract Peptone-galactose plates to induce DSBs as a consequence of HO endonuclease expression. B, role of NHEJ in the ligation of linearized plasmid DNA after transformation into yeast cells. EcoRV-cleaved and circular plasmid DNAs were transformed into SLY1A (WT), SLY1A ΔDNL4 (Δdnl4), SLY1A ΔLIF1 (Δlif1), and SLY1A ΔNEJ1 (Δnej1) (upper panel) and derivatives of SLY1A containing the empty expression vector (WT pVector) and derivatives of SLY1A ΔNEJ1-harboring plasmids expressing the indicated tagged versions of tagged Nej1 (Δnej1 pFlag-Nej1, pCBP-Nej1, and pNej1-V5) (lower panel). Results are presented as relative transformation efficiencies (ratios of cut versus uncut plasmid) from three independent experiments. Error bars indicate mean ± S.D. C, kinetics of recruitment of V5-tagged Nej1 to a site-specific DSB in SLY1A (WT), SLY1A ΔDNL4 (dnl4-del), SLY1A ΔLIF1 (lif1-del), and SLY1A ΔyKU70 (ku-del) strains were measured by chromatin immunoprecipitation as described under “Experimental Procedures.” Data represent the mean ± S.D. of three or more independent experiments.
FIGURE 2.
Role of Nej1 in the binding of yKu to an in vivo DSB: effect of genetic inactivation of NEJ1 on the recruitment of Rad51 to an in vivo DSB and sensitivity to phleomycin. The kinetics of recruitment of yKu to a site-specific DSB in SLY1A (WT), SLY1A ΔLIF1 (lif1), and SLY1A ΔNEJ1 (nej1) strains were measured by chromatin immunoprecipitation with (A) and without (B) formaldehyde cross-linking. C, kinetics of recruitment of Rad51 to a site-specific DSB in SLY1A (WT), SLY1A ΔLIF1 (lif1), SLY1A ΔyKU70 (yku), and SLY1A ΔNEJ1 (nej1) strains. D, growth of wild-type SLY1A (WT) and derivatives with the indicated gene deletions was compared on Yeast Extract Peptone-glucose plates containing increasing concentrations of phleomycin by spotting dilutions of the cultures onto plates.
Because the binding of yKu to in vivo DSBs inhibits end resection (19, 40), we examined the binding of Rad51 to the single-strand regions generated by 5′- to 3′-resection at a DSB using ChIP assays (19). As expected (19), Rad51 binding was significantly increased in the absence of functional yKu (Fig. 2C). There was also increased Rad51 binding in the absence of Nej1 that was greater than that observed in the lif1 strain but less than that in the yKu strain (Fig. 2C). Inactivation of yKu results in increased resistance to DSB-inducing agents such as phleomycin (19) because end binding by yKu suppresses the repair of DSBs by homologous recombination. To provide evidence that the stabilization of yKu binding to in vivo DSBs by Nej1 contributes to the yKu-dependent inhibition of homologous recombination, we examined the effect of inactivating Nej1 on sensitivity to phleomycin. As was observed in strains lacking either a functional yKu complex or a functional Dnl4-Lif1 complex (19), Nej1 inactivation resulted in increased resistance to phleomycin (Fig. 2D). Together, these results demonstrate that in addition to Dnl4-Lif1 (19), Nej1 contributes to the stable binding of yKu to in vivo DSBs and the channeling of these lesions into the NHEJ repair pathway.
Direct Physical Interaction between Ku and Nej1
Next, we asked whether there is a direct interaction between yKu and Nej1 that may underlie the yKu-dependent recruitment of Nej1 to in vivo DSBs and subsequent stabilization of yKu binding. In initial studies, tagged versions of Nej1 in yeast cell extracts were specifically retained by yKu beads (Fig. 3A, upper panel) in a reaction that was dependent upon the N-terminal domain of Nej1 (middle and lower panels). To determine whether Nej1 interacts directly with yKu, a CBP-tagged version of Nej1 was purified after expression in yeast (Fig. 3B). Purified yKu was specifically retained by CBP-Nej1 beads (Fig. 3C), and in the reciprocal experiment, purified CBP-Nej1 was specifically retained by yKu beads (Fig. 3D). Neither the presence of ethidium bromide nor incubation with DNase disrupted the interaction between the purified proteins (data not shown).
FIGURE 3.
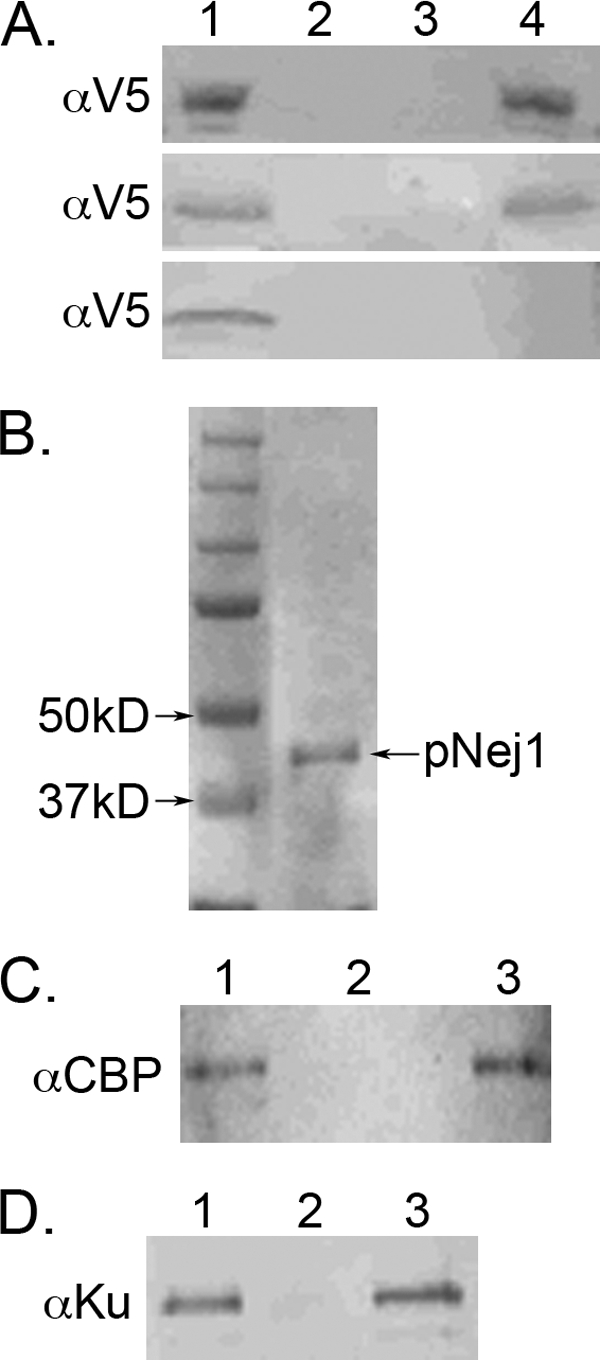
Physical interaction between Nej1 and yKu is dependent upon the N-terminal domain of Nej1. A, lane 1, immunoblots of extracts (20 μg) from yeast cells expressing V5-tagged wild-type Nej1 (upper panel), V5-tagged Nej1 lacking the C-terminal domain (middle panel), and V5-tagged Nej1 lacking the N-terminal domain (lower panel). Extracts (200 μg) were incubated with either nickel beads (lane 2) or nickel beads liganded by His-tagged yKu (lane 4). Lane 3, nickel beads liganded by His-tagged Ku incubated with extract expressing the V5 epitope. Epitope-tagged proteins retained by the beads were detected by immunoblotting. B, Coomassie Blue-stained gel showing molecular mass standards (left lane) and purified CBP-Nej1 (300 ng; right lane, pNej1). C, lane 1, immunoblot of purified CBP-Nej1 (100 ng). Purified CBP-Nej1 (1 μg) was incubated with either nickel beads (lane 2) or nickel beads liganded by yKu (lane 3). D, lane 1, immunoblot of purified yKu (100 ng). Purified yKu (1 μg) was incubated without (lane 2) or with (lane 3) purified CBP-Nej1 prior to the addition of protein G-Sepharose beads liganded by anti-CBP antibody. Proteins were detected by immunoblotting with the indicated antibody.
DNA Binding by Nej1
Because it had been shown previously that DNA binding by XLF and XRCC4 is dependent upon the length of the DNA duplex (31, 41), we examined the binding of Nej1 to linear blunt-ended DNA duplexes ranging in size from 0.2 to 5 kb in EMSAs. Nej1 did preferentially bind to the 5-kb substrate, forming discrete DNA-protein complexes, whereas interactions with the shorter DNA substrates resulted in less distinct DNA-protein complexes (data not shown). In similar assays, yKu formed discrete DNA-protein complexes with one end or both ends bound by yKu with all three DNA substrates (data not shown). Using surface plasmon resonance, we measured the binding of Nej1 and yKu to a 600-bp blunt-ended linear DNA immobilized on a sensor chip. Calculated affinities for Nej1 and yKu were 3 × 10−9 and 3 × 10−10 m, respectively. The higher affinity binding by yKu was due to its much slower rate of dissociation (9 × 10−3 1 per second) compared with Nej1 (9 × 10−2 1 per second).
Effect of yKu on DNA Binding by Nej1
The higher affinity binding of yKu to DNA prompted us to determine the effects of yKu on DNA binding by Nej1. As expected (19), incubation of yKu with a 200-bp linear blunt-ended DNA substrate resulted in the formation of a DNA-protein complex in which yKu was bound at one end of the linear DNA (Fig. 4A, lane 2). The addition of increasing amounts of Nej1 resulted in increasing amounts of a DNA-protein complex similar in size to that formed by yKu alone and the appearance of a DNA-protein complex with reduced electrophoretic mobility (Fig. 4A, compare lane 2 with lanes 3–5). In contrast, no DNA-protein complexes were detected in comparable reactions with Nej1 in the absence of yKu (Fig. 4A, lanes 6–8). To identify the proteins within the DNA-protein complexes, we performed supershift experiments with anti-yKu and anti-Nej1 antibodies. As expected, both DNA-protein complexes formed by coincubating yKu and Nej1 were shifted by anti-yKu antibody (Fig. 4B, compare lanes 2 and 3). In similar experiments with anti-Nej1 antibody, the two DNA-protein complexes formed by coincubating yKu and Nej1 were also shifted (Fig. 4B, compare lanes 6 and 7), whereas the DNA-protein complex formed with yKu alone was not (lane 8). Thus, Nej1 forms a stable complex with DNA ends bound by yKu.
FIGURE 4.
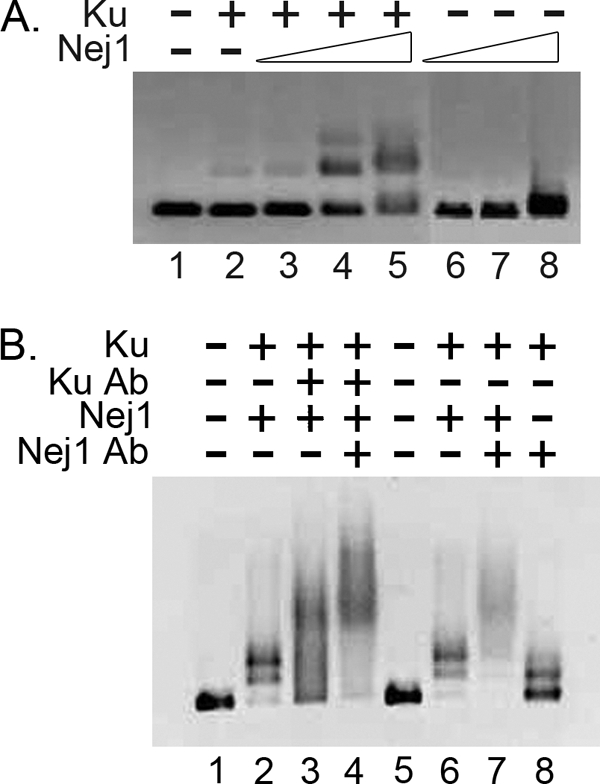
DNA-protein complexes formed by yKu and Nej1 in EMSAs. Formation of DNA-protein complexes by yKu and Nej1 with a 200-bp DNA duplex with blunt ends (38 nm) was detected in EMSAs as described under “Experimental Procedures.” A, lane 1, no protein; lane 2, 3 nm yKu; lane 3, 3 nm yKu and 6 nm Nej1; lane 4, 3 nm yKu and 12 nm Nej1; lane 5, 3 nm yKu and 25 nm Nej1; lane 6, 6 nm Nej1; lane 7, 12 nm Nej1; lane 8, 25 nm Nej1. B, DNA-protein complexes formed by yKu (3 nm; lanes 2–4 and 6–8) and Nej1 (25 nm; lanes 2–4, 6, and 7), either alone or in combination, were incubated with anti-yKu (lanes 3 and 4) or anti-Nej1 (anti-CBP, lanes 7 and 8) antibodies as indicated. Lanes 1 and 5, no protein.
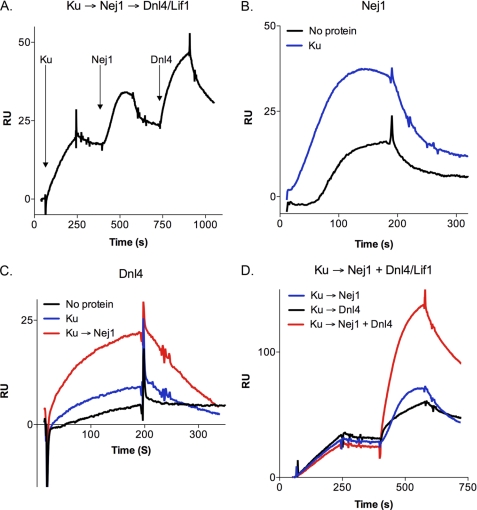
Assembly of DNA-Protein Complexes Containing yKu, Nej1, and Dnl4-Lif1
Based on our studies identifying an interaction between Nej1 and yKu and published studies describing interactions between yKu and Dnl4-Lif1 (16, 19) and between Dnl4-Lif1 and Nej1 (20–23), it appears that these factors are linked by pairwise interactions that presumably contribute to their assembly into a DNA-protein complex at DNA ends. To provide evidence for this model, we examined the assembly of DNA-protein complexes by surface plasmon resonance. In initial studies, we sequentially added Nej1 and Dnl4-Lif1 to DNA preloaded with yKu (Fig. 5A). In accord with the EMSA results, the presence of yKu on the DNA enhanced binding by Nej1 (Fig. 5B). The binding response observed with 1.25 nm Nej1 and yKu-bound DNA was similar to that observed with 5 nm Nej1 and DNA in the absence of yKu (data not shown), indicating that the presence of yKu enhances the binding of Nej1 to DNA by 3–4-fold. As expected (19), the presence of yKu on the DNA enhanced binding by Dnl4-Lif1 (Fig. 5C). A much larger increase in binding by Dnl4-Lif1 was observed when yKu and then Nej1 were preloaded onto the DNA (Fig. 5C), suggesting that interactions with both yKu and Nej1 contribute to enhanced DNA binding by Dnl4-Lif1. Our data and other published studies (19, 24) are consistent with a model in which yKu initiates binding at a DSB and then independently recruits Nej1 and Dnl4-Lif1. To mimic this scenario, we examined the effect of mixing Nej1 and Dnl4-Lif1 prior to loading onto DNA bound by yKu. The binding response of the mixture was higher than the sum of the binding responses of the individual factors (Fig. 5D), suggesting that the interaction between Nej1 and Dnl4-Lif1 contributes to their enhanced association with yKu at DNA ends.
FIGURE 5.
Interaction of yKu, Nej1, and Dnl4-Lif1 with DNA visualized by surface plasmon resonance. A, representative sensorgram generated by sequential injection of Ku (1 nm), Nej1 (1 nm), and Dnl4-Lif1 (10 nm) onto a sensor chip coated with 669-bp blunt-ended duplex DNA. B, representative sensorgrams showing the profile of binding of Nej1 (1.25 nm) to the DNA chip that had either been preloaded with yKu (2 nm; blue line, Ku) or not (black line, No protein). C, representative sensorgrams showing the profile of binding of Dnl4-Lif1 (10 nm) to the DNA chip (black line, No protein) and the effect of preloading of the DNA chip with yKu (1 nm; blue line, Ku) or yKu followed by Nej1 (1 nm yKu and 1 nm Nej1; red line, Ku → Nej1). D, representative sensorgrams showing protein binding to the DNA chip preloaded with yKu followed by Nej1 alone (1 nm; blue line, Ku → Nej1), Dnl4-Lif1 alone (10 nm; black line, Ku → Dnl4), and a mixture of Nej1 and Dnl4-Lif1 (1 nm Nej1 and 10 nm Dnl4-Lif1, red line, Ku → Nej1 + Dnl4). RU, resonance units.
Nej1 Stimulates DNA Joining by Dnl4-Lif1 by ATP-independent and ATP-dependent Mechanisms
The DNA ligase IV family is distinct from the other families of eukaryotic DNA ligases in that the purified enzyme is not only predominantly preadenylated but also appears to be capable of completing only a single ligation reaction (32, 42–45). Because XLF stimulates DNA joining by DNA ligase IV-XRCC4 (31–34, 46), we asked whether Nej1 modulates the catalytic activity of Dnl4-Lif1. In initial studies, we examined the effect of Nej1 on adenylation and deadenylation of Dnl4. When Dnl4-Lif1 was incubated with [α-32P] ATP, only ∼5% of the Dnl4 molecules in the purified Dnl4-Lif1 fraction formed the labeled Dnl4-AMP intermediate (Fig. 6A, lanes 1 and 4). Coincubation with Nej1 resulted in an ∼2-fold increase in the amount of the labeled Dnl4-AMP intermediate (Fig. 6A, lanes 2 and 3). These results indicate that at least 10% of the Dnl4 molecules in the purified Dnl4-Lif1 fraction are not adenylated and that about half of these molecules require Nej1 for adenylation.
FIGURE 6.
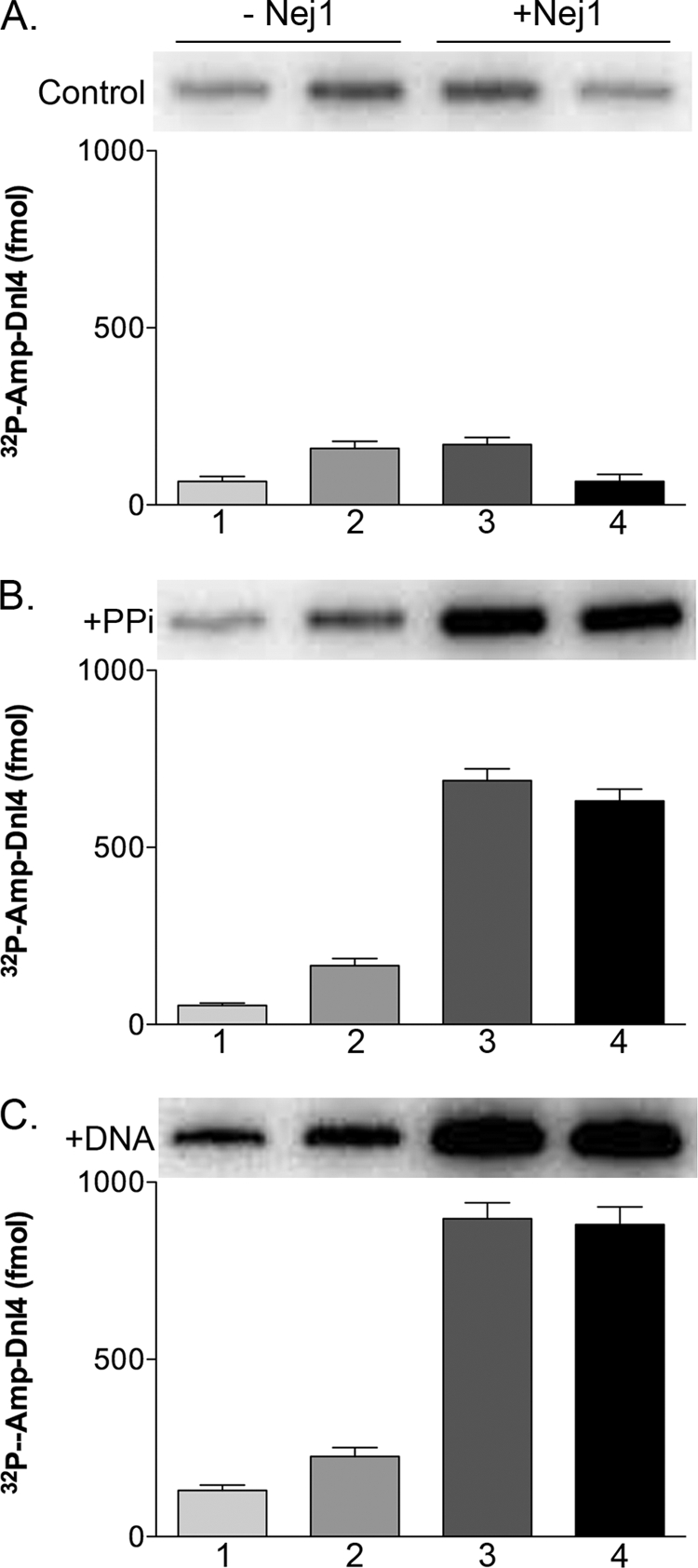
Effect of Nej1 on Dnl4 deadenylation and readenylation. A, His-tagged Dnl4-Lif1 (1 pmol) was preincubated without (lanes 1 and 2) or with (2 pmol; lanes 3 and 4) Nej1. B, His-tagged Dnl4-Lif1 (1 pmol) was preincubated without (lanes 1 and 2) or with (2 pmol; lanes 3 and 4) Nej1 in the presence of 10 mm pyrophosphate. C, His-tagged Dnl4-Lif1 (1 pmol) was preincubated without (lanes 1 and 2) or with (2 pmol; lanes 3 and 4) Nej1 in the presence of linear DNA oligonucleotide duplex with cohesive ligatable ends (100 pmol). After collection on nickel beads, Dnl4-Lif1 was incubated with [α-32P]ATP in the absence (lanes 1 and 4) or presence (2 pmol; lanes 2 and 3). of Nej1 Proteins were separated by SDS-PAGE and labeled. Dnl4 was detected and quantitated by PhosphorImager analysis. The results of three independent experiments are shown graphically. Error bars indicate mean ± S.D.
To determine the effect of Nej1 on deadenylation and readenylation, Dnl4-Lif1 was preincubated with pyrophosphate to reverse the first step of the ligation reaction, in either the presence or absence of Nej1. After collection on nickel beads and washing, Dnl4-Lif1 was incubated with [α-32P] ATP in either the presence or absence of Nej1 (Fig. 6B). Notably, preincubation with both Nej1 and pyrophosphate resulted in a 5–10-fold increase in the amount of the labeled Dnl4-AMP intermediate (Fig. 6B, compare lanes 1 and 2 with lanes 3 and 4), whereas the subsequent inclusion of Nej1 with [α-32P]ATP had no significant effect on the amount of labeled Dnl4-AMP intermediate (compare lanes 3 and 4). Dnl4 can also be deadenylated as a consequence of transfer of the AMP group to the DNA substrate during the ligation reaction. To determine the effect of Nej1 on the nucleotidyl transfer step of the ligation reaction, Dnl4-Lif1 was preincubated with DNA molecules with cohesive ligatable ends prior to incubation with [α-32P] ATP (Fig. 6C). The addition of Nej1 to the reactions with the DNA substrate resulted in a 5–10-fold increase in the amount of the labeled Dnl4-AMP intermediate (Fig. 6C, compare lanes 1 and 2 with lanes 3 and 4), but as was observed in the reactions with pyrophosphate, Nej1 did not significantly increase readenylation. Thus, under these reaction conditions, Nej1 has a major effect on deadenylation rather than adenylation of Dnl4.
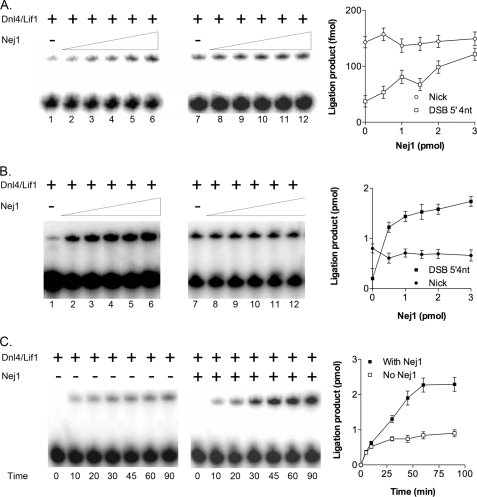
Next, we examined the effect of Nej1 on DNA joining by Dnl4-Lif1. In assays carried out in the absence of ATP, the intermolecular joining of linear cohesive-ended DNA molecules was less efficient than nick joining by Dnl4-Lif1. The addition of Nej1 stimulated intermolecular joining but not nick joining under these single-turnover conditions (Fig. 7A). When similar assays were carried out in the presence of ATP, Nej1 again stimulated intermolecular ligation but not nick ligation (Fig. 7B). With the nicked DNA substrate, the amount of ligated product was less than the number of Dnl4 molecules, suggesting that the Dnl4 molecules were carrying out only a single ligation event. In contrast, the addition of Nej1 to intermolecular joining reactions resulted in more ligation events than Dnl4 molecules, demonstrating that Nej1 enables Dnl4-Lif1 to carry out more than one catalytic cycle. Under these conditions, Nej1 did not significantly alter the initial reaction rate (Fig. 7C), indicating that the increased ligation results from Nej1 inducing the turnover of Dnl4-Lif1 molecules.
FIGURE 7.
Effect of Nej1 on DNA joining by Dnl4-Lif1 under single- and multiple-turnover reaction conditions. A, ligation of cohesive-ended duplex DNA (lanes 1–6; DSB 5′ 4nt; 1 pmol; open squares) and nicked DNA (lanes 7–12; Nick; 1 pmol; open circles) in the absence of ATP. B, ligation of cohesive-ended duplex DNA (lanes 1–6; DSB 5′ 4nt; 10 pmol; filled squares) and nicked DNA (lanes 7–12; Nick; 10 pmol; filled circles) in the presence of ATP. The reaction contained Dnl4-Lif1 (1 pmol) alone (lanes 1 and 7) and increasing amounts of Nej1 (lanes 2 and 8, 0.5 pmol; lanes 3 and 9, 1 pmol; lanes 4 and 10, 1.5 pmol; lanes 5 and 11, 2 pmol; lanes 6 and 12, 3 pmol). C, kinetics of joining of cohesive-ended duplex DNA (10 pmol) by Dnl4-Lif1 (1 pmol) in the absence (No Nej1; open squares) or presence (With Nej1; 2 pmol; filled squares) of Nej1 in reactions containing ATP (1 mm). The results of three independent experiments are shown graphically. Error bars indicate mean ± S.D.
DISCUSSION
Yeast Nej1 and its mammalian ortholog XLF are the most recently identified core components of the major eukaryotic NHEJ pathway (20–23, 25, 26). These proteins are structurally similar to and interact with the yeast Lif1 and human XRCC4 subunit of the DNA ligase IV complex (20–23, 25, 28, 30, 35). In addition, XLF enhances joining by DNA ligase IV-XRCC4 of both matched and mismatched DNA ends (31–34, 46). Using a combination of molecular genetic and biochemical approaches, we have demonstrated that Nej1 not only contributes to the final ligation step of NHEJ but also plays a critical role in the initiation of the NHEJ pathway.
Previously, live cell imaging studies had shown that the recruitment of XLF to DSBs is dependent upon Ku but not XRCC4 (27). Similarly, we found that the recruitment of Nej1 to an in vivo site-specific DSB measured by ChIP was dependent upon yKu but not Lif1 or Dnl4. Notably, our ChIP studies revealed that in addition to Dnl4-Lif1 (19), Nej1 is required for the stable association of yKu at in vivo DSBs. This stabilization of yKu binding suppresses homologous recombination by inhibiting end resection (19) and Rad51 recruitment, thereby sequestering DNA ends into the NHEJ repair pathway. Consistent with the Ku-dependent recruitment of XLF to DSBs (27) and our ChIP results, there is a direct physical interaction between yKu and Nej1 that occurs in both the absence and presence of DNA ends. Similar to Dnl4-Lif1 (19), Nej1 preferentially interacts with DNA ends bound by yKu. Furthermore, the binding of a mixture of Dnl4-Lif1 and Nej1 to DNA ends bound by yKu is greater than the sum of the binding of Dnl4-Lif1 and Nej1 alone, suggesting that the interaction between Dnl4-Lif1 and Nej1 contributes to the assembly of complexes with yKu at DNA ends.
The complex formed by yKu, Nej1, and Dnl4-Lif1 proteins at DNA ends plays a critical role in channeling DNA ends into the NHEJ pathway by preventing resection (19). In mammalian NHEJ, the stability of Ku binding to DNA ends is also influenced by DNA-PKcs, which causes the Ku ring to translocate inward along the DNA duplex during formation of the DNA-PK complex (47). Because the recruitment of XLF and DNA ligase IV-XRCC4 to in vivo DSBs is dependent upon Ku but independent of DNA-PKcs (27, 48), XLF, DNA ligase IV-XRCC4, and DNA-PKcs may all contribute to the stable association of Ku with DNA ends. Indeed, we suggest that the increased contribution of NHEJ to DSB repair in mammals is a consequence of DNA-PKcs greatly enhancing the stability of Ku at DNA ends.
An unusual feature of purified DNA ligase IV family proteins is that they appear to catalyze only a single ligation event because of inefficient readenylation following ligation (32, 42, 45). Recently, it has been shown that XLF promotes readenylation of DNA ligase IV-XRCC4, and it was suggested that this readenylation may enable DNA ligase IV-XRCC4 to join both strands at a ligatable DSB (32). Here, we have found that although Nej1 does enhance adenylation of Dnl4-Lif1, it has a much greater effect on deadenylation, enhancing pyrophosphate-driven reversal of Step 1 of the ligation reaction and Step 2 of the ligation reaction, nucleotidyl transfer from Dnl4 to DNA. Importantly, we have demonstrated that Dnl4-Lif1 molecules catalyze more than one ligation event in the presence of Nej1 but only during intermolecular ligation. Because only one strand of the aligned DNA ends was ligatable, the increased ligation reflects more intermolecular ligation rather than enhanced ligation of the second strand at aligned DNA ends. This reactivation of Dnl4 by Nej1 has important implications for the repair capacity of the NHEJ pathway because, without such a mechanism, the repair of DSBs by NHEJ will be limited by the number of Dnl4 molecules.
In summary, our molecular genetic and biochemical studies have provided novel mechanistic insights into the role of Nej1 in the repair of DSBs by NHEJ. Notably, this protein is a key component of the initial NHEJ complex assembled at the DSB and also appears to be involved in the turnover of NHEJ factors after repair is completed. In addition to participating in NHEJ, Nej1 is also involved in the repair of DSBs by yKu-independent microhomology-mediated end joining and single-strand annealing (49, 50). Further studies are needed to determine how Nej1 contributes to these repair pathways.
Acknowledgments
We thank Tom Wilson and Sang Eun Lee for plasmids and strains.
This work was supported, in whole or in part, by National Institutes of Health Grant GM47251 (to A. E. T.).
- DSB
- double-strand break
- NHEJ
- nonhomologous end joining
- yKu
- yeast Ku
- PKcs
- PK catalytic subunit
- CBP
- calmodulin-binding peptide.
REFERENCES
- 1. San Filippo J., Sung P., Klein H. (2008) Annu. Rev. Biochem. 77, 229–257 [DOI] [PubMed] [Google Scholar]
- 2. Weterings E., Chen D. J. (2008) Cell Res. 18, 114–124 [DOI] [PubMed] [Google Scholar]
- 3. Hefferin M. L., Tomkinson A. E. (2005) DNA Repair 4, 639–648 [DOI] [PubMed] [Google Scholar]
- 4. Critchlow S. E., Jackson S. P. (1998) Trends Biochem. Sci. 23, 394–398 [DOI] [PubMed] [Google Scholar]
- 5. Gottlieb T. M., Jackson S. P. (1993) Cell 72, 131–142 [DOI] [PubMed] [Google Scholar]
- 6. Chan D. W., Chen B. P., Prithivirajsingh S., Kurimasa A., Story M. D., Qin J., Chen D. J. (2002) Genes Dev. 16, 2333–2338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Huang Y., Giblin W., Kubec M., Westfield G., St Charles J., Chadde L., Kraftson S., Sekiguchi J. (2009) J. Exp. Med. 206, 893–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. DeFazio L. G., Stansel R. M., Griffith J. D., Chu G. (2002) EMBO J. 21, 3192–3200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen L., Trujillo K., Ramos W., Sung P., Tomkinson A. E. (2001) Mol. Cell 8, 1105–1115 [DOI] [PubMed] [Google Scholar]
- 10. Boulton S. J., Jackson S. P. (1996) Nucleic Acids Res. 24, 4639–4648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Boulton S. J., Jackson S. P. (1998) EMBO J. 17, 1819–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Herrmann G., Lindahl T., Schär P. (1998) EMBO J. 17, 4188–4198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schär P., Herrmann G., Daly G., Lindahl T. (1997) Genes Dev. 11, 1912–1924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Teo S. H., Jackson S. P. (1997) EMBO J. 16, 4788–4795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Grawunder U., Wilm M., Wu X., Kulesza P., Wilson T. E., Mann M., Lieber M. R. (1997) Nature 388, 492–495 [DOI] [PubMed] [Google Scholar]
- 16. Palmbos P. L., Daley J. M., Wilson T. E. (2005) Mol. Cell. Biol. 25, 10782–10790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Palmbos P. L., Wu D., Daley J. M., Wilson T. E. (2008) Genetics 180, 1809–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Teo S. H., Jackson S. P. (2000) Curr. Biol. 10, 165–168 [DOI] [PubMed] [Google Scholar]
- 19. Zhang Y., Hefferin M. L., Chen L., Shim E. Y., Tseng H. M., Kwon Y., Sung P., Lee S. E., Tomkinson A. E. (2007) Nat. Struct. Mol. Biol. 14, 639–646 [DOI] [PubMed] [Google Scholar]
- 20. Frank-Vaillant M., Marcand S. (2001) Genes Dev. 15, 3005–3012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kegel A., Sjöstrand J. O., Aström S. U. (2001) Curr. Biol. 11, 1611–1617 [DOI] [PubMed] [Google Scholar]
- 22. Ooi S. L., Shoemaker D. D., Boeke J. D. (2001) Science 294, 2552–2556 [DOI] [PubMed] [Google Scholar]
- 23. Valencia M., Bentele M., Vaze M. B., Herrmann G., Kraus E., Lee S. E., Schär P., Haber J. E. (2001) Nature 414, 666–669 [DOI] [PubMed] [Google Scholar]
- 24. Wu D., Topper L. M., Wilson T. E. (2008) Genetics 178, 1237–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ahnesorg P., Smith P., Jackson S. P. (2006) Cell 124, 301–313 [DOI] [PubMed] [Google Scholar]
- 26. Buck D., Malivert L., de Chasseval R., Barraud A., Fondanèche M. C., Sanal O., Plebani A., Stéphan J. L., Hufnagel M., le Deist F., Fischer A., Durandy A., de Villartay J. P., Revy P. (2006) Cell 124, 287–299 [DOI] [PubMed] [Google Scholar]
- 27. Yano K., Morotomi-Yano K., Wang S. Y., Uematsu N., Lee K. J., Asaithamby A., Weterings E., Chen D. J. (2008) EMBO Rep. 9, 91–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Andres S. N., Modesti M., Tsai C. J., Chu G., Junop M. S. (2007) Mol. Cell 28, 1093–1101 [DOI] [PubMed] [Google Scholar]
- 29. Doré A. S., Furnham N., Davies O. R., Sibanda B. L., Chirgadze D. Y., Jackson S. P., Pellegrini L., Blundell T. L. (2006) DNA Repair 5, 362–368 [DOI] [PubMed] [Google Scholar]
- 30. Li Y., Chirgadze D. Y., Bolanos-Garcia V. M., Sibanda B. L., Davies O. R., Ahnesorg P., Jackson S. P., Blundell T. L. (2008) EMBO J. 27, 290–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lu H., Pannicke U., Schwarz K., Lieber M. R. (2007) J. Biol. Chem. 282, 11155–11162 [DOI] [PubMed] [Google Scholar]
- 32. Riballo E., Woodbine L., Stiff T., Walker S. A., Goodarzi A. A., Jeggo P. A. (2009) Nucleic Acids Res. 37, 482–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tsai C. J., Kim S. A., Chu G. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 7851–7856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gu J., Lu H., Tippin B., Shimazaki N., Goodman M. F., Lieber M. R. (2007) EMBO J. 26, 1010–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Deshpande R. A., Wilson T. E. (2007) DNA Repair 6, 1507–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lee S. E., Moore J. K., Holmes A., Umezu K., Kolodner R. D., Haber J. E. (1998) Cell 94, 399–409 [DOI] [PubMed] [Google Scholar]
- 37. Miller C. A., 3rd, Martinat M. A., Hyman L. E. (1998) Nucleic Acids Res. 26, 3577–3583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tseng H. M., Tomkinson A. E. (2002) J. Biol. Chem. 277, 45630–45637 [DOI] [PubMed] [Google Scholar]
- 39. Chen X., Pascal J., Vijayakumar S., Wilson G. M., Ellenberger T., Tomkinson A. E. (2006) Methods Enzymol. 409, 39–52 [DOI] [PubMed] [Google Scholar]
- 40. Clikeman J. A., Khalsa G. J., Barton S. L., Nickoloff J. A. (2001) Genetics 157, 579–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Modesti M., Junop M. S., Ghirlando R., van de Rakt M., Gellert M., Yang W., Kanaar R. (2003) J. Mol. Biol. 334, 215–228 [DOI] [PubMed] [Google Scholar]
- 42. Chen X., Ballin J. D., Della-Maria J., Tsai M. S., White E. J., Tomkinson A. E., Wilson G. M. (2009) DNA Repair 8, 961–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Riballo E., Doherty A. J., Dai Y., Stiff T., Oettinger M. A., Jeggo P. A., Kysela B. (2001) J. Biol. Chem. 276, 31124–31132 [DOI] [PubMed] [Google Scholar]
- 44. Robins P., Lindahl T. (1996) J. Biol. Chem. 271, 24257–24261 [DOI] [PubMed] [Google Scholar]
- 45. Wang Y., Lamarche B. J., Tsai M. D. (2007) Biochemistry 46, 4962–4976 [DOI] [PubMed] [Google Scholar]
- 46. Hentges P., Ahnesorg P., Pitcher R. S., Bruce C. K., Kysela B., Green A. J., Bianchi J., Wilson T. E., Jackson S. P., Doherty A. J. (2006) J. Biol. Chem. 281, 37517–37526 [DOI] [PubMed] [Google Scholar]
- 47. Dynan W. S., Yoo S. (1998) Nucleic Acids Res. 26, 1551–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mari P. O., Florea B. I., Persengiev S. P., Verkaik N. S., Brüggenwirth H. T., Modesti M., Giglia-Mari G., Bezstarosti K., Demmers J. A., Luider T. M., Houtsmuller A. B., van Gent D. C. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 18597–18602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Carter S. D., Vigasová D., Chen J., Chovanec M., Aström S. U. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 12037–12042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lee K., Lee S. E. (2007) Genetics 176, 2003–2014 [DOI] [PMC free article] [PubMed] [Google Scholar]