Abstract
The functions of a gene are traditionally annotated textually using either free text (Gene Reference Into Function or GeneRIF) or controlled vocabularies (e.g., Gene Ontology or Disease Ontology). Inspired by the latest word cloud tools developed by the Information Visualization Group at IBM Research, we have prototyped a visual system for capturing gene annotations, which we named Gene Graph Into Function or GeneGIF. Fully developing the GeneGIF system would be a significant effort. To justify the necessity and to specify the design requirements of GeneGIF, we first surveyed the end-user preferences. From 53 responses, we found that a majority (64%, p < 0.05) of the users were either positive or neutral toward using GeneGIF in their daily work (acceptance); in terms of preference, a slight majority (51%, p > 0.05) of the users favored visual presentation of information (GeneGIF) compared to textual (GeneRIF) information. The results of this study indicate that a visual presentation tool, such as GeneGIF, can complement standard textual presentation of gene annotations. Moreover, the survey participants provided many constructive comments that will specify the development of a phase-two project (http://128.248.174.241/) to visually annotate each gene in the human genome.
Keywords: Gene function, Social networking, Visualization, Word cloud
79.1 Introduction
Genes in the human genome have been predominantly annotated using unstructured text. For example, the Gene Reference Into Function (GeneRIF) provides a tool to include one or more 255-character-long “gene function” statements that couple a specific publication with a gene [4, 6]. An example GeneRIF annotation of the human Kruppel-like factor 4 (KLF4, GeneID:9314) gene is shown in Fig. 79.1. For genes with more than about ten GeneRIFs, it is time-consuming to review the knowledge present in GeneRIFs.
Fig. 79.1.
GeneRIF annotation of gene KLF4 (human). Each GeneRIF is a statement up to 255-character long. Note that only 9 out of the 49 GeneRIFs are presented
Gene Ontology annotations [3] and Disease Ontology annotations [5] of a gene are more compact and the ontological structure makes these annotations much easier for a human reader to parse, in addition to the advantages of these ontologies for semantic reasoning and inference. However, these ontological systems require training to use consistently and accurately, and require a significant investment in curatorial time to build the ontological structure.
We investigated a different approach to present the genome annotation data. Research in human cognition has suggested that visual presentation can facilitate human learning and knowledge acquisition [2, 7, 10]. New semantic web tools, such as word clouds, appear to be ideally suited for helping people rapidly parse large amounts of textual data. Thus, we explored the impact of using a word cloud visual presentation of gene annotation information using the latest visualization tools developed by the Information Visualization Group at IBM Research (http://manyeyes.alphaworks.ibm.com/manyeyes). We call this visual annotation of a gene a “Gene Graph Into Function (GeneGIF).” Results from the user survey suggest that the visual presentation (GeneGIF) is complementary to the raw text presentation (GeneRIF) in current use.
79.2 Results
79.2.1 Word Clouds: A Direct Application of Existing Visualization Tools
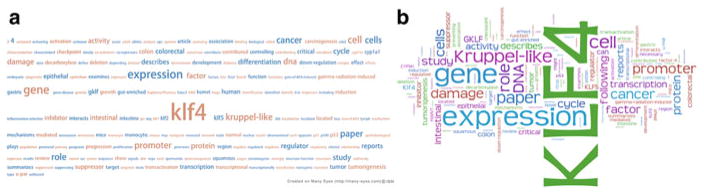
A word cloud is a visual display of a set of words, where the font, size, color, or even movement can represent some underlying information. When a reader is in the process of acquiring new information by examining evidences in a cursory manner, a word cloud can be very effective. Our first attempt was to apply the existing “word clould” tools of “tag-cloud” and “Wordle” from the “Many Eyes” project of IBM Research to display the words with their fonts proportional to their frequencies in the GeneRIFs of a gene, and the colors were added to enhance readability. Compared with the tag cloud tool that presents the words alphabetically (Fig. 79.2a) the Wordle tool utilizes the screen space more effectively (Fig. 79.2b).
Fig. 79.2.
Word clouds for the gene annotation of KLF4. (a) Using the tag cloud algorithm (b) using the Wordle algorithm
79.2.2 Incorporating Domain-Specific Knowledge to Improve Wordle
Although Fig. 79.2b effectively summarizes the functions of KLF4, such as its role in DNA damage, cell cycle, promoter activity, and cancer, a careful examination of Fig. 79.2b suggests the following problems:
Self-referencing: The gene symbol, “KLF4,” is a self-reference. Although it appears as the most frequent word, it carries little extra information. Thus, the self-referencing words should be removed.
Inflected words: The plural form of “cells” carries redundant information with the single form of “cell.” Other examples include “KLF4” and “klf4” (upper vs. lower case). Thus, word stemming should be included in the algorithm.
Stopwords: Although the common English stopwords are removed, some domain-specific stopwords, such as “gene,” “paper,” or “study,” should also be removed.
Phrases: Currently, the Wordle algorithm cannot identify phrases such as “cell cycle.”
To count the word frequency more accurately, we applied the Porter Stemming Algorithm [9] to reduce words to word stems: for example, the inflected words “stimulates,” “stimulated,” and “stimulating” are all reduced to the stem form of “stimulate.” In addition, we use the vocabulary list from the Gene Ontology [1] to identify phrases such as “cell cycle” and count them as a unit.
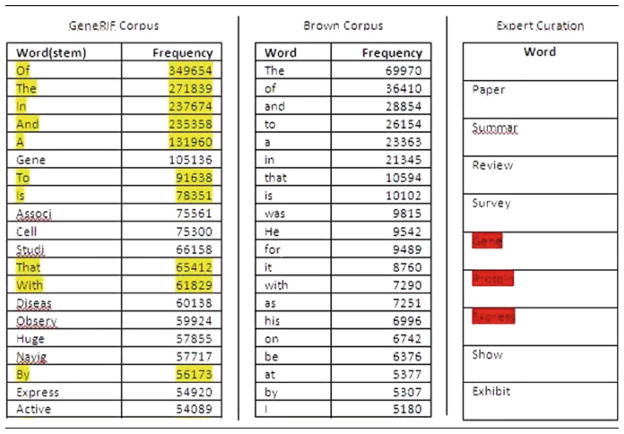
As noted, most word cloud generation engines remove stopwords from the input set before generating the cloud. Stopwords are overrepresented words or phrases which constitute parts of speech which occur frequently but convey nonspecific information. In text of a general nature, it suffices to remove definite and indefinite articles, prepositions, pronouns, and so on. However, in the application specific sense, there can be a large set of words which are redundant. In examining GeneRIFs, for example, common biological terms such as “gene” or “protein” will occur frequently and were added to a list of GeneGIF stopwords. To do this more formally, we used the entire GeneRIF as a corpus to identify the 50 most frequently occurring words (top 20 shown in Table 79.1). In contrast with the common English stopwords identified from the Brown corpus [8], we call the domain-specific stop-words “bio-stopwords.” We combined the three lists from (Table 79.1) to remove the stopwords in GeneRIF.
Table 79.1.
Stopwords. The 20 most frequently occurring words (after stemming) in the entire GeneRIF dataset (left) and the Brown Corpus (center) are shown, and the overlaps between the two lists are highlighted. On the right is a list of manually identified stopwords, and its overlaps with the GeneRIF-corpus-list are highlighted
 |
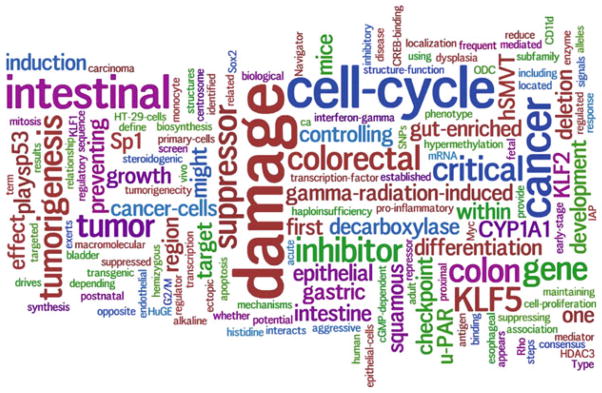
The final visual presentation of KLF4 is shown in Fig. 79.3. We call this improved visual annotation of a gene “Gene Graph Into Function” (GeneGIF). The GeneGIF of KLF4 quickly summarizes the major functions of KLF4 from 49 entries of GeneRIF by displaying the more frequent keywords in bigger font: KLF4 is a cell-cycle checkpoint protein that prevents mitosis after DNA damage, and is thought to function as a tumor suppressor. KLF4 plays an important role in the tumorigenesis of intestinal cancers, especially colorectal adenocarcinomas. Decreased expression of KLF4 has been demonstrated in surgically resected colorectal cancers. The normal function of KLF4 seems to require the wild type p53 protein. (The underscored words above are the keywords identified by GeneGIF.)
Fig. 79.3.
Improvements over the Wordle algorithm. We removed all of the self-references, stopwords (English stopwords and bio-stopwords) and hyphenate the phrases (for example, cell-cycle, interferon-gamma, and cancer-cells are identified)
79.3 User Survey
To test the utility of GeneGIF, we surveyed the users of gene annotations. We sent a survey to participants of the MAQC project who are experts on genomic data analysis using microarrays. Prospective participants were informed with the purpose, procedure, and handling of the survey. Since it was an anonymous survey, signatures for the consent were waived. 53 responses were collected. Statistical tests of the survey results suggest that in terms of acceptance, a majority (point estimate: 64%; 95% confidence interval: 50–77%) of the users were either positive or neutral toward using GeneGIF in their daily work; in terms of preference, a slight majority (51%, not statistically significant) of the users favored visual (GeneGIF) information compared to textual (GeneRIF) information. An Analysis of Variance (ANOVA) suggests no significant association of the outcome (GeneGIF vs. GeneRIF) with either gender, age, field of study, English as first language, or education level.
79.4 Discussion
The state of knowledge about a given gene changes, and these changes are reflected in the literature. Although the GeneRIF provides a mechanism to keep the functional annotation of a gene up-to-date, reading through GeneRIF entries to identify significant and recurring points is not easy when there are dozens or even hundreds of GeneRIFs.
For the first time, we have prototyped a visual system of gene annotation (GeneGIF) by summarizing the phrases used in a collection of GeneRIFs. As the user comments indicate, GeneGIF is much more effective in getting a rough overview of the gene’s major functions while GeneRIF can provide more detailed and precise information. Therefore, GeneGIFs are complementary to the raw textual display of GeneRIFs. The MAQC respondents also pointed out that the current prototype of GeneGIF is very primitive. For instance, we can make the GeneGIF clickable and directly linked it to individual GeneRIF items with the keyword highlighted. Based on these positive feedbacks, we have begun a phase II project to use GeneGIF to annotate each gene in the human genome (http://128.248.174.241/).
We have also found that the same visual representation can be used for more than just single genes. We have used gene lists from gene expression experiments to build word clouds that are based on a collection of GeneRIF collections. This is a rapid way to identify functional pathways that are affected in the collection. Another application is to directly graph gene expression data into the cloud structure. For example, position can be used to define whether the expression was negative or positive (right to left, respectively), the size of the term for expression magnitude, colored grouping for related biomarkers (e.g., common pathway), and even some degree of movement (vibration) to express the noise/discrepancy. These various types of data are all amenable to word cloud visualization.
79.5 Materials and Methods
Gene annotations were downloaded from the NCBI Entrez database in January 2009. The word cloud was created using the Wordle algorithm from the “Many Eyes” project of IBM Research (http://manyeyes.alphaworks.ibm.com/manyeyes/). The survey was designed using the django-survey application (http://code.google.com/p/django-survey/). Other programs were written in Python. Normal approximation was used to estimate the 95% confidence interval for the proportion of users who were positive or neutral toward using GeneGIF in their daily work. A one sample z-test was used to test users’ preference of using GeneGIF to GeneRIF, i.e., the proportion of users who prefer using GeneGIF is greater than 50%.
Acknowledgments
The authors would like to thank Martin Wattenberg, Matthew M Mckeon, and Jonathan Feinberg at IBM Research for helpful discussion of Wordle and comments on this manuscript. The authors would also like to thank Rhett Sutphin at NUCATS for exploring the application programing interface to Wordle.
References
- 1.Ashburner M, Ball CA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature Genetics. 2000;25(1):25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Childers TL, Houston MJ, et al. Measurement of individual-differences in visual versus verbal information-processing. Journal of Consumer Research. 1985;12(2):125–134. [Google Scholar]
- 3.Harris MA, Clark J, et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Research. 2004;32(Database issue):D258–61. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maglott D, Ostell J, et al. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Research. 2007;35(Database issue):D26–31. doi: 10.1093/nar/gkl993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Osborne JD, Flatow J, et al. Annotating the human genome with disease ontology. BMC Genomics. 2009;10(Suppl 1):S6. doi: 10.1186/1471-2164-10-S1-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Osborne JD, Lin S, et al. Other riffs on cooperation are already showing how well a wiki could work. Nature. 2007;446(7138):856. doi: 10.1038/446856a. [DOI] [PubMed] [Google Scholar]
- 7.Plass JL, Chun DM, et al. Supporting visual and verbal learning preferences in a second-language multimedia learning environment. Journal of Educational Psychology. 1998;90(1):25–36. [Google Scholar]
- 8.Weiss SM. Text mining : predictive methods for analyzing unstructured information. New York: Springer; 2005. [Google Scholar]
- 9.Willett P. The Porter stemming algorithm: then and now. Program-Electronic Library and Information Systems. 2006;40(3):219–223. [Google Scholar]
- 10.Wyer RS, Jiang YW, et al. Visual and verbal information processing in a consumer context: Further considerations. Journal of Consumer Psychology. 2008;18(4):276–280. [Google Scholar]