Abstract
Mango malformation is the most serious disease of mango causing considerable damage to the mango orchards worldwide. It is a major threat for mango cultivation in north Indian belt. In recent years, Fusarium sp. is finding wide acceptability in scientific community as a causal agent of this disease. However, little information is known about the variability in Fusarium isolates from malformed mango tissues. Therefore, the major objective of present study was the identification and analysis of genetic diversity among Fusarium isolates collected from malformed mango tissues. Two texon selective primers, ITS-Fu-f and ITS-Fu-r, were used for quick identification of Fusarium spp. The fungal genomic DNA was extracted from using CTAB method and was utilized as template for PCR amplification. Total 224 bands were amplified by 18 RAPD primers at an average of 12.44 bands per primer. The size of the obtained amplicons ranged from 0.264 kb (minimum) to 3.624 kb (maximum). Data scored from 25 isolates of Fusarium sp. with 18 RAPD primers were used to generate similarity coefficients. The similarity coefficient ranged from 0.17 to 0.945. Based on DNA fingerprints, all isolates were categorized into two major clusters. This study indicated a wide variability among different isolates of Fusarium.
1. Introduction
Malformation, arguably the most important disease of mango (Mangifera indica L.) globally, is of growing concern not only because of its widespread and destructive nature but also because its etiology and control are not well understood. Mango malformation was reported for the first time by an expert mango grower from Darbhanga district in Bihar in 1891 [1]. Malformation is not only well known in India but has also been confirmed in most mango growing countries like Pakistan, Egypt, South Africa, Brazil, Israel, Central America, Mexico, and USA [2]. There is a lot of confusion in the literature about the etiology of this malady because research efforts made hitherto have not been able to ascertain its etiology. The complexity of the disorder is attributed by many factors like mites, fungal, viral, and physiological factors. However, in recent years, Fusarium spp. are finding wide acceptability in scientific community as a causal agent of this disease.
All the disease management strategies based on host resistance require the knowledge of variability in pathogens, that is why the objective of this study was to develop a polymerase chain reaction (PCR) assay to examine genetic variation in a larger collection of the pathogen. Also, DNA-based genetic markers provide a genetic diagnostic tool that permits direct identification of pathotypes in any developmental stage in environment-independent manner [3]. The two texon selective primers, ITS-Fu-f and ITS-Fu-r, were used for quick identification of Fusarium spp. [4]. Arbitrary 18 primers were used in RAPD to produce characteristic profiles of amplified products.
2. Material and Method
2.1. Isolates of Fusarium spp
Twenty-five samples of malformed panicles and seedlings were collected from various orchards from different locations at Pantnagar. Infected samples were cut into small pieces and surface sterilized with 0.2% sodium hypochlorite solution for 2 minutes. Thereafter, the samples were washed with sterilized distilled water before placing them in Petriplates containing Potato Dextrose Agar (PDA) medium. The sealed plates were kept in a BOD incubator at 28°C ± 2°C for 4 days until the fungus growth appeared. Fresh fungal growth from the plated samples was then transferred on PDA. Finally, every isolate was further purified by single-spore culture on PDA. potato dextrose broth (PDB) medium was used for the harvesting of mycelium for DNA extraction.
3. DNA Extraction
Fungal genomic DNA was extracted for molecular characterization studies. Total DNA was extracted by using the CTAB (Hexa-decyl tri-methyl ammonium bromide) method of [5]. For the extraction of DNA, 1 g of freshly harvested mycelium was ground in liquid nitrogen with a mortar pestle into a very fine powder. Powder was suspended in 10 ml of DNA extraction buffer (50 mM Tris Buffer pH 8.0, 100 mM EDTA, 150 mM NaCl). After proper shaking, 1 ml of 10% SDS was added and incubated for 1 h at 37°C. 1.5 ml of 5 M NaCl and 1.25 ml of CTAB solution (10% CTAB and 0.7 M NaCl) were added and incubated at 65°C for 20 minutes in an incubator shaker at 60 rev. per minute. DNA was extracted by adding an equal volume of Chloroform: Isoamyl Alcohol (24 : 1 V/V) and mixed thoroughly but gently and then centrifuged at 10000 rpm for 12 min at 10°C. Aqueous viscous supernatant was removed to a fresh tube and precipitated with 0.6 volumes of ice-cold isopropanol and 0.1 volume sodium acetate and left overnight in the freezer at −20°C. The mixture was centrifuged at 10000 rpm for 10 min at 10°C. Pellet was washed with 70% ethanol, dried completely, and dissolved in minimum amount of TE buffer. DNA was purified by RNAse treatment and quantified by UV Spectrophotometer.
4. Primers, PCR Amplification, and Gel Electrophoresis
PCR amplification with arbitrary primers for RAPD was carried out in 25 μl reaction containing 2 μl dNTP (250 μM each dNTP), 1 μl primer (20 ng/μl), 1 μl template DNA (30 ng/μl), 2.5 μl reaction buffer (10X), 0.5 μl Taq DNA polymerase (3 U/μl), and deionized water 18.0 μl. PCR amplification for ribosomal DNA regions was carried out in 50 μl reaction containing 2 μl dNTP (250 μM each dNTP), 1 μl primer reverse and forward (20 ng/μl each), 1 μl template DNA (30 ng/μl), 2.5 μl reaction buffer (10X), 2.5 U Taq DNA polymerase, and remaining deionized water. PCR reactions were performed with PTC-200 peltier thermal cycler (MJ research inc., Watertown, MS, USA) for both ribosomal amplification with 30 cycles (1 min denaturing at 94°C, 30 sec annealing at 54°C, and 1 min polymerization at 72°C) and RAPD with 35 cycles (1 min denaturing at 94°C, 1 min annealing at 44°C, and 2 min polymerization at 72°C).
After completion of amplifications, 3 μl of gel loading dye was added to each sample, and 25 μl total volumes were resolved on 1.8% (Ribosomal amplified) and 1.5% (RAPD) agarose gel in 0.5X TBE buffer. The size of amplified DNA fragments was estimated with 100 bp ladders (Bangalore Genei Pvt. Ltd., India). Detail of arbitrary primers, synthesized from SIGMA (Sigma-Aldrich, St. Louis, USA), is given in Table 1.
Table 1.
Primers and their codes used for PCR amplification of 25 isolates of Fusarium spp.
| Primers code used in present study | Primers sequence | GC (%) |
|---|---|---|
| PP1 | 5′ ACC GCG AAG G 3′ | 70 |
| PP2 | 5′ GGA CCC AAC C 3′ | 70 |
| PP3 | 5′ GTC GCC GTC A 3′ | 70 |
| PP4 | 5′ GTC TGC CCC A 3′ | 70 |
| PP5 | 5′ AGA TGC AGC C 3′ | 60 |
| PP6 | 5′ GTT TCG CTC C 3′ | 60 |
| PP7 | 5′ GTG AGG CGT C 3′ | 70 |
| PP8 | 5′ GTG ACA TGC C 3′ | 60 |
| PP9 | 5′ ACTCAGCCAC 3′ | 60 |
| PP10 | 5′ CGTAGTGGTG 3′ | 60 |
| PP11 | 5′ CGGTTTGGTC 3′ | 60 |
| PP12 | 5′ GGACGATTCG 3′ | 60 |
| PP13 | 5′ GGGGGTTAGG 3′ | 70 |
| PP14 | 5′ GAGGAGGAGGAGGAG 3′ | 66.6 |
| PP15 | 5′ CATCATCATCATCAT 3′ | 33.3 |
| PP16 | 5′ TCTGGTGACC 3′ | 60 |
| PP17 | 5′ CCGCATCCTA 3′ | 60 |
| PP18 | 5′ CAGGCCCTTC 3′ | 70 |
5. Data Analysis
DNA fingerprints were scored for the presence (1) or absence (0) of bands of various molecular weight sizes in the form of binary matrix. Data were analyzed to obtain Jaccard's coefficients among the isolates by using NTSYS-pc (version 2.11V; Exeter Biological Software, Setauket, NY). Jaccard's coefficients were clustered to generate dendrograms by using the SHAN clustering programme, selecting the unweighted pair-group method with arithmetic average (UPGMA) algorithm version 2.11v in NTSYS-PC computer package (Exter software, NY; [6]).
6. Results and Discussion
The present work deals with molecular characterization of Fusarium sp. isolated from malformed mango tissues. The molecular studies were carried out with the optimization or standardization of DNA extraction procedures from mycelium of the fungus, and evaluation of polymerase chain reaction to explore the variability among different isolates of Fusarium sp. has emerged as most likely cause of malformation disease in mango ([2]; Freeman et al.; U. S. Singh, personnel communication). However, little information is available on variability in Fusarium sp. isolated from malformed mango tissues. Knowledge of genetic mechanisms underlying the variability in pathogen (Fusarium sp.) is almost invariably achieved through the use of molecular markers, that is, molecules which serve to distinguish one species or isolate it from another.
The dominant marker RAPD was used to calculate the genetic distance between 25 Fusarium isolates using Jaccard's similarity coefficient which takes into account the presence or absence of bands.
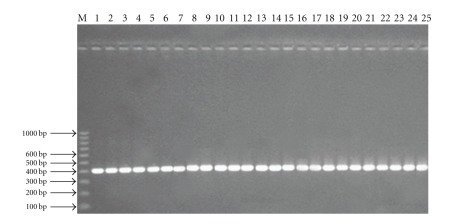
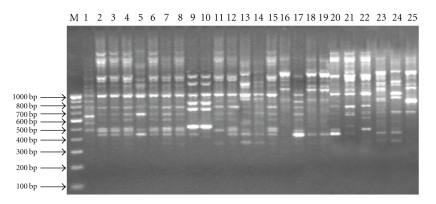
All twenty-five isolates of Fusarium sp. were isolated from malformed panicles and bunchy top seedlings of mango. They were purified by single-spore isolation and subjected to identification by using two Fusarium selective primers which gives approximately 410 bp band (Figure 1). These isolates were compared and categorized at molecular level by using a powerful and extremely sensitive technique RAPD-PCR. The conditions for PCR amplification of fungal DNA were optimized for determining the template DNA concentration and primer suitability. Thirty primers were used to characterize the genetic diversity present among 25 isolates. Eighteen of these primers showed a total of 224 reproducible bands with 12.44 bands per primer. Each of the primers varied greatly in their ability to resolve variability among the genotypes. All primers were able to give high polymorphism among the isolates (Figure 2). The size of the obtained amplicons ranged from 0.264 kb (minimum) to 3.624 (maximum) while the similarity coefficient ranged from 0.17 to 0.94.5 (Table 2).
Figure 1.
Amplification of 410 bp amplicon using Fusarium-specific primers.
Figure 2.
Amplification profile of 25 isolates of Fusarium spp. obtained using PP1 primer.
Table 2.
Similarity matrix of 25 isolates of Fusarium sp. isolated from mango malformed tissues of mango.
| F1 | F2 | F3 | F4 | F5 | F6 | F7 | F8 | F9 | F10 | F11 | F12 | F13 | F14 | F15 | F16 | F17 | F18 | F19 | F20 | F21 | F22 | F23 | F24 | F25 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| F1 | 1.000 | ||||||||||||||||||||||||
| F2 | 0.664 | 1.000 | |||||||||||||||||||||||
| F3 | 0.629 | 0.868 | 1.000 | ||||||||||||||||||||||
| F4 | 0.664 | 0.872 | 0.895 | 1.000 | |||||||||||||||||||||
| F5 | 0.452 | 0.500 | 0.525 | 0.547 | 1.000 | ||||||||||||||||||||
| F6 | 0.635 | 0.835 | 0.842 | 0.924 | 0.546 | 1.000 | |||||||||||||||||||
| F7 | 0.600 | 0.712 | 0.744 | 0.211 | 0.525 | 0.831 | 1.000 | ||||||||||||||||||
| F8 | 0.606 | 0.798 | 0.805 | 0.839 | 0.521 | 0.863 | 0.869 | 1.000 | |||||||||||||||||
| F9 | 0.400 | 0.446 | 0.439 | 0.460 | 0.346 | 0.451 | 0.413 | 0.429 | 1.000 | ||||||||||||||||
| F10 | 0.396 | 0.442 | 0.435 | 0.446 | 0.325 | 0.447 | 0.400 | 0.426 | 0.945 | 1.000 | |||||||||||||||
| F11 | 0.547 | 0.639 | 0.657 | 0.676 | 0.510 | 0.698 | 0.664 | 0.669 | 0.448 | 0.454 | 1.000 | ||||||||||||||
| F12 | 0.563 | 0.676 | 0.669 | 0.737 | 0.547 | 0.746 | 0.750 | 0.716 | 0.404 | 0.391 | 0.704 | 1.000 | |||||||||||||
| F13 | 0.353 | 0.371 | 0.365 | 0.402 | 0.562 | 0.421 | 0.411 | 0.392 | 0.371 | 0.350 | 0.455 | 0.472 | 1.000 | ||||||||||||
| F14 | 0.314 | 0.278 | 0.280 | 0.326 | 0.443 | 0.338 | 0.356 | 0.326 | 0.259 | 0.244 | 0.316 | 0.369 | 0.526 | 1.000 | |||||||||||
| F15 | 0.557 | 0.696 | 0.652 | 0.733 | 0.510 | 0.768 | 0.695 | 0.737 | 0.449 | 0.437 | 0.608 | 0.710 | 0.421 | 0.360 | 1.000 | ||||||||||
| F16 | 0.303 | 0.351 | 0.354 | 0.357 | 0.289 | 0.359 | 0.331 | 0.339 | 0.351 | 0.329 | 0.366 | 0.377 | 0.342 | 0.228 | 0.352 | 1.000 | |||||||||
| F17 | 0.431 | 0.459 | 0.485 | 0.507 | 0.440 | 0.507 | 0.542 | 0.515 | 0.340 | 0.335 | 0.503 | 0.532 | 0.340 | 0.307 | 0.472 | 0.289 | 1.000 | ||||||||
| F18 | 0.254 | 0.234 | 0.227 | 0.226 | 0.206 | 0.230 | 0.227 | 0.229 | 0.278 | 0.255 | 0.253 | 0.269 | 0.228 | 0.174 | 0.243 | 0.386 | 0.265 | 1.000 | |||||||
| F19 | 0.230 | 0.221 | 0.223 | 0.214 | 0.227 | 0.210 | 0.206 | 0.209 | 0.291 | 0.268 | 0.273 | 0.264 | 0.240 | 0.200 | 0.247 | 0.388 | 0.231 | 0.815 | 1.000 | ||||||
| F20 | 0.368 | 0.450 | 0.453 | 0.454 | 0.378 | 0.464 | 0.427 | 0.481 | 0.386 | 0.382 | 0.490 | 0.460 | 0.403 | 0.259 | 0.472 | 0.348 | 0.464 | 0.268 | 0.263 | 1.000 | |||||
| F21 | 0.343 | 0.415 | 0.439 | 0.431 | 0.375 | 0.411 | 0.371 | 0.418 | 0.335 | 0.340 | 0.398 | 0.400 | 0.335 | 0.256 | 0.383 | 0.383 | 0.349 | 0.238 | 0.242 | 0.507 | 1.000 | ||||
| F22 | 0.381 | 0.381 | 0.412 | 0.405 | 0.363 | 0.388 | 0.396 | 0.413 | 0.327 | 0.322 | 0.395 | 0.405 | 0.327 | 0.262 | 0.371 | 0.400 | 0.338 | 0.323 | 0.305 | 0.368 | 0.353 | 1.000 | |||
| F23 | 0.369 | 0.406 | 0.438 | 0.430 | 0.371 | 0.431 | 0.393 | 0.428 | 0.388 | 0.384 | 0.486 | 0.437 | 0.378 | 0.284 | 0.439 | 0.349 | 0.376 | 0.275 | 0.306 | 0.510 | 0.480 | 0.333 | 1.000 | ||
| F24 | 0.343 | 0.336 | 0.347 | 0.369 | 0.384 | 0.372 | 0.389 | 0.369 | 0.283 | 0.278 | 0.378 | 0.408 | 0.392 | 0.324 | 0.373 | 0.321 | 0.369 | 0.240 | 0.244 | 0.351 | 0.344 | 0.363 | 0.422 | 1.000 | |
| F25 | 0.315 | 0.292 | 0.311 | 0.308 | 0.317 | 0.310 | 0.299 | 0.308 | 0.301 | 0.305 | 0.316 | 0.346 | 0.301 | 0.220 | 0.305 | 0.391 | 0.291 | 0.389 | 0.403 | 0.316 | 0.375 | 0.392 | 0.420 | 0.323 | 1.000 |
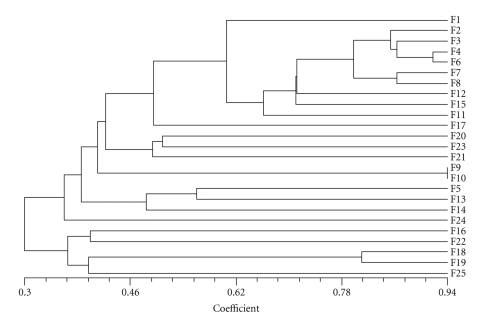
Association among 18 genotypes revealed by unweighted pair-group methods with Arithmetic mean (UPGMA) cluster analysis was presented in Figure 3.
Figure 3.
Combined phenogram of 25 isolates of Fusarium species using 18 primers constructed by NTsys PC.
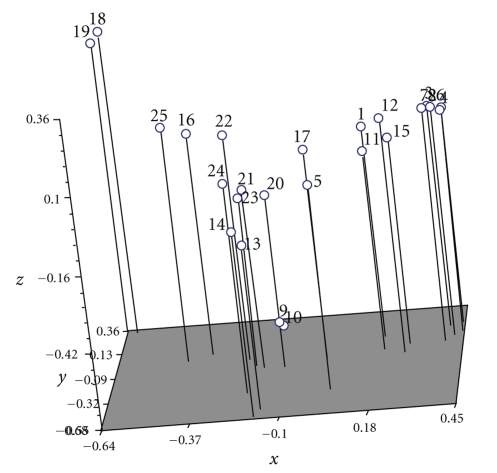
A total of 18 primers having GC content ranging from 33.3 to 70% were used in the study. The UPGMA cluster analysis clearly grouped these isolates into two major clusters and established their relationship of similarity. The cluster analysis comprising the 25 isolates showed 17.4% (isolates 18 and 14) to 94.5% similarity in RAPD analysis. Cluster I has 20 isolates F1, F2, F3, F4, F5, F6, F7, F8, F9, F10, F11, F12, F13, F14, F15, F17, F20, F21, F23, and F24 and Cluster II has 5 isolates F16, F18, F19, F22, and F25. A relatively high level of variability (17.4 to 94.5%) was recorded among different isolates of Fusarium sp. Similarly Zheng and Ploetz [7] recorded wide variation in RAPD profiles of 74 isolates of Fusarium from mango using 10-mer primers. Further studies are needed to confirm this fact by morphological studies. A number of Fusarium spp. have already been isolated from malformed mango tissues [1]. However, F. subglutinans (Wollenw. and Reink) P. E. Nelson, T. A. Toussoun & Marasas (= F. moniliforme var. subglutinans, Wollenw. and Reink) is reported to be most commonly associated species with both floral and vegetative malformation. However, taxonomy and nomenclature of this species have recently been in flux [7]. Based on molecular characterization of 95 isolates of Fusarium sp. from malformed mango tissues, it can be concluded that all mango isolates belong to a new species of Fusarium. The pattern of cluster analysis is further confirmed by principal component analysis (Figure 4). The numbers plotted represent individual isolates of Fusarium spp.
Figure 4.
Three-dimensional view of score plot resulted from principle coordinate analysis of RAPD data.
The Matrix correlation (r) value of this marker is 0.93842, indicating very good fit between distance or similarity matrix and dendogram obtained. The results for RAPD marker are presented in two- and three-dimensional score plots (Figure 4). In this case, the cluster as well as the score plots of principal component analysis was found similar. The result of pairwise combinations indicated highest similarity (coefficient 0.945) between isolates 9 and 10. Isolates 4 and 6 also exhibited high degree of similarity (92.4%). The use of single or combination of two or more primers differentiated most of the genotypes. By using the 18 primers, all the genotypes could be distinguished from each other. So this study is helpful to understand the actual cause as well as the causal organism of the disease and can further support and strengthen the fact that Fusarium sp. is the actual causal organism of this disease.
References
- 1.Kumar J, Beniwal SPS. Role of Fusarium species in the etiology of mango malformation. In: Proceedings of the the 4th International Mango Symposium; July 1992; Miami, Fla, USA. p. 133. [Google Scholar]
- 2.Kumar J, Singh US, Beniwal SPS. Mango malformation: one hundred years of research. Annual Review of Phytopathology. 1993;31:217–232. [Google Scholar]
- 3.Powell W, Morgante M, Andre C, et al. The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis. Molecular Breeding. 1996;2(3):225–238. [Google Scholar]
- 4.Abd-Elsalam KA, Aly IN, Abdel-Satar MA, Khalil MS, Verreet JA. PCR identification of Fusarium genus based on nuclear ribosomal-DNA sequence data. African Journal of Biotechnology. 2003;2(4):96–103. [Google Scholar]
- 5.Lee SB, Taylor JW. Isolation of DNA from fungal mycelia and single spores. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. The PCR Protocols: A Guide to Methods and Applications. New York, NY, USA: Academic Press; 1990. pp. 282–314. [Google Scholar]
- 6.Rohlf FJ. NT-SYS-pc: Numerical Taxonomy and Multivariate Analysis System. Version 2.11V. Setauket, NY, USA: Exteer Software; 1993. [Google Scholar]
- 7.Zheng Q, Ploetz R. Genetic diversity in the mango malformation pathogen and development of a PCR assay. Plant Pathology. 2002;51(2):208–216. [Google Scholar]