Abstract
BK virus (BKV) is the causative agent for polyomavirus-associated nephropathy, a severe disease found in renal transplant patients due to reactivation of a persistent BKV infection. BKV replication relies on the interactions of BKV with many nuclear components, and subnuclear structures such as promyelocytic leukemia nuclear bodies (PML-NBs) are known to play regulatory roles during a number of DNA virus infections. In this study, we investigated the relationship between PML-NBs and BKV during infection of primary human renal proximal tubule epithelial (RPTE) cells. While the levels of the major PML-NB protein components remained unchanged, BKV infection of RPTE cells resulted in dramatic alterations in both the number and the size of PML-NBs. Furthermore, two normally constitutive components of PML-NBs, Sp100 and hDaxx, became dispersed from PML-NBs. To define the viral factors responsible for this reorganization, we examined the cellular localization of the BKV large tumor antigen (TAg) and viral DNA. TAg colocalized with PML-NBs during early infection, while a number of BKV chromosomes were adjacent to PML-NBs during late infection. We demonstrated that TAg alone was not sufficient to reorganize PML-NBs and that active viral DNA replication is required. Knockdown of PML protein did not dramatically affect BKV growth in culture. BKV infection, however, was able to rescue the growth of an ICP0-null herpes simplex virus 1 mutant whose growth defect was partially due to its inability to disrupt PML-NBs. We hypothesize that the antiviral functions of PML-NBs are inactivated through reorganization during normal BKV infection.
IMPORTANCE
BK virus (BKV) is a human pathogen that causes severe diseases, including polyomavirus-associated nephropathy in kidney transplant patients and hemorrhagic cystitis in bone marrow transplant recipients. How BKV replication is regulated and the effects of a lytic BKV infection on host cells at the molecular level are not well understood. Currently, there is no specific antiviral treatment for BKV-associated disease, and a better understanding of the complete life cycle of the virus is necessary. Here, we report the interplay between BKV and one of the regulatory structures in the host cell nucleus, promyelocytic leukemia nuclear bodies (PML-NBs). Our results show that BKV infection reorganizes PML-NBs as a strategy to inactivate the negative functions of PML-NBs.
INTRODUCTION
BK virus (BKV) is a ubiquitous human polyomavirus that normally persists in the urinary tract of healthy individuals without causing any disease. Under immunosuppressed conditions, particularly in kidney transplant patients, BKV can reactivate from a persistent to a lytic infection in renal proximal tubule epithelial (RPTE) cells (1). This reactivation can cause severe disease, including polyomavirus-associated nephropathy (PVAN); ~90% of patients who develop PVAN will lose their graft (2). BKV-related disease has become a serious concern due to an increase in the number of transplants performed and improvements in the immunosuppressive regimens used in these procedures (3). Unfortunately, little is known about how BKV switches from a persistent to a lytic infection in these cells, and there is currently no specific antiviral treatment available (4). Our laboratory has developed a cell culture system using primary human RPTE cells to analyze BKV infection (5). BKV grows lytically in these cells, and the morphology of BKV-infected cells is similar to that seen in PVAN patient kidneys. This system provides a first step towards gaining a complete picture of the BKV life cycle.
BKV begins its infectious life cycle by binding to cellular ganglioside receptors, followed by internalization and intracellular trafficking prior to delivery of its genome to the nucleus (6–8). Once inside the nucleus, viral early proteins are expressed and interact with various nuclear components to create an optimal environment conducive to viral replication. For example, the large tumor antigen (TAg) interacts with members of the retinoblastoma susceptibility protein (pRb) family to alleviate E2F-mediated transcription inhibition. TAg also binds to p53 and disrupts the ability of p53 to activate transcription of its target genes (reviewed in reference 9). The result of these interactions is to drive the cells into and keep the cells in S phase to provide the host DNA synthetic machinery that is necessary for BKV genome replication, and to prevent apoptosis to maximize the yield of progeny virus. Small t antigen contributes to BKV replication by orchestrating cell cycle progression through inhibiting the enzymatic activity of protein phosphatase 2A, increasing the transcription of cyclin D1 and cyclin A, and downregulating the cyclin kinase inhibitor p27 (10, 11). Simian virus 40 (SV40) TAg also manipulates the ubiquitin-proteasome pathway by inhibiting Cul7 and SCFFbw7 E3 ligase activities (12). The interactions of TAg with E3 ligases, however, were examined in transformation studies, and the implication of these interactions in productive viral infection remains to be determined.
Promyelocytic leukemia nuclear bodies (PML-NBs), also known as nuclear domains 10 (ND10), nuclear dots, or PML oncogenic domains (PODs), represent a potential obstacle for BKV replication. PML-NBs were identified almost 20 years ago (13), and a diverse array of functions have been assigned to these dynamic, small, punctate subnuclear structures, including transcription regulation, apoptosis, DNA damage response, oncogenesis, and chromatin remodeling (reviewed in references 14 and 15). Their name is derived from the major component of this structure, the PML protein, which was discovered as a fusion protein with the retinoic acid receptor through a chromosomal translocation event in patients with acute promyelocytic leukemia (16, 17). PML-NBs are complicated multiprotein complexes that are present in almost all cell types examined so far (13). They have a dynamic composition that changes upon various stress conditions, such as heat shock and DNA damage, and the list of proteins that are found to be present in or associated with PML-NBs is growing (reviewed in references 14 and 18). Several proteins, including the transcriptional regulators hDaxx and Sp100, are constitutively present in PML-NBs, while others are found only under certain stress conditions (19). The identification of a functional link between PML-NBs and viral infections began with the discoveries that herpes simplex virus 1 (HSV-1) infection destroys PML-NBs (20) and that PML-NBs are the nuclear deposition and replication site for a number of other DNA viruses (21–23). Later, it was demonstrated that PML-NBs possess both intrinsic and interferon (IFN)-mediated antiviral effects against several DNA and RNA viruses (15, 18, 24). There is also limited evidence that certain PML isoforms may facilitate bovine papillomavirus gene expression (25).
The roles that PML-NBs play in polyomavirus infection are less well characterized, but PML-NBs seem to be associated with all stages of the viral life cycle. In SV40-infected HEp-2 epidermoid carcinoma cells, some intense TAg areas are found juxtaposed to PML-NBs (21). In addition, in situ hybridization revealed that most of the DNA replication centers are adjacent to PML-NBs; however, there is no disruption of PML-NBs in these cells. Using a plasmid-based β-galactosidase expression system, Tang et al. demonstrated that the origin of SV40 DNA replication and TAg expression are required for targeting of β-galactosidase transcripts to PML-NBs (26). The accumulation of transcripts at PML-NBs can be disrupted by inhibiting DNA replication, which led the authors to conclude that TAg-directed DNA replication rather than transcription occurs in the vicinity of PML-NBs (26). For BKV, it has been shown by BrdU pulse-labeling that PML-NBs associate with newly synthesized viral DNA and that a portion of the DNA within these PML-NBs is single stranded (27). The authors hypothesized that the single-stranded DNA may represent recombination intermediates that accumulate during a replication-induced DNA repair process. Depleting PML protein using small interfering RNA (siRNA) had little effect on viral DNA replication (27). These studies, however, were conducted with primary brain cells and breast cancer cell lines, which are not the natural host cells for BKV; therefore, the biological significance of these results is unclear. For another human polyomavirus, JC virus (JCV), infection also does not cause degradation or relocalization of PML in the human glial cell line SVG-A, and short hairpin RNA (shRNA) knockdown of PML does not have significant effects on JCV replication (28). The inhibition of JCV by IFN-β, however, can be reversed by arsenite treatment, which degrades PML protein (28). Finally, the major and minor capsid proteins of JCV have been found to accumulate at PML-NBs in both transfected cells and oligodendrocytes derived from patients with progressive multifocal leukoencephalopathy, a disease caused by JCV (29). Both capsid proteins and virus-like particles were seen at discrete locations along the inner nuclear periphery, leading to the conclusion that PML-NBs may serve as a scaffold for JCV assembly (29, 30).
In this study, we characterized the effects of BKV infection on PML-NBs in its natural host, RPTE cells. Our results show that BKV infection caused a dramatic reorganization of PML-NBs, with a decrease in PML-NB number and an increase in size. In addition, both hDaxx and Sp100 were found to be dispersed from PML-NBs. This reorganization required BKV DNA replication; consistent with this, BKV DNA foci were found adjacent to PML-NBs during late infection. More importantly, we present evidence that the reorganization of PML-NBs is a strategy that BKV uses to inactivate intrinsic antiviral functions of PML-NBs.
RESULTS
BKV infection in RPTE cells alters both the number and the size of PML-NBs.
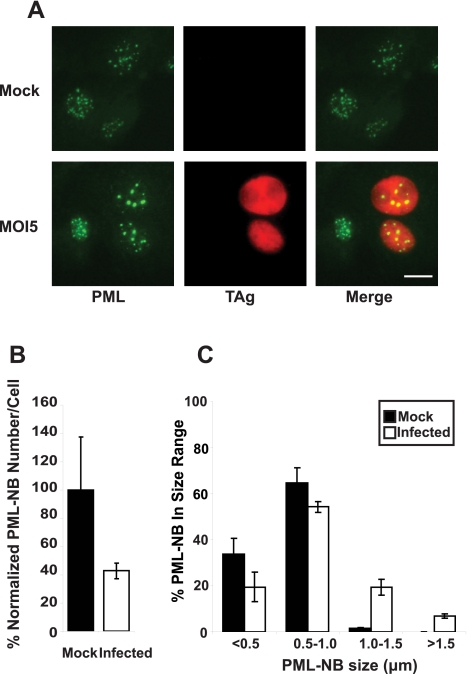
Various DNA virus infections have been shown to cause changes to PML-NBs. For example, PML-NBs are reorganized into track-like structures upon adenovirus (Ad) infection (23). This is accomplished by the early viral protein E4 ORF3 (23, 31). The HSV-1 regulatory protein ICP0 induces the degradation of PML in a proteasome-dependent manner (20, 32). In the case of human cytomegalovirus (HCMV), PML-NBs are dispersed into a diffuse nucleoplasmic form by the major immediate-early protein IE1, which interferes with the SUMOylation of PML (33, 34). To begin our characterization of the interplay between BKV and PML-NBs, we first examined the effects of BKV infection of RPTE cells on the morphology of PML-NBs. We visualized PML-NBs at 2 days postinfection (dpi) by immunostaining for the major defining component of PML-NBs, the PML protein (Fig. 1A). The viral early protein TAg was used as a marker for infected cells. In mock-infected cells, PML-NBs displayed a small, punctate, nuclear staining pattern. In TAg-positive cells, however, there was a dramatic difference in the appearance of PML-NBs. Overall, there were fewer PML-NBs in each nucleus, and some of the PML-NBs were larger than the ones in the mock-infected cells.
FIG 1 .
PML-NBs are altered morphologically during BKV infection. (A) RPTE cells were mock infected or infected with BKV at an MOI of 5 IU/cell for 48 h. The cells were fixed and immunostained for PML protein (green) and TAg (red). Images were obtained with a 40× objective on an Olympus BX41 fluorescence microscope. Scale bar, 10 µm. The number (B) and the size distribution (C) of PML-NBs were quantified as described in Materials and Methods. For panel B, the average number of PML-NBs in the mock-infected cells was set to 100%, and P was 0.06. At least 50 cells (B) or 200 PML-NBs (C) were counted or measured, respectively, in each sample. Each bar represents the average of results from three independent experiments, and the error bars represent the standard deviation (SD) values.
We quantified both the number (Fig. 1B) and the size distribution (Fig. 1C) of PML-NBs in both mock- and BKV-infected RPTE cells. There was an approximately 60% reduction in the average number of PML-NBs in the cells that were positive for TAg. To graph the size distribution of PML-NBs, we grouped PML-NBs based on their diameter measurements. PML-NBs that were larger than 1 µm in diameter were found exclusively in the BKV-infected samples. Cells that were assayed at 3 dpi showed a slightly more dramatic change in both PML-NB number and size than at 2 dpi (data not shown).
Levels and relative localization of PML-NB components during BKV infection.
To determine whether the decrease in PML-NB number was due to a decrease in PML protein level, we harvested total cell proteins from mock- and BKV-infected RPTE cells over a 3-day time course. The proteins were immunoblotted for three of the major components of PML-NBs: PML, hDaxx, and Sp100. Both PML and Sp100 have multiple isoforms produced from alternative mRNA splicing (35, 36), and the antibodies that we used recognize all the isoforms. TAg expression was also examined as a marker for the progression of the infection. No significant differences were seen in the levels of any PML-NB proteins between mock- and BKV-infected samples, although there was a reproducible slight increase of hDaxx in the BKV-infected samples compared to the level in the mock-infected samples at 2 and 3 dpi (see Fig. S1 in the supplemental material).
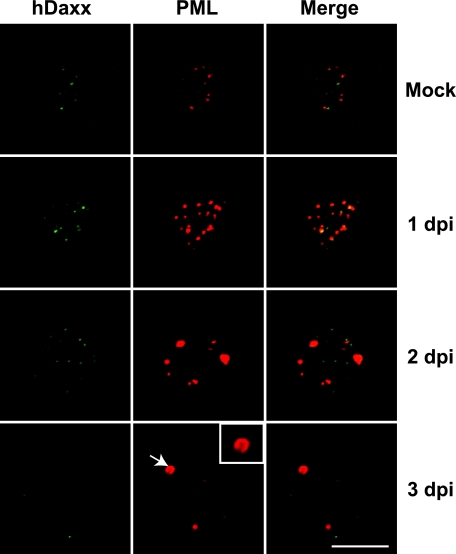
We then determined whether the enlarged PML-NBs in the infected cells have components similar to those of normal PML-NBs. We first examined the relative localizations of PML and hDaxx over the course of BKV infection, since hDaxx is considered a constitutive component of PML-NBs under normal growth conditions (37). In mock-infected cells, both hDaxx and PML displayed similar small punctate patterns, and most of the signals from these two proteins are colocalized with or adjacent to each other (Fig. 2). RPTE cells that were infected with BKV for 1 day showed a staining pattern similar to that of the mock-infected samples. By 2 dpi, enlarged PML-NBs started to appear, and these PML-NBs became more evident by 3 dpi, with some PML-NBs displaying a ring-like structure (Fig. 2 and 3A, arrows and insets). Intriguingly, as the PML-NBs enlarged, hDaxx began to disperse from the PML. There was still partial colocalization of PML and hDaxx in the 2-dpi samples, but by 3 dpi, the percentage of hDaxx foci that were colocalized with or adjacent to PML foci significantly decreased compared to the level for mock-infected cells (see Fig. S2A in the supplemental material). Similarly, Sp100 was also found to separate from PML by 3 dpi (Fig. S2B). SUMO-1, however, remains associated with PML throughout the course of infection (Fig. S2C). Due to the fact that hDaxx and Sp100 separate from PML, these results suggest that the enlargement of PML-NBs is not simply the fusion of preexisting PML-NBs.
FIG 2 .
PML and hDaxx localization during BKV infection. RPTE cells were mock infected or infected with BKV as described in the legend to Fig. 1. Confocal images of cells that were fixed at the indicated time points postinfection and immunostained for hDaxx (green) and PML protein (red) are shown. The arrow points to an enlarged ring-like PML-NB, which is magnified 2-fold in the inset. Scale bar, 10 µm.
FIG 3 .
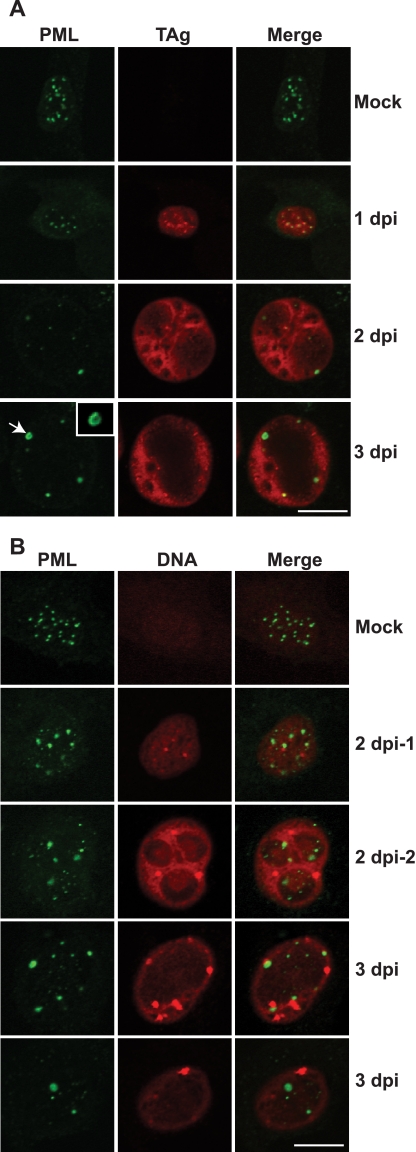
PML protein localization with BKV TAg and DNA. RPTE cells were mock infected or infected with BKV as described in the legend to Fig. 1 and fixed at the indicated time points postinfection. Samples were either coimmunostained for PML (green) and TAg (red) (A) or stained for PML (green) and BKV viral DNA using FISH (red) (B), and images were taken using a confocal microscope. The arrow in panel A points to an enlarged ring-like PML-NB, which is magnified 2-fold in the inset. Scale bar, 10 µm. In panel B, “2 dpi-1” and “2 dpi-2” represent early- and late-infected cells, respectively, at 2 dpi. Two different fields for 3 dpi are shown.
Dynamic localization of PML, TAg, and BKV DNA.
We then determined the localization of PML and viral factors to obtain insight into how the reorganization of PML-NBs occurs. Many DNA virus regulatory proteins are targeted to PML-NBs before they disrupt or reorganize these nuclear structures (reviewed in reference 19). We therefore first examined the relative localizations of PML and the BKV master regulatory protein TAg by immunostaining (Fig. 3A). TAg sets up the host environment for BKV replication by inactivating pRb and p53 and also participates in the initiation of viral DNA replication (9). During early infection (1 dpi), TAg appeared as small nuclear foci, many of which were colocalized with PML nuclear foci. Starting from 2 dpi, the majority of the TAg signals became diffused throughout the nucleus and there were several nuclear exclusion areas that were devoid of TAg. The TAg-excluded areas continued to increase in size to 3 dpi. PML localization did not appear to be random relative to the TAg staining; most of PML-NBs were found at the periphery of these TAg-excluded areas (Fig. 3A, arrow). We also examined the relative localizations of hDaxx and TAg (see Fig. S3 in the supplemental material); hDaxx colocalized with small TAg foci throughout the infection; no obvious association, however, was seen between hDaxx and the diffusely staining TAg observed during late infection.
Since the timing of PML-NB enlargement (2 dpi) coincides with the detection of BKV DNA replication (5), we also investigated the relative localizations between PML and viral DNA as visualized by fluorescent in situ hybridization (FISH) (Fig. 3B). We could not consistently detect viral DNA signals by FISH at 1 dpi, which is prior to the onset of DNA replication (5). Therefore, the FISH signals that we detected at 2 and 3 dpi were likely mostly newly replicated viral DNA. No FISH signals were seen in DNase-treated or nondenatured samples (data not shown), confirming that the signals were not from detection of viral RNA. A dynamic pattern was also observed between PML and viral DNA. In some of the cells at 2 dpi, viral DNA was present as distinct nuclear foci (Fig. 3B, 2 dpi-1). These foci were found to be adjacent to, but not colocalized with, PML foci. In some infected cells at 2 dpi that had progressed to a later point in the infection, as judged by nuclear enlargement (5), the DNA staining was more intense and was localized in a pattern that was similar to that of TAg, with areas of apparent nuclear exclusion (compare Fig. 3B, 2 dpi-2, and 3A, 2 dpi). By 3 dpi, most of the DNA signals were in several bright, large foci. Some, but not all, of these foci were found to be close to enlarged PML foci (Fig. 3B, 3 dpi). Taken together, PML foci were seen close to both TAg and viral DNA, albeit at different times during the infection.
Reorganization of PML-NBs requires BKV DNA replication.
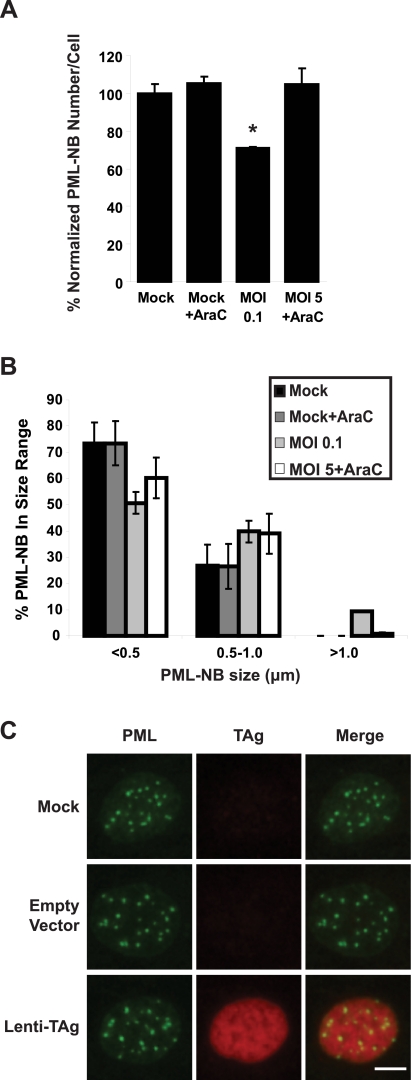
Given the timing of the PML-NB reorganization, we determined whether this reorganization required BKV DNA replication. We treated cells with the DNA replication inhibitor AraC, which blocks replication by interfering with topoisomerase I. This drug effectively inhibited BKV DNA replication as assayed by real-time PCR (see Fig. S4A in the supplemental material) and did not cause significant cytotoxicity (>80% of the cells were viable compared to the untreated controls). To ensure that DNA replication was the only variable, we adjusted the multiplicities of infection (MOIs) to keep the levels of TAg protein similar between treated and untreated cells, because otherwise the TAg level would be higher in untreated cells due to genome amplification. Western blotting confirmed that the levels of TAg were equivalent between cells that were infected with a low MOI and had active viral DNA replication and cells that were infected with a high MOI with DNA replication blocked (Fig. S4A, inset).
Both the number and the size distribution of PML-NBs were quantified in these samples (Fig. 4A and B). AraC treatment itself did not affect PML-NBs. In cells that were infected with an MOI of 0.1 infectious units (IU)/cell, there was ~30% reduction in the average number of PML-NBs. The change was not as dramatic as that shown in Fig. 1, probably due to the lower MOI used here. This decrease, however, was abolished when viral DNA replication was inhibited (Fig. 4A). Moreover, enlarged PML-NBs (diameter, >1 µm) were present only in cells in which BKV DNA replicated (Fig. 4B). Similar results were obtained when we treated the cells with a different DNA replication inhibitor, aphidicolin, which inhibits DNA polymerase (data not shown). Finally, we expressed TAg alone in RPTE cells by use of a lentivirus-based transduction method, and no difference was seen in either the number or the size of PML-NBs compared to mock- or empty-vector-transduced cells (Fig. 4C). Together, these results suggest that TAg alone is not sufficient to reorganize PML-NBs in RPTE cells and that active BKV DNA replication is necessary for this process.
FIG 4 .
PML-NB reorganization is dependent on viral DNA replication. RPTE cells were mock infected or infected with BKV at the indicated MOIs. At 24 hpi, 40 µg/ml AraC was added to the indicated samples. Average number (A) and size distribution (B) of PML-NBs in cells treated or not treated with AraC. At least 30 cells (A) or 130 PML-NBs (B) were counted or measured in each sample. Each bar represents the average of results from three independent experiments, and the error bars represent the SD values. *, P < 0.01 for comparison to all the other samples. (C) RPTE cells were mock transduced or transduced with empty-lentivirus control or with lentivirus expressing TAg. Cells were fixed at 3 days posttransduction and immunostained for PML (green) and TAg (red). Images were obtained as described in the legend to Fig. 1. Scale bar, 5 µm.
PML-NBs are not required for BKV growth.
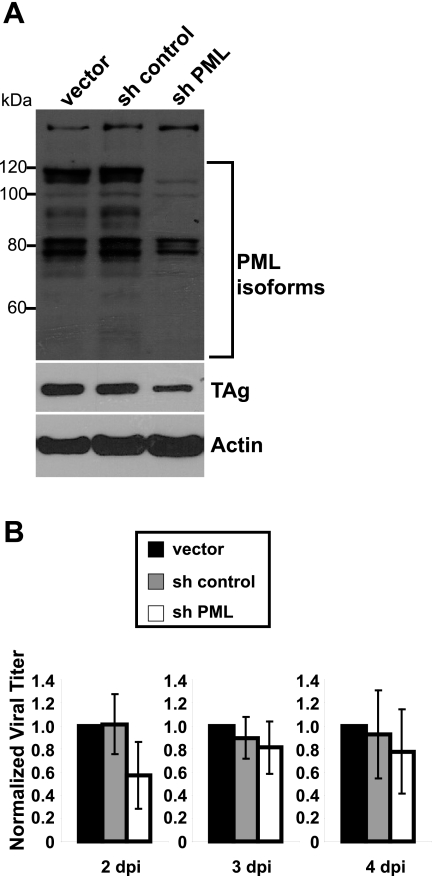
To gain a better understanding of the functional significance of PML-NB reorganization, we transduced RPTE cells with a retrovirus expressing shRNA against PML and assessed the effects on BKV infection. The PML shRNA and control vectors have previously been used in primary human fibroblasts to define the functions of PML in HCMV infection (38). After puromycin selection of transduced RPTE cells, there was a downregulation of a majority of the isoforms of PML (Fig. 5A). Although two isoforms of PML migrating at ~80 kDa were still present in the PML knockdown samples, most cells examined by an immunofluorescence analysis (IFA) did not contain detectable PML-NBs (see Fig. S5A in the supplemental material; also data not shown). In the PML knockdown samples, hDaxx had a more speckled nuclear staining pattern than in the control samples (Fig. S5A), as previously reported (38).
FIG 5 .
PML knockdown does not affect BKV replication. (A) RPTE cells were transduced with the indicated retroviruses (vector, shRNA control, and PML shRNA) and infected with BKV at an MOI of 0.5 IU/cell. Total proteins were harvested at 2 dpi and probed for PML, TAg, and actin. (B) Retrovirally transduced RPTE cells were infected as described for panel A, viral lysates were harvested at 2 to 4 dpi, and progeny production was determined by an IU assay. Data were normalized to protein concentration, and the values for the empty-vector samples were set to 1 for each time point. Each bar represents the average of results from at least four independent experiments, and the error bars represent the SD values. The P values are 0.02, 0.49, and 0.60 for 2, 3, and 4 dpi, respectively (for comparison of PML knockdown sample to shRNA control sample).
We infected these cells with BKV and harvested total cell proteins at 2 dpi. A lower MOI was used because it has been shown that the effect of PML knockdown on HCMV infection is more pronounced when cells were infected at low MOIs (38). Western blotting demonstrated that there was ~50% reduction of TAg in the PML knockdown cells (Fig. 5A; see also Fig. S5B in the supplemental material). We observed a similar reduction of TAg when we used pooled siRNA to knock down PML, during which the two isoforms of PML at ~80 kDa were downregulated (data not shown). PML knockdown also did not affect BKV growth: there was only a 2-fold reduction of progeny virions in knockdown cells at 2 dpi, and no differences were detected at 3 and 4 dpi (Fig. 5B).
BKV infection is able to rescue an ICP0-null mutant of HSV-1.
PML-NBs have intrinsic antiviral activities and are therefore targeted by many DNA viruses to facilitate the progression of their infections (15, 18). If the reorganization of PML-NBs is a way for BKV to inactivate the antiviral functions of PML-NBs, we would expect to see no major effect of PML knockdown on BKV infection, since the PML-NBs have already been disrupted by the virus. To test the hypothesis that BKV inhibits PML-NB function, therefore, we examined whether BKV infection was able to rescue the growth defect of an ICP0-null HSV-1 mutant. During HSV-1 infection, ICP0 is targeted to PML-NBs and induces PML degradation (37, 39). An ICP0-null mutant is not able to degrade PML, and hence, viral gene expression is repressed (40, 41). Depletion of PML, however, can partially complement the growth defects of an ICP0-null mutant, suggesting that PML has antiviral effects that are counteracted by ICP0 during normal infection (42). Therefore, we reasoned that the rescue of the ICP0-null mutant could be used as an indicator of PML-NB inactivation by BKV.
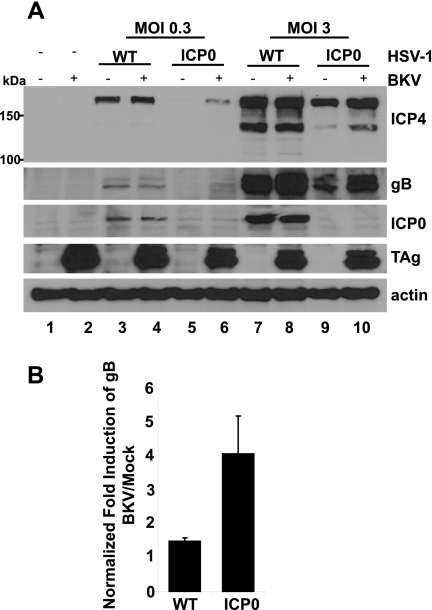
Because HSV-1 replicates more rapidly than BKV, RPTE cells were first infected with BKV for 3 days and subsequently superinfected with wild-type (WT) or ICP0-null mutant HSV-1 at two different MOIs. Total cell proteins were collected 8 h after HSV infection and immunoblotted for ICP4 (an immediate-early HSV-1 gene product), gB (a late HSV-1 gene product), ICP0, TAg, and actin (Fig. 6A). The ICP0 blot confirmed the identities of the two herpesviruses. Examining ICP4 revealed that BKV infection did not have any noticeable effect on WT HSV-1 at either MOI (Fig. 6A, lane 4 versus lane 3 and lane 8 versus lane 7). However, there were significant increases in ICP4 in the ICP0-null mutant at both MOIs (Fig. 6A, lane 6 versus lane 5 and lane 10 versus lane 9). In the case of gB, BKV infection caused a slight increase in WT HSV-1 at the high MOI (Fig. 6A, lane 8 versus lane 7, and B), but the induction of gB with BKV coinfection in the ICP0-null mutant was much stronger (Fig. 6B). Using IFA, we confirmed that PML was degraded in the WT-HSV-1-superinfected samples but remained as enlarged structures in the ICP0-null-mutant-superinfected samples (data not shown). These results corroborated our hypothesis that the reorganization of PML-NBs during BKV infection is a novel form of functional inactivation of these structures.
FIG 6 .
BKV infection partially complements an ICP0-null HSV-1 mutant. (A) RPTE cells were mock infected or infected with BKV at an MOI of 5 IU/cell for 3 days. The cells were subsequently superinfected with WT or ICP0-null HSV-1 at the indicated MOIs. Total cell proteins were harvested 8 h after HSV-1 infection and immunoblotted for ICP4, gB, ICP0, TAg, and actin. (B) Quantitation of gB induction by BKV infection. Western blots of samples infected with an MOI of 3 PFU/cell HSV-1 were probed for gB and GAPDH and visualized using the Odyssey imaging system. The fold induction of gB caused by BKV infection was quantified for both WT and ICP0-null HSV-1s and normalized to the level for the GAPDH loading control. Each bar represents the average of results from three independent experiments, and the error bars represent the SD values (P = 0.02).
DISCUSSION
To establish an optimal environment for replication, viruses must utilize host components that are beneficial to their propagation; at the same time, inhibitory host factors need to be inactivated or destroyed to ensure the progression of the replication process. We investigated the effects that BKV infection has on PML-NBs and the roles that PML-NBs might play in a productive BKV infection. PML-NBs are multiprotein structures that are able to produce antiviral effects against both DNA and RNA viruses through various mechanisms (18). We show here that, similar to other DNA viruses, BKV causes PML-NBs to reorganize in RPTE cells upon infection, albeit in a novel manner. Instead of being degraded or reshaped into track-like structures, PML-NBs in BKV-infected cells became reduced in number, larger in size, and altered in composition, although the levels of PML components were unchanged. The reorganization was accompanied by a functional change in PML-NBs, as BKV-infected cells were able to complement the growth defects of an ICP0-null mutant of HSV-1 whose deficiencies lie partially in its inability to destroy the detrimental PML-NBs. These results suggest that antiviral roles of PML-NBs are disrupted by BKV during its infection. While it is possible that functions of ICP0 other than disruption of PML-NBs are supplied by BKV, our preliminary results show that TAg alone is not sufficient to complement the ICP0-null mutant (data not shown).
PML knockdown, which disrupts PML-NBs, does not seem to have a major impact on WT BKV growth. These results are consistent with the observations that JCV, another human polyomavirus, does not require PML to undergo a productive infection (28). We hypothesize that PML-NBs are inactivated by BKV and therefore that PML knockdown does not significantly affect BKV infection. This is similar to inactivation of pRb by SV40 TAg: the absence of pRb does not enhance TAg-dependent DNA replication (43). Our data, however, do not rule out the possibility that low levels of some PML isoforms that persist in the PML knockdown cells may have additional roles in BKV infection.
PML-NBs were colocalized with or adjacent to BKV components, and the association occurs in a temporally controlled manner. During the early phase of infection, PML-NBs colocalized with TAg foci. It is possible that incoming polyomavirus genomes get deposited at PML-NBs, although unfortunately we could not consistently detect input viral genomes. PML-NBs may provide an initial microenvironment that is conducive to early transcription. This phenomenon has been documented for HCMV, whose immediate-early transcripts have been found close to PML-NBs along with the spliceosome assembly factor SC35 and two other transcription factors (44). In PML knockdown cells infected with BKV, we detected a 2-fold decrease in TAg levels compared to the level for control cells. This small decrease was still present in cells treated with AraC, which rules out the effects of genome amplification (data not shown), suggesting that PML may contribute to the optimal transcription of the TAg gene.
Later during BKV infection, PML-NB reorganization occurred and PML-NBs were detected adjacent to intense BKV DNA foci. Furthermore, we show here that, unlike for other viruses, BKV DNA replication was necessary for this process. How viral replication elicits reorganization is not known, and we are currently isolating BKV DNA replication mutants to investigate this process. It has been suggested that newly replicated single-stranded DNA accumulates in PML-NBs in BKV-infected cells (27), and therefore the reorganization of PML-NBs may be coupled to postreplication DNA-processing events such as recombination to repair breaks that are generated during replication. We think it is unlikely that BKV late proteins play a major role in PML-NB reorganization, because AraC treatment has only a slight effect on VP1 levels (see Fig. S4B in the supplemental material) and it does not inhibit SV40 late gene expression (45). During late infection, we observe that TAg is excluded from certain areas of the nucleus. We currently do not know what these TAg-excluded areas represent, but there is limited electron microscopy and IFA staining evidence indicating that these are the areas where viral capsid proteins and newly produced BKV virions accumulate (data not shown and reference 46).
As BKV infection progressed in RPTE cells, the PML-NBs continued to enlarge to the point that they sometimes formed ring-like structures. Similar ring-like structures have been reported to occur in several other cases. For example, HSV-1 genomes in quiescently infected cells are found sequestered in PML-NBs with ring-like structures (47). Unlike for BKV infection, these ring structures contain hDaxx along with heterochromatin protein HP1 and conjugated ubiquitin. In addition, the PML-NB ring surrounds, instead of being juxtaposed to, the HSV-1 DNA (47). The functional significance of this association remains unknown. Enlarged ring-like PML-NBs are also found in telomerase-negative tumor cells that maintain their telomeres using the recombination-mediated alternative-lengthening-of-telomeres (ALT) mechanism (48). These PML-NBs are called ALT-associated PML-NBs and contain telomeric DNA, telomere binding proteins, and proteins involved in DNA recombination, such as RAD51 and RAD52. Furthermore, a similar PML-NB ring structure has been described to occur in lymphocytes of patients suffering immunodeficiency, centromeric instability, and facial dysmorphy syndrome (49). Interestingly, the authors discovered that these PML-NBs also contain DNA and may function in the reestablishment of the condensed heterochromatic state on the enclosed DNA. High-resolution 4Pi fluorescence microscopy has revealed that PML-NBs have a distinct outer shell and may surround core structures such as telomeric DNA (50). Taken together, the common features of the enlarged, ring-like PML-NBs are that they are all associated with DNA and that DNA recombination or chromatin remodeling processes may occur within or adjacent to them. It is possible that the ring-like PML-NBs detected in BKV infection also represent sites that allow viral DNA to undergo postreplication recombination events or chromatin modification.
It is intriguing that hDaxx and Sp100 separate from PML during BKV infection at a time that correlates with viral DNA replication and PML-NB enlargement. During Ad infection, other PML-NB components, Sp100 and NDP55, relocate from PML-NBs to viral replication domains (23). Mutational analyses have revealed that Ad DNA replication may be required for the redistribution of these proteins and that formation of PML-NB tracks may be required only for early replication. There are two major questions remaining with regard to the redistribution of PML and hDaxx/Sp100 during BKV infection. First, what is the functional significance of this event? It is possible that hDaxx/Sp100 play an inhibitory role to combat BKV replication and that the detachment of these proteins from PML-NBs counters those negative effects. Alternatively, the dissociated hDaxx/Sp100 may play a positive role during BKV late infection. The second question seeks to determine how the separation of PML and hDaxx/Sp100 occurs. It is known that SUMOylation of PML is required for formation of PML-NBs and for recruitment of hDaxx to PML-NBs through interactions with a SUMO-interacting motif in hDaxx (37, 51). Both HCMV and Epstein-Barr virus have the capacity to regulate the SUMOylation of PML, disrupting inhibitory PML-NBs (34, 52, 53). BKV infection did not seem to cause a dramatic change in the SUMOylation status of PML (see Fig. S1 in the supplemental material); therefore, additional studies are required to determine how the infection causes PML and hDaxx/Sp100 to dissociate, which might help explain how reorganization leads to functional inactivation of PML-NBs.
MATERIALS AND METHODS
Cell culture, viruses, and plasmids.
Primary human RPTE cells were maintained in renal epithelial cell growth medium (REGM) as previously described (54). Vero cells and 293T cells were maintained in Dulbecco’s modified Eagle medium (Gibco) containing 10% fetal bovine serum (HyClone), 100 U/ml penicillin, and 100 µg/ml streptomycin (Cambrex). 293TT cells (a gift from Chris Buck, NIH) were maintained in the same medium with 400 µg/ml hygromycin (55). All cells were grown at 37°C with 5% CO2 in a humidified incubator.
BKV (Dunlop) was grown and purified and its titer determined using an infectious unit (IU) assay as previously described (8). Wild-type (WT) and ICP0-null mutant HSV-1s were obtained from Neal DeLuca, University of Pittsburgh. shRNA empty plasmid (pSIREN-RetroQ), shRNA control plasmid (psiC), and PML shRNA plasmid (psiPML2) were described previously (56). The retrovirus packaging plasmid pLP/VSVG was from Invitrogen, and pUMVC3-gag-pol was obtained from Alice Telesnisky, University of Michigan. Lentivirus packaging plasmids pMDLg-pRRE, pRSV-Rev, and pCI-VSVG as well as the lentiviral expression plasmid pLentilox-IRES-PURO (pLL-Puro) were obtained from the Vector Core, University of Michigan.
The BKV TAg cDNA was cloned using fusion PCR. The 5′ untranslated region (5′-UTR) and exon 1 of TAg were PCR amplified using primers BKV-TAg-ext-F-new (5′ GGCTGCATTCCATGGGTAAGCAGCT 3′) and BK-TAg-int-R (5′ AGGTTGGCACCTCTGAGCTACTCCAGGT 3′). Exon 2 and the 3′-UTR were PCR amplified using primers BK-TAg-int-F (5′ GTAGCTCAGAGGTGCCAACCTATGGAACA 3′) and BK-TAg-ext-R (5′ CCAGGGGACCCAGATATGATAAGA 3′). The two PCR products were fused together in a separate PCR using primers F-XhoI-5UTR (5′ GACTCGAGGCCTCCACCCTTTCTCTCAA 3′) and BK-TAg-R-BamHI (5′ CAGGATCCTTATTTTGGGGGTGGTGT 3′). The final PCR product was cloned into the pCR2.1-TOPO vector (Invitrogen) and subcloned into pLL-Puro via the XhoI and BamHI restriction sites.
Antibodies.
The following antibodies and concentrations were used unless otherwise stated: anti-PML (sc-5621; Santa Cruz) for immunofluorescence analyses (IFAs; 1:200) and Western blotting (WB; 1:500), anti-PML (sc-966; Santa Cruz) for IFAs (1:200), anti-Daxx (sc-7152; Santa Cruz) for IFAs (1:50), anti-Daxx (no. 1094-1; Epitomics) for WB (1:5,000), anti-SP100 (H00006672-D01P; Novus Biologicals) for IFAs (1:300) and for WB (1:1,000), anti-SUMO-1 (small ubiquitin-like modifier 1) (sc-5308; Santa Cruz) for IFAs (1:100), anti-TAg pAb416 (57) for IFAs (1:200) and WB (1:3,000), anti-VP1 (8) for WB (1:5,000), anti-GAPDH (anti-glyceraldehyde-3-phosphate dehydrogenase; ab9484; Abcam) for WB (1:10,000), anti-β-actin (no. 4967; Cell Signaling) for WB (1:5,000), anti-HSV-1 ICP4 for WB (1:1,000) and ICP0 for WB (1:1,000), anti-HSV-1 gB for WB (1:500); horseradish peroxidase-conjugated sheep anti-mouse and donkey anti-rabbit IgG (Amersham) for WB (1:2,000-15,000), and Alexa Fluor 488 goat anti-rabbit and Alexa Fluor 594 goat anti-mouse IgG(H+L) (Invitrogen) for IFAs (1:200). Anti-gB serum was a gift from Oveta Fuller (University of Michigan), and anti-HSV-1 antibodies were gifts from Neal DeLuca.
Infections and drug treatments.
To synchronize infections, 70% confluent RPTE cells were prechilled for 15 min at 4°C. The cells were then infected with purified BKV diluted in REGM at the indicated multiplicities of infection (MOIs) and incubated for 1 h at 4°C. The viral inoculum was removed, and the cells were washed once with cold medium. Infection was initiated by adding prewarmed REGM and transferring the cells to 37°C. Total cell proteins and viral lysates were harvested as previously described (54). For the HSV-1 superinfection experiment, RPTE cells were first infected with BKV for 3 days. Subsequently, the cells were infected with HSV-1 (WT or ICP0-null) at the indicated MOIs and incubated for 1 h at 37°C. The viral inoculum was then replaced with fresh REGM, and total cell proteins were harvested at 8 h postinfection (hpi).
Cytosine β-d-arabinofuranoside (AraC; Sigma) was reconstituted according to the manufacturer’s recommendations and was added at 24 hpi at 40 µg/ml and present until the time of the assays. The AraC treatment did not cause significant cytotoxic effects, according to a cell metabolism WST-1 assay (Roche; data not shown).
Western blotting and quantification.
Total cell proteins were harvested, quantified, and immunoblotted as previously described (8). For quantitative blots using the Odyssey infrared imaging system, the membrane was processed according to the manufacturer’s instructions (LI-COR). The following antibodies and concentrations were used: gB, 1:1,000; GAPDH, 1:5,000; and goat anti-rabbit IRDye 680 and goat anti-mouse IRDye 800CW, both at 1:10,000. The membrane was scanned using the Odyssey infrared imaging system. The relevant bands were quantified using the Odyssey software program.
Microscopy and PML-NB measurements.
At the indicated time points postinfection, RPTE cells grown on coverslips (Fisher) were fixed in ice-cold 95% ethanol and 5% acetic acid for 5 min. Following three washes with phosphate-buffered saline (PBS), the samples were blocked with 5% goat serum (Vector Laboratories, Inc.) for 1 h at room temperature (RT). After being blocked, the samples were incubated with primary antibodies and the corresponding secondary antibodies sequentially in 5% goat serum for 1 h each at RT, with three PBS washes in between. Slides were mounted with ProLong gold antifade reagent with DAPI (4′,6-diamidino-2-phenylindole; Invitrogen). For standard fluorescence microscopy, slides were examined using an Olympus BX41 microscope with a Plan 40×/0.65 objective or a Plan 100×/1.25 oil objective. For laser-scanning confocal microscopy, all images were obtained using a Zeiss LSM 510 confocal microscope with a 63×/1.2 objective and a 1-µm optical section. Images were analyzed and processed using an LSM image browser (Zeiss).
All PML-NB measurements were obtained using an Olympus BX41 microscope. To determine the number of PML-NBs per cell, images were taken using a 40× objective from randomly distributed fields on the slide, and the number of PML-NBs per cell was counted. To measure the average size of PML-NBs, images were obtained using a 100× objective. A ruler was imprinted onto each image using the DP Controller software program (Olympus). The images were then changed to 8-bit format by use of ImageJ software (NIH). The previously imprinted ruler was used to set a pixel/µm scale, and the line tool was used to measure the longest dimension for each PML-NB. All statistical analyses were performed using the Student t test.
Fluorescent in situ hybridization (FISH).
The hybridization probe was prepared using the pBR322-Dunlop plasmid (54). DNA was labeled with Cy3-dCTP using a nick translation kit (GE Healthcare) according to the manufacturer’s instructions, and the unincorporated dye was removed with a Micro Bio-Spin 30 spin column (Bio-Rad). Cells were fixed and washed with PBS as described above. Samples were treated with 5 µg/ml DNase-free RNase at 37°C for 1 h. After three washes with PBS, the cells were incubated with prehybridization buffer (50% formamide, 10% dextran sulfate, 4× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate]) at 37°C for 30 min. Labeled probe was diluted in the prehybridization buffer with 50 µg/ml salmon sperm DNA (Invitrogen) to 1 µg/ml. Both the probes and the samples were denatured at 95°C for 2 min. The cells were hybridized with the probe overnight at 37°C in a humidified chamber. After hybridization, the samples were washed with 2× SSC, once at 60°C for 5 min and then once at RT for 5 min. The samples were then subjected to immunostaining and confocal microscopy as described above.
Retrovirus/lentivirus production and shRNA knockdown.
To produce replication-deficient Moloney murine leukemia virus-based retroviruses, 293T cells were cotransfected with pSIREN-RetroQ plasmids along with packaging plasmids pLP/VSVG and pUMVC3-gag-pol at a ratio of 1.5:1.5:1, with a total of 12 µg DNA per 10-cm dish, using TransIT-LT1 reagents (Mirus) according to the manufacturer’s recommendations. Lentivirus expressing TAg was produced similarly by transfecting 293TT cells with plasmids pMDLg-pRRE, pRSV-Rev, pCI-VSVG, and pLL-puro at a ratio of 1:1:1:2. Retrovirus- or lentivirus-containing supernatants were harvested at 3 days posttransfection and concentrated 100× using a Lenti-X concentrator (Clontech) according to the manufacturer’s instructions.
RPTE cells were infected with retroviruses or lentiviruses in the presence of 8 µg/ml Polybrene (10 µl concentrated virus per well of a 12-well plate) for 6 h at 37°C. The viral inoculum was replaced with fresh REGM, and the cells were selected with 3.1 µg/ml puromycin beginning at 2 dpi. The cells were infected with BKV 3 days after puromycin selection. Lentivirus-infected cells were fixed at 3 dpi and immunostained for PML and TAg as described above.
Real-time PCR.
To quantify the viral DNA load in cells, low-molecular-weight DNA was isolated using a modified Hirt protocol (58) in which the viral DNA was further purified using a QIAquick PCR purification kit (Qiagen) after removal of the chromosomal DNA. Primers were designed to amplify 152- and 164-base-pair fragments of the TAg (TAg-Forward, 5′ AAGGAAAGGCTGGATTCTGA 3′; TAg-Reverse, 5′ TGTGATTGGGATTCAGTGCT 3′) and mitochondrial 16s rRNA (Mitochondrial 16s rRNA-Forward, 5′ GAGGAACAGCTCTTTGGACA 3′; Mitochondrial 16s rRNA-Reverse, 5′ CAATTGGGTGTGAGGAGTTC 3′), respectively. Real-time PCRs were performed in a total volume of 25 µl using 12.5 µl Power SYBR green PCR master mix (Applied Biosystems), 2.5 µl DNA template, and 300 nM each primer. Amplification was performed in 96-well PCR plates using the iCycler iQ5 real-time detection system (Bio-Rad) with the following program: 2 min at 50°C, 10 min at 95°C, 40 cycles of denaturation at 95°C for 15 s, and annealing and extension at 58°C for 30 s. Results are presented as the relative BKV DNA levels, with the levels in samples infected with an MOI of 0.1 IU/cell at 2 dpi arbitrarily set to 1. Results were normalized to the levels of mitochondrial DNA present using the threshold cycle (2–ΔΔCT; Livak) method (54).
SUPPLEMENTAL MATERIAL
ACKNOWLEDGMENTS
We thank the members of the Imperiale laboratory for help and discussion with this work, Justin Namm for technical assistance, and Christiane Wobus, Kathy Spindler, and Akira Ono for critical reading of the manuscript. We are extremely grateful to Alice Telesnisky, Neal DeLuca, Oveta Fuller, and Chris Buck for providing us with the necessary reagents for this study. We also thank the Microscopy & Image Analysis Laboratory at the University of Michigan for microscopy assistance.
This work was supported by AI060584, awarded to M.J.I. from the NIH, and in part by CA046592, awarded to the University of Michigan Cancer Center from the NIH. M.J. was supported by American Heart Association Postdoctoral Fellowship 0825806G.
Footnotes
Citation Jiang, M., P. Entezami, M. Gamez, T. Stamminger, and M. J. Imperiale. 2011. Functional reorganization of promyelocytic leukemia nuclear bodies during BK virus infection. mBio 2(1):e00281-10. doi:10.1128/mBio.00281-10.
REFERENCES
- 1. Nickeleit V., Hirsch H. H., Binet I. F., Gudat F., Prince O., Dalquen P., Thiel G., Mihatsch M. J. 1999. Polyomavirus infection of renal allograft recipients: from latent infection to manifest disease. J. Am. Soc. Nephrol. 10:1080–1089 [DOI] [PubMed] [Google Scholar]
- 2. Jiang M., Abend J. R., Johnson S. F., Imperiale M. J. 2009. The role of polyomaviruses in human disease. Virology 384:266–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bonvoisin C., Weekers L., Xhignesse P., Grosch S., Milicevic M., Krzesinski J. M. 2008. Polyomavirus in renal transplantation: a hot problem. Transplantation 85:S42–S48 [DOI] [PubMed] [Google Scholar]
- 4. Hilton R., Tong C. Y. 2008. Antiviral therapy for polyomavirus-associated nephropathy after renal transplantation. J. Antimicrob. Chemother. 62:855–859 [DOI] [PubMed] [Google Scholar]
- 5. Low J., Humes H. D., Szczypka M., Imperiale M. 2004. BKV and SV40 infection of human kidney tubular epithelial cells in vitro. Virology 323:182–188 [DOI] [PubMed] [Google Scholar]
- 6. Low J. A., Magnuson B., Tsai B., Imperiale M. J. 2006. Identification of gangliosides GD1b and GT1b as receptors for BK virus. J. Virol. 80:1361–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Moriyama T., Sorokin A. 2008. Intracellular trafficking pathway of BK Virus in human renal proximal tubular epithelial cells. Virology 371:336–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jiang M., Abend J. R., Tsai B., Imperiale M. J. 2009. Early events during BK virus entry and disassembly. J. Virol. 83:1350–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Imperiale M. J., Major E. O. 2007. Polyomaviruses, p. 2263–2298 In Knipe D. M., Howley P. M. Fields virology, 5th ed., vol. 2 Lippincott Williams & Wilkins, Philadelphia, PA. : [Google Scholar]
- 10. Yang S. I., Lickteig R. L., Estes R., Rundell K., Walter G., Mumby M. C. 1991. Control of protein phosphatase 2A by simian virus 40 small-t antigen. Mol. Cell. Biol. 11:1988–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Skoczylas C., Fahrbach K. M., Rundell K. 2004. Cellular targets of the SV40 small-t antigen in human cell transformation. Cell Cycle 3:606–610 [PubMed] [Google Scholar]
- 12. Blanchette P., Branton P. E. 2009. Manipulation of the ubiquitin-proteasome pathway by small DNA tumor viruses. Virology 384:317–323 [DOI] [PubMed] [Google Scholar]
- 13. Ascoli C. A., Maul G. G. 1991. Identification of a novel nuclear domain. J. Cell Biol. 112:785–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bernardi R., Pandolfi P. P. 2007. Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat. Rev. Mol. Cell Biol. 8:1006–1016 [DOI] [PubMed] [Google Scholar]
- 15. Everett R. D., Chelbi-Alix M. K. 2007. PML and PML nuclear bodies: implications in antiviral defence. Biochimie 89:819–830 [DOI] [PubMed] [Google Scholar]
- 16. de The H., Lavau C., Marchio A., Chomienne C., Degos L., Dejean A. 1991. The PML-RAR alpha fusion mRNA generated by the t(15;17) translocation in acute promyelocytic leukemia encodes a functionally altered RAR. Cell 66:675–684 [DOI] [PubMed] [Google Scholar]
- 17. Kakizuka A., Miller W. H., Jr., Umesono K., Warrell R. P., Jr., Frankel S. R., Murty V. V., Dmitrovsky E., Evans R. M. 1991. Chromosomal translocation t(15;17) in human acute promyelocytic leukemia fuses RAR alpha with a novel putative transcription factor, PML. Cell 66:663–674 [DOI] [PubMed] [Google Scholar]
- 18. Tavalai N., Stamminger T. 2008. New insights into the role of the subnuclear structure ND10 for viral infection. Biochim. Biophys. Acta 1783:2207–2221 [DOI] [PubMed] [Google Scholar]
- 19. Everett R. D. 2006. Interactions between DNA viruses, ND10 and the DNA damage response. Cell. Microbiol. 8:365–374 [DOI] [PubMed] [Google Scholar]
- 20. Maul G. G., Guldner H. H., Spivack J. G. 1993. Modification of discrete nuclear domains induced by herpes simplex virus type 1 immediate early gene 1 product (ICP0). J. Gen. Virol. 74(Pt. 12):2679–2690 [DOI] [PubMed] [Google Scholar]
- 21. Ishov A. M., Maul G. G. 1996. The periphery of nuclear domain 10 (ND10) as site of DNA virus deposition. J. Cell Biol. 134:815–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Maul G. G., Ishov A. M., Everett R. D. 1996. Nuclear domain 10 as preexisting potential replication start sites of herpes simplex virus type-1. Virology 217:67–75 [DOI] [PubMed] [Google Scholar]
- 23. Doucas V., Ishov A. M., Romo A., Juguilon H., Weitzman M. D., Evans R. M., Maul G. G. 1996. Adenovirus replication is coupled with the dynamic properties of the PML nuclear structure. Genes Dev. 10:196–207 [DOI] [PubMed] [Google Scholar]
- 24. Regad T., Chelbi-Alix M. K. 2001. Role and fate of PML nuclear bodies in response to interferon and viral infections. Oncogene 20:7274–7286 [DOI] [PubMed] [Google Scholar]
- 25. Day P. M., Baker C. C., Lowy D. R., Schiller J. T. 2004. Establishment of papillomavirus infection is enhanced by promyelocytic leukemia protein (PML) expression. Proc. Natl. Acad. Sci. U. S. A. 101:14252–14257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tang Q., Bell P., Tegtmeyer P., Maul G. G. 2000. Replication but not transcription of simian virus 40 DNA is dependent on nuclear domain 10. J. Virol. 74:9694–9700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jul-Larsen A., Visted T., Karlsen B. O., Rinaldo C. H., Bjerkvig R., Lonning P. E., Boe S. O. 2004. PML-nuclear bodies accumulate DNA in response to polyomavirus BK and simian virus 40 replication. Exp. Cell Res. 298:58–73 [DOI] [PubMed] [Google Scholar]
- 28. Gasparovic M. L., Maginnis M. S., O’Hara B. A., Dugan A. S., Atwood W. J. 2009. Modulation of PML protein expression regulates JCV infection. Virology 390:279–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shishido-Hara Y., Ichinose S., Higuchi K., Hara Y., Yasui K. 2004. Major and minor capsid proteins of human polyomavirus JC cooperatively accumulate to nuclear domain 10 for assembly into virions. J. Virol. 78:9890–9903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shishido-Hara Y., Higuchi K., Ohara S., Duyckaerts C., Hauw J. J., Uchihara T. 2008. Promyelocytic leukemia nuclear bodies provide a scaffold for human polyomavirus JC replication and are disrupted after development of viral inclusions in progressive multifocal leukoencephalopathy. J. Neuropathol. Exp. Neurol. 67:299–308 [DOI] [PubMed] [Google Scholar]
- 31. Carvalho T., Seeler J. S., Ohman K., Jordan P., Pettersson U., Akusjarvi G., Carmo-Fonseca M., Dejean A. 1995. Targeting of adenovirus E1A and E4-ORF3 proteins to nuclear matrix-associated PML bodies. J. Cell Biol. 131:45–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Boutell C., Sadis S., Everett R. D. 2002. Herpes simplex virus type 1 immediate-early protein ICP0 and its isolated RING finger domain act as ubiquitin E3 ligases in vitro. J. Virol. 76:841–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ahn J. H., Brignole E. J., III, Hayward G. S. 1998. Disruption of PML subnuclear domains by the acidic IE1 protein of human cytomegalovirus is mediated through interaction with PML and may modulate a RING finger-dependent cryptic transactivator function of PML. Mol. Cell. Biol. 18:4899–4913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lee H. R., Kim D. J., Lee J. M., Choi C. Y., Ahn B. Y., Hayward G. S., Ahn J. H. 2004. Ability of the human cytomegalovirus IE1 protein to modulate sumoylation of PML correlates with its functional activities in transcriptional regulation and infectivity in cultured fibroblast cells. J. Virol. 78:6527–6542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jensen K., Shiels C., Freemont P. S. 2001. PML protein isoforms and the RBCC/TRIM motif. Oncogene 20:7223–7233 [DOI] [PubMed] [Google Scholar]
- 36. Guldner H. H., Szostecki C., Schroder P., Matschl U., Jensen K., Luders C., Will H., Sternsdorf T. 1999. Splice variants of the nuclear dot-associated Sp100 protein contain homologies to HMG-1 and a human nuclear phosphoprotein-box motif. J. Cell Sci. 112(Pt. 5):733–747 [DOI] [PubMed] [Google Scholar]
- 37. Ishov A. M., Sotnikov A. G., Negorev D., Vladimirova O. V., Neff N., Kamitani T., Yeh E. T., Strauss J. F., III, Maul G. G. 1999. PML is critical for ND10 formation and recruits the PML-interacting protein daxx to this nuclear structure when modified by SUMO-1. J. Cell Biol. 147:221–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tavalai N., Papior P., Rechter S., Leis M., Stamminger T. 2006. Evidence for a role of the cellular ND10 protein PML in mediating intrinsic immunity against human cytomegalovirus infections. J. Virol. 80:8006–8018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Everett R. D., Freemont P., Saitoh H., Dasso M., Orr A., Kathoria M., Parkinson J. 1998. The disruption of ND10 during herpes simplex virus infection correlates with the Vmw110- and proteasome-dependent loss of several PML isoforms. J. Virol. 72:6581–6591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Everett R. D., Boutell C., Orr A. 2004. Phenotype of a herpes simplex virus type 1 mutant that fails to express immediate-early regulatory protein ICP0. J. Virol. 78:1763–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hagglund R., Roizman B. 2004. Role of ICP0 in the strategy of conquest of the host cell by herpes simplex virus 1. J. Virol. 78:2169–2178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Everett R. D., Rechter S., Papior P., Tavalai N., Stamminger T., Orr A. 2006. PML contributes to a cellular mechanism of repression of herpes simplex virus type 1 infection that is inactivated by ICP0. J. Virol. 80:7995–8005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Trifillis P., Picardi J., Alwine J. C. 1990. Simian virus 40 T antigen can transcriptionally activate and mediate viral DNA replication in cells which lack the retinoblastoma susceptibility gene product. J. Virol. 64:1345–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ishov A. M., Stenberg R. M., Maul G. G. 1997. Human cytomegalovirus immediate early interaction with host nuclear structures: definition of an immediate transcript environment. J. Cell Biol. 138:5–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Keller J. M., Alwine J. C. 1984. Activation of the SV40 late promoter: direct effects of T antigen in the absence of viral DNA replication. Cell 36:381–389 [DOI] [PubMed] [Google Scholar]
- 46. Bernhoff E., Tylden G. D., Kjerpeseth L. J., Gutteberg T. J., Hirsch H. H., Rinaldo C. H. 2010. Leflunomide inhibition of BK virus replication in renal tubular epithelial cells. J. Virol. 84:2150–2156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Everett R. D., Murray J., Orr A., Preston C. M. 2007. Herpes simplex virus type 1 genomes are associated with ND10 nuclear substructures in quiescently infected human fibroblasts. J. Virol. 81:10991–11004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yeager T. R., Neumann A. A., Englezou A., Huschtscha L. I., Noble J. R., Reddel R. R. 1999. Telomerase-negative immortalized human cells contain a novel type of promyelocytic leukemia (PML) body. Cancer Res. 59:4175–4179 [PubMed] [Google Scholar]
- 49. Luciani J. J., Depetris D., Usson Y., Metzler-Guillemain C., Mignon-Ravix C., Mitchell M. J., Megarbane A., Sarda P., Sirma H., Moncla A., Feunteun J., Mattei M. G. 2006. PML nuclear bodies are highly organised DNA-protein structures with a function in heterochromatin remodelling at the G2 phase. J. Cell Sci. 119:2518–2531 [DOI] [PubMed] [Google Scholar]
- 50. Lang M., Jegou T., Chung I., Richter K., Munch S., Udvarhelyi A., Cremer C., Hemmerich P., Engelhardt J., Hell S. W., Rippe K. 2010. Three-dimensional organization of promyelocytic leukemia nuclear bodies. J. Cell Sci. 123:392–400 [DOI] [PubMed] [Google Scholar]
- 51. Lin D. Y., Huang Y. S., Jeng J. C., Kuo H. Y., Chang C. C., Chao T. T., Ho C. C., Chen Y. C., Lin T. P., Fang H. I., Hung C. C., Suen C. S., Hwang M. J., Chang K. S., Maul G. G., Shih H. M. 2006. Role of SUMO-interacting motif in Daxx SUMO modification, subnuclear localization, and repression of sumoylated transcription factors. Mol. Cell 24:341–354 [DOI] [PubMed] [Google Scholar]
- 52. Bell P., Lieberman P. M., Maul G. G. 2000. Lytic but not latent replication of Epstein-Barr virus is associated with PML and induces sequential release of nuclear domain 10 proteins. J. Virol. 74:11800–11810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Adamson A. L., Kenney S. 2001. Epstein-Barr virus immediate-early protein BZLF1 is SUMO-1 modified and disrupts promyelocytic leukemia bodies. J. Virol. 75:2388–2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Abend J. R., Low J. A., Imperiale M. J. 2007. Inhibitory effect of gamma interferon on BK virus gene expression and replication. J. Virol. 81:272–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Buck C. B., Pastrana D. V., Lowy D. R., Schiller J. T. 2004. Efficient intracellular assembly of papillomaviral vectors. J. Virol. 78:751–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tavalai N., Papior P., Rechter S., Stamminger T. 2008. Nuclear domain 10 components promyelocytic leukemia protein and hDaxx independently contribute to an intrinsic antiviral defense against human cytomegalovirus infection. J. Virol. 82:126–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Harlow E., Crawford L. V., Pim D. C., Williamson N. M. 1981. Monoclonal antibodies specific for simian virus 40 tumor antigens. J. Virol. 39:861–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hirt B. 1967. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 26:365–369 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.