Abstract
Listeria monocytogenes (Lm) is known to induce strong cellular immune responses. We constructed a live-attenuated Lm vector, Lmdd-BdopSIVgag, which encodes SIVmac239 gag. Intragastric (i.g.) administration of 3×1012 bacteria to rhesus macaques was safe and induced anti-Gag cellular but no humoral immune responses. Boosting of Gag-specific cellular responses was observed after i.g. administration of Lmdd-BdopSIVgag to previously vaccinated RM despite preexisting anti-Lm immunity shown by lymphoproliferative responses. Surprisingly, anti-Lm cellular responses were also detected in non-vaccinated controls, which may reflect the fact that Lm is a ubiquitous bacterium. The novel, live-attenuated Lmdd-BdopSIVgag may be an attractive platform for oral vaccine delivery.
Keywords: Live attenuated Listeria monocytogenes, live oral vaccine, HIV/AIDS vaccine, cellular immune responses, anti-vector immunity
1. Introduction
According to recent UNAIDS (www.UNAIDS.org) global estimates, 2.7 million new HIV-1 infections occur each year, yet a safe and effective HIV-1/AIDS vaccine has remained elusive. The encouraging results of the recent RV144 Phase III Thai AIDS vaccine trial suggest that a vaccine strategy seeking to generate cellular immune responses may need to be combined with one designed to induce humoral responses in order to gain substantial vaccine protection against HIV-1. Although neutralizing human monoclonal antibodies provided complete protection from acquisition of neutralization-sensitive simian-human immunodeficiency virus (SHIV) strains by passive immunization [1–4], induction of antibodies capable of neutralizing diverse HIV-1 strains has not been achieved with the current experimental immunogens. Primate model studies [5, 6] suggest that vaccine-induced cellular responses may limit lentiviral replication during the acute phase of infection; lowering of peak and steady-state viremia is associated with better clinical outcome. In a recent SIV/rhesus macaque model study, vaccine-induced cellular responses offered protection against persistent systemic infection [7]. Development of immunogens that can induce potent cellular responses is ongoing in several laboratories.
Among vaccination strategies to stimulate adaptive immunity, an important niche is occupied by live bacterial vectors. Live intracellular bacteria carrying foreign genes [8–11] are attractive candidates for the induction of cellular immune responses. Among the intracellular bacteria, the ubiquitous gram positive bacterium Listeria monocytogenes (Lm) has been studied in animal models as a vector for candidate cancer vaccines [12–20]; and was recently used in a Phase I human clinical trial among cervical carcinoma patients [21]. Several features of Lm, such as infection of antigen-presenting cells and the mucosal route of infection, make it an attractive vector for AIDS vaccine development (reviewed in [22]). Since most HIV-1 infections occur at mucosal surfaces, induction of immune responses at mucosal sites is crucial and Lm is a potential tool to achieve this goal. Lm has been used to induce HIV-1 Gag-specific immunity in murine [23–25] and non-human primate models [26]. Oral immunization of mice with Lm expressing HIV-1 Gag induced strong mucosal Gag-specific T-cell responses and protected against vaginal challenge with recombinant vaccinia virus expressing HIV-1 gag [25, 27, 28]. Lm induces multiple effector mechanisms, including antigen presentation via MHC class I and II pathways as well as induction of innate immune responses (reviewed in [29, 30]). Lm is able to survive in macrophages [31], in antigen presenting cells (APC), and also in dendritic cells (DC) [32], by escaping from the phagosomic compartment into the cytosol of target cells [33]. Lm can replicate in mammalian cells that are normally non-phagocytic, such as epithelial gastrointestinal cells, by actively translocating across the intestinal barrier and entering organs via the lymphatic system (reviewed in [34–36]). Of particular interest is the fact that Lm is naturally transmitted by the oral route. Thus, Lm-based vaccine vectors may be potential oral vaccines, which may be attractive for use in developing countries.
The use of Lm also presents major concerns. Lm is a food-borne pathogen that causes listeriosis, which can be manifest clinically by signs ranging from gastroenteritis to encephalitis and meningitis, which could be fatal (reviewed in [37]). Listeriosis mostly affects vulnerable individuals, such as immune compromised hosts, neonates, and pregnant women [37–39]. Thus, a number of strategies have been considered to attenuate this bacterium [11, 40, 41].
Another concern for using Lm vectors as vaccine platform is pre-existing immunity against Listeria, a ubiquitous bacterium. Although few studies has have shown that pre-existing immunity against Lm did not preclude the generation of immunity to foreign antigens expressed by the Listeria vector in feline [42] and murine [43, 44] models, pre-existing immunity against the vector is a widely discussed concern.
Earlier, we reported the hyperattenuated strain of Listeria, Lmdd, which lacked alanine racemase (dal) and D-amino acid aminotransferase (dat), the genes responsible for the synthesis of D-alanine, an essential component of the bacterial cell wall. Consequently, Lmdd critically depended on an exogenous supply of this amino acid [45]. While Lmdd encoding HIV gag was safe in rhesus monkeys (RM), the immunogenicity was not as potent as in mouse models [26], a difference we ascribed in part to the difficulties in co-administering the live-attenuated bacteria with D-alanine. Therefore, we further modified Lmdd to eliminate the strict D-alanine dependence. The new strain, Lmdd-Bdop, was constructed by allelic recombination between the Lmdd and the shuttle vector pKSV-7 carrying a Bacillus subtilis dal gene [27, 46].
Here, we present safety and immunogenicity data on the recombinant Lmdd-BdopSIVgag, which encodes Lm-codon-optimized SIV gag, in Indian-origin rhesus macaques. In parallel, we tested the parental vector, Lmdd-Bdop. Furthermore, we addressed the issue of boosting cellular immune responses to the vector transgene in monkeys with known prior Lm exposure.
2. Methods
2.1. Construction of recombinant Lmdd-BdopSIVgag
The hyperattenuated, double-deletion Lm mutant, Lmdd, lacks alanine racemase (dal) and D-amino acid aminotransferase (dat), the genes responsible for the synthesis of D-alanine (an essential component of the bacterial cell wall) and hence required exogenous D-alanine for its replication [45]. Lmdd-BdopSIVgag was constructed by allelic recombination between Lmdd and the shuttle vector pKSV-7 carrying a B. subtilis dal gene [27, 46] and a Lm-codon-optimized SIV gag; the Lmdd-Bdop control vector did not contain any SIV insert. Both Lmdd-Bdop and Lmdd-BdopSIVgag can grow transiently in infected mammalian cells without exogenous D-alanine. The final recombinant bacteria carry neither plasmids nor plasmid-encoded drug-resistance genes (Fig. 1A), but retain a significant degree of attenuation, with a 50% lethal dose (LD50) for the intravenous (i.v.) route in BALB/c mice of about 108 colony-forming units (CFU) versus 104 CFU for wild-type Lm.
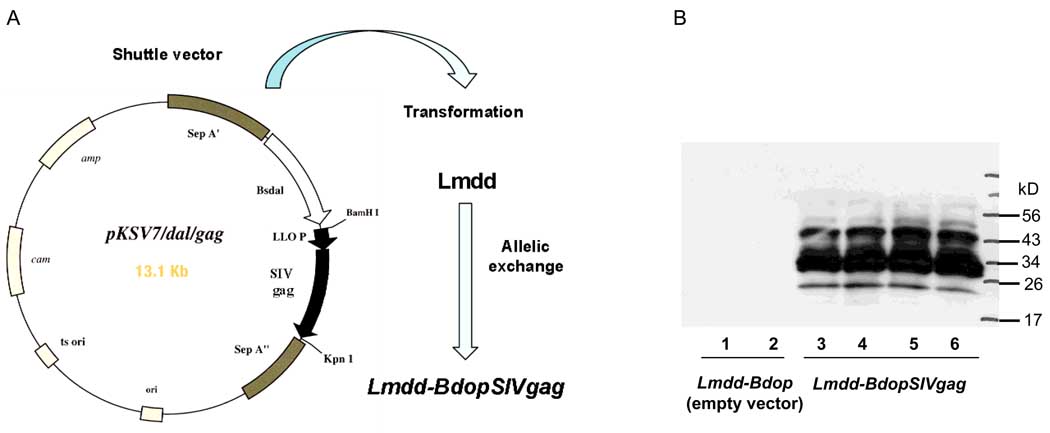
Fig. 1.
Construction of Lmdd-BdopSIVgag and analysis of SIV gag expression by Lmdd-BdopSIVgag. The recombinant Listeria monocytogenes (Lm) vector Lmdd-BdopSIVgag was obtained by allelic exchange between the Lm dal dat vector and pKSV-7 carrying a Bacillus subtilis dal gene and Lm-codon-optimized SIV gag (A). Culture supernatants from control Lmdd-Bdop (lanes 1, 2) and from gag-expressing Lmdd-BdopSIVgag (lanes 3–6) were analyzed by Western blot. SIV Gag was detected using the anti-SIV Gag monoclonal antibody 2F12 (AIDS Research & Reference Reagent Program, NIH). Cultures contained approximately 7 ng/ml p27 antigen as determined by ELISA. The multiple bands reflect proteolytic processing of Gag within the Lm and following secretion into the medium.
Lmdd-BdopSIVgag and the corresponding empty vector were grown in Bacto Brain Heart Infusion broth (DIFCO, BD Diagnostic, Sparks, MD) overnight at 30°C and stored at −80°C. The expression of SIV Gag by Lmdd-BdopSIVgag was confirmed by the Western blot analysis of bacterial culture supernatants (Fig. 1B).
2.2. Animals
Indian-origin rhesus monkeys (Macaca mulatta) were housed at the Yerkes National Primate Research Center (YNPRC), Atlanta, Georgia, USA. YNPRC is fully accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International (AAALAC). Approval for all procedures was received from the Institutional Animal and Care and Use Committees of Emory University and the Dana-Farber Cancer Institute. All animals were MHC typed for the Mamu-A*001 allele (Table 1).
Table 1.
Immunization schedule of rhesus monkeys with Lmdd-BdopSIVgag and empty vector control
| Vaccination Phase | Groups (status at start) |
Monkey name | Immunogen | Dose (CFU) | Schedule |
|---|---|---|---|---|---|
| Phase I | A (naive) | RHq-9 | Lmdd-Bdop | 1×1012 | Days 0, 2, 4, and 6 at weeks 0 and 13 |
| RUo-9 | |||||
| RVr-9 | |||||
| RYv-9 | |||||
| RYw-9 | |||||
| B (naive) | RCg-10* | Lmdd-BdopSIVgag | 3×1012 | Days 0, 2, 4, and 6 at weeks 0 and 13 | |
| RFa-10* | |||||
| RKs-9* | |||||
| RQe-10* | |||||
| RWy-9* | |||||
| Phase II | B (re-enrolled from Phase I) | RCg-10* | Lmdd-BdopSIVgag | 3×1012 | Days 0, 1, and 2 at weeks 0 and 6 |
| RFa-10* | |||||
| RKs-9* | |||||
| RQe-10* | |||||
| RWy-9* | |||||
| C (naive) | RFq-10* | Lmdd-BdopSIVgag | 3×1012 | Days 0, 1, and 2 at weeks 0 and 6 | |
| RIq-10* | |||||
| RKh-10 | |||||
| RLu-10 | |||||
| RTj-10* | |||||
| D (naive) | RAq-10 | Lmdd-Bdop | 1×1012 | Days 0, 1, and 2 at weeks 0 and 6 | |
| RDm-10 | |||||
| RQp-10 | |||||
| RTn-10 | |||||
| RWu-10 | |||||
Animals positive for the Mamu-A*001 allele
2.3. Immunization regimen
Immunizations were performed in two phases (Table 1). During Phase I, five RM were given the empty vector Lmdd-Bdop (Group A, Phase I controls) and five RM received Lmdd-BdopSIVgag. In Phase II, two new groups of five RM (Groups C and D) were enrolled. Group C RM were given Lmdd-BdopSIVgag, whereas Group D received the empty vector Lmdd-Bdop (Phase II controls). Group B (re-enrolled from Phase I) was the only group to be continued for the Phase II and again received two sets of Lmdd-BdopSIVgag immunizations. Fifteen min before the vector administration, each animal was anesthetized with ketamine and pretreated with a saturated sodium bicarbonate solution given by nasogastric (NG) tube to neutralize gastric acid. During Phases I and II, the immunizations differed in the number of doses and schedules of administration as described in Table 1.
2.4. Sample collection
Peripheral blood was collected in CPT tubes (Becton Dickinson, Franklin Lakes, NJ) containing sodium heparin as an anticoagulant. Peripheral blood mononuclear cells (PBMC) and plasma were separated and used for various assays. Remaining cells were cryopreserved and plasma aliquots were stored at −80°C.
In Phases I and II, rectal pinch biopsies were collected after 3 and 2 weeks following each immunization, respectively. Mononuclear cells were obtained from the biopsies as described earlier [47]. Briefly, the tissue was washed with Hanks balanced salt solution (HBSS), digested by mixture of DNase (Roche, Indianapolis, IN, USA) and collagenase IV (Sigma, St. Louis, MO, USA). The digest was passed through needles of different sizes and filtered to obtain single cell suspension.
2.5. Monitoring of fecal shedding of Lm-based vaccine vectors
To monitor shedding of live organisms, rectal swabs or fecal samples were collected following each immunization. These specimens were plated directly on selective media and incubated at 35°C for 24 h. Biochemical tests were performed to confirm colonies of Lm and the colony counts were documented (Tables 2 and 3).
Table 2.
Shedding of live Lm in feces after vaccination during Phase I.
| Bacterial colonies (CFU) | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Groups | RM | 1st set of four immunizations | 2nd set of four immunizations | ||||||||||||||||||
| D0 | D1 | D2 | D3a | D4 | D5 | D6 | D7 | D8 | D9 | D0 | D1 | D2 | D3 | D4 | D5 | D6 | D7 | D8 | D9 | ||
| A | RHq-9 | − | +++ | − | + | + | − | +++ | − | − | − | ++++ | − | ++++ | − | ++++ | ++ | +++ | − | − | |
| RUo-9 | − | + | − | − | ++ | − | + | + | − | − | ++ | − | +++ | − | +++ | − | +++ | − | − | ||
| RVr-9 | − | ++ | + | ++ | − | − | ++ | + | − | − | +++ | ++ | +++ | − | +++ | ++ | ++++ | − | − | ||
| RYv-9 | − | +++ | − | + | +++ | − | + | − | − | − | ++++ | ++ | +++ | − | ++++ | + | ++++ | − | − | ||
| RYw-9 | − | ++ | − | + | + | − | ++ | − | − | − | ++ | − | +++ | − | ++++ | − | − | − | − | ||
| B | RCg-0 | − | ++++ | + | + | + | + | +++ | ++ | − | − | ++++ | − | +++ | − | +++ | + | +++ | − | − | |
| RFa-10 | − | ++ | − | ++ | + | − | ++ | ++ | − | − | +++ | − | − | − | − | + | ++ | − | − | ||
| RKs-9 | − | + | ++ | ++ | − | − | ++ | + | − | − | +++ | − | +++ | − | − | − | ++ | − | − | ||
| RQe-10 | − | +++ | + | + | + | − | + | − | − | − | +++ | − | +++ | + | ++++ | − | ++ | − | − | ||
| RWy-9 | − | + | − | − | + | − | ++ | − | − | − | ++ | − | + | − | − | − | ++ | − | − | ||
++++, > 100 CFU;
+++, 50 to 100 CFU;
++, 10 to 50 CFU;
+, < 10 CFU;
−, no CFU;
no samples available
Table 3.
Shedding of live Lm in feces after vaccination during Phase II.
| Bacterial colonies (CFU) | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Groups | RM | 1st set of three immunizations | 2nd set of three immunizations | ||||||||||||
| D0 | D1 | D2 | D3 | D4 | D5 | D6 | D0 | D1 | D2 | D3 | D4 | D5 | D6 | ||
| B | RCg-10 | − | +++ | +++ | +++ | + | − | − | − | +++ | ++ | ++ | − | − | − |
| RFa-10 | − | ++ | ++ | ++++ | − | − | − | − | +++ | +++ | +++ | − | − | − | |
| RKs-9 | − | ++ | + | + | ++ | − | − | − | +++ | ++ | + | − | − | − | |
| RQe-10 | − | +++ | +++ | +++ | + | − | − | − | ++ | + | ++ | − | − | − | |
| RWy-9 | − | +++ | ++ | ++ | − | − | − | − | + | ++ | ++ | + | − | − | |
| C | RFq-10 | − | ++ | ++ | +++ | + | + | + | − | ++++ | +++ | ++ | − | − | − |
| RIq-10 | − | ++ | ++ | +++ | + | − | − | − | +++ | ++ | +++ | + | + | − | |
| RKh-10 | − | +++ | ++ | +++ | ++ | − | − | − | ++ | +++ | +++ | − | − | − | |
| RLu-10 | − | ++ | + | ++++ | +++ | − | − | − | +++ | ++ | ++ | − | − | − | |
| RTj-10 | − | +++ | + | ++++ | + | + | − | − | ++++ | ++ | +++ | − | − | − | |
| D | RAq-10 | − | +++ | +++ | +++ | − | − | − | − | +++ | +++ | − | +++ | − | − |
| RDm-10 | − | ++ | +++ | +++ | − | − | − | − | +++ | +++ | ++ | − | − | − | |
| RQp-10 | − | +++ | + | +++ | − | − | − | − | +++ | +++ | +++ | − | − | − | |
| RTn-10 | − | + | +++ | +++ | − | − | − | − | ++++ | ++ | +++ | +++ | − | + | |
| RWu-10 | − | ++++ | ++ | +++ | ++ | + | + | − | +++ | +++ | +++ | − | − | − | |
++++, > 100 CFU;
+++, 50 to 100 CFU;
++, 10 to 50 CFU;
+, < 10 CFU;
−, no CFU;
2.6. ELISPOT assay
The assay was performed as described earlier [48]. Multiscreen-IP plates (Millipore, Billerica, MA, USA) were coated with mouse monoclonal anti-human interferon (IFN)-γ antibody (clone B27, BD Pharmingen, San Jose, CA, USA) overnight at 4°C, then blocked with 10% heat-inactivated fetal bovine serum (FBS) in RPMI-1640 (R-10) for 2 h at room temperature. PBMC were isolated from whole blood by centrifugation in heparinized CPT tubes (BD Pharmingen) and washed three times. For antigen-specific T-cell stimulation, SIVmac239 Gag peptides (obtained through the AIDS Research & Reference Reagent Program (ARRRP), Division of AIDS, NIAID, NIH) were used; the set of consisted 15-mers with 11-amino acid overlaps between sequential peptides and represented the complete protein sequence. Three pools of peptides were prepared (pools #1, #2 and #3 consisting of peptides 1–42, 43–84 and 85–125, respectively). Cells were incubated overnight at 1×105 cells/well in R-10 medium containing 100 U/ml penicillin, 100 µg/ml streptomycin and 0.02 mM β-mercaptoethanol along with a pool of peptides (each peptide at 2 µg/ml). After washing the plate, biotinylated mouse anti-human IFN-γ antibody (clone 7-B6-1, Mabtech, Cincinnati, OH, USA) was added. Horseradish peroxidase-conjugated streptavidin (BD Biosciences, San Jose, CA, USA) and AEC chromogen substrate (BD Biosciences) was used to develop the spots. IFN-γ-secreting cells were enumerated using an Immunospot ELISPOT reader (CTL, Cleveland, OH, USA). Assays were done in duplicates and background counts with no peptide stimulation were subtracted. The results were expressed as the average number of spot-forming unit (SFU) per million PBMC. Cells of each sample were also stimulated with a mixture of phorbol myristic acid (PMA) and ionomycin (0.01 and 0.5 µg/ml, respectively) as positive control. The assay also included cells obtained from a naïve animal as well as from a RM with known strong SIV Gag-specific cellular responses (from a different study [48, 49]) as negative and positive controls, respectively.
2.7. Tetramer staining
Indian rhesus macaques display a high frequency of MHC class I allele Mamu-A*001 [50], which binds SIV Gag peptide p11C, the immunodominant epitope for this allele. Using this tetramer-peptide complex, we monitored frequency of p11C-specific, CD8+ T cells for all Mamu-A*001+ RM. Fresh PBMC or rectal mononuclear cells (1 × 106 cells) were stained at room temperature in the dark for 30 min with a mixture of anti-human CD3-Alexa Fluor 700 (SP34-2; BD Pharmingen), anti-human CD8-APC-Cy7 (RPA-T8; BD Pharmingen) antibodies and Gag p11C–Mamu-A*001 tetrameric complex-APC (kindly provided by Dr. Norman Letvin, Beth Israel Deaconess Medical Center, Boston, MA). Cells were washed twice with FACS buffer (PBS with 2% FBS) and fixed in 2% paraformaldehyde. PBMC from a Mamu-A*001-positive macaque with known SIV Gag-specific cellular response and a Mamu-A*001-negative macaque were used as positive and negative controls, respectively. Cells were analyzed using flow cytometer (LSR II; BD Biosciences) and FlowJo 6.0 (Tree Star) software. Cells were considered to be tetramer positive if they represented at least 0.03% of the total CD3+CD8+ T lymphocytes, and if the cell cluster was clearly separated from the tetramer-negative cell population.
2.8. Intracellular Cytokine staining (ICS)
PBMC collected at weeks 0, 2, 4, 6, 8 and 10 during Phase II of immunization were used for ICS. Cells (1 × 106) were suspended in 100 µl R-10 containing anti-CD49d (1 µg/ml, clone 9F10, BD Biosciences) and anti-CD28 (20 µl, clone CD28.2, eBioscience, San Diego, CA, USA) antibodies. The cells were stimulated with a mixture of all three Gag SIVmac239 peptide pools. To allow cytokine accumulation in the Golgi complex, GolgiPlug (BD Pharmingen) was added according to the manufacturer’s instructions after 1.5 h. Cells without peptide stimulation were used to measure background cytokine production. Cells stimulated with a mixture of PMA and ionomycin (0.01 and 0.5 µg/ml, respectively) served as positive controls. After overnight incubation, cells were washed with PBS containing 2% FBS and surface-stained with anti-human CD3-FITC (SP34, BD Pharmingen), anti-human CD4PerCP-Cy5.5 (L200, BD Pharmingen), anti-human CD8-PE (RPA-T8, BD Pharmingen). After surface staining, cells were fixed/permeabilized with BD Cytofix/Cytoperm solution (BD Cytofix/Cytoperm Plus Fixation/Permeabilization kit, BD Bioscience). The cells were then intracellularly stained with the following monoclonal antibodies (mAbs): anti-human IFN-γ-APC Cy7 (4S.B3, eBioscience, San Diego, CA), anti-human IL-2-APC (MQ1-17H12, BD Pharmingen) and anti-human TNF-α-Alexa Fluor 700 (mAb11, BD Pharmingen). The cells were then washed and fixed in 2% paraformaldehyde (PFA). Lymphocytes (based on forward and side scatter) were acquired by flow cytometry (LSR-II; BD Immunocytometry Systems) and analyzed using FACSDiva 6.0 software. The percentage of CD3+CD4+ or CD3+CD8+ T cells producing IFN-γ, IL-2 or TNF-α was determined after subtracting the corresponding percentages from unstimulated cells (cytokine production background).
2.9. Anti-Lm and anti-SIV Gag antibody ELISA
Antibodies against SIV Gag or against whole Lm in the plasma of immunized animals were measured by ELISA. To determine the presence of IgG against whole Lm, a suspension of heat-inactivated bacteria was prepared as follows. An inoculum of Lm (monkey clinical isolate 12443, bacterial strain RM3102, serotype 1/2a) [51] was grown overnight in Bacto Brain Heart Infusion broth (DIFCO); the concentration of bacteria was evaluated spectrophotometrically at 550 nm. After three washes in PBS, the bacterial suspension was heat-inactivated for 1 h at 65°C and resuspended in PBS to a concentration of 108 CFU/ml. Nunc-Immuno Maxi-Sorp 96-well ELISA plates (Nalgene, Nunc International, Rochester, NY, USA) were coated overnight at 4°C with 50 µl/well of Lm suspension; plates were washed with PBS/0.05% Tween 20 (Sigma), and blocked for 2 h at 37°C with the same buffer containing also 2% FBS. The plates were washed and incubated 2 h at 37°C with 100 µl/well of plasma samples diluted 1/100 in washing buffer. After additional washing, plates were incubated for 1.5 h at 37°C with goat HRP-anti-monkey IgG Fc (1:4000) (Accurate/Nordic, Westbury, NY, USA), washed, and finally developed by ABTS substrate solution (2,2’-azinobis (3-ethylbenzthiazoline-6-sulfonic acid-diammonium salt dissolved in stable peroxide substrate buffer 1X) (Pierce, Rockford, IL, USA) for 30 min at room temperature. The absorbance was measured at 410 nm. Negative controls consisted of six plasma samples obtained from naïve monkeys; positive control was plasma from a monkey previously vaccinated with Lmdd-HIVgag, known to have high anti-Lm antibody titers [26]. A cut-off value for the OD was obtained from plasma of six naïve animals, and was expressed as the mean OD410 + 3 SD. Antibody titers are expressed as the dilution at which the sample OD410 value was equal or above the cut-off OD410 value.
Anti-SIV Gag antibodies were assessed in the same samples. Plates were coated with 3 µg/ml SIVmac251 p55 Gag (ARRRP) in Coating Buffer B (Invitrogen, Carlsbad, CA, USA) and incubated overnight at 4°C. The same protocol as described above was followed, but samples were diluted 1/50, and plasma from an animal with high antibody titers against SIV Gag [48] was used as a positive control for the assay.
2.10. Lymphocyte proliferation assay
PBMC (5×106) were suspended in PBS (1×106/ml) containing 0.1% BSA and 5 µM CFSE (CellTrace™ CFSE Cell Proliferation Kit, Invitrogen, Camarillo, CA, USA) and incubated at 37°C for 10 min. To quench the staining, ice-cold R-10 media was added and the cell suspension was incubated on ice for 5 min. The cells were washed twice, resuspended in R-10, seeded at 1×106 cells/well in a 48-well plate and stimulated with either SIVmac251 Gag proteins (2 µg/ml; ARRRP) or 10 µl/well of heat-inactivated Lm suspension. Positive controls were stimulated with Con-A (5 µg/ml); cells without any stimuli were used to determine background proliferation. The cells were incubated at 37°C for 5 d then stained with anti-human CD3-Alexa Fluor 700 (SP34-2; BD Pharmingen), anti-human CD4PerCP-Cy5.5 (L200, BD Pharmingen) and anti-human CD8-APC-Cy 7 (RPA-T8; BD Pharmingen). After fixation (2% formaldehyde in PBS), at least 10,000 lymphocytes (based on forward and side scatter) were acquired by flow cytometry and data analyzed using FACSDiva (BD Biosciences) software. The percentages of antigen–specific CD3+CD4+ and CD3+CD8+ proliferating cells were determined by measuring CFSE fluorescence intensity for each subset; background proliferation (no stimulation) was subtracted.
2.11. Statistical analysis
Statistical analysis of data was performed by GraphPad Prism 4. The difference in frequency of SIV Gag-specific, IFN-γ-producing cells (SFU / 106 PBMC; measured by ELISPOT assay) between Groups B and C (during Phase II of immunization) was assessed by the Mann-Whitney U test. A P value < 0.05 was considered statistically significant.
3. Results
3.1. Safety of the new vector, Lmdd-BdopSIVgag
A major objective of this study was to test the safety of the new, live attenuated Lm vector in primates. In Phase I, five RM received 1 × 1012 of Lmdd-Bdop organisms (Group A) at weeks 0 and 13 (Table 1); the four doses were given on alternate days (days 0, 2, 4 and 6). An equivalent dose of Lmdd-BdopSIVgag (3 × 1012 organisms) was given to another five RM (Group B) using the same schedule of i.g. vaccination. The higher dose of Lmdd-BdopSIVgag compared to that of the parental vector accounted for the higher degree of attenuation of the former as tested in mice by LD50 (chromosomal integration of the transgene further attenuates Lm organisms). The animals were monitored for adverse clinical reactions and shedding of live Lm organisms in feces, which was detected within 24 h of inoculation. Twenty-two of 40 samples tested 48 h after each vaccine administration during the first set of four immunizations and 32 of 40 samples tested after the same time interval during second set of four immunizations were fecal culture negative for Lm (Table 2).
In Phase II, ten additional monkeys were enrolled (Groups C and D) that received three doses on consecutive days of either Lmdd-Bdop or Lmdd-BdopSIVgag at weeks 0 and 6 (Table 1). Group B was re-enrolled from Phase I and received additional Lmdd-BdopSIVgag according to the administration schedule followed for Groups C and D. These animals also cleared shedding of Listeria; 6 of 15 fecal samples tested 48 h after last dose of the first set of three immunizations and 11 of 15 samples tested at same time interval after last dose of the second set of immunizations were culture negative for Lm (Table 3).
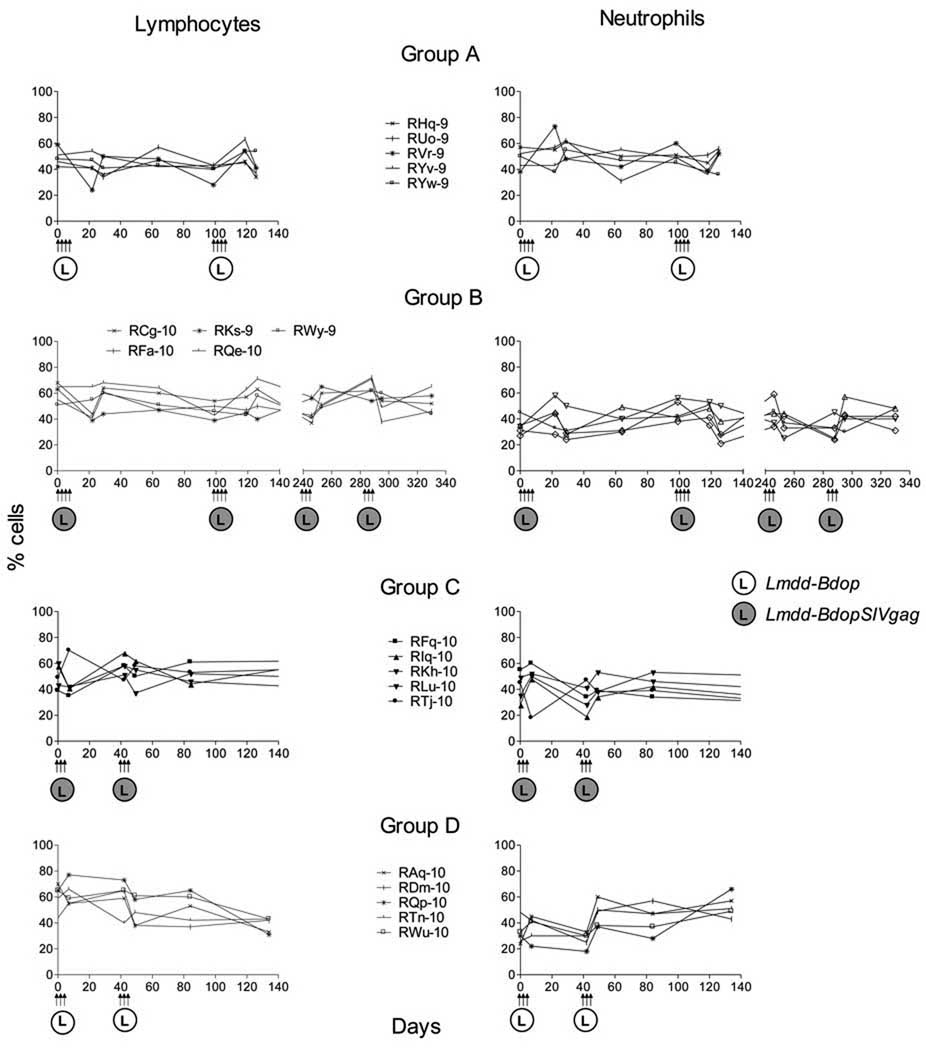
Administration of a single dose of 1 × 109 wild-type Lm in monkeys caused illness that included septicemia, irritability, loss of appetite and occasional diarrhea; the animals shed Listeria in feces for about 21 days [52]. These animals also showed lymphopenia and neutrophilia within 48 h after Lm administration. We monitored all animals for 4 months after the last dose of Lm administration. Routine physical exams, complete blood count (CBC) and liver function tests were performed periodically. After administration of high doses of the live attenuated Lmdd-Bdop (1 × 1012 CFU) and Lmdd-BdopSIVgag (3 × 1012 CFU), none of the macaques showed any of the above mentioned clinical signs and cleared fecal shedding of Lm within 48 to 72 h (Tables 2 and 3). Fig. 2 shows lymphocyte and neutrophil counts after Lm administration. Our data indicate that the new, live attenuated Lmdd-Bdop and Lmdd-BdopSIVgag vectors were safe and well tolerated by the rhesus monkeys.
Fig. 2.
Hematological profiles of RM following Lm administration. The percent lymphocytes (left panels) and neutrophils (right panels) are shown. No neutrophilia was seen; one Group C RM had an absolute neutrophil count on day 4 after the last dose of Lmdd-BdopSIVgag of 14,280 cells/mm3 (normal range for RM, 0.2–14.6 × 103 cells/mm3) without bands or concomitant changes in the percent neutrophils and lymphocytes. This RM also did not show any clinical signs of illness. The normal percentages of lymphocytes and neutrophils range between 8% to 92% and 5% to 88%, respectively, as wide fluctuations have been reported.
3.2. Lmdd-BdopSIVgag induced anti-SIV Gag cellular but not humoral immune responses after i.g. immunization
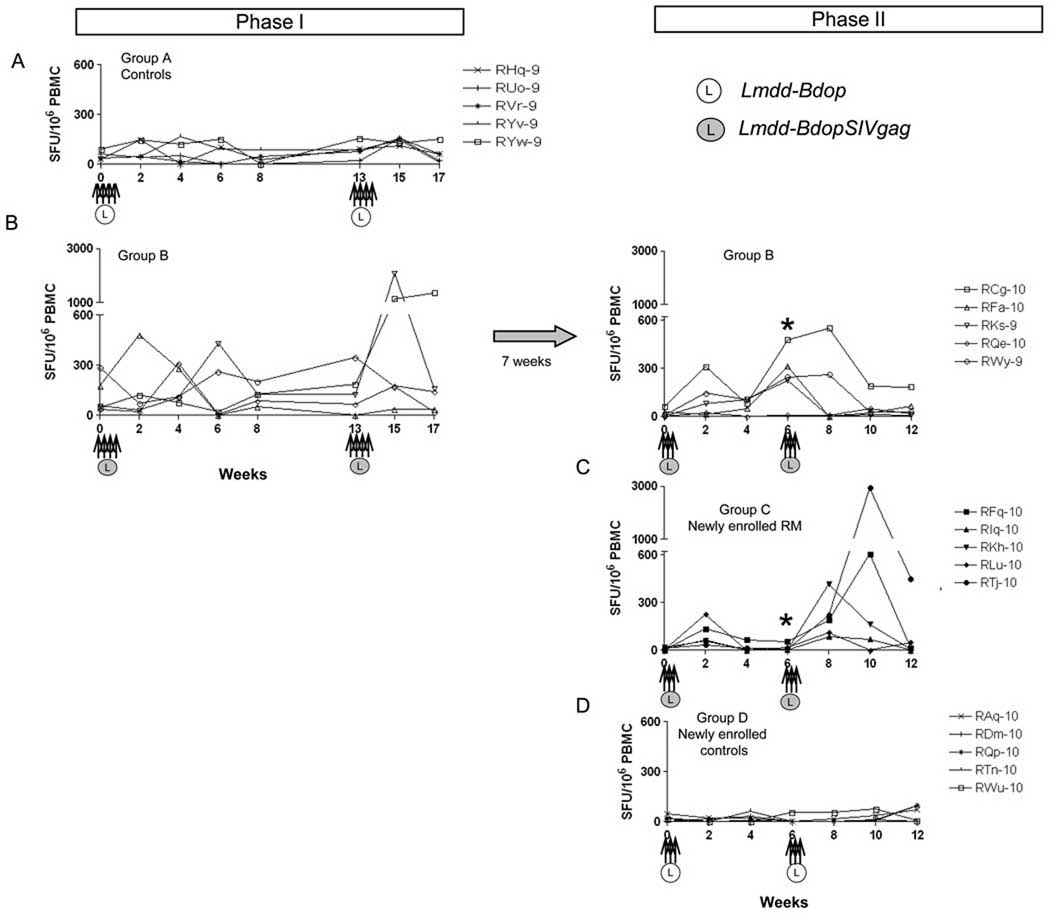
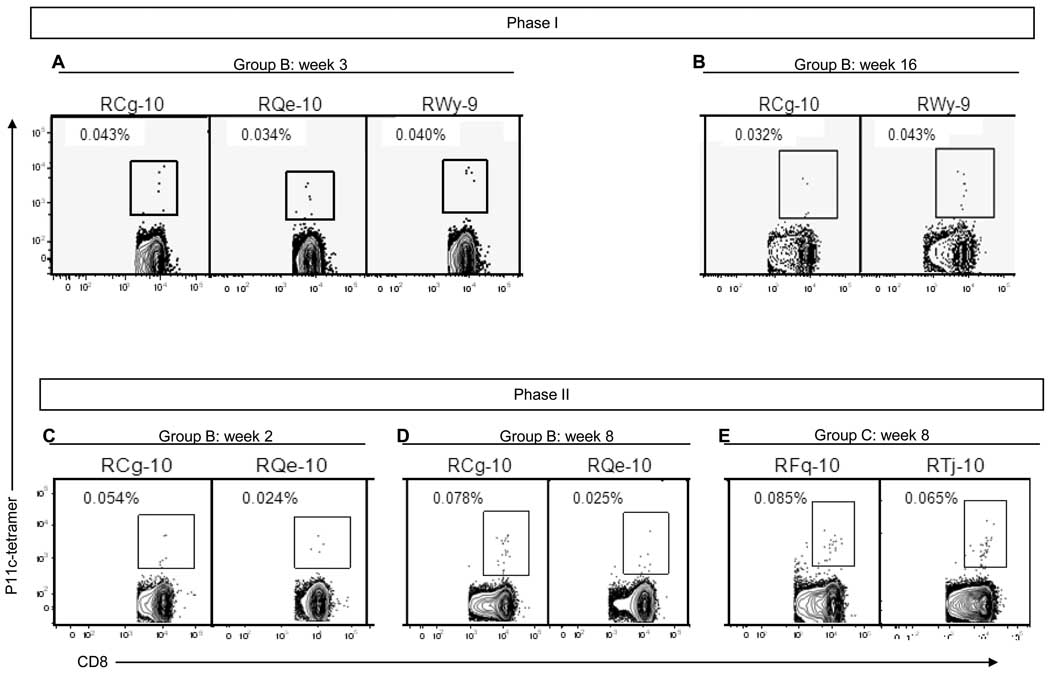
Next, we evaluated the immunogenicity of Lmdd-BdopSIVgag after i.g. administration of the vaccine vector. During Phase I, RM were exposed to Lmdd-bdop (Group A) or Lmdd-BdopSIVgag (Group B) on weeks 0 and 13 on alternate days for a total of four doses (two sets of immunization). The frequency of SIV Gag-specific, IFN-γ secreting cells was monitored by ELISPOT assay. Low-level cellular responses were first detected among Group B animals between weeks 2 to 6 (Fig. 3A and 3B) after the initial priming. p11C (SIV Gag epitope)-specific CD8+ T cells were also detected in PBMC among 3 out of 5 macaques 3 weeks after first set of Lmdd-BdopSIVgag administration (Fig. 4A). Two weeks after boosting with the second set of exposures to Lmdd-BdopSIVgag, two RM showed strong SIV Gag-specific responses (>1000 SFU/ 106 PBMC; Fig. 3B). Tetramer+, p11C-specific CD8+ T cells were detected in the peripheral blood of two RM (Fig. 4B). These data indicate that i.g. priming / boosting induced strong anti-SIV Gag cellular immune responses in two of the five vaccinated RM. No anti-Gag antibody responses were observed in any vaccinees (data not shown).
Fig. 3.
The frequency of SIV Gag-specific cells measured by IFN-γ ELISPOT assay at the time points indicated. Arrows, intragastric administration of either Lmdd-BdopSIVgag (L in dark circle) or Lmdd-Bdop (L in open circle); *, statistically significant difference (P = 0.031) between Groups B and C at week 6 in Phase II.
Fig. 4.
Frequency of CD3+CD8+ Gag p11C tetramer+ cells among Mamu-A*001+ animals during Phases I and II. The results are shown only for responder RM. Results were considered positive if >0.03%, a cut-off that was determined from analysis of naïve monkeys. No positive cells were detected among Mamu-A*001 negative vaccinated monkeys.
3.3. Pre-existing anti-Lm immunity was detected among all RM
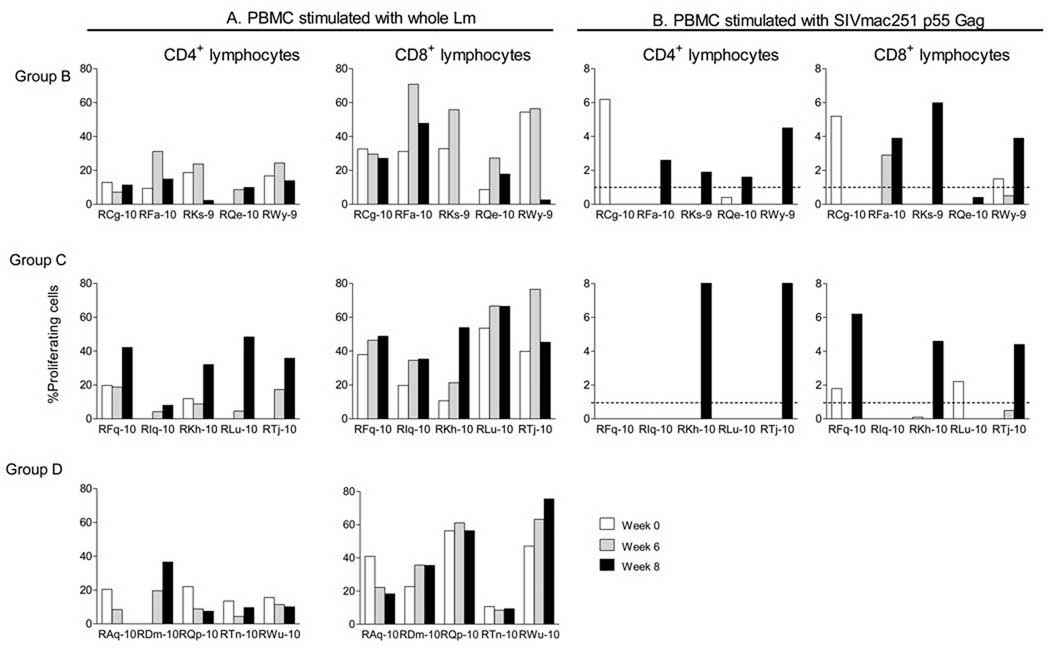
Next, we sought to test whether the known exposure of Group B RM during the Phase I immunogenicity testing had induced anti-Lm immune responses, and whether they would preclude recall responses to the transgene. To do this, we re-enrolled Group B monkeys into Phase II immunogenicity testing. We recruited two more groups of five RM each. Group C consisted of naïve monkeys that would receive the same vaccine, Lmdd-BdopSIVgag, as the animals in Group B. The difference between Groups B and C was prior exposure of Group B monkeys to the vaccine. Group D animals were enrolled as recipients of the empty vector, Lmdd-Bdop. Eleven weeks after the last immunization during Phase I, we tested Group B animals as well as the newly enrolled Groups C and D for anti-Lm humoral and cellular immune responses. None of the animals had measurable anti-Lm antibodies (data not shown). However, all animals of all three groups had proliferative responses among the CD4+ T cells, and the fraction of CD8+ T cells proliferating in response to whole Lm stimulation was surprisingly high for all three groups (Fig. 5A). It should be noted that all RM had been pre-screened prior to enrollment for antibody responses to Lm (all had been negative). We ascribe the proliferative responses of all monkeys to whole bacteria – with or without the additional vaccination with Lmdd-BdopSIVgag – to pre-existing Lm antigen-specific memory T cells, given that Listeria is a ubiquitous bacterium present in soil.
Fig. 5.
Proliferation of PBMC after stimulation with whole heat-inactivated Lm (A) or SIV Gag protein (B). Percent CD4+ (left panels) and CD8+ (right panels) proliferating cells are shown. Dashed line, mean frequency of proliferating cells shown by control (Group D) animals.
3.4. Prior immunization with Lmdd-BdopSIVgag did not prevent anamnestic responses to SIV Gag
Next, we sought to test whether the previously vaccinated Group B monkeys would show increases in anti-SIV Gag ELISPOT responses upon further exposure to the vaccine vector. Phase II re-vaccination was performed in two sets of three immunizations given on three consecutive days (Fig. 3, panels on right). At week 6, prior to the second set of immunization in Phase II, four out of the five RM of Group B had measurable ELISPOT responses (Fig. 3B, right panel); intracellular cytokine staining showed low and variable positivity (data not shown), and p11C-specific CD8+ T cells were detected in the peripheral blood of one out of five RM (monkey RCg-10, Fig. 4C). Overall, four out of five monkeys of Group B showed positive anti-SIV Gag responses by ELISPOT assay after only one set of immunization during Phase II, which is consistent with an anamnestic anti-Gag response.
Next, we compared the anti-Gag cellular responses of the re-exposed vaccine recipients of Group B with those of Group C animals that had never seen the vaccine vector before. As expected, the single set of immunization (priming) given at week 0 did not induce high ELISPOT responses (Fig. 3C), and at week 6, the latter were similar to those of monkeys of control Group D. When compared to Group B (re-exposed) monkeys, ELISPOT responses at week 6 in Group C were significantly lower (P = 0.031). This indicates that the re-exposed Group B animals mounted significant anamnestic response, whereas Group C monkeys needed boosting with antigen to develop anti-Gag ELISPOT responses, which peaked 2 to 4 weeks after the boost given at week 6 (Fig. 3C).
After second set of immunizations during Phase II, we also observed proliferation of CD4 as well as CD8 T cells in response to SIV Gag protein (Fig. 4B), suggesting induction of SIV Gag-specific memory T cells. Although this vaccination was performed i.g., we did not see p11C-specific T cells in rectal mucosal tissues after staining of mononuclear cells from biopsies (data not shown).
3.5 Lmdd-BdopSIVgag induced equivalent anti-Gag immune responses among animals immunized according to schedules yielding persistent versus non-persistent antigen exposure
Next, we compared the immunogenicity of the alternate day dose-schedule with daily vaccine administration schedule for three consecutive days. In this pilot study, we compared the anti-Gag ELISPOT responses of Group B during Phase I with that of the animals in Group C. Phase I data of Listeria shedding in feces suggest that the bacteria did not persist in the animals’ gut until the administration of next dose given at 48 h later. Therefore in Phase II, the dose schedule was modified and the oral dose of Listeria vector was given on 3 consecutive days (days 0, 1, 2). After this modification, all animals showed persistent shedding of Listeria from day 1 to 3 (Table 3), implying that continuous antigen exposure was achieved during this set of immunization, with which we sought to mimic the course of natural infection. In this pilot study, we compared the anti-Gag ELISPOT responses of Group B during Phase I with those of the RM in Group C. Anti-Gag ELISPOT responses did not differ much following vaccination according to the two different dose-schedules. After the initial prime/boost immunizations at weeks 0 and 13, two out of five RM in Group B (Phase I) versus three out of five RM in Group C (Phase II) were clearly positive. Given the small group size, this difference was not significant.
3.6. Lmdd-BdopSIVgag did not induce anti-Lm or anti-SIV Gag humoral responses
Although our vaccination induced SIV Gag-specific T-cell responses, we did not detect humoral responses to Gag or whole Lm. ELISA results were negative even after multiple i.g. Lm exposures of Group B animals (data not shown). The exclusive activation of cellular immunity without humoral counterpart is characteristic of the intracellular survival and proliferation of Listeria and these results are in accordance with our earlier study [26].
4. Discussion
The intracellular bacterium Lm has been considered as live vaccine vector to deliver foreign antigens to elicit antigen-specific cellular immune responses. However, Lm is a food-borne pathogen that causes listeriosis and hence, attempts are directed toward attenuating bacterial pathogenicity while retaining immunogenicity. We developed new live attenuated strain of Lm, Lmdd-BdopSIVgag, to induce SIV Gag-specific cellular immune responses.
We found that our new Listeria vector was safe and well tolerated by all RM. No adverse reaction or illnesses were induced even after administration of multiple high doses (3 × 1012 organisms). In contrast, a single oral dose of 1×109 wild-type Lm caused severe listeriosis in non-human primates [52]. Shedding of Lmdd-BdopSIVgag was observed only for the limited duration following i.g. administration. Hence, we conclude that our new vaccine vector was safe among non-human primates. The vector also induced SIV Gag-specific cellular immune responses that were higher than those induced by our earlier live attenuated strain, based upon the Lmdd vector [26]. Thus, Lmdd-BdopSIVgag was more immunogenic than the earlier version and was also easier to administer since it did not require exogenous D-alanine for its replication.
Besides safety, another major concern associated with the use of vectored vaccines is the issue of preexisting anti-vector immunity [53], which has gained more attention after the failed STEP trial. Despite initially encouraging data from preclinical and clinical studies, the efficacy of human adenoviral vector serotype 5 (AdHu5) was hampered by a strong preexisting anti-vector immunity among vaccinated macaques, in which transgene-specific T cells homed to different organs in the presence of anti-vector immunity [54]. Lower vaccine-vector transgene immunogenicity has been reported for the adeno-associated virus (AAV) vector in animal models in the presence of anti-vector nAbs [55]. Furthermore, prior exposure to wild-type Salmonella also negatively affected the use of this bacterium as vaccine vector [56, 57]. Since Lm is a ubiquitous bacterium, anti-Lm immune responses are likely to be present among the majority of individuals. The issue of preexisting immunity in the context of development of Listeria as vaccine vector has been addressed only in murine [43, 44] and feline [42] systems. These studies showed that despite previous exposure to Lm, induction of immune responses to the new antigens carried by Listeria was not hampered. Our current findings are in agreement with these earlier reports. Although we detected strong lymphocyte proliferation to whole Lm before the administration of the Lmdd-BdopSIVgag vaccine vector, SIV Gag-specific cellular responses were induced among the vaccinated monkeys. Moreover, we also found that secondary anamnestic cellular responses were generated after re-exposure to this vaccine strain, suggesting that tolerance was not induced despite repeated oral administrations of live attenuated Lmdd-BdopSIVgag.
We did not detect any anti-Lm antibodies despite multiple Lm exposures in our primate studies, a finding that is in agreement with earlier reports. Infection of mice with sublethal doses of wild-type Lm did not induce anti-Lm antibodies [58]. In a monkey model of listeriosis during pregnancy, highly invasive Lm infection was required to induce high-titer anti-Lm antibodies, which in turn were associated with placental invasion and stillbirths [51]. Similarly, in our earlier study with the live attenuated Lm vector, Lmdd-HIVgag, anti-Lm antibodies were not detected after oral priming and boosting but were detected after oral priming followed by intramuscular boosting in macaques [26]. In humans, characterization of Lm-specific cellular and humoral immune responses among healthy individuals showed that while 60% of individuals had Lm-specific cellular responses, only 16% of the individuals showed low-titer anti-Lm antibodies [43].
The present study shows that our newly developed, live attenuated Lm strain, Lmdd-BdopSIVgag, was safe and immunogenic in non-human primates despite preexisting anti-Lm immunity shown by proliferative responses. This new strain induced SIV Gag-specific cellular immune responses after i.g. administration, and repeated mucosal administrations did not lead to induction of tolerance.
Acknowledgements
We thank Elizabeth Samit for assistance with the preparation of this manuscript, Stephanie Ehnert for the coordination of the primate studies, and Dr. Richard Raybourne (formerly of the Food and Drug Administration) for assistance and protocols to assess anti-Lm immune responses. This study was supported by NIH grant P01 AI054558 to FF and RMR.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Baba TW, Liska V, Hofmann-Lehmann R, Vlasak J, Xu W, Ayehunie S, et al. Human neutralizing monoclonal antibodies of the IgG1 subtype protect against mucosal simian-human immunodeficiency virus infection. Nat Med. 2000 Feb;6(2):200–206. doi: 10.1038/72309. [DOI] [PubMed] [Google Scholar]
- 2.Mascola JR, Lewis MG, Stiegler G, Harris D, VanCott TC, Hayes D, et al. Protection of Macaques against pathogenic simian/human immunodeficiency virus 89.6PD by passive transfer of neutralizing antibodies. J Virol. 1999 May;73(5):4009–4018. doi: 10.1128/jvi.73.5.4009-4018.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parren PW, Marx PA, Hessell AJ, Luckay A, Harouse J, Cheng-Mayer C, et al. Antibody protects macaques against vaginal challenge with a pathogenic R5 simian/human immunodeficiency virus at serum levels giving complete neutralization in vitro. J Virol. 2001 Sep;75(17):8340–8347. doi: 10.1128/JVI.75.17.8340-8347.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mascola JR, Stiegler G, VanCott TC, Katinger H, Carpenter CB, Hanson CE, et al. Protection of macaques against vaginal transmission of a pathogenic HIV-1/SIV chimeric virus by passive infusion of neutralizing antibodies. Nat Med. 2000 Feb;6(2):207–210. doi: 10.1038/72318. [DOI] [PubMed] [Google Scholar]
- 5.Wilson NA, Keele BF, Reed JS, Piaskowski SM, MacNair CE, Bett AJ, et al. Vaccine-induced cellular responses control simian immunodeficiency virus replication after heterologous challenge. J Virol. 2009 Jul;83(13):6508–6521. doi: 10.1128/JVI.00272-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu J, O'Brien KL, Lynch DM, Simmons NL, La Porte A, Riggs AM, et al. Immune control of an SIV challenge by a T-cell-based vaccine in rhesus monkeys. Nature. 2009 Jan 1;457(7225):87–91. doi: 10.1038/nature07469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hansen SG, Vieville C, Whizin N, Coyne-Johnson L, Siess DC, Drummond DD, et al. Effector memory T cell responses are associated with protection of rhesus monkeys from mucosal simian immunodeficiency virus challenge. Nat Med. 2009 Mar;15(3):293–299. doi: 10.1038/nm.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stover CK, de la Cruz VF, Fuerst TR, Burlein JE, Benson LA, Bennett LT, et al. New use of BCG for recombinant vaccines. Nature. 1991 Jun 6;351(6326):456–460. doi: 10.1038/351456a0. [DOI] [PubMed] [Google Scholar]
- 9.Weiss S. Transfer of eukaryotic expression plasmids to mammalian hosts by attenuated Salmonella spp. Int J Med Microbiol. 2003 Apr;293(1):95–106. doi: 10.1078/1438-4221-00248. [DOI] [PubMed] [Google Scholar]
- 10.Cayabyab MJ, Hovav AH, Hsu T, Krivulka GR, Lifton MA, Gorgone DA, et al. Generation of CD8+ T-cell responses by a recombinant nonpathogenic Mycobacterium smegmatis vaccine vector expressing human immunodeficiency virus type 1 Env. J Virol. 2006 Feb;80(4):1645–1652. doi: 10.1128/JVI.80.4.1645-1652.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brockstedt DG, Giedlin MA, Leong ML, Bahjat KS, Gao Y, Luckett W, et al. Listeria-based cancer vaccines that segregate immunogenicity from toxicity. Proc Natl Acad Sci U S A. 2004 Sep 21;101(38):13832–13837. doi: 10.1073/pnas.0406035101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jensen ER, Selvakumar R, Shen H, Ahmed R, Wettstein FO, Miller JF. Recombinant Listeria monocytogenes vaccination eliminates papillomavirus-induced tumors and prevents papilloma formation from viral DNA. J Virol. 1997 Nov;71(11):8467–8474. doi: 10.1128/jvi.71.11.8467-8474.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Souders NC, Verch T, Paterson Y. In vivo bactofection: listeria can function as a DNA-cancer vaccine. DNA Cell Biol. 2006 Mar;25(3):142–151. doi: 10.1089/dna.2006.25.142. [DOI] [PubMed] [Google Scholar]
- 14.Singh R, Paterson Y. Listeria monocytogenes as a vector for tumor-associated antigens for cancer immunotherapy. Expert Rev Vaccines. 2006 Aug;5(4):541–552. doi: 10.1586/14760584.5.4.541. [DOI] [PubMed] [Google Scholar]
- 15.Wallecha A, Maciag PC, Rivera S, Paterson Y, Shahabi V. Construction and characterization of an attenuated Listeria monocytogenes strain for clinical use in cancer immunotherapy. Clin Vaccine Immunol. 2009 Jan;16(1):96–103. doi: 10.1128/CVI.00274-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shahabi V, Reyes-Reyes M, Wallecha A, Rivera S, Paterson Y, Maciag P. Development of a Listeria monocytogenes based vaccine against prostate cancer. Cancer Immunol Immunother. 2008 Sep;57(9):1301–1313. doi: 10.1007/s00262-008-0463-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peng X, Hussain SF, Paterson Y. The ability of two Listeria monocytogenes vaccines targeting human papillomavirus-16 E7 to induce an antitumor response correlates with myeloid dendritic cell function. J Immunol. 2004 May 15;172(10):6030–6038. doi: 10.4049/jimmunol.172.10.6030. [DOI] [PubMed] [Google Scholar]
- 18.Sewell DA, Pan ZK, Paterson Y. Listeria-based HPV-16 E7 vaccines limit autochthonous tumor growth in a transgenic mouse model for HPV-16 transformed tumors. Vaccine. 2008 Sep 26;26(41):5315–5320. doi: 10.1016/j.vaccine.2008.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wood LM, Guirnalda PD, Seavey MM, Paterson Y. Cancer immunotherapy using Listeria monocytogenes and listerial virulence factors. Immunol Res. 2008;42(1–3):233–245. doi: 10.1007/s12026-008-8087-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tangney M, Gahan CG. Listeria monocytogenes as a vector for anticancer therapies. Curr Gene Ther. 2010 Feb;10(1):46–55. doi: 10.2174/156652310790945539. [DOI] [PubMed] [Google Scholar]
- 21.Maciag PC, Radulovic S, Rothman J. The first clinical use of a live-attenuated Listeria monocytogenes vaccine: a Phase I safety study of Lm-LLO-E7 in patients with advanced carcinoma of the cervix. Vaccine. 2009 Jun 19;27(30):3975–3983. doi: 10.1016/j.vaccine.2009.04.041. [DOI] [PubMed] [Google Scholar]
- 22.Lieberman J, Frankel FR. Engineered Listeria monocytogenes as an AIDS vaccine. Vaccine. 2002 May 6;20(15):2007–2010. doi: 10.1016/s0264-410x(02)00088-9. [DOI] [PubMed] [Google Scholar]
- 23.Frankel FR, Hegde S, Lieberman J, Paterson Y. Induction of cell-mediated immune responses to human immunodeficiency virus type 1 Gag protein by using Listeria monocytogenes as a live vaccine vector. J Immunol. 1995 Nov 15;155(10):4775–4782. [PubMed] [Google Scholar]
- 24.Peters C, Peng X, Douven D, Pan ZK, Paterson Y. The induction of HIV Gag-specific CD8+ T cells in the spleen and gut-associated lymphoid tissue by parenteral or mucosal immunization with recombinant Listeria monocytogenes HIV Gag. J Immunol. 2003 May 15;170(10):5176–5187. doi: 10.4049/jimmunol.170.10.5176. [DOI] [PubMed] [Google Scholar]
- 25.Zhao X, Zhang M, Li Z, Frankel FR. Vaginal protection and immunity after oral immunization of mice with a novel vaccine strain of Listeria monocytogenes expressing human immunodeficiency virus type 1 gag. J Virol. 2006 Sep;80(18):8880–8890. doi: 10.1128/JVI.00894-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang S, Rasmussen RA, Nolan KM, Frankel FR, Lieberman J, McClure HM, et al. Live attenuated Listeria monocytogenes expressing HIV Gag: immunogenicity in rhesus monkeys. Vaccine. 2007 Oct 16;25(42):7470–7479. doi: 10.1016/j.vaccine.2007.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Z, Zhang M, Zhou C, Zhao X, Iijima N, Frankel FR. Novel vaccination protocol with two live mucosal vectors elicits strong cell-mediated immunity in the vagina and protects against vaginal virus challenge. J Immunol. 2008 Feb 15;180(4):2504–2513. doi: 10.4049/jimmunol.180.4.2504. [DOI] [PubMed] [Google Scholar]
- 28.Rayevskaya MV, Frankel FR. Systemic immunity and mucosal immunity are induced against human immunodeficiency virus Gag protein in mice by a new hyperattenuated strain of Listeria monocytogenes. J Virol. 2001 Mar;75(6):2786–2791. doi: 10.1128/JVI.75.6.2786-2791.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wallecha A, Carroll KD, Maciag PC, Rivera S, Shahabi V, Paterson Y. Multiple effector mechanisms induced by recombinant Listeria monocytogenes anticancer immunotherapeutics. Adv Appl Microbiol. 2009;66:1–27. doi: 10.1016/S0065-2164(08)00801-0. [DOI] [PubMed] [Google Scholar]
- 30.Paterson Y, Maciag PC. Listeria-based vaccines for cancer treatment. Curr Opin Mol Ther. 2005 Oct;7(5):454–460. [PubMed] [Google Scholar]
- 31.Birmingham CL, Canadien V, Kaniuk NA, Steinberg BE, Higgins DE, Brumell JH. Listeriolysin O allows Listeria monocytogenes replication in macrophage vacuoles. Nature. 2008 Jan 17;451(7176):350–354. doi: 10.1038/nature06479. [DOI] [PubMed] [Google Scholar]
- 32.Brzoza KL, Rockel AB, Hiltbold EM. Cytoplasmic entry of Listeria monocytogenes enhances dendritic cell maturation and T cell differentiation and function. J Immunol. 2004 Aug 15;173(4):2641–2651. doi: 10.4049/jimmunol.173.4.2641. [DOI] [PubMed] [Google Scholar]
- 33.Webster P. Early intracellular events during internalization of Listeria monocytogenes by J774 cells. J Histochem Cytochem. 2002 Apr;50(4):503–518. doi: 10.1177/002215540205000407. [DOI] [PubMed] [Google Scholar]
- 34.Cossart P, Toledo-Arana A. Listeria monocytogenes, a unique model in infection biology: an overview. Microbes Infect. 2008 Jul;10(9):1041–1050. doi: 10.1016/j.micinf.2008.07.043. [DOI] [PubMed] [Google Scholar]
- 35.Lecuit M. Understanding how Listeria monocytogenes targets and crosses host barriers. Clin Microbiol Infect. 2005 Jun;11(6):430–436. doi: 10.1111/j.1469-0691.2005.01146.x. [DOI] [PubMed] [Google Scholar]
- 36.Ireton K. Entry of the bacterial pathogen Listeria monocytogenes into mammalian cells. Cell Microbiol. 2007 Jun;9(6):1365–1375. doi: 10.1111/j.1462-5822.2007.00933.x. [DOI] [PubMed] [Google Scholar]
- 37.Drevets DA, Bronze MS. Listeria monocytogenes: epidemiology, human disease, and mechanisms of brain invasion. FEMS Immunol Med Microbiol. 2008 Jul;53(2):151–165. doi: 10.1111/j.1574-695X.2008.00404.x. [DOI] [PubMed] [Google Scholar]
- 38.Williams D, Castleman J, Lee CC, Mote B, Smith MA. Risk of fetal mortality after exposure to Listeria monocytogenes based on dose-response data from pregnant guinea pigs and primates. Risk Anal. 2009 Nov;29(11):1495–1505. doi: 10.1111/j.1539-6924.2009.01308.x. [DOI] [PubMed] [Google Scholar]
- 39.Swaminathan B, Gerner-Smidt P. The epidemiology of human listeriosis. Microbes Infect. 2007 Aug;9(10):1236–1243. doi: 10.1016/j.micinf.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 40.Brockstedt DG, Bahjat KS, Giedlin MA, Liu W, Leong M, Luckett W, et al. Killed but metabolically active microbes: a new vaccine paradigm for eliciting effector T-cell responses and protective immunity. Nat Med. 2005 Aug;11(8):853–860. doi: 10.1038/nm1276. [DOI] [PubMed] [Google Scholar]
- 41.Rudnicka W, Kaczmarek M, Szeliga J, Germann T, Wieckowska M, Rozalska B. The host response to Listeria monocytogenes mutants defective in genes encoding phospholipases C (plcA, plcB) and actin assembly (actA) Microbiol Immunol. 1997;41(11):847–853. doi: 10.1111/j.1348-0421.1997.tb01939.x. [DOI] [PubMed] [Google Scholar]
- 42.Stevens R, Lavoy A, Nordone S, Burkhard M, Dean GA. Pre-existing immunity to pathogenic Listeria monocytogenes does not prevent induction of immune responses to feline immunodeficiency virus by a novel recombinant Listeria monocytogenes vaccine. Vaccine. 2005 Feb 10;23(12):1479–1490. doi: 10.1016/j.vaccine.2004.09.033. [DOI] [PubMed] [Google Scholar]
- 43.Leong ML, Hampl J, Liu W, Mathur S, Bahjat KS, Luckett W, et al. Impact of preexisting vector-specific immunity on vaccine potency: characterization of listeria monocytogenes-specific humoral and cellular immunity in humans and modeling studies using recombinant vaccines in mice. Infect Immun. 2009 Sep;77(9):3958–3968. doi: 10.1128/IAI.01274-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Starks H, Bruhn KW, Shen H, Barry RA, Dubensky TW, Brockstedt D, et al. Listeria monocytogenes as a vaccine vector: virulence attenuation or existing antivector immunity does not diminish therapeutic efficacy. J Immunol. 2004 Jul 1;173(1):420–427. doi: 10.4049/jimmunol.173.1.420. [DOI] [PubMed] [Google Scholar]
- 45.Thompson RJ, Bouwer HG, Portnoy DA, Frankel FR. Pathogenicity and immunogenicity of a Listeria monocytogenes strain that requires D-alanine for growth. Infect Immun. 1998 Aug;66(8):3552–3561. doi: 10.1128/iai.66.8.3552-3561.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Z, Zhao X, Zhou C, Gu B, Frankel FR. A truncated Bacillus subtilis dal gene with a 3' ssrA gene tag regulates the growth and virulence of racemase-deficient Listeria monocytogenes. Microbiology. 2006 Oct;152(Pt 10):3091–3102. doi: 10.1099/mic.0.28994-0. [DOI] [PubMed] [Google Scholar]
- 47.Siddappa NB, Song R, Kramer VG, Chenine AL, Velu V, Ong H, et al. Neutralization-sensitive R5-tropic simian-human immunodeficiency virus SHIV-2873Nip, which carries env isolated from an infant with a recent HIV clade C infection. J Virol. 2009 Feb;83(3):1422–1432. doi: 10.1128/JVI.02066-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rasmussen RA, Ong H, Song R, Chenine AL, Ayash-Rashkovsky M, Hu SL, et al. Efficacy of a multigenic protein vaccine containing multimeric HIV gp160 against heterologous SHIV clade C challenges. AIDS. 2007 Sep 12;21(14):1841–1848. doi: 10.1097/QAD.0b013e32828684ea. [DOI] [PubMed] [Google Scholar]
- 49.Rasmussen RA, Lakhashe SK, Ruprecht RM. Bimodal AIDS vaccine approach: induction of cellular as well as humoral immunity can protect from systemic infection. Vaccine. 2010 May 26;28 Suppl 2:B25–B31. doi: 10.1016/j.vaccine.2009.10.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dzuris JL, Sidney J, Appella E, Chesnut RW, Watkins DI, Sette A. Conserved MHC class I peptide binding motif between humans and rhesus macaques. J Immunol. 2000 Jan 1;164(1):283–291. doi: 10.4049/jimmunol.164.1.283. [DOI] [PubMed] [Google Scholar]
- 51.Smith MA, Takeuchi K, Brackett RE, McClure HM, Raybourne RB, Williams KM, et al. Nonhuman primate model for Listeria monocytogenes-induced stillbirths. Infect Immun. 2003 Mar;71(3):1574–1579. doi: 10.1128/IAI.71.3.1574-1579.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Farber JM, Daley E, Coates F, Beausoleil N, Fournier J. Feeding trials of Listeria monocytogenes with a nonhuman primate model. J Clin Microbiol. 1991 Nov;29(11):2606–2608. doi: 10.1128/jcm.29.11.2606-2608.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cohen P. Immunity's yin and yang. A successful vaccine must first avoid being eliminated by pre-existing immunity before it can promote a protective immune response. IAVI Rep. 2006 Jan–Feb;10(1):1–5. [PubMed] [Google Scholar]
- 54.McCoy K, Tatsis N, Korioth-Schmitz B, Lasaro MO, Hensley SE, Lin SW, et al. Effect of preexisting immunity to adenovirus human serotype 5 antigens on the immune responses of nonhuman primates to vaccine regimens based on human- or chimpanzee-derived adenovirus vectors. J Virol. 2007 Jun;81(12):6594–6604. doi: 10.1128/JVI.02497-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lin J, Calcedo R, Vandenberghe LH, Figueredo JM, Wilson JM. Impact of preexisting vector immunity on the efficacy of adeno-associated virus-based HIV-1 Gag vaccines. Hum Gene Ther. 2008 Jul;19(7):663–669. doi: 10.1089/hum.2008.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vindurampulle CJ, Attridge SR. Impact of vector priming on the immunogenicity of recombinant Salmonella vaccines. Infect Immun. 2003 Jan;71(1):287–297. doi: 10.1128/IAI.71.1.287-297.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gahan ME, Webster DE, Wijburg OL, Wesselingh SL, Strugnell RA. Impact of prior immunological exposure on vaccine delivery by Salmonella enterica serovar Typhimurium. Vaccine. 2008 Nov 18;26(49):6212–6220. doi: 10.1016/j.vaccine.2008.09.060. [DOI] [PubMed] [Google Scholar]
- 58.Hage-Chahine CM, Del Giudice G, Lambert PH, Pechere JC. Hemolysin-producing Listeria monocytogenes affects the immune response to T-cell-dependent and T-cell-independent antigens. Infect Immun. 1992 Apr;60(4):1415–1421. doi: 10.1128/iai.60.4.1415-1421.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]