Summary
We previously showed that mutations in the genes encoding the two main biosynthetic enzymes responsible for polyamine production, arginine decarboxylase (SpeA) and ornithine decarboxylase (SpeC), cause a loss of biofilm formation in Yersinia pestis. In Y. pestis the development of a biofilm is dependent on 6 Hms (hemin storage) proteins (HmsH, F, R, S, T, and P) grouped into 3 operons; hmsHFRS, hmsT, and hmsP. In this article we show that polyamines are necessary to maintain the levels of key Hms proteins. In the absence of polyamines there is an ~93%, ~43%, and ~90% reduction in protein levels of HmsR, HmsS, and HmsT respectively. Over-expression of hmsR and hmsT from plasmids alone can restore biofilm formation to a SpeA− SpeC− mutant. Addition of exogenous putrescine also restores normal levels of HmsR, HmsS, HmsT, and biofilm production. Analyses using transcriptional reporters and quantitative RT-PCR indicate that the initiation of transcription and mRNA stability are not reduced by polyamine deficiency. Instead, translational reporters indicate that polyamines function at least in part by modulating the translation of HmsR and HmsT. Although construction of a consensus Shine-dalgarno sequence upstream of hmsT modestly reduced the stimulation of translation by putrescine, additional mechanisms likely contribute to the polyamine-dependent expression of HmsT. Finally, we have shown that polyamines play a role in bubonic plague.
Keywords: Y. pestis, Polyamines, Biofilm, hms, GGDEF, c-di-GMP
Introduction
Biofilms are complex bacterial communities that congregate within an exopolysacchride matrix often at liquid-surface interfaces. Members of these biofilm communities are afforded protection from a number of environmental stresses like changes in pH and UV exposure as well as from some host defenses and medical treatments (Mah and O’Toole, 2001; Hall-Stoodley et al., 2004). Biofilm production is a dynamic event which can be stimulated by environmental cues or by the presence of small signaling molecules. Extracellular molecules include autoinducers which are quorum sensing molecules released by bacteria to give feedback on population densities. The release and detection of autoinducers leads to changes in gene expression of a number of important processes including biofilm production (Williams et al., 2007). External conditions can also affect the production of intracellular signaling molecules, in particular 3′,5′-cyclic diguanylic acid (c-di-GMP). C-di-GMP levels are regulated by diguanylate cyclases, which produce c-di-GMP from two molecules of GTP, and phosphodiesterases which cleave c-di-GMP into a non-functional linear form. C-di-GMP is a ubiquitous molecule that has been identified as a positive regulator of biofilm formation in a wide array of bacteria (Jefferson, 2004; Camilli and Bassler, 2006; Jenal and Malone, 2006; Karatan and Watnick, 2009).
In Yersinia pestis, biofilm formation has been associated with the ability of the bacteria to create a blockage of the proventriculus valve of infected fleas contributing to one form of transmission of plague to mammals (Hinnebusch et al., 1996; Jarrett et al., 2004). While biofilm development plays a role in transmission from fleas to mammals it is not required for the virulence of bubonic or pneumonic plague in mice (Lillard et al., 1999; Abu Khweek et al., 2010) or Y. pestis infection of Caenorhabditis elegans (Styer et al., 2005). Biofilm development in Y. pestis involves 6 hms (hemin storage) genes. Four of these genes, hmsHFRS, form a polycistronic operon located within the 102-kb pigmentation (pgm) locus. The pgm locus is spontaneously deleted at high frequency which results in the elimination of the biofilm phenotype (Fetherston et al., 1992). HmsH and HmsF are outer membrane (OM) proteins with a predicted β-barrel structure and a deacetylase domain, respectively, whereas HmsR and HmsS are inner membrane (IM) proteins with a putative glycosyltransferase domain and no currently recognized domains, respectively (Lillard et al., 1997; Perry et al., 2004). The HmsHFRS proteins show high to moderate similarities to the pgaABCD (formerly ycdSRQP) gene products (Jones et al., 1999); PgaABCD and HmsHFRS are necessary for the biosynthesis of the biofilm polysaccharide poly-β-1,6-N-acetyl-D-glucosamine in Escherichia coli and Y. pestis, respectively (Wang et al., 2004; Itoh et al., 2005; Bobrov et al., 2008).
Located outside of the pgm locus are two genes, hmsT and hmsP, necessary for biofilm regulation. HmsT is an IM protein characterized by a GGDEF domain that is associated with diguanylate cyclase activity responsible for the formation of c-di-GMP in Y. pestis (Jones et al., 1999; Kirillina et al., 2004; Simm et al., 2005) and a number of other bacteria (Cotter and Stibitz, 2007). When the entire gene is deleted or when single substitutions are made in the GGEE residues the biofilm phenotype is lost (Kirillina et al., 2004). HmsP is an IM protein that contains an EAL domain that exhibits phosphodiesterase activity which likely degrades c-di-GMP and therefore is necessary for the negative regulation of biofilm formation (Kirillina et al., 2004; Bobrov et al., 2005).
In Y. pestis, biofilm formation is characterized by the absorption of large amounts of hemin or Congo red (CR) during growth at 26°C but not at 37°C indicating that biofilm formation is temperature sensitive. Transcriptional reporters and RNA dot blots revealed no significant temperature regulation at the RNA level. Degradation of HmsR, HmsS, and HmsT appear to be the primary mechanism for temperature-dependent biofilm development (Jackson and Burrows, 1956; Surgalla and Beesley, 1969; Jones et al., 1999; Perry et al., 2004).
Previously we have shown that polyamines are necessary for biofilm formation in Y. pestis (Patel et al., 2006). Polyamines are small, polycationic molecules that are thought to exist in virtually all living organisms. HPLC analysis showed that when the genes encoding two key biosynthetic enzymes necessary for polyamine production, arginine decarboxylase (ADC; SpeA) and ornithine decarboxylase (ODC; SpeC), were mutated, the two main polyamines in bacteria, putrescine and spermidine, were reduced to levels below detectable limits. This reduction in polyamines, especially putrescine, correlated with a loss of biofilm formation. The loss of biofilm caused by the undetectable levels of internal polyamines was reversed by the addition of exogenous putrescine (Patel et al., 2006).
In this article, we present evidence which demonstrates that the polyamine requirement for Y. pestis biofilm results from its effect on the levels of key Hms proteins. Modulation of the translation of HmsR and HmsT appears to be a primary effect of polyamine deprivation.
Results
Uptake of polyamines by Y. pestis
Since exogenous putrescine but not spermidine restored biofilm development in the Y. pestis ΔspeA ΔspeC mutant (KIM6-2112.1+), we examined the ability of Y. pestis to accumulate these two polyamines. In E. coli, the PotABCD system is considered spermidine preferential because it transports spermidine but is also able to transport putrescine. PotFGHI is specific for uptake of putrescine (Igarashi and Kashiwagi, 1999). BLAST analysis of the putrescine-specific PotFGHI proteins identified highly homologous open reading frames (ORFs) in Y. pestis. Each of the four Y. pestis ORFs (Y2847-Y2849 and Y2851) that comprise this system had a percent identity of at least 73%. BLAST analysis of each of the spermidine-preferential PotABCD proteins identified only Y. pestis ORFs with low similarity. The ATP-binding protein PotA (Y1388) had the highest similarity, 62%. The lowest similarity found between the E.coli and Y. pestis transport systems was for the polyamine-binding protein PotD (Y1391) which had only a 44% similarity.
Both Y. pestis KIM6+ and E. coli K12 accumulated 14C-putrescine to similar levels over a 30 min period in an energy-dependent manner (Fig. 1A and data not shown). However, E. coli but not Y. pestis was also able to transport 14C-spermidine from an exogenous concentration of 10 μM (Fig. 1B). The level of Y. pestis cell-associated spermidine binding was energy-independent since cells poisoned by CCCP had similar levels of cell-associated 14C-spermidine (data not shown). These results indicate that Y. pestis KIM6+ transports exogenous putrescine but not spermidine into the bacterial cell in an energy dependent manner.
Fig. 1.
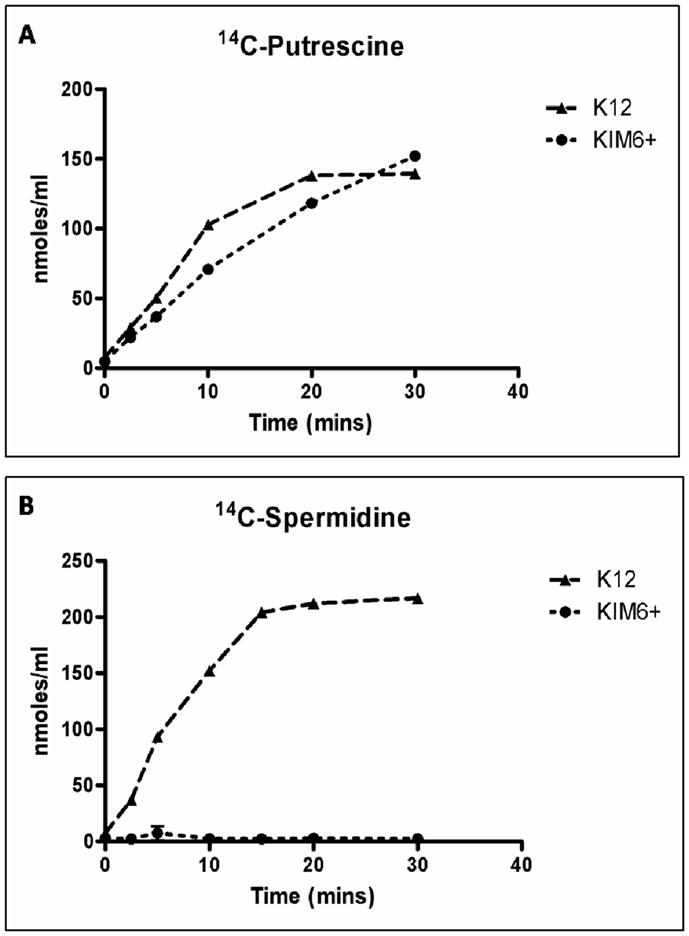
Putrescine and spermidine transport by Y. pestis and E. coli. Uptake of 14C-putrescine (A) or 14C-spermidine (B) into Y. pestis KIM6+ and E. coli K12 cells. Cell-associated substrate accumulation is reported as nmol ml−1 of culture at an OD620 of 0.4.
Polyamines and c-di-GMP metabolism are independently essential for biofilm formation
The hmsHFRS and hmsT operons are both necessary to form biofilm in Y. pestis. When any of these genes are mutated loss of biofilm development results; when either operon is over-expressed, an over abundance of biofilm is produced at 26°C and 37°C (Jones et al., 1999; Kirillina et al., 2004). Fig. 2 shows that over-expression of hmsT in a SpeA−SpeC− mutant (Fig. 2A), the addition of 1 mM exogenous putrescine to an hmsT::kan mutant, or over-expression of SpeA in a hmsT::kan mutant (Fig. 2B) all fail to restore biofilm formation. Thus exogenous putrescine cannot compensate for the lack of HmsT and HmsT alone cannot compensate for polyamine deprivation indicating that both pathways are necessary for biofilm production.
Fig. 2.
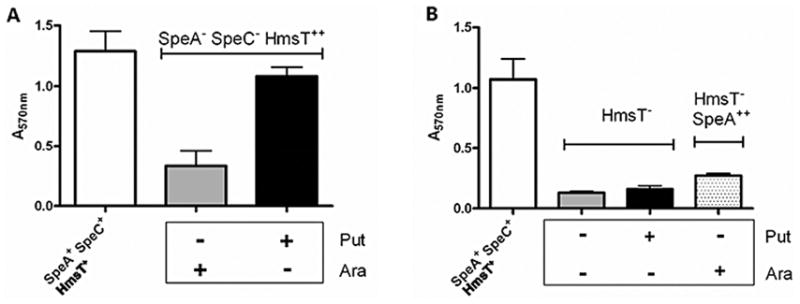
Cross complementation does not restore biofilm formation in Y. pestis SpeA−SpeC− or HmsT− mutants. Panels show CV staining (A570nm), as a measure of biofilm. A) Y. pestis SpeA−SpeC− cells expressing HmsT from an inducible plasmid[SpeA−SpeC− HmsT++; KIM6-2112.1(pBADhmsT6xH)+] B) Y. pestis HmsT− mutant (HmsT−; KIM6-2051+) alone and with SpeA expressed from a plasmid [HmsT− SpeA++; KIM6-2051(pBAD/HisbADC)+]. Cells grown with (+) or without (−) exogenous 1 mM putrescine (Put) or 0.2% (w/v) arabinose (Ara). Expression of hmsT from pBADHmsT6xH and speA from pBAD/HisbADC is arabinose inducible. Parent strain, KIM6+ (SpeA+SpeC+ HmsT+), is shown as a control. Averages for CV staining were calculated from duplicate samples from two or more independent experiments with error bars indicating standard deviations.
Effects of polyamines on protein expression of Hms proteins
To determine whether the lack of polyamines affects the levels of Hms proteins, extracts collected from the Y. pestis SpeA+SpeC+ parent strain (KIM6+) and the Y. pestis SpeA−SpeC− mutant (KIM6-2112+) grown in the absence of polyamines and the same mutant grown with 1 mM putrescine were analyzed by Western blot. The levels of HmsH and HmsF are nearly unchanged while the level of HmsR was reduced by ~93% and HmsS had an ~43% reduction in its protein level. HmsT, encoded outside of the pgm locus, also showed a dramatic decrease in protein level with a reduction of ~90% while HmsP showed little change (Fig. 3A). In each instance where there was a significant reduction in the amount of protein, addition of 1 mM putrescine to the medium during growth resulted in full recovery of protein levels to that of the parent strain. In contrast, exogenous putrescine did not increase the levels of HmsH, HmsF, or HmsP (Fig. 3A) and the addition of exogenous putrescine to the SpeA+SpeC+ parent did not increase any Hms protein level (data not shown). Putrescine had no effect on the level of an unrelated control protein, YfeA (a periplasmic-binding protein for an ABC transporter for iron and manganese; Fig. 3B) (Bearden and Perry, 1999; Perry et al., 2007).
Fig. 3.
Western blot analysis of Hms proteins in the Y. pestis SpeA−SpeC− mutant (KIM6-2112.1+) and parent strain (SpeA+ SpeC+ KIM6+). Equal concentrations of whole-cell lysates were separated by SDS-PAGE, blotted and reacted with anti-serum against the indicated protein. The SpeA− SpeC− mutant was grown with or without 1 mM exogenous putrescine (Put). The average percent reduction (± standard deviation) in the level of the YfeA control protein and each Hms protein in SpeA+ SpeC+ cells compared to SpeA−SpeC− cells, as quantified by the image processing software ImageJ, is shown. Average percentages and standard deviations were calculated from three individual cultures and blots. Arrows identify HmsT and HmsP from cross-reacting bands.
To determine whether biofilm production can be restored by increasing the levels of Hms proteins that were reduced by polyamine deficiency, SpeA−SpeC− cells were transformed with plasmids expressing the affected hms genes (Fig. 4). We were unable to obtain SpeA−SpeC− cells expressing all three hms genes affected by polyamines. However, complementation of the SpeA−SpeC− mutant with plasmids expressing HmsR and HmsT restored biofilm to near wild-type levels without the addition of exogenous putrescine (Fig. 4A). Fig. 4B shows that the levels of HmsR and HmsT are increased in the complemented strain, relative to the vector control. These results indicate that lower levels of these Hms proteins is the primary cause for loss of biofilm formation in the polyamine mutant.
Fig. 4.
Biofilm production is restored by over-expressing HmsT and HmsR in Y. pestis SpeA−SpeC− cells. (A) CV staining as a measure of biofilm production in SpeA− SpeC− cells carrying both pBAD30 and pACYC184 vectors and the same vectors expressing HmsR (pBADHmsR) and HmsT (pAHMS8) (HmsT++ HmsR++) are shown. The SpeA+ SpeC+ strain is included for comparison. CV staining values are the averages of three independent experiments with error bars indicating standard deviations. (B) Western blot analysis comparing protein levels of HmsT and HmsR in the control Y. pestis SpeA+ SpeC+ and SpeA− SpeC− cells carrying vectors alone or plasmids expressing HmsR and HmsT.
Exogenous putrescine does not affect transcription of hms genes or mRNA levels
A systematic approach was taken to determine how polyamines affected the production of the Hms proteins. First, the transcription of hms genes was examined using hmsT::lacZ (pEUHmsT-Pro) or hmsH::lacZ (pEUHmsH-Pro) transcriptional reporters. Fig. 5 shows that the promoter activity from the hmsHFRS and hmsT reporters is not reduced in a SpeA−SpeC− mutant grown in the absence of putrescine. These results indicate that polyamine deficiency did not lower proteins levels (see Fig. 3) by reducing the level of transcription of the relevant genes.
Fig. 5.
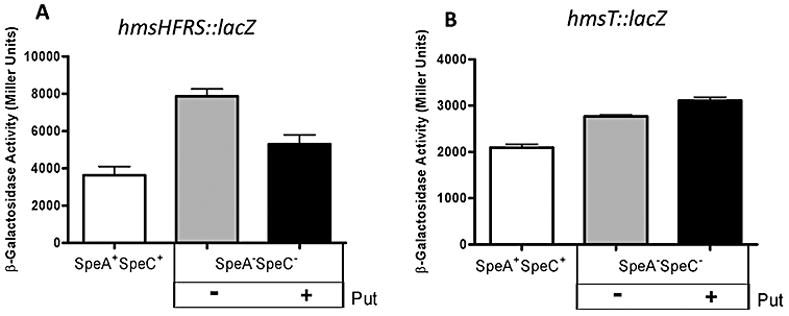
The effect of polyamines on hms::lacZ transcriptional reporters. β-galactosidase activities from hmsHFRS::lacZ (A) and hmsT::lacZ (B) transcriptional reporters in Y. pestis SpeA−SpeC− cells grown with (+) or without (−) 1mM putrescine (Put) are shown. SpeA+SpeC+ cells are shown for comparison. β-galactosidase values are the averages of duplicate samples from two or more independent experiments with error bars indicating standard deviations.
Due to their cationic nature, large amounts of cellular polyamines bind RNA; the vast majority of spermidine and ~50% of putrescine are found complexed with RNA (Miyamoto et al., 1993) This suggest that polyamines play an important role in both RNA stability and function. Therefore quantitative RT-PCR (qRT-PCR) was used to investigate whether the lack of polyamines could reduce protein levels by modulating mRNA levels. The amount of hmsHFRS and hmsT RNA in the parent strain (SpeA+SpeC+) and the polyamine deficient mutant (SpeA−SpeC−) was normalized relative to an internal standard, gyrA, encoding DNA gyrase subunit A (Fig. 6). No significant changes that correlate with the large differences in protein levels seen in Fig. 3 were found in mRNA levels for the Hms proteins (Fig. 6).
Fig. 6.
Messenger RNA ratios of hms genes. Values represent mean fold-expression differences (± standard deviation) between Y. pestis SpeA+ SpeC+ and SpeA− SpeC− cells grown in PMH2, a polyamine-deficient medium. Messenger levels were quantified from DNase treated RNA collected at mid-log phase from three independent cultures. RNA was run in duplicate using a Roche one step Lightcycler RNA Master SYBR Green I mix and quantified using the Pfaffl analysis method.
Post-transcriptional effects of polyamines
Previously we have shown that HmsS interacts with HmsR and is less stable in the absence of HmsR (Wang et al., 2004; Itoh et al., 2005; Bobrov et al., 2008). To determine whether the moderately lower HmsS level in the SpeA−SpeC− mutant was simply due to the lower HmsR level, we over-expressed HmsR in the SpeA−SpeC− mutant. Western blot analysis shows the expected drop in the HmsS level in the SpeA−SpeC− mutant compared to SpeA+SpeC+ KIM6+ (Fig. 7, lanes 1 and 2). Over-expression of HmsR in the SpeA−SpeC− mutant increased the level of HmsS to that found in KIM6+ over-expressing HmsR (Fig. 7, lanes 3 and 4). Thus the primary effect of polyamines on HmsS is indirect acting through HmsR.
Fig. 7.

Western blot analysis of HmsS levels in the Y. pestis SpeA−SpeC− mutant (KIM6-2112.1+) and parent strain (SpeA+ SpeC+ KIM6+) with and without over-expression of HmsR. Equal concentrations of whole-cell lysates were separated by SDS-PAGE, blotted and reacted with anti-serum against HmsS. The label identifies HmsS from cross-reacting bands. KIM6-2112.1+ and KIM6+ strains are indicated by the plus and minus signs, respectively, in the SpeAC row. These strains carried either the pBAD30 vector or the HmsR over-expression vector pBADHmsR (plus and double plus signs, repectively, in the HmsR row). Western blot analysis with HmsR anti-sera showed increased HmsR levels in strains carrying pBADHmsR compared to those carrying pBAD30 (data not shown).
Translation reporters were used to assess the effect of putrescine on translation of hmsP (negative control), hmsR, and hmsT. Plasmid translational reporters for HmsT (HmsT::β-gal; pRMCD70hmsT-P390) and HmsP (HmsP::β-gal; pRMCD70hmsP-H647) include putative promoter regions, ribosome binding sites and start codons (Wang et al., 2004; Itoh et al., 2005; Bobrov et al., 2008). Because hmsR is the third gene of a four gene operon, a chromosomally-integrated fusion was used (HmsR::β-gal; KIM6-2118.5; Table 1) to avoid using an artificial promoter for expression from a plasmid. Suicide vectors were used to construct a polyamine deficient background in the HmsR integrated reporter strain (KIM6-2118.6; Table 1). β-galactosidase activities of SpeA+SpeC+ and SpeA−SpeC− cells carrying these translational reporters were analyzed after growth with and without 1 mM putrescine. Fig. 8A and 8B shows that the activities of HmsR and HmsT translational reporters in the SpeA−SpeC− cells were decreased ~2-fold, ~1.5-fold, respectively compared to the SpeA+SpeC+ strain. With both reporters in the SpeA−SpeC− strain, the addition of exogenous putrescine increased expression of the β-gal fusion proteins (Fig. 8A and 8B). Since HmsP levels in the SpeA−SpeC− cells are not affected by the presence of putrescine (Fig. 3), an HmsP::β-gal fusion was used a control. Fig. 8C shows that the activity of the translational reporter for HmsP in the polyamine deficient cells was not significantly reduced compared to the parent strain. For each of the translational reporters the addition of exogenous putrescine had no effect on β-galactosidase activity in the SpeA+SpeC+ cells (Fig. 8).
Table 1.
Bacterial strains, plasmids, and primers used in this study.
| Strains* | Characteristics | Reference or source |
|---|---|---|
| KIM6+ | Pgm+ Pla+ Lcr−SpeA+ SpeC+ | (Fetherston et al., 1992) |
| KIM6-2051+ | Pgm+ Pla+ Lcr−Kmr hmsT2051::mini-kan | (Jones et al., 1999) |
| KIM6-2112 (pKD46)+ | Pgm+ Pla+ Lcr−SpeA− (ΔspeA::cam2111) SpeC− (ΔspeC::kan2010) Apr Cmr Kmr | (Patel et al., 2006) |
| KIM6-2112.1+ | Pgm+ Pla+ Lcr−SpeA− (ΔspeA2111.2) SpeC− (ΔspeC2010.2) | This Study |
| KIM5(pCD1Ap)+ | Pgm+ Pla+ Lcr+SpeA+ SpeC+ Apr | (Gong et al., 2001) |
| KIM5-2112.1 (pCD1Ap)+ | Pgm+ Pla+ Lcr+SpeA− (ΔspeA2111.2) SpeC− (ΔspeC2010.2) Apr | This Study |
| KIM6-2118.5 | Pgm+ Pla+ Lcr−SpeA+ SpeC+ Hms− (ΔhmsRS::lacZ)ΔlacZ2084 β-Gal+ (hmsR::lacZ translational fusion) Kmr | A. Bobrov and O. Kirillina |
| KIM6-2118.6 | Pgm+ Pla+ Lcr−SpeA− (ΔspeA2111.2) SpeC− (ΔspeC2010.2) (ΔhmsRS::lacZ) Kmr | This Study |
| KIM6-2181+ | Pgm+ Pla+ Lcr−SpeA− (ΔspeA2111) SpeC− (ΔspeC2010) (altered hmsT SD) | This Study |
| Plasmids | characteristics | Reference or source |
| pBAD30 | 4.9 kb, Apr, araC+, arabinose-inducible promoter upstream of multiple-cloning site | (Guzman et al., 1995) |
| pBADHmsT6xH | 6.1 kb, Apr, arabinose-inducible hmsT with C-terminal His tag; 1.2-kbSmaI-EcoRI PCR product from pAHMS16 ligated into pBAD30 | (Simm et al., 2005) |
| pBADHmsR | 6.3 kb, Apr, araC+, arabinose-inducible hmsR 1.4 kb EcoRI-XbaI PCR product ligated into pBAD30 | (Forman et al., 2006) |
| pBAD/Hisb | 4.1 kb, Apr, six-histidine-tagged fusion protein plasmid | Invitrogen |
| pBAD/HisbADC | 6.1kb, Apr, His-ADC expression vector, 2-kb speA PCR product ligated into SacI-HindIII sites of pBAD/Hisb | (Patel et al., 2006) |
| pACYC184 | 4.2kb, Cmr Tcr, moderate-copy-number cloning vector | (Chang and Cohen, 1978) |
| pAHMS8 | 10.5 kb, Cmr, hmsT+, 6.3 kb BamHI fragment from pAHMS496 ligated into pACYC184 | (Jones et al., 1999) |
| pEU730 | 15.2 kb, Spcr, low-copy-number vector with promoterless lacZ | (Froehlich et al., 1994) |
| pEUHmsT-Pro | 15.5 kb, Spcr, low-copy-number vector with hmsT::lacZ reporter; 340 bp KpnI and PacI fragment ligated into pEU730 | This Study |
| pEUHmsH-Pro | 15.5 kb, Spcr, low-copy-number vector with hmsH::lacZ reporter; 314 bp KpnI and PmeI fragment ligated into pEU730 | This Study |
| pRMCD70hmsP- H647 | 2106 bp (hmsP) insert (aa 1–646) cloned into XbaI/PstI of pRMCD70 | (Wang et al., 2004; Itoh et al., 2005; Bobrov et al., 2008) |
| pRMCD70hmsT- P390 | Apr, HmsT::β-gal, 1411 bp (hmsT) insert (aa 1-390) into pRMCD70 | (Wang et al., 2004; Itoh et al., 2005; Bobrov et al., 2008) |
| pNEB193 | 2.7 kb cloning vector; Apr | New England Biolabs |
| pKNG101 | 6.8 kb suicide vector, sacB+, R6K origin, Smr | (Kaniga et al., 1991) |
| pKNG_ADC | 1026 bp insert cloned from KIM6-2112.1+ into BamHI/SmaI of pKNG101 | This Study |
| pKNG_ODC | 1369 bp insert cloned from KIM6-2112.1+ into BamHI/SmaI of pKNG101 | This Study |
| pKNG_hmsTSD | 784 bp insert cloned into SmaI/XbaI of pKNG101 | This Study |
| Primers | Sequence | use |
| T-ProF | 5′-TTAATTAACGGTGATCCCCCACGAACG-3′ | hmsT Transcriptional reporter |
| T-ProR | 5′-GGTACCCGTGCTGTCAGTAGACTAAT-3′ | hmsT Transcriptional reporter |
| H-ProF | 5′-GCTTTGTTTAAACAGAACACTGTATCGCAGCAT-3′ | hmsH Transcriptional reporter |
| H-ProR | 5′-GGGGTACCTATATAACCCTTAAGCCAGC-3′ | hmsH Transcriptional reporter |
| SpeA-pKNGF | 5′-CGGGATCCCTGCGCGTAGCGAGCAAATCA-3′ | SpeA Suicide Vector |
| SpeA-pKNGR | 5′-TGCTACTCGCCATCGCCATCCA-3′ | SpeA Suicide Vector |
| ODC-pKNGF | 5′-CGGGATCCCAGGCGATCAACTGCTCTCTTTTCAA3′ | SpeC Suicide Vector |
| ODC-pKNGR | 5′-CCGGCCAACAGCAGGCCAATTA-3′ | SpeC Suicide Vector |
| hmsT_SVF1 | 5′-CGGGATCCCATCCAAGCTGTTACTTACTGTATTACCTAC-3′ | hmsT SD Insertion |
| hmsT_SVR3 | 5′-CTCTGCATAATATCCTCCTGTCAGTAGACTAA-3′ | hmsT SD Insertion |
| hmsT_SV4 | 5′-TTAGTCTACTGACAGGAGGATATTATGCAGAG-3′ | hmsT SD Insertion |
| hmsT_SV2 | 5′-GCTCTAGAGATTATACAAACTGGTCAGAGGGTCTATCATGC-3′ | hmsT SD Insertion |
| hmsH-RTf | 5′-TTCGTGCCAGAGCCGGTGAT-3′ | RT-PCR |
| hmsH-RTr | 5′-CCAGCGCTTTTCGTTGCGGT-3′ | RT-PCR |
| hmsF-RTf | 5′-GCGCCGAATGGTACGGTGCT-3′ | RT-PCR |
| hmsF-RTr | 5′-ATCAACATGCGCCACCCGCA3′ | RT-PCR |
| hmsR-RTf | 5′-TGGCTCCAGTGACGATACCGCAC-3′ | RT-PCR |
| hmsR-RTr | 5′-TCGAAGAGAACTCCCCGACCTG-3′ | RT-PCR |
| hmsS-RTf | 5′-ATGGGGCCGATACCGTTGAGGA-3′ | RT-PCR |
| hmsS-RTr | 5′-GCGCCATGCTGCTTGCCAAT-3′ | RT-PCR |
| hmsT-RTf | 5′-TTCGCGATGCTGTGCGTTCA-3′ | RT-PCR |
| hmsT-RTr | 5′-AGCGCTTCATCCGCGGCTTT-3′ | RT-PCR |
| gyrA-RTf | 5′-AGACGGCATGCGCATCGTGA-3′ | RT-PCR |
| gyrA-RTr | 5′-TGGGCGCGATCACGTGCTTT-3′ | RT-PCR |
A plus sign indicates an intact chromosomal 102-kb pgm locus in Y. pestis. All other Y. pestis strains have a mutation within this locus or a deletion of the entire locus.
Fig. 8.
The effect of polyamines on Hms::β-gal translational reporters. β-galactosidase enzyme assays were performed on permeabilized Y. pestis cells carrying the appropriate reporter fusion plasmids grown without (−) and with (+) 1 mM putrescine. The values are averages from at least two independent cultures with enzyme assays performed in duplicate. Error bars indicate standard deviations.
Effects of a consensus Shine-dalgarno (SD) sequence on HmsT expression
Using microarray analysis of a polyamine deficient E. coli strain, it was found that 309 genes were up-regulated by the addition of exogenous polyamines (Yoshida et al., 2004; Igarashi and Kashiwagi, 2006). Up-regulation of these 300+ genes was not a direct result of polyamine stimulation but an indirect effect through the enhanced translation of a smaller group of genes encoding transcriptional regulators. Of these genes, four lack a consensus SD sequence which is important in the recruitment of the 30S ribosomal unit to the initiation site for translation (Shine and Dalgarno, 1975). When a consensus SD sequence is placed upstream of the start codon in these genes increased translation is observed in the absence of polyamines compared to that of the wt gene – indicating an expected, general increase in translation efficiency. More significantly, a canonical SD sequence also diminishes polyamine-stimulation of these genes as judged by comparing the ratio of protein synthesis with and without polyamines in the wild type gene relative to the recombinant gene with a consensus SD sequence. Thus the absence of a canonical SD in genes whose translation is stimulated by polyamines is one mechanism for polyamine control of translation (Yoshida et al., 2004; Higashi et al., 2006; Terui et al., 2007; Terui et al., 2009). Of the five hms genes necessary for biofilm production, hmsR and hmsT lack a consensus SD sequence (Fig. 9A). Because the protein levels expressed from these two genes are most affected by polyamines it seemed likely that the insertion of a consensus SD sequence might diminish or eliminate the polyamine control of translation of hmsT and hmsR.
Fig. 9.

Shine-dalgarno (SD) sequences of hms genes and polyamine-dependency. A) Sequences upstream of the predicted start codon for the hms genes required for biofilm production. Consensus Shine-dalgarno (SD) sequences are in bold text and underlined; sequences end at the predicted ATG start (bold text). B) Upstream sequence of hmsT before and after the construction of a consensus SD sequence. The two residues changed are underlined in bold text. The predicted ATG starts are shown in bold text. C) CV staining as a measure of biofilm production in Y. pestis SpeA+ SpeC+ cells, and SpeA− SpeC− cells with (+) and without (−) an altered SD sequence (hmsT_SD+) and grown with (+) and without (−) 1mM putrescine (Put). D) Western blot analysis comparing protein levels of HmsT under the same conditions as those for the CV assay. Percentages represent the relative protein level of HmsT with respect to the parent strain (SpeA+ SpeC+).
Of the two genes lacking an SD sequence, hmsT was chosen for genetic manipulation because it lies outside the four gene hmsHFRS operon. Using a two step PCR procedure in combination with suicide vectors, single point mutations were made in the chromosomal sequence upstream of hmsT in the SpeA−SpeC− mutant to create a consensus SD sequence (hmsT_SD+) (Fig. 9B). As expected, biofilm levels correlated with HmsT protein levels (Fig. 9C and 9D). If it follows the E. coli example, the hmsT_SD+ alteration should decrease the ratio of HmsT protein levels in the presence vs the absence of exogenous putrescine in the polyamine-deficient mutant. Fig. 9D shows that the ratio of HmsT protein levels in the presence vs absence of putrescine was 9.5 and 7.8 in the strains carrying wt hmsT and hmsT_SD+, respectively. Thus introduction of an SD for hmsT caused an ~18% reduction in polyamine stimulation of translation.
The polyamine-deficient mutant is attenuated in a mouse model of bubonic plague
To determine the effect of polyamine deficiency on the virulence of Y. pestis for mammals, we tested SpeA+ SpeC+ KIM5(pCD1Ap)+ and SpeA− SpeC− KIM5-2112.1(pCD1Ap)+ cells for virulence in both subcutaneous and intranasal routes of infection. The non-mutated parent strain had an LD50 of 329 ± 105 and 23 ± 14 for intranasal and subcutaneous infection routes, respectively, while the polyamine deficient mutant had an LD50 of 186 ± 88 and 12,109 ± 1,766 respectively. Thus, the polyamine deficient mutant had an ~500-fold loss of virulence in this model of bubonic plague but no significant affect on virulence in the pneumonic plague model.
Discussion
In Y. pestis, we have previously shown that polyamine deficiency, through mutation of the two genes encoding the key biosynthetic enzymes necessary for polyamine production, causes a loss of biofilm production. In contrast to E. coli, the effect of the SpeA− SpeC− mutations on growth was minimal showing that biofilm loss was not simply due to a lower bacterial population. Exogenous putrescine, but not spermidine, or over-expression of SpeA restored biofilm formation (Patel et al., 2006).
Polyamines also affect biofilm development in Vibrio cholerae. Exogenous spermidine inhibits while exogenous norspermidine enhances biofilm formation possibly by acting via an NspS/MbaA signaling pathway. In addition, norpsermidine biosynthesis is essential for biofilm development (Karatan et al., 2005; Karatan and Watnick, 2009; Lee et al., 2009; McGinnis et al., 2009).
Here we show that exogenous putrescine but not spermidine was transported into the bacterial cell. This suggests that polyamine uptake is necessary for the effect on biofilm formation. The energy-dependent uptake of putrescine by Y. pestis (Fig. 1) could be due to either (or both) the PotFGHI ABC transporter or the putrescine/ornithine antiporter PotE; both show high similarities to the E. coli systems which are specific for putrescine. The putative periplasmic binding protein, PotF (Y2851), has the highest similarity to its E. coli counterpart where seven residues (Trp-37, Tyr-40, Ser-85, Glu-185, Trp-244, Asp-278, and Tyr-314) are critical for putrescine binding (Vassylyev et al., 1998; Igarashi and Kashiwagi, 1999). Using the SIM protein alignment tool (ExPASy), a comparison of E. coli PotF and the Y. pestis PotF homologue (Y2851) revealed that each of these residues is conserved.
In E. coli, the seven critical residues necessary for putrescine binding by PotF are also conserved in PotD which transports spermidine and putrescine; of these, four (Trp-37, Trp-229, Asp-257, Tyr-293) are important for spermidine binding as well (Igarashi and Kashiwagi, 1999). However, a multiple sequence alignment of E. coli PotF, PotD, and the Y. pestis homologue of PotD, Y1391, found that none of the important conserved residues for putrescine/spermidine binding are apparent in Y1391. Thus it is unlikely that Y. pestis has a functional PotABCD transport system. Thus the lack of spermidine uptake may explain the inability of exogenous spermidine to restore biofilm development to the Y. pestis ΔspeA ΔspeC mutant. However, once putrescine is taken into these cells, it can be converted into spermidine. Thus it is still possible that intracellular spermidine may play a role in biofilm development.
A recent study found that y1392 and y1391 mRNA levels were increased in the flea relative to in vitro flow cell growth (Vadyvaloo et al., 2007). While the apparent regulation is intriguing, our bioinformatic analysis suggests that this PotABCD-like transport system may not be functional for putrescine or spermidine uptake. The level of putrescine in plasma, has been measured at ~0.23 μM (Shipe et al., 1979; Teti et al., 2002). Assuming this is equivalent to the concentration in the flea gut following a blood meal, this alone is unlikely to be enough to promote biofilm formation since our polyamine-deficient strain requires 1 mM exogenous putrescine for maximal biofilm production (Patel et al., 2006). Rather endogenously synthesized polyamines alone or in combination with that supplied by the putative transporters PotFGHI and/or PotE may provide sufficient putrescine for biofilm formation by the Y. pestis y1390-y1392 deletion mutant.
Here we have also identified the mechanism for polyamine control of biofilm development – polyamines are necessary for maintaining the levels of three key Hms proteins required for the production of biofilm: HmsR, HmsS, and HmsT. Transcriptional reporters and qRT-PCR demonstrated that polyamines are not affecting transcription of the hms genes or their mRNA stability. The effect on HmsS occurs through HmsR; HmsS is less stable in the absence of HmsR. Since HmsR levels are lower in the SpeA−SpeC− mutant, this causes lower HmsS levels. We have shown that over expression of HmsR in our polyamine-deficient mutant, even in the absence of exogenous putrescine, increases the HmsS protein level (Fig. 7). In contrast, translational lacZ fusions indicate that polyamines enhance the translation of HmsT and HmsR mRNAs.
Due to their polycationic nature polyamines have been implicated in a number of cellular mechanisms. Many of the functions associated with polyamines are thought to occur because their polycationic charge acts as a counterion to nucleic acids. In E. coli, putrescine and spermidine, respectively, are estimated to be 50% and 90% bound in polyamine-RNA complexes with an additional 9.3% and 5.1% bound to DNA (Miyamoto et al., 1993). Due to these interactions, polyamines can affect protein production on several levels. In E. coli, lacZ fusions reveal that antizyme (Az), the protein which inhibits polyamine biosynthesis, is regulated by polyamines in a concentration-dependent manner at the transcriptional level (Filippou et al., 2007). In Pseudomonas aeruginosa, exogenous putrescine can increase transcription of several loci including the spuABCDEFGH operon which is involved in polyamine uptake and utilization (Lu et al., 2002).
Because polyamines are often found in complexes with RNA they also have the ability to influence protein expression through translation. In E. coli several of the genes which comprise the polyamine modulon, including rpoN, fecI, oppA, hns, and fis, are characterized by the absence of an SD sequence, a weak SD sequence, or suboptimal spacing for an SD. The translational stimulation by polyamines of these genes ranged from 5- to 2-fold. Introduction of a consensus SD (for fecI, fis, and hns) or a spacing change to an optimal distance (rpoN) significantly reduces the stimulation of translation by polyamines (Yoshida et al., 2004; Higashi et al., 2006; Terui et al., 2007). The two hms genes most affected by the lack of polyamines, hmsR and hmsT, also appear to lack a consensus SD sequence. Introduction of a consensus SD sequence upstream of hmsT did cause an ~18% reduction in polyamine-stimulation of HmsT protein levels. However this reduction does not abolish polyamine stimulation (Fig. 8) which indicates that polyamines are affecting the levels of, at least HmsT, through additional mechanism(s). These results suggest that polyamines stimulate of translation of hmsT (and possibly hmsR) independent of a canonical SD.
Polyamines can bind to and stabilize DNA (Balasundaram and Tyagi, 1991; Lindemose et al., 2005; Pastre et al., 2006) as well as RNA. In the case of E. coli oppA, the addition of polyamines induces a structural change in the oppA RNA which aids in the formation of an initiation complex and increases the rate of translation (Igarashi et al., 1997; Yoshida et al., 1999). This mechanism may also contribute to enhanced levels of HmsT (and possibly HmsR) in the presence of polyamines.
The reduced level of translation of HmsR and HmsT in the absence of polyamines does not directly correspond to the protein levels observed by Western blot analysis (compare Fig. 3 and Fig. 8) raising the possibility of additional affects due to polyamine deficiency. Perhaps the stability of HmsR, HmsS and HmsT are also reduced due to alterations in the Hms complex or changes in protease levels or activity. We have recently shown that HmsP interacts with HmsT and HmsR while HmsR interacts with HmsS. In addition HmsS is unstable in the absence of HmsR (Wang et al., 2004; Itoh et al., 2005; Bobrov et al., 2008) which causes the reduced protein levels of HmsS in the SpeA− SpeC−. These protein interactions may also affect the stability of HmsR and HmsT as well – amplifying the translational affects of polyamines. Creating fusions protein with β-galactosidase may also affect the stability of the chimeric protein in the absence of polyamines and therefore may be partially masking the effect of polyamine deficiency.
Our polyamine-deficient mutant was significantly less virulent in a mouse model of bubonic plague but fully virulent in a mouse model of pneumonic plague. This suggests that either 1) polyamines are more readily obtained from the mammalian host during a lung infection than during systemic spread via the blood to internal organs or 2) some Y. pestis proteins dependent upon polyamines for full expression, perhaps in a translational control mechanism, are important for the virulence of bubonic but not pneumonic plague.
Since it has been previously demonstrated that hmsR or hmsH mutations that prevent biofilm development have no effect on the virulence of bubonic plague in mice, (Lillard et al., 1999; Abu Khweek et al., 2010), the attenuated virulence of our polyamine-deficient mutant is not due to lack of biofilm formation but to other effects of polyamines in Y. pestis. Further studies will be required to elucidate any additional putative polyamine functions in other important biological processes.
Experimental Procedures
Bacterial strains and cultivation
All bacterial strains used in this study are listed in Table 1. The Y. pestis KIM6-2112(pKD46)+ mutant was cured of pKD46 and the incorporated antibiotic cassettes as previously described (Forman et al., 2006) and designated KIM6-2112.1+. Y. pestis cells were streaked onto CR (Surgalla and Beesley, 1969) plates from fresh cultures made from buffered glycerol stocks stored at −80°C and incubated at 30°C for 48 h. For most experiments, individual colonies picked from agar plates were inoculated onto slants made from PMH2, a chemically defined medium lacking polyamines (Gong et al., 2001). Cells were washed off slants with PMH2 and grown overnight with aeration at the appropriate temperature. Where indicated, experiments that characterized yfeA gene products were grown in the same manner except that the PMH2 media was deferrated by Chelex 100 extraction (Staggs and Perry, 1991) to create iron-deficient conditions that increase the expression of the yfeABCD operon. The glassware used in the iron-deficient experiments was treated with Scotclean (Owl Scientific, Inc.) to remove contaminating iron. Where appropriate, ampicillin (Ap) (100 μg ml−1), kanamycin (Km) (40 μg ml−1), or chloramphenicol (Cm) (15 μg ml−1) was added to cultures. Cell growth was monitored at an optical density (OD) of 620 nm with a Spectronic Genesys5 spectrophotometer. E. coli cells were grown in Luria broth (LB) or M0 medium.
Polyamines uptake assay
Y. pestis KIM6+ cells were inoculated onto PMH2 slants, and E. coli K12 cells were inoculated onto M9 medium slants and both cultures incubated at 37°C overnight. Cells were washed from the corresponding slant with PMH2 or M9 media and a 5 ml culture was inoculated at an OD of 0.1 and grown to OD620 of ~0.6. The transport reaction was started with the addition of 50 μl of 14C-putrescine or a mixture of 14C-spermidine and unlabeled spermidine to a final concentration of 10 μM. The cells were incubated at 37°C with cells in 0.5 ml of culture collected at regular intervals by vacuum filtration through membrane filters (0.45 μm, MF-Millipore) pre-soaked in PMH2 or M9 containing 10 μm spermidine or putrescine. Filters were washed twice with a total of 8 ml of the appropriate medium and suspended in Bio-Safe II counting cocktail (Research Products International). Cell associated radioactivity in each sample was measured on a Beckman LS6500 liquid scintillation spectrometer. Unfiltered samples determined the total radioisotope content of cultures in each experiment. To demonstrate energy-dependent uptake control cultures were poisoned metabolically with 100 μM carbonyl cyanide m-chlorophenylhydrazone (CCCP) 10 min before the addition of isotope.
Crystal violet (CV) staining
Cells attached to glass test tubes were detected with CV staining essentially as described previously (O’Toole et al., 1999). Briefly, cells grown overnight on PMH2 slants were diluted into fresh PMH2 to an OD620 of 0.1 and grown for 16 to 18 h with shaking at room temperature. The cultures were incubated with 0.1% CV for 15 to 20 min before draining the liquid and washing the test tubes three times with water. CV retained by attached bacterial cells was solubilized with a mixture of 80% ethanol-20% acetone. The amount of dye bound, representing the mass of attached bacterial cells, was monitored by measuring the absorbance at 570 nm on a Spectronic Genesys 5 spectophotometer.
β-Galactosidase assays
Y. pestis cells carrying β-galactosidase constructs were grown overnight at 30°C, back- diluted to an OD620 of 0.1 and incubated at 30°C to an OD of ~0.5–0.6. The β-galactosidase activity in cell lysates were measured spectrophotometrically with a Genesys 5 spectrophotometer (Spectronic, Rochester, NY) following previously described enzymatic assays (Manoil, 1991; Miller, 1992).
Western blot analysis
Individual Y. pestis colonies were picked from CR agar plates and inoculated onto slants made from PMH2 and incubated for 48hrs at 30°C. From the slants, cultures were inoculated to an OD of 0.1 and grown overnight in PMH2 media in the presence or absence of 1mM putrescine. One ml of the overnight culture was centrifuged and resuspended in SDS loading buffer at a concentration of 0.01 OD/μl. Bacterial cells were homogenized for Western blot analysis using 0.1μM silica beads (RPI Corp). Equal amounts of protein, as confirmed by Comaisse blue staining, were separated on polyacrylamide gels containing SDS and immunoblotted to polyvinylidene fluoride membranes (Immobilon P; Millipore). The blots were processed using a modified procedure of Towbin et al. (Towbin et al., 1979). Briefly, the proteins were fixed with methanol and the membranes were blocked with 5% non-fat dry milk in 10mM Tris-HCL, pH 7.6, 137mM NaCl (TBS) with 0.1% Tween 20 (TBST). The blots were incubated with anti-serum diluted in TBS plus 1% gelatin and washed in TBST. Blots were incubated with horseradish peroxidase-conjugated protein A and processed with the ECL Western blot detection reagent (GE Healthcare). Immunoreative proteins were visualized on Kodak Biomax Light film. Immunoblots from three independent cultures were quantified using ImageJ (Abramoff et al., 2004).
Plasmids and recombinant DNA techniques
Plasmids were purified by alkaline lysis (Birnboim and Doly, 1979) from cultures grown overnight. Standard cloning and recombinant DNA techniques (Sambrook and Russell, 2001) were used to construct the plasmids listed in Table 1. Y. pestis cells were transformed by electroporation as previously described (Fetherston et al., 1995). Synthetic oligonucleotide primers were purchased from Integrated DNA Technologies (Table 1).
qRT-PCR
Y. pestis cells were grown at 30°C in PMH2 for a total of two transfers (~6–8 generations). Cells at an OD of 0.6 were combined with Bacterial RNA protect (Qiagen) and stored at −20°C according to the manufacture’s protocol. RNA was extracted from cell pellets using RNeasy Mini Kits (Qiagen) and treated with Turbo DNA free DNaseI (Ambion). PCR reactions, using the purified RNA as template, were run to confirm the absence of contaminating genomic DNA. RT-PCR primers for genes hmsH-F-S-T and gyrA (Table 1) were designed using mPrimer3 (Rozen and Skaletsky, 2000). Reactions were performed on a Roche Light cycler 2.0 using the one step Light cycler RNA Master SYBR Green I mix (Roche) and contained 100 ng of DNase-treated RNA and 0.2 μM primers in a total volume of 20 μl. For each target gene, PCR conditions were optimized and PCR efficiency was determined. Melt curve analysis was performed to ensure that a single product was amplified. Target gene expression was normalized to the house keeping gene DNA gyrase subunit A (gyrA), chosen because its expression was determined not to vary between the two strains under these growth conditions, and corrected according to the PCR efficiency value (Pfaffl, 2001). RNA from three independent cultures was run in duplicate and quantified using the comparative threshold-cycle method (Livak and Schmittgen, 2001).
Construction of transcriptional reporters
The hmsT::lacZ reporter strain was constructed by PCR amplification of a 340-bp fragment upstream of the predicted start codon of the hmsT gene using primers T-ProF and T-ProR (Table 1). The product was digested with KpnI and PacI and cloned into the corresponding sites upstream of the promoterless lacZ of pEU730. The hmsH::lacZ reporter was constructed by PCR amplification of a 314-bp fragment upstream of the predicted start codon of hmsH using primers H-ProF and H-ProR (Table 1). The product was digested with KpnI and PmeI and cloned into the corresponding sites upstream of the promoterless lacZ of pEU730. Constructs were confirmed by DNA sequencing (Davis Sequencing).
Construction of a SpeA− SpeC− HmsR translational reporter
A chromosomally integrated translational HmsR::β-gal fusion reporter strain, KIM6-2118.5, was kindly provided by A. Bobrov and O. Kirillina. In this strain β-galactosidase replaces the C-terminal sequences of hmsR starting at the predicted M162 residue and all of hmsS. Starting with the HmsR::β-gal integrated reporter strain, a polyamine deficient background was constructed. Using the SpeA− SpeC− mutant (KIM6-2112.1+) as the template, primers were designed (SpeA-pKNGF/SpeA-pKNGR; SpeC-pKNGF/SpeC-pKNGR; Table 1) to sequences ~500–600bp upstream and downstream of the FRT sites that remained from lambda red recombinase mutagenesis of SpeA and SpeC, respectively. The products were digested with BamHI and cloned into the BamHI and HincII sites of pNEB193. The pNEB193 plasmid containing the insert was then digested with BamHI and PmeI and ligated into the BamHI and SmaI sites of the suicide vector pKNG101. The suicide vector constructs containing either speA or speC deletion sequences were electroporated into the HmsR::β-gal integrated reporter strain and selected for sucrose resistance. Clones were checked for positive recombination and gene elimination by PCR. Once one mutation was achieved the process was repeated with the other vector plus insert of the other gene to achieve the double mutant.
Construction of an altered hmsT Shine-Dalgarno (SD) sequence
Construction of a consensus SD sequence upstream of hmsT involved the two-step PCR cloning scheme as described previously (Forman et al., 2006) using two outside primers and two overlapping interior primers carrying the desired mutation. Briefly, two smaller PCR products were generated using primers hmsT_SVF1/hmsT_SVR3 and hmsT_SVF4/hmsT_SVR2, overlapping at the region of interest by primers carrying the desired mutation(hmsT_SVR3 and hmsT_SVF4; Table 2). To produce a full-length, double-stranded template, the products of the first PCR reaction underwent 2 cycles of PCR fill in using the conditions 94°C for 2 min, 94°C for 30 s, 55°C for 30 s, and 72°C for 2 min. Next, outside primers (hmsT_SVF1, hmsTSVR2; Table 2) were added to the reaction and it was allowed to continue to run under the same conditions for an addition 35 cycles to complete the second PCR and thus generate a gene fragment carrying the desired mutation. The resulting PCR products were digested with BamHI and XbaI and ligated into the same sites of pNEB193. Clones with the desired SD sequence were confirmed by sequencing using PCR products generated by the hmsT_SVF1 and hmsT_SVR2 primers. One plasmid was selected and digested with EcoRV and XbaI. The insert DNA fragment was ligated into the SmaI and XbaI sites of the pKNG101 suicide vector. The vector plus insert was electroporated into competent KIM6-2112.1+ (SpeA− SpeC−) cells and screened for sucrose resistance.
Virulence testing of the polyamine-deficient mutant
Construction of potentially virulent strains and virulence testing was performed in a CDC-approved BSL3 laboratory following Select Agent regulations using procedures approved by the University of Kentucky Institutional Biosafety Committee. Y. pestis strains were transformed with the virulence plasmid, pCD1Ap, by electroporation (Gong et al., 2001; Forman et al., 2007) and plated on TBA plates containing Ap (50 μg ml−1). The plasmid profile of transformants was analyzed as well as their phenotype on CR agar (Surgalla and Beesley, 1969) and magnesium-oxalate plates (Higuchi and Smith, 1961). Supernatants from cultures grown at 37°C in the absence of CaCl2 were tested for the secretion of LcrV by Western blot analysis using polyclonal antisera against histidine-tagged LcrV (Fields and Straley, 1999; Forman et al., 2007). For subcutaneous infections, overnight cultures of Y. pestis cells grown in heart infusion broth (HIB) at 26°C, were diluted to an OD of 0.1 at 620 nm and incubated in HIB at 26°C until they reached mid-logarithmic phase (OD of ~0.5). Samples were harvested and diluted in mouse isotonic PBS (149 mM NaCl, 16 mM Na2HPO4, 4 mM NaH2PO4). Groups of four 6- to 8- week old female Swiss Webster (Hsd::ND4) mice were injected subcutaneously with 0.1 ml of 10-fold serially diluted bacterial suspensions ranging from 100 to 105 cfu ml−1. Cells used for intranasal infections were grown at 37°C in HIB containing 4 mM CaCl2 to prevent full induction of Lcr in vitro and were similarly diluted in mouse isotonic PBS. Twenty microliters of the bacterial suspension was administered to the nares of mice sedated with 100 μg of ketamine and 10 mg of xylazine/KG. The actual administered bacterial doses were determined by plating aliquots of serially diluted suspensions of each dose, in duplicate, onto TBA plates containing Ap (50 μg ml−1). The colonies were counted on plates incubated at 30°C for 2 days. Mice were observed daily for 2 weeks and LD50 values were calculated according to the method of Reed and Muench (Reed, 1938). All animal care and experimental procedures were conducted in accordance with the Animal Welfare Act, Guide for the Care and Use of Laboratory Animals, PHS Policy and the U.S. Government Principals for the Utilization of and Care for Vertebrate Animals in Teaching, Research, and Training and approved by the University of Kentucky Institutional Animal Care and Use Committee. The University of Kentucky Animal Care Program first achieved accreditation by the Association for the Assessment and Accreditation of Laboratory Animal Care, Inc. (AAALAC) in 1966 and has maintained full accreditation continuously since that time.
Acknowledgments
This project was supported by Public Health Services grant AI25098 from the U.S. National Institutes of Health. BWW was supported in part by the Department of Pharmaceutical Sciences, University of Kentucky. MAO is supported by EARDA grant 5G11HD052388-03. We thank Alex Bobrov and Olga Kirillina for providing Y. pestis strain KIM6-2118.5 and Jennifer Abney for her assistance with some experiments.
References
- Abramoff MD, Magelhaes PJ, Ram SJ. Image processing with ImageJ. Biophotonics Intl. 2004;11:36–42. [Google Scholar]
- Abu Khweek A, Fetherston JD, Perry RD. Analysis of HmsH and its role in plague biofilm formation. Microbiology. 2010 doi: 10.1099/mic.0.036640-0. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasundaram D, Tyagi AK. Polyamine--DNA nexus: structural ramifications and biological implications. Mol Cell Biochem. 1991;100:129–140. doi: 10.1007/BF00234162. [DOI] [PubMed] [Google Scholar]
- Bearden SW, Perry RD. The Yfe system of Yersinia pestis transports iron and manganese and is required for full virulence of plague. Mol Microbiol. 1999;32:403–414. doi: 10.1046/j.1365-2958.1999.01360.x. [DOI] [PubMed] [Google Scholar]
- Birnboim HC, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobrov AG, Kirillina O, Perry RD. The phosphodiesterase activity of the HmsP EAL domain is required for negative regulation of biofilm formation in Yersinia pestis. FEMS Microbiol Lett. 2005;247:123–130. doi: 10.1016/j.femsle.2005.04.036. [DOI] [PubMed] [Google Scholar]
- Bobrov AG, Kirillina O, Forman S, Mack D, Perry RD. Insights into Yersinia pestis biofilm development: topology and co-interaction of Hms inner membrane proteins involved in exopolysaccharide production. Environ Microbiol. 2008;108:1419–1432. doi: 10.1111/j.1462-2920.2007.01554.x. [DOI] [PubMed] [Google Scholar]
- Camilli A, Bassler BL. Bacterial small-molecule signaling pathways. Science. 2006;311:1113–1116. doi: 10.1126/science.1121357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang AC, Cohen SN. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotter PA, Stibitz S. c-di-GMP-mediated regulation of virulence and biofilm formation. Curr Opin Microbiol. 2007;10:17–23. doi: 10.1016/j.mib.2006.12.006. [DOI] [PubMed] [Google Scholar]
- Fetherston JD, Schuetze P, Perry RD. Loss of the pigmentation phenotype in Yersinia pestis is due to the spontaneous deletion of 102 kb of chromosomal DNA which is flanked by a repetitive element. Mol Microbiol. 1992;6:2693–2704. doi: 10.1111/j.1365-2958.1992.tb01446.x. [DOI] [PubMed] [Google Scholar]
- Fetherston JD, Lillard JW, Jr, Perry RD. Analysis of the pesticin receptor from Yersinia pestis: role in iron-deficient growth and possible regulation by its siderophore. J Bacteriol. 1995;177:1824–1833. doi: 10.1128/jb.177.7.1824-1833.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields KA, Straley SC. LcrV of Yersinia pestis enters infected eukaryotic cells by a virulence plasmid-independent mechanism. Infect Immun. 1999;67:4801–4813. doi: 10.1128/iai.67.9.4801-4813.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippou PS, Lioliou EE, Panagiotidis CA, Athanassopoulos CM, Garnelis T, Papaioannou D, Kyriakidis DA. Effect of polyamines and synthetic polyamine-analogues on the expression of antizyme (AtoC) and its regulatory genes. BMC Biochem. 2007;8:1. doi: 10.1186/1471-2091-8-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forman S, Nagiec MJ, Abney J, Perry RD, Fetherston JD. Analysis of the aerobactin and ferric hydroxamate uptake systems of Yersinia pestis. Microbiology. 2007;153:2332–2341. doi: 10.1099/mic.0.2006/004275-0. [DOI] [PubMed] [Google Scholar]
- Forman S, Bobrov AG, Kirillina O, Craig SK, Abney J, Fetherston JD, Perry RD. Identification of critical amino acid residues in the plague biofilm Hms proteins. Microbiology. 2006;152:3399–3410. doi: 10.1099/mic.0.29224-0. [DOI] [PubMed] [Google Scholar]
- Froehlich B, Husmann L, Caron J, Scott JR. Regulation of rns, a positive regulatory factor for pili of enterotoxigenic Escherichia coli. J Bacteriol. 1994;176:5385–5392. doi: 10.1128/jb.176.17.5385-5392.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong S, Bearden SW, Geoffroy VA, Fetherston JD, Perry RD. Characterization of the Yersinia pestis Yfu ABC inorganic iron transport system. Infect Immun. 2001;69:2829–2837. doi: 10.1128/IAI.67.5.2829-2837.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall-Stoodley L, Costerton JW, Stoodley P. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol. 2004;2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- Higashi K, Kashiwagi K, Taniguchi S, Terui Y, Yamamoto K, Ishihama A, Igarashi K. Enhancement of +1 frameshift by polyamines during translation of polypeptide release factor 2 in Escherichia coli. J Biol Chem. 2006;281:9527–9537. doi: 10.1074/jbc.M513752200. [DOI] [PubMed] [Google Scholar]
- Higuchi K, Smith JL. Studies on the nutrition and physiology of Pasteurella pestis. VI. A differential plating medium for the estimation of the mutation rate to avirulence. J Bacteriol. 1961;81:605–608. doi: 10.1128/jb.81.4.605-608.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch BJ, Perry RD, Schwan TG. Role of the Yersinia pestis hemin storage (hms) locus in the transmission of plague by fleas. Science. 1996;273:367–370. doi: 10.1126/science.273.5273.367. [DOI] [PubMed] [Google Scholar]
- Igarashi K, Kashiwagi K. Polyamine transport in bacteria and yeast. Biochem J. 1999;344:633–642. [PMC free article] [PubMed] [Google Scholar]
- Igarashi K, Kashiwagi K. Polyamine Modulon in Escherichia coli: genes involved in the stimulation of cell growth by polyamines. J Biochem. 2006;139:11–16. doi: 10.1093/jb/mvj020. [DOI] [PubMed] [Google Scholar]
- Igarashi K, Saisho T, Yuguchi M, Kashiwagi K. Molecular mechanism of polyamine stimulation of the synthesis of oligopeptide-binding protein. J Biol Chem. 1997;272:4058–4064. doi: 10.1074/jbc.272.7.4058. [DOI] [PubMed] [Google Scholar]
- Itoh Y, Wang X, Hinnebusch BJ, Preston JF, III, Romeo T. Depolymerization of s-1,6-N-acetyl-D-glucosamine disrupts the integrity of diverse bacterial biofilms. J Bacteriol. 2005;187:382–387. doi: 10.1128/JB.187.1.382-387.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson S, Burrows TW. The pigmentation of Pasteurella pestis on a defined medium containing haemin. Br J Exp Pathol. 1956;37:570–576. [PMC free article] [PubMed] [Google Scholar]
- Jarrett CO, Deak E, Isherwood KE, Oyston PC, Fischer ER, Whitney AR, et al. Transmission of Yersinia pestis from an infectious biofilm in the flea vector. J Infect Dis. 2004;190:783–792. doi: 10.1086/422695. [DOI] [PubMed] [Google Scholar]
- Jefferson KK. What drives bacteria to produce a biofilm? FEMS Microbiol Lett. 2004;236:163–173. doi: 10.1016/j.femsle.2004.06.005. [DOI] [PubMed] [Google Scholar]
- Jenal U, Malone J. Mechanisms of cyclic-di-GMP signaling in bacteria. Annu Rev Genet. 2006;40:385–407. doi: 10.1146/annurev.genet.40.110405.090423. [DOI] [PubMed] [Google Scholar]
- Jones HA, Lillard JW, Jr, Perry RD. HmsT, a protein essential for expression of the haemin storage (Hms+) phenotype of Yersinia pestis. Microbiology. 1999;145:2117–2128. doi: 10.1099/13500872-145-8-2117. [DOI] [PubMed] [Google Scholar]
- Kaniga K, Delor I, Cornelis GR. A wide-host-range suicide vector for improving reverse genetics in gram-negative bacteria: inactivation of the blaA gene of Yersinia enterocolitica. Gene. 1991;109:137–141. doi: 10.1016/0378-1119(91)90599-7. [DOI] [PubMed] [Google Scholar]
- Karatan E, Watnick P. Signals, regulatory networks, and materials that build and break bacterial biofilms. Microbiol Mol Biol Rev. 2009;73:310–347. doi: 10.1128/MMBR.00041-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karatan E, Duncan TR, Watnick PI. NspS, a predicted polyamine sensor, mediates activation of Vibrio cholerae biofilm formation by norspermidine. J Bacteriol. 2005;187:7434–7443. doi: 10.1128/JB.187.21.7434-7443.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirillina O, Fetherston JD, Bobrov AG, Abney J, Perry RD. HmsP, a putative phosphodiesterase, and HmsT, a putative diguanylate cyclase, control Hms-dependent biofilm formation in Yersinia pestis. Mol Microbiol. 2004;54:75–88. doi: 10.1111/j.1365-2958.2004.04253.x. [DOI] [PubMed] [Google Scholar]
- Lee J, Sperandio V, Frantz DE, Longgood J, Camilli A, Phillips MA, Michael AJ. An alternative polyamine biosynthetic pathway is widespread in bacteria and essential for biofilm formation in Vibrio cholerae. Journal of Biological Chemistry. 2009;284:9899–9907. doi: 10.1074/jbc.M900110200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillard JW, Jr, Bearden SW, Fetherston JD, Perry RD. The haemin storage (Hms+) phenotype of Yersinia pestis is not essential for the pathogenesis of bubonic plague in mammals. Microbiology. 1999;145:197–209. doi: 10.1099/13500872-145-1-197. [DOI] [PubMed] [Google Scholar]
- Lillard JW, Jr, Fetherston JD, Pedersen L, Pendrak ML, Perry RD. Sequence and genetic analysis of the hemin storage (hms) system of Yersinia pestis. Gene. 1997;193:13–21. doi: 10.1016/s0378-1119(97)00071-1. [DOI] [PubMed] [Google Scholar]
- Lindemose S, Nielsen PE, Mollegaard NE. Polyamines preferentially interact with bent adenine tracts in double-stranded DNA. Nucleic Acids Res. 2005;33:1790–1803. doi: 10.1093/nar/gki319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lu CD, Itoh Y, Nakada Y, Jiang Y. Functional analysis and regulation of the divergent spuABCDEFGH-spuI operons for polyamine uptake and utilization in Pseudomonas aeruginosa PAO1. J Bacteriol. 2002;184:3765–3773. doi: 10.1128/JB.184.14.3765-3773.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mah TF, O’Toole GA. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 2001;9:34–39. doi: 10.1016/s0966-842x(00)01913-2. [DOI] [PubMed] [Google Scholar]
- Manoil C. Analysis of membrane protein topology using alkaline phosphatase and beta-galactosidase gene fusions. Methods Cell Biol. 1991;34:61–75. doi: 10.1016/s0091-679x(08)61676-3. [DOI] [PubMed] [Google Scholar]
- McGinnis MW, Parker ZM, Walter NE, Rutkovsky AC, Cartaya-Marin C, Karatan E. Spermidine regulates Vibrio cholerae biofilm formation via transport and signaling pathways. FEMS Microbiology Letters. 2009;299:166–174. doi: 10.1111/j.1574-6968.2009.01744.x. [DOI] [PubMed] [Google Scholar]
- Miller JH. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- Miyamoto S, Kashiwagi K, Ito K, Watanabe S, Igarashi K. Estimation of polyamine distribution and polyamine stimulation of protein synthesis in Escherichia coli. Arch Biochem Biophys. 1993;300:63–68. doi: 10.1006/abbi.1993.1009. [DOI] [PubMed] [Google Scholar]
- O’Toole GA, Pratt LA, Watnick PI, Newman DK, Weaver VB, Kolter R. Genetic approaches to study of biofilms. Methods Enzymol. 1999;310:91–109. doi: 10.1016/s0076-6879(99)10008-9. [DOI] [PubMed] [Google Scholar]
- Pastre D, Pietrement O, Landousy F, Hamon L, Sorel I, David MO, et al. A new approach to DNA bending by polyamines and its implication in DNA condensation. Eur Biophys J. 2006;35:214–223. doi: 10.1007/s00249-005-0025-7. [DOI] [PubMed] [Google Scholar]
- Patel CN, Wortham BW, Lines JL, Fetherston JD, Perry RD, Oliveira MA. Polyamines are essential for the formation of plague biofilm. J Bacteriol. 2006;188:2355–2363. doi: 10.1128/JB.188.7.2355-2363.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry RD, Mier I, Jr, Fetherston JD. Roles of the Yfe and Feo transporters of Yersinia pestis in iron uptake and intracellular growth. Biometals. 2007;20:699–703. doi: 10.1007/s10534-006-9051-x. [DOI] [PubMed] [Google Scholar]
- Perry RD, Bobrov AG, Kirillina O, Jones HA, Pedersen L, Abney J, Fetherston JD. Temperature regulation of the hemin storage (Hms+) phenotype of Yersinia pestis is posttranscriptional. J Bacteriol. 2004;186:1638–1647. doi: 10.1128/JB.186.6.1638-1647.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed LJ, Muench H. A simple method of estimating fifty percent endpoints. The American Journal of Hygiene. 1938;27:493–497. [Google Scholar]
- Rozen S, Skaletsky H. Primer 3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular cloning, a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Shine J, Dalgarno L. Determinant of cistron specificity in bacterial ribosomes. Nature. 1975;254:34–38. doi: 10.1038/254034a0. [DOI] [PubMed] [Google Scholar]
- Shipe JR, Jr, Hunt DF, Savory J. Plasma polyamines determined by negative-ion chemical ionization/mass spectrometry. Clin Chem. 1979;25:1564–1571. [PubMed] [Google Scholar]
- Simm R, Fetherston JD, Kader A, Romling U, Perry RD. Phenotypic convergence mediated by GGDEF-domain-containing proteins. J Bacteriol. 2005;187:6816–6823. doi: 10.1128/JB.187.19.6816-6823.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staggs TM, Perry RD. Identification and cloning of a fur regulatory gene in Yersinia pestis. J Bacteriol. 1991;173:417–425. doi: 10.1128/jb.173.2.417-425.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Styer KL, Hopkins GW, Bartra SS, Plano GV, Frothingham R, Aballay A. Yersinia pestis kills Caenorhabditis elegans by a biofilm-independent process that involves novel virulence factors. EMBO Rep. 2005;6:992–997. doi: 10.1038/sj.embor.7400516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surgalla MJ, Beesley ED. Congo red-agar plating medium for detecting pigmentation in Pasteurella pestis. Appl Microbiol. 1969;18:834–837. doi: 10.1128/am.18.5.834-837.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terui Y, Higashi K, Tabei Y, Tomitori H, Yamamoto K, Ishihama A, et al. Enhancement of the synthesis of RpoE and StpA by polyamines at the level of translation in Escherichia coli under heat shock conditions. J Bacteriol. 2009;191:5348–5357. doi: 10.1128/JB.00387-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terui Y, Higashi K, Taniguchi S, Shigemasa A, Nishimura K, Yamamoto K, et al. Enhancement of the synthesis of RpoN, Cra, and H-NS by polyamines at the level of translation in Escherichia coli cultured with glucose and glutamate. J Bacteriol. 2007;189:2359–2368. doi: 10.1128/JB.01562-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teti D, Visalli M, McNair H. Analysis of polyamines as markers of (patho)physiological conditions. Journal of Chromatography B. 2002;781:107–149. doi: 10.1016/s1570-0232(02)00669-4. [DOI] [PubMed] [Google Scholar]
- Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadyvaloo V, Jarrett C, Sturdevant D, Sebbane F, Hinnebusch BJ. Analysis of Yersinia pestis gene expression in the flea vector. Adv Exp Med Biol. 2007;603:192–200. doi: 10.1007/978-0-387-72124-8_16. [DOI] [PubMed] [Google Scholar]
- Vassylyev DG, Tomitori H, Kashiwagi K, Morikawa K, Igarashi K. Crystal Structure and Mutational Analysis of the Escherichia coli putrescine receptor. Structural basis for substrate specificity. J Biol Chem. 1998;273:17604–17609. doi: 10.1074/jbc.273.28.17604. [DOI] [PubMed] [Google Scholar]
- Wang X, Preston JF, 3rd, Romeo T. The pgaABCD locus of Escherichia coli promotes the synthesis of a polysaccharide adhesin required for biofilm formation. J Bacteriol. 2004;186:2724–2734. doi: 10.1128/JB.186.9.2724-2734.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams P, Winzer K, Chan WC, Camara M. Look who’s talking: communication and quorum sensing in the bacterial world. Philos Trans R Soc Lond B Biol Sci. 2007;362:1119–1134. doi: 10.1098/rstb.2007.2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida M, Meksuriyen D, Kashiwagi K, Kawai G, Igarashi K. Polyamine stimulation of the synthesis of oligopeptide-binding protein (OppA). Involvement of a structural change of the Shine-Dalgarno sequence and the initiation codon aug in oppa mRNA. J Biol Chem. 1999;274:22723–22728. doi: 10.1074/jbc.274.32.22723. [DOI] [PubMed] [Google Scholar]
- Yoshida M, Kashiwagi K, Shigemasa A, Taniguchi S, Yamamoto K, Makinoshima H, et al. A unifying model for the role of polyamines in bacterial cell growth, the polyamine modulon. J Biol Chem. 2004;279:46008–46013. doi: 10.1074/jbc.M404393200. [DOI] [PubMed] [Google Scholar]


