Abstract
The emergence of multidrug-resistant Mycobacterium tuberculosis strains has made many of the currently available anti-tuberculosis (TB) drugs ineffective. Accordingly, there is a pressing need to identify new drug targets. Filamentous temperature-sensitive protein Z (FtsZ), a bacterial tubulin homologue, is an essential cell-division protein that polymerizes in a GTP-dependent manner, forming a highly dynamic cytokinetic ring, designated as the Z ring, at the septum site. Other cell-division proteins are recruited to the Z ring and, upon resolution of the septum, two daughter cells are produced. Since inactivation of FtsZ or alteration of FtsZ assembly results in the inhibition of Z-ring and septum formation, FtsZ is a very promising target for novel antimicrobial drug development. This review describes the function and dynamic behaviors of FtsZ and the recent development of FtsZ inhibitors as potential anti-TB agents.
Tuberculosis, caused by Mycobacterium tuberculosis (Mtb) is a leading cause of death worldwide [1]. Indeed, recent statistics from WHO estimate that there are approximately 9.2 million new tuberculosis (TB) cases every year with a global mortality rate of 23% [301]. The primary site of infection is the lungs, followed by dissemination via the circulatory system and lymphatic system to secondary sites including the CNS, bones, joints, liver and spleen. It has been estimated that 90% of those who are exposed to or infected with TB remain latent, with only 10% progressing to active disease. However, with the emergence of HIV/AIDS in the last couple of decades, TB has become the most common opportunistic infection for HIV/AIDS patients [301]. There are 700,000 HIV-positive people infected with TB, contributing to 200,000 deaths worldwide [301]. Furthermore, poor patient compliance and inadequate control programs have lead to the emergence of multidrug-resistant (MDR) strains of Mtb [2]. The WHO estimates that up to 490,000 cases of MDR-TB emerge every year, leading to more than 110,000 deaths worldwide. Treatment of MDR-TB requires longer treatment times and is much more expensive, as treatment takes up to 2 years and cost US$250,000 per patient in the USA. Bacterial resistance to three or more ‘second-line’ antibiotics is classified as extremely drug-resistant TB (XDR-TB). Recent findings by WHO from 2000 to 2004 suggested that 4% of MDR-TB cases meet the criteria for XDR-TB [301]. Therefore, there is an urgent need for the development of new anti-TB drugs with novel mechanism of action(s), which are active against drug-resistant as well as drug-sensitive Mtb stains.
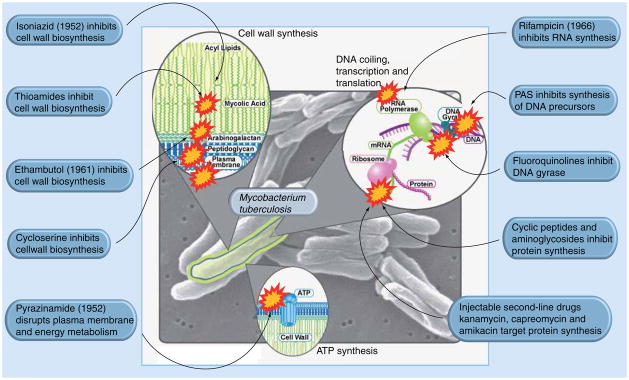
Despite extensive research in the last 40 years, the treatment of TB is limited to a cocktail of drugs, including isoniazid, ethambutol, pyrazinamide and rifampicin, which target cell-wall biosynthesis and RNA synthesis (Figure 1). Unlike eukaryotic cells, cytokinesis remains largely unexploited for the development of novel bacterial therapeutics purposes [3]. Filamentous temperature-sensitive protein Z (FtsZ), a tubulin homolog, is the most abundant bacterial cell-division protein. Owing to its vital importance in bacterial cell division, FtsZ has recently attracted considerable interest as an attractive drug target. In the presence of GTP, FtsZ polymerizes bidirectionally at the center of the cell on the inner membrane to form a highly dynamic helical structure known as the ‘Z-ring’ [4–8]. The recruitment of several other cell-division proteins leads to Z-ring contraction, resulting in septum formation and eventually cell division [9]. It was hypothesized that the inhibition of the proper FtsZ assembly would cause an absence of septum formation, leading to bacterial cell division arrest. The bacterial cell continues to elongate, resulting in a filamentation, which ultimately leads to cell death [10–11]. Owing to the central role that FtsZ plays in cell division, it is a very promising target for the development of new anti-TB drugs active against drug-resistant Mtb stains.
Figure 1. Current drugs and their targets.
Adapted and reorganized with permission from the National Institute of Allergy and Infectious Diseases’ website [302].
Filamentous temperature-sensitive protein Z
Biological role of the Z-ring
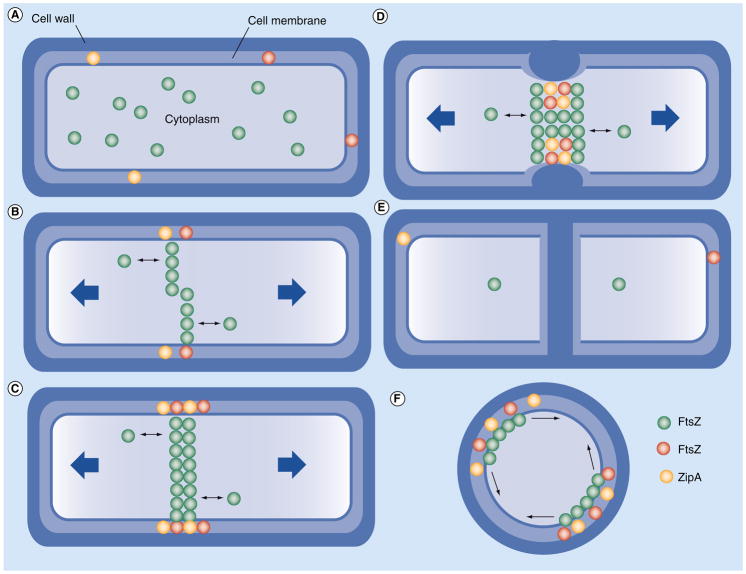
Prokaryotic cell division is a dynamic process that requires a concentration-dependent, temporal and spatial septation of the cell membrane and cell wall [12–14]. This process involves cytoplasmic protein, FtsZ, an essential GTPase that dynamically polymerizes to form a macro-molecular contractile ‘Z-ring’ [15]. This Z-ring structure is essential in initiating invagination of the cytoplasmic membrane and guiding the biosynthesis of septal peptidoglycan, which eventually leads to the formation of two daughter cells (Figure 2) [14].
Figure 2. Prokaryotic cell division.
(A) A normal cell, ready to divide with sufficient GTP-FtsZ, initiates polymerization.
(B) Polymerization of FtsZ proteins mid-cell and coordination of membrane-bound FtsA and ZipA, forming the MinC gradient.
(C) Steady-state turnover equilibrium reached with GTP-FtsZ in the cytoplasm and the Z-ring formed. (D) Contraction of the Z-ring and invagination of the cell membrane as well as cell wall. (E) Full septation of the cell and formation of two daughter cells, as well as the depolymerization of the Z-ring. (F) Radial view of a prokaryotic cell, showing bidirectional growth of FtsZ polymers along the cell membrane mid-cell.
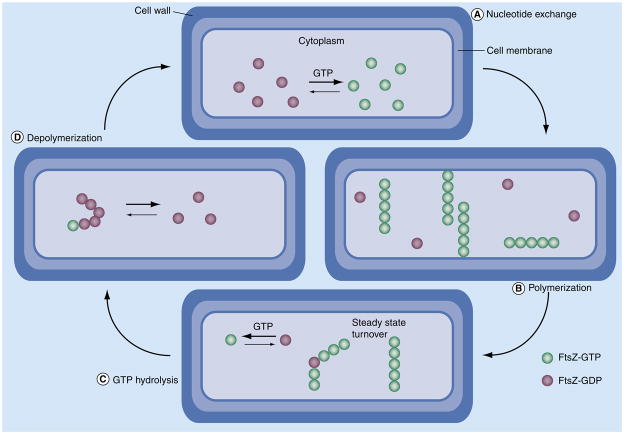
FtsZ is a homologue of tubulin but shares only 10% of its sequence identity at the protein level. Like tubulin, FtsZ undergoes dynamic polymerization, but major differences in this process make it distinct (Figure 3) [14]. In particular, FtsZ differs substantially in structural associations and dynamic nature. Polymerization experiments with isolated FtsZ protein have yielded no evidence of ‘treadmilling’ or ‘dynamic instability’, despite its critical role in microtubule dynamics. Dynamic instability encompasses the buildup of stored energy in the form of strained helical structures with several lateral interactions and GTP cap. Instead, what is observed is that individual subunits of hydrolyzed GDP-bound FtsZ can be exchanged independently with GTP-bound FtsZ in the cytoplasm. These dynamics, termed ‘steady-state turnover’, indicate an alternative model for Z-ring formation and contraction [16].
Figure 3. Dynamic polymerization of FtsZ.
(A) First expression levels of suitable quantities of FtsZ protein in the cytoplasm eliciting nucleotide exchange with rapid equilibration in favor of GTP bound FtsZ. (B) After a critical concentration of GTP-bound FtsZ is achieved, polymerization begins and long-strait protofilaments begin to form. (C) During polymerization, GTP hydrolysis is in continuous competition with protofilament growth in a process referred to ‘steady-state turnover’. (D) After bacterial cell division occurs, regulation of GTP yields distorted GDP-bound FtsZ-dominated protofilaments that revert to the monomer form.
Regulation of FtsZ assembly & septum formation
While many of the proteins involved in chromosomal segregation and cell division are conserved across taxa, others appear limited to Gram-negative or Gram-positive bacteria. Interestingly, many of the genes that encode proteins involved in regulation of septum formation and cell division are not annotated in Mtb [11]. This indicates that the division apparatus of Mtb consists of either fewer proteins than model systems such as Escherichia. coli and Bacillus subtilis or nonorthologous but functional cognates are used in Mtb.
Much of what is known about regulation of septum formation is derived from studies in model organisms, which provides evidence for the presence of a complex regulatory network consisting of a two-component signal transduction master regulator, accessory kinases, subordinate transcriptional regulators and FtsZ assembly modulating proteins [17]. Activation of master two-component regulators relies on a complex network consisting of multiple two-component systems resulting in phosphor-relays that altered phosphorylation states of the master response regulator (e.g., CtrA) producing expression changes in other regulatory elements and genes encoding cell cycle processes and components such as cell division [17]. Mtb encodes numerous two-component systems, but one such system MtrAB (rv3246c, rv3245c) is an essential two-component system that has been mapped as a group I regulator in Mtb studies, shown to target the origin of regulation, involved in cytokinetic control, cell-wall homeostasis, and modulation of proliferation [11,18–20]. Accordingly, this two-component system has been compared with the global regulator CtrA and is considered a candidate for coordination of DNA replication and cell division, and cell-cycle regulation [17].
The nontranscriptional mechanisms of septum regulation involve modulating FtsZ polymerization through protein–protein interaction. Three such, perhaps even redundant mechanisms have been described in other bacteria; the min-system, nucleoid occlusion and recently the spatial regulator MipZ [21–23]. In Mtb proteins known to interact with FtsZ and govern septum formation (MinC, MinD, MinE, EzrA, MipZ, SulA, SlmA and Noc) are not annotated [11]. The min-system directs division to the center of the cell. In the Gram-negative bacteria E. coli, the min-system consists of the proteins MinC, MinD and MinE, while in Gram-positive bacteria B. subtilis, the min-system consist of MinC, MinD and DivIVA. MinC and MinD form a complex that directly inhibits Z-ring formation whereas the role of MinE and DivIVA is for topological specificity [24]. The min-system is not essential for viability, but when absent aberrant division has been observed [24]. Bioinformatics modeling and transcriptional mapping have identified two open reading frames rv3660c and rv1708 that encode putative MinD orthologs [11]. The MinD-like proteins Rv1708 and Rv3660c have been annotated as a putative initiation inhibition protein and putative septum site-determining protein, respectively. However, other min-system components and the role of these putative mycobacterial cell-division components have not been experimentally demonstrated. While, nucleoid occlusion and the Z-ring regulator MipZ have not been identified in the mycobacterial genome, often these systems co-exist and are thought to cooperate in directing the location of Z-ring formation to mid-cell and coordinating cell division with the completion of DNA replication and segregation [23,25]. Therefore, it remains possible that the putative MinD-like proteins guide the location of septum formation along with another poorly defined mechanism such as nucleoid occlusion.
The cell-division proteins FtsA, ZapA and ZipA that promote Z-ring assembly have also not been annotated in Mtbs [11]. ZipA and FtsA accessory proteins provide both a scaffold and stimulus for bidirectional Z-ring growth. FtsA and ZipA are known to colocalize and interact with FtsZ [26–27]. These interactions provide substantial guiding forces during the formation of the Z-ring. The role of FtsA as the principal component of anchoring the Z-ring is supported by the identification of a conserved membrane targeting sequence in FtsA and the fact that FtsA is widely conserved among prokaryotes, except mycobacteria [11]. However, it has been demonstrated that both FtsA and ZipA are essential for cell division and involved in (although maybe not required for) recruitment of many of the remaining proteins [5]. Transcriptional mapping studies and bioinformatics have lead to the identification of two open reading frames, rv2345 and rv3835, which encode putative ZipA orthologs [11]. However, the role of these ZipA-like proteins in septum formation has not been experimentally substatiated. The necessary role and confusion about FtsA and ZipA arises because they can be bypassed, thus these proteins cannot be solely responsible for recruiting other cell division proteins through direct interaction [28–29]. Rather, these proteins may enhance the recruitment activity of the Z-ring by preventing disassembly [14].
Septum formation consists of independent assembly of proteins into subcomplexes that are then assembled to form the complete divisome [30–31]. Accordingly, in this model, three protein complexes are preassembled then recruited to mid-cell. The first proteins to interact are FtsZ, FtsA and ZipA to form a proto-ring on the cytoplasmic side of the plasma membrane. As discussed previously, Mtb does not encode the typical FtsA cell-division component as determined by bioinformatics and may encode two putative ZipA-like proteins. Thus, the assembly of the Z-ring on the cytoplasmic side of the plasma membrane in mycobacteria may employ proteins encoded by rv2345 and rv3835 or is in inherently stable once formed. In model organisms the proteins FtsQ, FtsL and FtsB assemble into what is known as the QLB complex, which is recruited to the septum [30]. Based on the fact that FtsQ, FtsL and FtsB contain cytoplasmic and membrane domains, this complex is thought to tether the Z-ring complex with proteins involved with peptidoglycan remodeling [30]. FtsQ and FtsL, protein components of the QLB complex are present in the mycobacterial genome, but bioinformatics has failed to identify the third protein of the complex, FtsB [11]. The next proteins recruited are the proteins of the PG complex that are involved in remodeling and synthesis of septal peptidoglycan [30]. The PG complex components are FtsW, FtsN and FtsI. FtsW is a putative transporter believed to play a role in shuttling precursors, and FtsI is a septation-specific transpeptidase [30–31]. FtsN is a late-assembly protein that needs all the other components to be present for recruitment, while both FtsK and FtsN have been shown to interact with FtsA [30–31]. Therefore, FtsK and FtsN are thought to promote association between the Z-ring complex and the QLB complex, as well as modulate septal constriction, respectively. Only FtsK and the PG complex members, FtsI and FtsW, are annotated in Mtb genome [11]. Overall, bioinformatics indicates that the Mtb genome does not encode the number of cell-division components as model Gram-positive or Gram-negative bacteria, and is specifically lacking prototypical components involved in chromosome segregation, site selection and Z-ring assembly and regulation.
FtsZ polymerization & interactions
In addition to controlling the spatial organization of the Z-ring by use of accessory proteins, bacteria can also control Z-ring formation by regulating intracellular FtsZ polymerization [15]. Bacteria accomplish this by closely regulating their intracellular GTP-FtsZ concentrations. In particular, this can be controlled by the rate of nucleotide turnover, which varies from 6.9 nmol mg−1 h−1 for Mtb to 30 μmol mg−1 h−1 for E. coli [32]. Despite having different rates of GTP hydrolysis, the GTPase activity of FtsZ has not been found essential in cell division [33]. It is not entirely conclusive how GTP functions in Z-ring dynamics, although GTP binding (specifically the γ-phosphate of GTP) has been found essential to promote protofilament stability [34]. The GTP-binding has effects on up to six ‘tubulin’ (T-1–T-6) loop regions, but has its most pronounced effects on the T-3 loop, containing the essential conservative ‘tubulin’ sequence GGGTGTG [6]. It should be noted that the GTP-binding region does not contain the specific amino acid residues required for nucleotide hydrolysis. Instead, the three residues responsible in hydrolysis are located in the T-7 loop on the opposing side of FtsZ. This has fortified the idea of ‘head-to-tail’ or longitudinal associations of individual FtsZ monomers. ‘Head-to-tail’ or longitudinal organization is also consistent with the crystal structure of Methanococcus jannaschii FtsZ (MjFtsZ [PDB:1W5A]; Figure 4) reported by Lowe et al. [35].
Figure 4. Dimerized Methanococcus jannacschii (protein data bank: 1W5A) resolution 2.40 Å in complex with GTP.
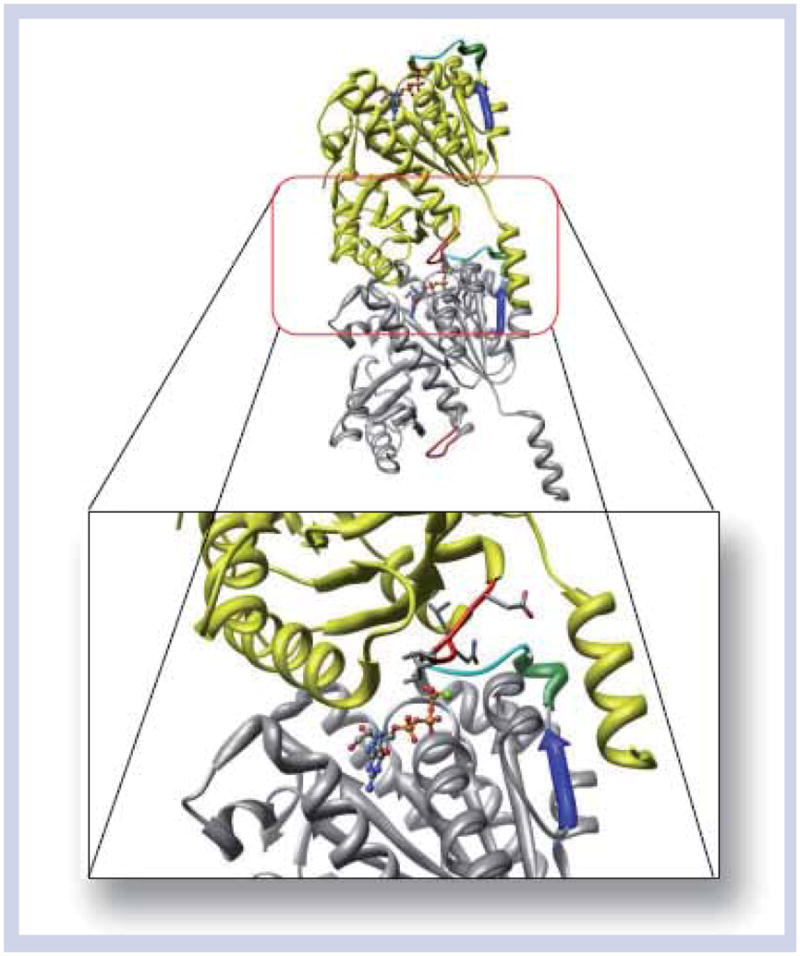
In yellow and grey are the individual monomer FtsZ proteins. In cyan is the T-3 loop. In green is the H-2 helix. In blue is the β-2 β-strand. In red is the T-7 catalytic loop. The light green ball is Mg2+, which is required for hydrolysis.
Adapted with permission from [35].
As reported by Diaz et al., hydrolysis of GTP results in the loss of hydrogen bonding responsible in stabilizing the T-3 loop [36]. As a consequence, this is believed to reduce protein–protein interactions longitudinally and destabilize polymerization. Alternatively, Leung et al. explain this with both longitudinal and lateral associations, as seen in their crystal structure of Mtb FtsZ (MtbFtsZ [PDB:1RLU]; Figure 5) [6]. Mutations in lateral amino acids have provided evidence that lateral interactions are critical to proper Z-ring formation and functionality [37].
Figure 5. Chain A (yellow) and chain B (grey) of the MtbFtsZ homodimer.
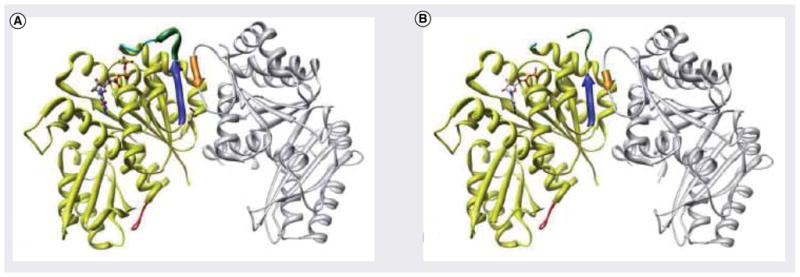
In cyan is the T-3 loop. In green is the H2 helix, if present. In blue is the β-2 β-sheet. In orange is the β-3 β– sheet in chain B. In red is the T-7 catalytic loop. (A) Crystal structure of homo-dimer MtbFtsZ bound to GTPγS (PDB:1RLU) at 2.08 Å resolution. Denoted is the presence of an H2 helix seen in green. (B) Crystal structure of homodimer MtbFtsZ bound to GDP (PDB:1RQ7) at 2.60 Å resolution. Denoted is the absence of an H2 helix seen in green.
Adapted with permission from [6].
The most critical element observed in the crystal structure of MtbFtsZ is the lateral nature of its homodimer. Although still believed to associate longitudinally, MtbFtsZ appears to involve substantial lateral interactions as well, on the basis of in-vitro experimentation and x-ray diffraction [6,32]. MtbFtsZ also can regulate these associations, utilizing two coupled conformational switches, switch I and switch II. Switch I is believed to be involved in the traditional G-protein α-to-β switch, while switch II corresponds to the well-documented T-3 loop ordering and disordering involved in the regulation of FtsZ polymerization. Despite having two switches, each is directly coupled to the phosphorylation state of the nucleotide. Switch I consists of the sH-2 helix stabilized by the γ-phosphate of GTP. The α-helix produced both extends and stabilizes β-3 in monomer A and increases the hydrogen bonding with β-2 in monomer B. It is believed that, upon hydrolysis of the nucleotide, sterics contribute to the disordering of sH-2 helix, which results in the reduction of lateral interactions.
Z-ring formation & contraction: a new target in antibiotics drug discovery
The exact mechanism of FtsZ polymerization and Z-ring contraction remains highly debated. Computational models have provided useful insights by using simple coarse-grain Monte-Carlo simulations based on physical observables [38–39]. In silico models have revealed a propensity of FtsZ polymers to condense and maximize bundling laterally. Although promising, these models deviate from the strained model, particularly in the source of energy required for contraction [40]. Despite these differences, each model requires FtsZ polymerization. Owing to the critical role of FtsZ in both models, FtsZ contraction is receiving attention as a logical and promising target for the development of new chemotherapeutics. Specifically, a high level of homology between FtsZ proteins of different species implies that FtsZ is an excellent target for broad-spectrum antimicrobials.
Mtb FtsZ inhibitors
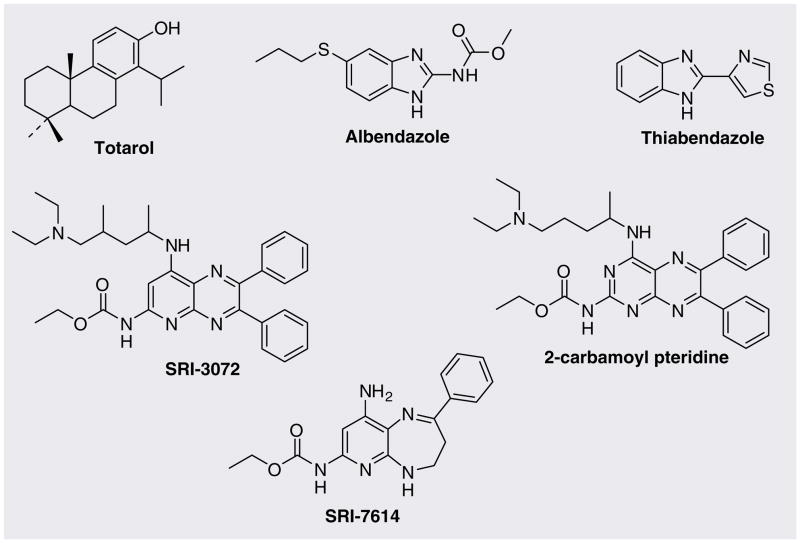
Inhibitors of Mtb FtsZ reported so far to date are summarized in Table 1 and their structures are shown in Figures 6 & 7. Characteristics of these inhibitors are discussed below.
Table 1.
Mycobacterium tuberculosis FtsZ inhibitors.
| Compound | Mode of action | Assays used | MIC | Ref. |
|---|---|---|---|---|
| Albendazole Thiabendazole | Filamentous phenotype response; up-regulation of gene encoding Z-ring formation | Cell-based assay; cell morphology; transcriptional profiling | Albendazole 61 μM, Thiabendazole 80 μM | [11,44] |
| Totarol | Dose-dependent inhibition of GTPase and FtsZ polymerization; reduced width and aggregation of FtsZ protofilaments | GTPase and FtsZ polymerization assay; TEM | [43] | |
| SRI-3072, SRI-7614, 2-carbamoyl- pteridine | Inhibition of FtsZ polymerization and GTPase activity | FtsZ GTPase and polymerization assay | SRI-3072 0.15 μg/ml SRI-7614 6.25 μg/ml 2-carbamoylpteridine 0.25 μg/ml |
[46–47] |
| Taxanes | FtsZ polymer stabilization | Real-time PCR-based assay, cell filamentation | 1.25–2.5 μM | [59] |
| Benzimidazoles | Inhibition of FtsZ polymerization; GTPase activity enhancement; budding and deformation of M. tuberculosis cells | FtsZ polymerization and GTPase assay; SEM; TEM; cell-based assay | 0.5–6 μg/ml | [Unpublished Data] |
MIC: Minimum inhibitory concentration; SEM: Scanning electron microscopy; TEM: Transmission electron microscopy.
Figure 6.
Mtb FtsZ inhibitors.
Figure 7.
Further Mtb FtsZ inhibitors: taxanes.
Totarol
Totarol is a naturally occurring diterpenoid phenol extracted from Podocarpus totara. Since it has been shown to inhibit the proliferation of several pathogenic Gram-positive bacteria such as Staphylococcus aureus and Mtb [41–42], Jaiswal et al. examined the effect of totarol on the assembly dynamics of Mtb FtsZ in vitro [43]. In the preliminary study they found that totarol increased the length of B. subtilis cells by several fold, indicating its inhibitory activity against cell division.
Totarol inhibited the polymerized mass of Mtb FtsZ by 27% at 50 μM concentration. GTPase activity was also reduced in a concentration dependent manner without affecting the binding of GTP to Mtb FtsZ. Transmission electron microscopy (TEM) images indicated that totarol significantly reduced the average width of FtsZ protofilaments and also induced aggregation in the absence of GTP. Totarol increased the fluorescence intensity of the FtsZ–ANS complex (ANS=1-anilinonaphthalene-8-sulfonic acid), whereas decreased that of PM-FtsZ (PM = N-[1-pyrene[maliemide), indicating that totarol induced conformational changes in FtsZ.
Albendazole & thiabendazole
Albendazole and thiabendazole (TBZ) are known fungicides and parasiticides, which cause degenerative alterations in the intestinal cells of the worm by binding to the colchicine-sensitive site of tubulin and inhibiting its polymerization, or assembly into microtubules. Sarcina et al. reported that the treatment of cyanobacterial and bacterial cultures with thiabendazole resulted in cell elongation [44]. This suggested that the cell-division cycle was arrested and therefore tubulin inhibitors may act in a similar manner on the FtsZ gene product. In addition, the increased amount of DNA in DAPI-DNA-stained TBZ-treated Synechococcus 7942 cells was observed, which indicated that DNA replication still occurred in the presence of thiabendazole. Slayden et al. examined the effect of albendazole and thiabendazole on Mtb cell growth [11]. MIC99 values of albendazole and thiabendazole were determined to be 61 and 80 μM, respectively. The cell morphology and transcriptional response indicated cell filamentation and gene regulation causing the inhibition of septum formation in Mtb cells. These results suggested that thiabendazole and albendazole interfered and delayed Mtb cytokinesis.
2-alkoxycarbonylaminopyridines & 2-carbamoylpteridine
Based on the premise that known tubulin inhibitors can inhibit FtsZ assembly [45], White, Reynolds and their co-workers screened a library of 200 2-alkoxycarbonylaminopyridines for antimicrobial activity against Mtb [46]. Two compounds, SRI-3072 and SRI-7614, were found to inhibit the growth of Mtb cells at 0.15 and 6.25 μg/ml, respectively. SRI-3072 and SRI-7614 inhibited Mtb FtsZ polymerization in a dose-dependent manner with ID50 values of 52 and 60 μM, respectively. These compounds also inhibited the GTPase activity by 20–25% at 100 μM concentration. In addition, these compounds were evaluated for their inhibitory activity against tubulin polymerization. SRI-3072 was specific to FtsZ and did not exhibit tubulin polymerization inhibition at 100 μM concentration, whereas SRI-7614 inhibited tubulin polymerization (ID50 4 μM). Furthermore, SRI-3072 reduced the growth of Mtb in mouse-derived macrophages. However, SRI-3072 had some issues in the synthesis and purification. Thus, optimization of these lead compounds was carried out to yield a more easily accessible and more potent (eightfold better MIC than SRI-3072) 2-carbamoylpteridine analogue of SRI-3072 [47].
Taxanes
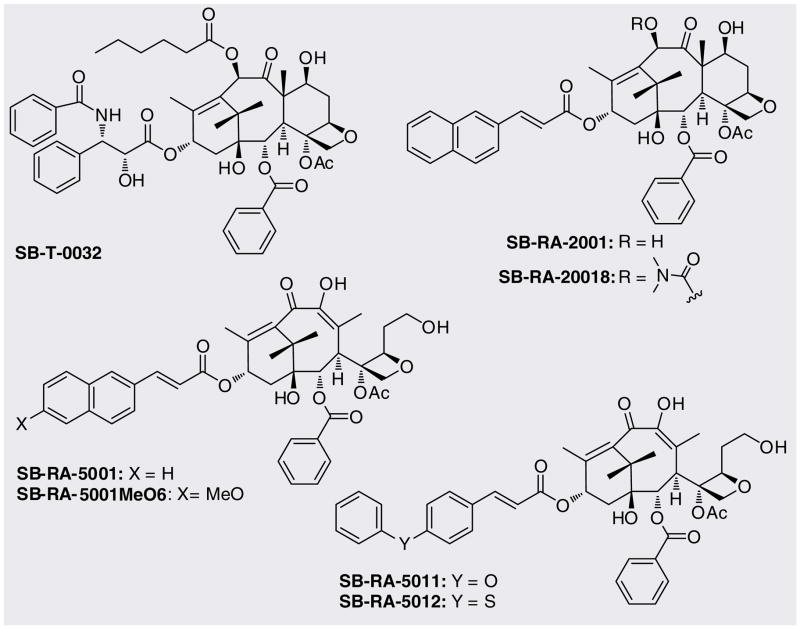
Paclitaxel (Taxol®), a microtubule-stabilizing anticancer agent, exhibits modest antibacterial activity against drug-sensitive and drug-resistant Mtb strains (MIC 40 μM), although its cytotoxicity against human cancer cell lines (a benchmark for activity against human host cells) is three orders of magnitude more potent (IC50 0.019–0.028 μM). Following the hypothesis that compounds that affect tubulin/microtubule would serve as leads for developing FtsZ inhibitors [44–46], a library of taxanes was designed and screened for antibacterial activity against Mtb cells. A real-time PCR-based assay was employed to screen taxanes, such as cytotoxic taxoids that stabilize microtubules [48–50] and noncytotoxic taxane-MDR reversal agents [51–58] that inhibit the efflux pumps of ATP-binding cassette transporters such as P-glycoprotein. Of the 120 taxanes screened, several compounds exhibited significant anti-TB activity [59]. From the MIC99 values and cytotoxicity assay, SB-T-0032 and SB-RA-2001 (Figure 7 & Table 2) were selected as lead compounds for further studies. SB-RA series of taxanes bearing a (E)-3-(naphtha-2-yl) acryloyl (2-NpCH=CHCO) group at the C-13 position exhibited MIC99 between 2.5 and 5 μM against drug-sensitive and drug-resistant Mtb strains. A new library of taxanes based on this SB-RA series was prepared through modification of 10-deacetylbaccatin III (DAB). These SB-RA-2001 analogs were found to have higher specificity to FtsZ than microtubules and exhibit the same level of anti-TB activity as that of SB-T-0032.
Table 2.
Antimicrobial activities of taxanes against drug-sensitive and multidrug-resistant Mycobacterium tuberculosis strains†.
| Taxanes | MIC M. tuberculosis (μM) | Cytotoxicity (IC50, μM) | ||
|---|---|---|---|---|
| H37Rv | IMCJ946.K2 | MCF79 | A549 | |
| Paclitaxel | 40 | 40 | 0.019 | 0.028 |
| SB-T-0032 | 5 | 1.25 | 0.65 | 0.65 |
| SB-RA-2001 | 5 | 2.5 | 4.5 | 15.7 |
| SB-RA-5001 | 2.5 | 1.25 | >80 | >80 |
| SB-RA-5001MeO6 | 2.5 | 2.5 | >80 | >80 |
| SB-RA-5011 | 2.5 | 1.25 | >80 | >80 |
| SB-RA-5012 | 2.5 | 1.25 | >80 | >80 |
Mycobacterium tuberculosis H37Rv is sensitive to all antibiotics tested. M. tuberculosis IMCJ946.K2 is resistant to nine drugs including INH, REF, EB, streptomycin, kanamycin, ethionamid, p-aminosalicilic acid, cycloserine and enviomycin. MCF7 and A549 cells: human breast and non-small-cell lung cancer cell lines, respectively.
MIC: Minimum inhibitory concentration.
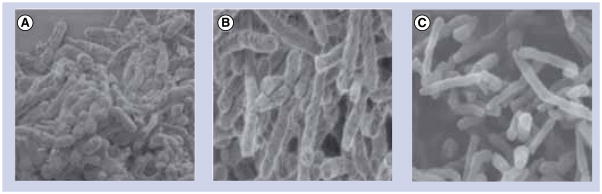
It has been shown that novel and effective anti-angiogenic taxoid, IDN5390 [60–61], bearing a C-seco-baccatin moiety possesses substantially less cytotoxicity than paclitaxel. Therefore, C-seco analogs of SB-RA-2001 (Figure 7) were designed, synthesized and evaluated. These novel C-seco-taxanes SB-RA-5001 and its congeners (Figure 7 & Table 2) were found to exhibit promising anti-TB activity (MIC99 1.25–2.5 μM) against both drug-sensitive and drug-resistant Mtb strains without appreciable cytotoxicity (IC50 > 80 μM). Thus, the specificity of these novel taxanes to microtubules as compared with FtsZ was completely reversed through systematic rational drug design. The scanning electron microscopy images of Mtb cells treated with SB-RA-20018 and SB-RA-5001 clearly show the substantial elongation and filamentation, a phenotypic response to FtsZ inactivation (Figure 8). In addition, a preliminary study on the effect of SB-RA-5001 on the polymerization–depolymerization, using the standard light-scattering assay exhibited a dose-dependent stabilization of FtsZ against depolymerization.
Figure 8. Electron micrograph of Mycobacterium tuberculosis cells.
(A) Control, (B) SB-RA-20018 and (C) SB-RA-5001.
Reprinted with permission from [59].
Benzimidazoles
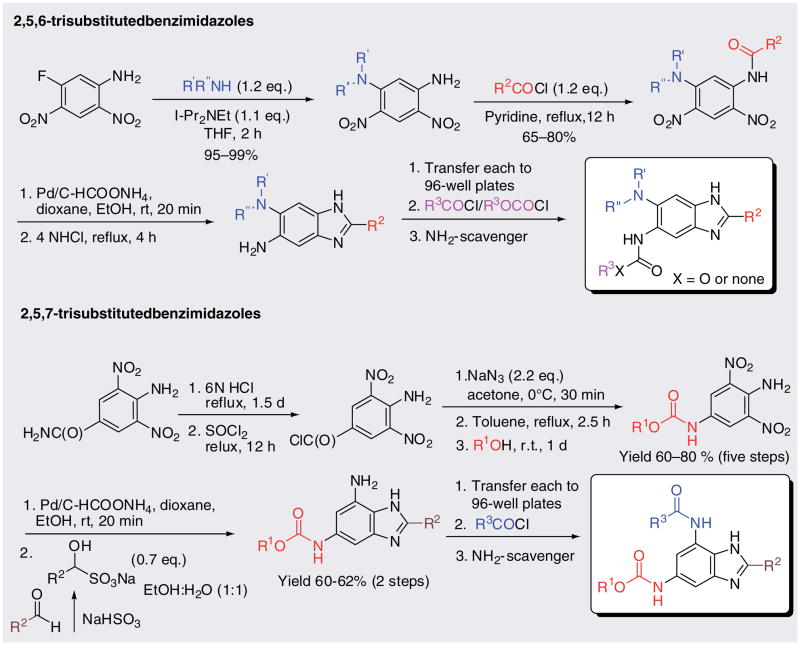
Based on the similarity of the benzimidazole moiety to the pyridopyrazine and pteridine pharamacophores in the FtsZ inhibitors identified by White, Reynolds and others [46–47], we hypothesized that the benzimidazole framework might be a promising starting point for the development of novel FtsZ inhibitors. This hypothesis was derived from the known ability of albendazole and thiabendazole to inhibit septum formation [11]. The benzimidazole skeleton is a privileged nitrogen heterocycle that represents an important pharmacophore that is present in a number of currently used drugs in clinic, for example, proton-pump inhibitors, antihistaminic drugs, antihypertensive drugs, anthelimintic drugs and antipsychotic drug. As such, the pharmacological, metabolic and toxicological profiles of the benzimidazole pharmacophore have been extensively studied. Several solid- or solution-phase synthetic methods for mono- or di-substituted benzimidazoles have been reported, but few deal with the synthesis of tri-substituted benzimidazoles. Owing to the difficulties in the synthesis of tri-substituted benzimidazoles, only 25 compounds are available from specialized commercial sources based on our thorough SciFinder search. Consequently, we selected trisubstituted benzimidazoles, especially 2,5,6- and 2,5,7- trisubstituted benzimidazoles, as the basic scaffold for the creation of novel benzimidazole libraries.
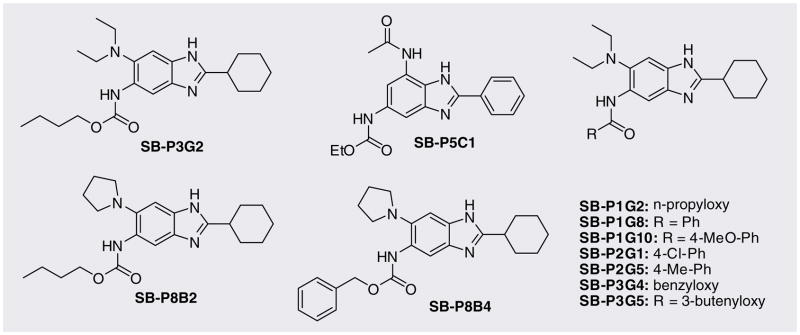
A library of 360 novel 2,5,6- and 2,5,7-tri-substituted benzimidazoles was synthesized as illustrated in Figure 9, and screened against Mtb H37Rv [201, 62,63]. From the assay, 27 compounds were identified that inhibited the Mtb cell growth with MIC99 values at most 5 μg/ml in triplicates. Furthermore, among the 27 hits, nine compounds showed MIC99 values at 0.56–6.1 μg/ml based on the more accurate Alamar blue assay (Table 3) (see Figure 10 for chemical structures) [64]. SB-P3G2 and SB-P5C1 exhibited the lowest MIC99 values of 0.78 μg/ml (2.0 μM) and 0.5 μg/ml (1.6 μM), respectively.
Figure 9.
Synthetic routes to the novel 2,5,6- and 2,5,7-trisubstituted benzimidazole libraries.
Table 3.
Antimicrobial activities of benzimidazole hits against Mycobacterium tuberculosis H37Rva.
| Benzimidazoles | MIC99 (μM) against H37Rv | Cytotoxicity (IC50, μM) against vero cells |
|---|---|---|
| SB-P1G8 | 7.9 | >400 |
| SB-P1G2 | 4.3 | >400 |
| SB-P1G10 | 7.4 | >400 |
| SB-P3G5 | 4.2 | >400 |
| SB-P2G1 | 14.1 | >400 |
| SB-P2G5 | 15.1 | >400 |
| SB-P3G2 | 2 | >400 |
| SB-P3G4 | 3.8 | >400 |
| SB-P8B2 | 1.3 | >400 |
| SB-P8B4 | 1.2 | >400 |
| SB-P5C1 | 1.6 | >100 |
MIC: Minimum inhibitory concentration.
Figure 10.
Further Mtb FtsZ inhibitors: benzimidazoles.
Preliminary structure–activity relationship studies indicated that cyclohexyl and diethyl amino groups at the 2- and 6-position, respectively, were critically important for antibacterial activity. Alkyl carbamates and benzamides showed better activity over alkyl or cycloalkyl-amides at the 5-position. The number of hits was more prevalent in the 2,5,6-tri-substituted series than the 2,5,7-tri-substituted series. Based on these findings, we designed and synthesized an additional 238 compound library of 2,5,6-tri-substituted benzimidazoles and screened them against Mtb H37Rv drug-sensitive strain [64]. Based on the assay, 54 compounds were identified, which inhibited the cell growth with MIC99 values at most 5 μg/ml in triplicates. Among the 54 hits, two compounds, SB-P8B2 and SB-P8B4, showed MIC99 values at most 0.5 μg/ml based on the Alamar blue assay [64].
SB-P3G2, SB-P5C1, SB-P8B2 and SB-P8B4 were advanced to in vivo efficacy evaluation in the rapid TB model in mice. Although the formulation of these lead compounds needs to be worked out, SB-P3G2 exhibited very promising activity via intraperitoneal injection [Unpublished Data]. Further optimization of these leads is actively in progress.
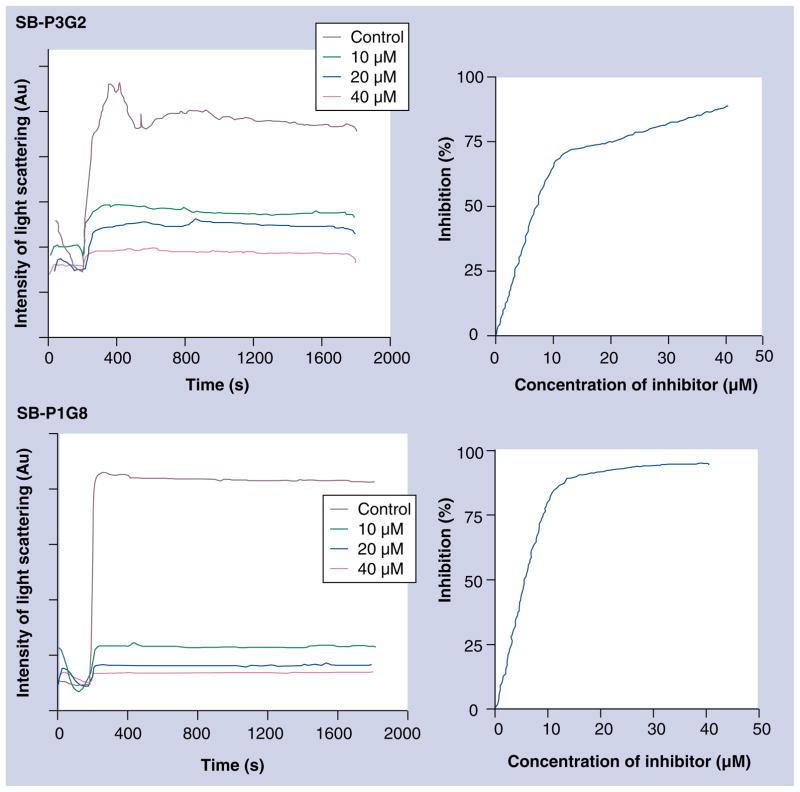
In order to confirm that these compounds inhibit bacterial growth by interfering with the FtsZ assembly, a light scattering experiment was carried out to investigate the effect of the lead compounds on FtsZ (de)polymerization [64]. Two representative results are shown in Figure 11 (FtsZ concentration = 15 μM) as examples. As anticipated, SB-P1G8 (MIC99 3.1 μg/ml, 7.9 μM) inhibited FtsZ polymerization in a dose-dependent manner and approximately 80% inhibition was observed at a concentration of 10 μM and the complete inhibition was achieved at 40 μM (IC50 6.21 μM). A similar result was obtained for SB-P3G2 (IC50 7.69 μM), as well as for other lead compounds [64].
Figure 11.
Inhibition of FtsZ polymerization by SB-P3G2 and SB-P1G8.
As the duration of steady state of FtsZ polymers is dependent on the rate of GTP hydrolysis, the effect of these lead compounds on GTPase activity was investigated. Unexpectedly, these compounds enhanced the GTPase activity by three- to fourfold. The result indicated that these novel benzimidazole leads decreased the stability of FtsZ protofilaments by increasing the rate of GTP hydrolysis, hence inhibiting FtsZ assembly [Unpublished Data], curcumin has also been reported to enhance the GTPase activity to disrupt the FtsZ assembly in B. subtilis and E. coli: vide infra [65]. Scanning electron micrograph images of Mtb cells treated with SB-P3G2 showed an absence of septum formation and slight cell elongation, indicative of FtsZ assembly inhibition [Unpublished Data]. The drastic reduction in the mass of FtsZ polymers and bundling of FtsZ protofilaments was observed by TEM analysis of Mtb FtsZ treated with SB-P3G2 [Unpublished Data]. These results will be published elsewhere in due course.
FtsZ inhibitors for other bacterial strains
Representative FtsZ inhibitors for bacterial strains other than Mtb are summarized in Tables 4 & 5. Characteristics of these inhibitors are discussed below. These inhibitors have relevance to the discovery and development of Mtb FtsZ inhibitors since FtsZ structure is well conserved in all bacterial species.
Table 4.
FtsZ inhibitors for bacteria other than Mycobacterium tuberculosis (1).
| Compound | Target organism | Mode of action | MIC/IC50 (cell growth inhibition) | Ref. |
|---|---|---|---|---|
| Viriditoxin | Escherichia coli, Staphylococcus aureus, MRSA, VRE, Enterococcus faecilus | Inhibits GTPase and FtsZ polymerization | 4–8 μg/ml (S. aureus and MRSA) 2–16 μg/ml (enterococcus faecalis and VRE) |
[68] |
| Zantrins | Mycobacterium tuberculosis, E. coli, MRSA, VRE | Inhibits GTPase, Z1, Z2, Z4 destabilizes FtsZ polymer, Z5 stabilizes FtsZ assembly | [3] | |
| GTP analogs: | [69,70,71] | |||
| ▪ BrGTP | E. coli | Inhibits GTPase and FtsZ polymerization assay in reversible competitive mode | ||
| ▪ GAL analogs | E. coli, S. aureus | Inhibits GTPase activity (E. coli FtsZ), Competitive inhibition of FtsZ polymerization and GTPase assay | 0.4–20 mg/ml (S. aureus) | |
| Sanguinarine | Bacillus subtilis, E. coli | Inhibits FtsZ assembly and protofilament bundling, thinner and shorter FtsZ protofilaments (E. coli) | [73] | |
| N-benzyl-3- sulfonamidopyrrolidine | E. coli | Lethal cell filamentation, moderate inhibition of GTPase activity, no effect on FtsZ polymerization | Compound 14–10 μM Compound 15–20 μM |
[74] |
| (±)-dichamanetin | S. aureus, B. subtilis, Mycobacterium smegmatis, E. coli | Inhibits GTPase activity of E. coli FtsZ | 1.7–3.4 μM (S. aureus, B. subtilis, M. smegmatis and E. coli) | [78] |
| (±)-2′-hydroxy-5′-benzylisouvarinol-B | S. aureus, B. subtilis, M. smegmatis, E. coli | Inhibits GTPase activity of E. coli FtsZ | 2.3–10.7 μM (S. aureus, B. subtilis, M. smegmatis and E. coli) | [78] |
MIC: Minimum inhibitory concentration; VRE: Vancomycin-resistant enterococcus.
Table 5.
FtsZ inhibitors for bacteria other than Mycobacterium tuberculosis (2).
| Compound | Target species | Mode of action | MIC/IC50 (cell growth inhibition) | Refs. |
|---|---|---|---|---|
| Cinnamaldehyde | Escherichia coli, Bacillus subtilis, MRSA | Inhibits GTPase activity, FtsZ polymerization and FtsZ protofilament bundling (E. coli); reduction in Z-ring/unit cell; conformational change blocking optimal contact with the GTP binding T1–T6 loop in a neighboring monomer | 1 μg/ml (E. coli) 0.5 μg/ml (B. subtilis) 0.25 μg/ml (MRSA) |
[79] |
| OTBA | E. coli, B. subtilis | Inhibition of GTPase activity and enhanced FtsZ polymerization (E. coli and B. subtilis); promoting assembly and stabilization of FtsZ polymer | 2 μM (B. subtilis) | [80] |
| Curcumin | E. coli, B. subtilis | Filamentation of B. subtilis 168 cells, inhibition of Z-ring formation without affecting organization of nucleoids; inhibition of FtsZ polymerization and enhancement of GTPase assay (E. coli) | 17 μM against (B. subtilis) 100 μM (E. coli) |
[65] |
| Berberine | E. coli, B. subtilis, Staphylococcus aureus B. subtilis, Staphylococcus aureus | Inhibits GTPase and FtsZ polymerization (E. coli); destabilization of FtsZ assembly-thin, short and curved FtsZ protofilaments; binding site in the vicinity of GTP binding site | [84] | |
| PC190723 | B. subtilis, S. aureus | Inhibits GTPase activity of S. aureus FtsZ by possible interaction with H7 (docking study); discrete foci throughout the elongated B. subtilis 168 cell, indicating mislocalization of Z-ring Competitive inhibition of GTPase and FtsZ polymerization (B. subtilis) | 0.5–1 μg/ml against B. subtilis, S. aureus, MRSA, MDRSA | [93,94] |
| PC58538 and PC170942 | B. subtilis, S. aureus, MRSA | Long aseptate filaments; inhibition of GTPase activity in a dose-dependent manner |
B. subtilis 168 PC58538 128 μg/ml PC170942 16 μg/ml S. aureus PC170942 64 μg/ml |
[91] |
| DAPI | E. coli | Enhancing FtsZ protofilament stability while inhibiting GTPase activity; bundling of FtsZ protofilaments | [90] | |
| Stathmin- derived I19L peptide | E. coli | Inhibition of FtsZ polymerization, bundling of FtsZ protofilaments; proposed binding site near GTP-binding pocket | [86] |
DAPI: 4′,6-diamidino-2-phenylindole; MIC: Minimum inhibitory concentration; MRSA: Methicillin-resistant Staphylococcus aureus; OBTA: 3-{5-[4-oxo-2-thioxo-3-(3-trifluoromethylphenyl)-thiazolidin-5-ylidenemethyl]-fu ran-2-yl}-benzoic acid.
Zantrins
A high-throughput (HTP) protein-based screening of 18,320 molecules lead to the identification of five structurally diverse compounds named ‘zantrins’, which inhibited E. coli FtsZ GTPase activity by 50% at 4–25 μM concentration [3]. Zantrins were also examined for their inhibitory activity againt a FtsZ ortholog from Mtb. The majority of zantrins inhibited the GTPase activity of Mtb FtsZ with IC50 values in the range of 50–70 μM. It was proposed that these zantrins may bind to a pocket between two FtsZ subunits to block the loop T7 of one monomer to come in optimal contact with GTP bound to the T-1–T-6 loops of the neighboring monomer, which is essential for polymerization [66–67]. In contrast, some zantrins appeared to stabilize the FtsZ assembly by contracting the movement of T3 loop that causes a bend in the filament upon GTP hydrolysis.
Viriditoxin
Viriditoxin, a potent E. coli FtsZ inhibitor, was identified from a pool of more than 100,000 extracts of microbial fermentation broths and plants using FtsZT65C-fluorescein polymerization assay [68]. Viriditoxin inhibited E. coli FtsZ polymerization (IC50 8.2 μg/ml) and GTPase activity (IC50 7 μg/ml). Furthermore, viriditoxin exhibited broad-spectrum antibacterial activity against clinically relevant Gram-positive as well as Gram-negative bacteria.
GTP analogs
Lappchen et al. designed a novel inhibitor of E. coli FtsZ, 8-bromogunosine-5′-triphosphate (BrGTP), based on the natural substrate GTP [69]. GTPase assay and polymerization assay indicated that BrGTP is a reversible competitive inhibitor of E. coli FtsZ. Subsequently, they designed various GTP analogues with small hydrophobic substituents at the C8 position and observed that these GTP analogues efficiently inhibited FtsZ polymerization [70]. The IC50 values of these GTP analogues in the polymerization as well as GTPase assay depended on the analogue:GTP ratio, suggesting competitive inhibition. The crystal structure of E. coli FtsZ/MorphGTP indicated that these C8-substituted GTP analogues bound to FtsZ in the same manner as that of GTP, which further supports the notion that these analogues are competitive inhibitors. Paradis-Bleau et al. synthesized a library of GTP analogues with core structures composed of a guanine-like moiety linked to an alanine side chain (GAL analogs) [71]. All the GAL analogues moderately inhibited the GTPase activity of Psuedomonas aeruginosa FtsZ (IC50 450 μM ~ 2.6 mM).
Sanguinarine
Sanguinarine, a benzophenanthridine alkaloid derived from the rhizomes of Sanguinaria canadensis, is known to have a broad range of antimicrobial activity [72]. Beuria et al. reported that sanguinarine inhibited cytokinesis in both Gram-positive and Gram-negative bacteria by arresting the E. coli FtsZ assembly [73]. Electron microscopic images of FtsZ polymer treated with sanguinarine indicated reduced thickness and significantly shorter lengths of FtsZ bundles.
N-benzyl-3-sulfonamidopyrrolidines
A high HTP screening of a library of compounds at the Broad Institute of Harvard University and MIT identified several compounds that caused lethal cell filamentation in E. coli without significantly inhibiting the GTPase activity of FtsZ [74]. For example, the lead compound, 534F6, displayed only weak inhibition (20%) of E. coli FtsZ GTPase activity. Based on this lead compound, an optimized library of 45 compounds was synthesized and evaluated against E. coli. Two of the compounds were found to be highly potent and caused extensive E. coli cell filamentation.
(±)-dichamanetin & (±)-2′-hydroxy-5′-benzylisouvarinol-B
Dichamanetin and 2′-hydroxy-5′-benzylisouvarinol-B, polyphenolic natural products, were isolated from U. chamae and X. afticana, respectively [75–76]. These compounds show a high level of activity against Gram-positive bacteria, such as S. aureus, B. subtilis, M. smegmatis and E. coli, with MIC values in the range of 1.7–3.4 μM for dichamanetin and 2.3–10.7 μM for hydroxybenzylisouvarinol-B [77]. Dichamanetin and hydroxybenzylisouvarinol-B were found trifluoromethylphenyl)- to be potent GTPase inhibitors of E. coli FtsZ (IC50 8–12 μM) [78].
Cinnamaldehyde
Cinnmaldehyde, a natural product, was shown to exhibit broad-spectrum antibacterial activity (MIC: 1 μg/ml for E. coli; 0.5 μg/ml for B. subtilis; 0.25 μg/ml for MRSA) [79]. E. coli FtsZ polymerization and GTPase activity were strongly inhibited with an IC50 values of 6.86 ± 2.2 and 5.81 ± 2.2 μM, respectively. Electron microscopy images and confocal imaging of live E. coli cells with GFP-tagged FtsZ further confirmed the reduction in Z-ring formation. Saturation transfer difference (STD)-NMR in conjunction with AutoDock computer modeling suggested that cinnamaldehyde binds around the T7 loop in a FtsZ monomer, inducing conformational changes that block optimal contact with the GTP-binding T1–T6 loop in a neighboring monomer.
3-{5-[4-oxo-2-thioxo-3-(3--5-ylidenemethyl]-furan-2-yl}-benzoic acid
Beuria et al. screened their library of 81 compounds with 29 diverse scaffolds using a sedimentation assay of E. coli FtsZ polymer, and found that 3-{5-[4-oxo-2-thioxo-3-(3-trifluoromethylphenyl)-thiazolidin-5-ylidenemethyl]-furan-2-yl}-benzoic acid (OTBA) promoted the E. coli FtsZ assembly in vitro and inhibited the B. subtilis cell proliferation, leading to filamentation (MIC = 2 μM) [80]. The light-scattering assay showed that OTBA enhanced the extent of E. coli FtsZ and B. subtilis FtsZ polymerization by threefold.
Curcumin
Curcumin is a naturally occurring polyphenolic compound that has been used as an important dietary ingredient for centuries. Curcumin has also shown antibacterial activity against a number of Gram-positive and Gram-negative bacteria. Rai et al. reported that curcumin induced the filamentation of B. subtilis 168 cells [65] and inhibited the cell proliferation of B. subtilis and E. coli (MIC = 17±3 μM for B. subtilis; 100 μM for E. coli). Curcumin was found to inhibit FtsZ polymerization, while enhancing GTPase activity [65]. The result indicated that curcumin decreased the stability of FtsZ protofilaments by increasing its GTPase activity.
Berberine
Berberine is a natural product isolated from various species of Berberis. It inhibits the overgrowth of pathogenic Gram-positive and -negative bacteria such as B. subtilis, Staphylococcus aureus, E. coli and MRSA [81–83]. Domadia et al. showed that berberine inhibited FtsZ assembly (IC50 10 ± 2.5 μM) and GTPase activity (IC50 16.01 ± 5 μM) [84]. Electron microscopy images further supported the destabilization of FtsZ protofilaments by berberine, wherein FtsZ filaments were found to be thin, short and curved. Fluorescence study of FtsZ-bound TNP-GTP (TNP = 2,4,6-trinitro-2,5-cyclohexadienylidene) implied that the binding site of berberine was in the vicinity of the GTP-binding pocket, overlapping with hydrophobic residues of the active site of FtsZ, which was supported by AutoDock and STD-NMR.
Stathmin-derived I19L peptide
Clement et al. showed that I19L, a peptide derived from the N-terminal region of stathmin, hampered tubulin polymerization [85]. Thus, the effect of I19L on FtsZ polymerization was also studied [86]. Sedimentation assay on E. coli FtsZ showed that I19L co-sedimented mainly with FtsZ bundles and thereby affected the bundling of FtsZ. A combination of docking studies and STD-NMR experiments indicated a possible binding site of I19L in E. coli FtsZ near the GTP-binding pocket.
4′,6-diamidino-2-phenylindole
Extending the study on tubulin–4′,6-diamidino-2-phenylindole (DAPI) binding [87–89], Nova et al. investigated the characterization of DAPI interaction with E. coli FtsZ [90]. Polymerization kinetics indicated that DAPI binding increased FtsZ polymer stability in a concentration-dependent manner. The GTPase assay illustrated that DAPI behaved as a non-competitive inhibitor of E. coli FtsZ with a Ki of 29.4 ± 0.3 μM. Anisotropy fluorescence measurements suggested that the binding of one mole of DAPI per mole of E. coli FtsZ was responsible for the bundling of the protofilaments and the inhibition of GTPase activity.
PC58538 & PC170942
Stokes et al. screened a library of 105,000 synthetic compounds against B. subtilis by adopting newly designed microtiter plate assay in combination with phase-contrast microscopy, and identified PC58538 [91]. The FtsZ GTPase assay showed that PC58538 inhibited FtsZ assembly in a dose-dependent manner (IC50 362 μM; Ki 82 μM). PC170942, an analog of PC58538, was found to be more potent than its parent molecule (IC50 44 μM; Ki 10μM). In addition, fluorescent imaging of GFP-FtsZ expressing B. subtilis, treated with PC58538 (128 μg/ml) lacked the band of FtsZ-GFP formed at the midpoint of the untreated cell.
PC190723
Ohashi et al. reported the effect of 3-methoxy-benzamide (3-MBA) on septation via inhibition of FtsZ cytokinesis [92]. Haydon et al. designed and screened more than 500 analogs of 3-MBA against various pathogenic bacterial strains and found that PC190723 exhibited potent antibacterial activity against B. subtilis, MRSA and MDRSA with MIC values in the range of 0.5–1 μg/ml [93]. PC190723 inhibited the GTPase activity of S. aureus FtsZ in a dose-dependent manner (IC50 55 ng/ml). The docking study of PC190723 suggested that the interaction of PC190723 with H7 could contribute to the inhibition of GTPase activity. Andreu et al. found that PC190723 induced B. subtilis FtsZ assembly into single-stranded coiled protofilaments, including bundles and toroids. PC190723 was also found to reduce the GTPase activity of FtsZ with the formation of straight bundles and ribbons [94].
Inhibition of the interaction of FtsZ with other cell-division proteins
The dynamic assembly of FtsZ is delicately balanced by FtsZ-interacting proteins, which include stabilizing auxiliary proteins such as ZapA, ZipA and FtsA, as well as destabilizing proteins such as SulA, EzrA and MinCD [5]. It has been shown that the C-terminal region of FtsZ is essential for interaction with several of these regulatory proteins and the deletion of this region hampers the bacterial cytokinesis [95–97]. Accordingly, the inhibition of FtsZ interaction with other regulatory proteins can be exploited to develop potential antibacterial agents. Among these interactions, FtsZ–ZipA interactions have generated a considerable interest. This could be very well attributed to the availability of crystal structure of the C-terminal region of E. coli ZipA with that of FtsZ [98,99].
Kenny et al. screened a library of 250,000 compounds and identified 29 genuine hit compounds with inhibition equal to or greater than 30% at 50 μg/ml [100]. The crystal structure of a promising small molecule, pyridylpyrimidine, bound to ZipA (KI of 12 μM) revealed that the compound binds to the same hydrophobic pocket where FtsZ binds. In a preliminary screening, Sutherland et al. identified weak inhibitors of the FtsZ–ZipA interaction (IC50 1,170–2,750 μM), which included indoles and the oxazole, binding to a largely distinct site. It was hypothesized that a hybrid of these indole- and oxazole-based inhibitors might exhibit enhanced activity. Then, carboxybiphenylindoles were studied, which displayed an improved inhibition of FtsZ–ZipA interaction (IC50 192 μM) [101].
Future perspective
Although the discovery and development of new anti-TB drugs with novel mechanisms of action, such as inhibition of the Mtb FtsZ assembly, leading to the disruption of the Z-ring formation and cell death, is still in the early stage, it appears that Mtb FtsZ has been proven to be a unique and promising target for drug discovery. We can already identify some promising lead compounds, as described in the review. Furthermore, the increasing interest and advancement in the drug discovery of broad-spectrum antimicrobials targeting FtsZ of various bacteria other than Mtb clearly suggests that this line of research will surely be translated into anti-TB drug discovery and development. We feel certain that new clinical candidates targeting Mtb FtsZ will emerge in the next couple of years and a number of drug candidates will go into human clinical trials via extensive preclinical evaluations.
Key Terms
- Cytokinesis
Process in which the cytoplasm of a single cell is divided to form two daughter cells
- Z-ring
During cell division, FtsZ monomers bind to GTP and polymerizes in a head-to-tail manner forming a ring at mid cell. This ring is called the Z-ring
- Filamentation
Anomalous growth of certain bacteria in which cells continue to elongate but do not divide
- Septation
Division or partitioning of parent cell into two parts by septum
- Protofilament
Filament formed by the aggregation of tubulin or FtsZ in the form of microtubules or FtsZ polymers; a stage in the development of the cytoplasmic skeleton
- Ortholog
One of two or more homologous gene sequences found in different species, which are direct evolutionary counterparts, (i.e., those genes related by descent from a common ancestor)
- Competitive inhibition
Form of enzyme inhibition where binding of the inhibitor to the active site on the enzyme prevents binding of the substrate and vice versa
- STD-NMR
Technique used to detect magnetization that is transferred from a receptor protein to a bound ligand
Footnotes
For reprint orders, please contact reprints@future-science.com
Financial & competing interests disclosure<br>Part of the research conducted in the authors’ laboratories was supported by grants from the National Institutes of Health (AI078251 to Iwao Ojima, AI055298 to Richard A Slayden) and the New York State Office of Science, Technology and Academic Research (NYSTAR Faculty Development Award to Iwao Ojima). The authors have no other relevant affiliation or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in this manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
No writing assistance was utilized in the production of this manuscript.
Bibliography
Papers of special note have been highlighted as:
▪ of interest
▪▪ of considerable interest
- 1.Bloom BR, Murray CJ. Tuberculosis: commentary on a reemergent killer. Science. 1992;257(5073):1055–1064. doi: 10.1126/science.257.5073.1055. [DOI] [PubMed] [Google Scholar]
- 2.Raviglione MC. Issues facing TB control (7). Multiple drug-resistant tuberculosis. Scott Med J. 2000;45(Suppl 5):52–55. doi: 10.1177/00369330000450S124. discussion 56. [DOI] [PubMed] [Google Scholar]
- 3.Margalit DN, Romberg L, Mets RB, et al. Targeting cell division: small-molecule inhibitors of FtsZ GTPase perturb cytokinetic ring assembly and induce bacterial lethality. [Erratum to document cited in CA141:271048] Proc Natl Acad Sci USA. 2004;101(38):13969. doi: 10.1073/pnas.0404439101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ben-Yehuda S, Losick R. Asymmetric cell division in B. subtilis involves a spiral-like intermediate of the cytokinetic protein FtsZ. Cell. 2002;109(2):257–266. doi: 10.1016/s0092-8674(02)00698-0. [DOI] [PubMed] [Google Scholar]
- 5.Goehring NW, Beckwith J. Diverse paths to midcell: assembly of the bacterial cell division machinery. Curr Biol. 2005;15(13):R514–R526. doi: 10.1016/j.cub.2005.06.038. [DOI] [PubMed] [Google Scholar]
- 6.Leung AKW, White EL, Ross LJ, Reynolds RC, DeVito JA, Borhani DW. Structure of Mycobacterium tuberculosis FtsZ reveals unexpected, G protein-like conformational switches. J Mol Biol. 2004;342(3):953–970. doi: 10.1016/j.jmb.2004.07.061. [DOI] [PubMed] [Google Scholar]
- 7.Moller-Jensen J, Loewe J. Increasing complexity of the bacterial cytoskeleton. Curr Opin Cell Biol. 2005;17(1):75–81. doi: 10.1016/j.ceb.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 8.Thanedar S, Margolin W. FtsZ exhibits rapid movement and oscillation waves in helix-like patterns in Escherichia coli. Curr Biol. 2004;14(13):1167–1173. doi: 10.1016/j.cub.2004.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Errington J, Daniel RA, Scheffers D-J. Cytokinesis in bacteria. Microbiol Mol Biol Rev. 2003;67(1):52–65. doi: 10.1128/MMBR.67.1.52-65.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Respicio L, Nair PA, Huang Q, et al. Characterizing septum inhibition in Mycobacterium tuberculosis for novel drug discovery. Tuberculosis. 2008;88(5):420–429. doi: 10.1016/j.tube.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 11▪▪.Slayden RA, Knudson DL, Belisle JT. Identification of cell cycle regulators in Mycobacterium tuberculosis by inhibition of septum formation and global transcriptional analysis. Microbiology. 2006;152(6):1789–1797. doi: 10.1099/mic.0.28762-0. Key manuscript that addresses global regulation in Mycobacterium tuberculosis and mapped regulatory elements to cell division and other cell cycle processes. [DOI] [PubMed] [Google Scholar]
- 12.Bramhill D. Bacterial cell division. Annu Rev Cell Dev Biol. 1997;13:395–424. doi: 10.1146/annurev.cellbio.13.1.395. [DOI] [PubMed] [Google Scholar]
- 13.Lutkenhaus J, Addinall SG. Baceterial cell division and the Z ring. Annu Rev Biochem. 1997;66:93–116. doi: 10.1146/annurev.biochem.66.1.93. [DOI] [PubMed] [Google Scholar]
- 14▪▪.Romberg L, Levin PA. Assembly dynamics of the bacterial cell division protein FtsZ: poised at the edge of stability. Annu Rev Microbiol. 2003;57:125–154. doi: 10.1146/annurev.micro.57.012903.074300. Excellent and thorough review on bacterial cell division and dynamics of FtsZ assembly. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Boer PCR, Rotherfield L. The essential bacterial cell-division protein FtsZ is a GTPase. Nature. 1992;359:254–256. doi: 10.1038/359254a0. [DOI] [PubMed] [Google Scholar]
- 16.Sticker JMP, Salmon ED, Erickson PH. Rapid assembly dynamics of the Escherichia coli. FtsZ-ring demonstrated by fluorescence recovery after photobleaching. Proc Natl Acad Sci USA. 2002;99:3171–3175. doi: 10.1073/pnas.052595099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McAdams HH, Shapiro L. System-level design of bacterial cell cycle control. FEBS Lett. 2009;583(24):3984–3991. doi: 10.1016/j.febslet.2009.09.030. Excellent review describing cell cycle regulation in Caulbacter, which serves as a model for other organisms. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nguyen HT, Wolff KA, Cartabuke RH, Ogwang S, Nguyen L. A lipoprotein modulates activity of the MtrAB two-component system to provide intrinsic multidrug resistance, cytokinetic control and cell wall homeostasis in Mycobacterium. Mol Microbiol. 2010;76(2):348–364. doi: 10.1111/j.1365-2958.2010.07110.x. [DOI] [PubMed] [Google Scholar]
- 19.Rajagopalan M, Dziedzic R, Al Zayer M, et al. Mycobacterium tuberculosis origin of replication and the promoter for immunodominant secreted antigen 85B are the targets of MtrA, the essential response regulator. J Biol Chem. 2010;285(21):15816–15827. doi: 10.1074/jbc.M109.040097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fol M, Chauhan A, Nair NK, et al. Modulation of Mycobacterium tuberculosis proliferation by MtrA, an essential two-component response regulator. Mol Microbiol. 2006;60(3):643–657. doi: 10.1111/j.1365-2958.2006.05137.x. [DOI] [PubMed] [Google Scholar]
- 21.Lutkenhaus J. Assembly dynamics of the bacterial MinCDE system and spatial regulation of the Z Ring. Annu Rev Biochem. 2007;76:539–562. doi: 10.1146/annurev.biochem.75.103004.142652. [DOI] [PubMed] [Google Scholar]
- 22.Mohl DA, Easter J, Jr, Gober JW. The chromosome partitioning protein, ParB, is required for cytokinesis in Caulobacter crescentus. Mol Microbiol. 2001;42(3):741–755. doi: 10.1046/j.1365-2958.2001.02643.x. [DOI] [PubMed] [Google Scholar]
- 23▪.Thanbichler M, Shapiro L. MipZ, a spatial regulator coordinating chromosome segregation with cell division in Caulobacter. Cell. 2006;126(1):147–162. doi: 10.1016/j.cell.2006.05.038. Important paper describing a spatial regulator with a dual role of positioning the Z-ring and delaying formation until chromosome segregation has initiated. [DOI] [PubMed] [Google Scholar]
- 24.Migocki MD, Freeman MK, Wake RG, Harry EJ. The Min system is not required for precise placement of the midcell Z ring in Bacillus subtilis. EMBO Rep. 2002;3(12):1163–1167. doi: 10.1093/embo-reports/kvf233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vicente M, Rico AI. The order of the ring: assembly of Escherichia coli cell division components. Mol Microbiol. 2006;61(1):5–8. doi: 10.1111/j.1365-2958.2006.05233.x. [DOI] [PubMed] [Google Scholar]
- 26.Hale CA, de Boer PA. Direct binding of FtsZ to ZipA, an essential component of the septal ring structure that mediates cell division in E. coli. Cell. 1997;88(2):175–185. doi: 10.1016/s0092-8674(00)81838-3. [DOI] [PubMed] [Google Scholar]
- 27.Hale CA, de Boer PA. ZipA is required for recruitment of FtsK, FtsQ, FtsL and FtsN to the septal ring in Escherichia coli. J Bacteriol. 2002;184(9):2552–2556. doi: 10.1128/JB.184.9.2552-2556.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Margolin W. FtsZ and the division of prokaryotic cells and organelles. Nat Rev Mol Cell Biol. 2005;6(11):862–871. doi: 10.1038/nrm1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Geissler B, Margolin W. Evidence for functional overlap among multiple bacterial cell-division proteins: compensating for the loss of FtsK. Mol Microbiol. 2005;58(2):596–612. doi: 10.1111/j.1365-2958.2005.04858.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30▪▪.Goehring NW, Gonzalez MD, Beckwith J. Premature targeting of cell-division proteins to midcell reveals hierarchies of protein interactions involved in divisome assembly. Mol Microbiol. 2006;61(1):33–45. doi: 10.1111/j.1365-2958.2006.05206.x. Key paper about how cell division components are assembled for the complete septal apparatus. [DOI] [PubMed] [Google Scholar]
- 31.Vicente M, Rico AI, Martinez-Arteaga R, Mingorance J. Septum enlightenment: assembly of bacterial division proteins. J Bacteriol. 2006;188(1):19–27. doi: 10.1128/JB.188.1.19-27.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32▪▪.White EL, Ross LJ, Reynolds RC, Seitz LE, Moore GD, Borhani DW. Slow polymerization of Mycobacterium tuberculosis FtsZ. J Bacteriol. 2000;182:4028–4034. doi: 10.1128/jb.182.14.4028-4034.2000. Describes in detail different factors that influence the Mycobacterium tuberculosis FtsZ polymerization. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mukherjee A, Saez C, Lutkenhaus J. Assembly of an FtsZ mutant deficient in GTPase activity has implications for FtsZ assembly and the role of the Z ring in cell division. J Bacteriol. 2001;183:7190–7197. doi: 10.1128/JB.183.24.7190-7197.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huecas S, Andreu JM. Polymerization of nucleotide-free, GDP- and GTP-bound cell division protein FtsZ: GDP makes the difference. FEBS Lett. 2004;569:43–48. doi: 10.1016/j.febslet.2004.05.048. [DOI] [PubMed] [Google Scholar]
- 35.Oliva MA, Trambaiolo D, Lowe J. Structural insights into the conformational variability of FtsZ. J Mol Biol. 2007;373:1229–1242. doi: 10.1016/j.jmb.2007.08.056. [DOI] [PubMed] [Google Scholar]
- 36.Diaz JF, Kralicek A, Mingorance J, Palacios JM, Vicente M, Andreu JM. Activation of cell division protein FtsZ. J Biol Chem. 2001;276:17307–17315. doi: 10.1074/jbc.M010920200. [DOI] [PubMed] [Google Scholar]
- 37.Lu C, Stricker J, Erickson HP. Site-specific mutations of FtsZ - effects on GTPase and in vitro assembly. BMC Microbiology. 2001;1(7) doi: 10.1186/1471-2180-1-7. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lan G, Daniels BR, Dobrowsky TM, Wirtz D, Sun SX. Conensation of FtsZ filaments can drive bacterial cell division. Proc Natl Acad Sci USA. 2009;106:121–126. doi: 10.1073/pnas.0807963106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paez A, Mateos-Gil P, Horger I, et al. Simple modeling of FtsZ polymers on flat and curved surfaces: correlation with experimental in vitro observations. PMC Biophysics. 2009;2(8) doi: 10.1186/1757-5036-2-8. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Erickson PH, Taylor DW, Taylor KA, Bramhill D. Bacterial cell division protein FtsZ assembles into protofilament sheets and minirings, structural homologs of tubulin polymers. Proc Natl Acad Sci USA. 1996;93:519–523. doi: 10.1073/pnas.93.1.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Constantine GH, Karchesy JJ, Franzblau SG, LaFleur LE. (+)-totarol from Chamaecyparis nootkatensis and activity against Mycobacterium tuberculosis. Fitoterapia. 2001;72(5):572–574. doi: 10.1016/s0367-326x(01)00272-6. [DOI] [PubMed] [Google Scholar]
- 42.Kubo I, Muroi H, Himejima M. Antibacterial activity of totarol and its potentiation. J Nat Prod. 1992;55(10):1436–1440. doi: 10.1021/np50088a008. [DOI] [PubMed] [Google Scholar]
- 43.Jaiswal R, Beuria TK, Mohan R, Mahajan SK, Panda D. Totarol inhibits bacterial cytokinesis by perturbing the assembly dynamics of FtsZ. Biochemistry. 2007;46(14):4211–4220. doi: 10.1021/bi602573e. [DOI] [PubMed] [Google Scholar]
- 44.Sarcina M, Mullineaux CW. Effects of tubulin assembly inhibitors on cell division in prokaryotes in vivo. FEMS Microbiol Lett. 2000;191(1):25–29. doi: 10.1111/j.1574-6968.2000.tb09314.x. [DOI] [PubMed] [Google Scholar]
- 45.de Pereda JM, Leynadier D, Evangelio JA, Chacon P, Andreu JM. Tubulin secondary structure analysis, limited proteolysis sites and homology to FtsZ. Biochemistry. 1996;35(45):14203–14215. doi: 10.1021/bi961357b. [DOI] [PubMed] [Google Scholar]
- 46▪.White EL, Suling WJ, Ross LJ, Seitz LE, Reynolds RC. 2-alkoxycarbonyl-aminopyridines: inhibitors of Mycobacterium tuberculosis FtsZ. J Antimicrob Chemother. 2002;50(1):111–114. doi: 10.1093/jac/dkf075. Describes the first examples of inhibitors of M. tuberculosis FtsZ, effective against TB. [DOI] [PubMed] [Google Scholar]
- 47.Reynolds RC, Srivastava S, Ross LJ, Suling WJ, White EL. A new 2-carbamoyl pteridine that inhibits mycobacterial FtsZ. Bioorg Med Chem Lett. 2004;14(12):3161–3164. doi: 10.1016/j.bmcl.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 48.Georg GI, Chen TT, Ojima I, Wyas DM, editors. ACS Symp Ser. Vol. 583. American Chemical Society; Washington DC, USA: 1995. Taxane Anticancer Agents: Basic Science and Current Status; p. 353. [Google Scholar]
- 49.Kingston DGI, Jagtap PG, Yuan H, Samala L. The chemistry of taxol and related taxoids. Prog Chem Org Nat Prod. 2002;84:53–225. doi: 10.1007/978-3-7091-6160-9_2. [DOI] [PubMed] [Google Scholar]
- 50.Ojima I, Kuduk SD, Chakravarty S. Recent advances in the medicinal chemistry of taxoid anticancer agents. Adv Med Chem. 1999;4:69–124. [Google Scholar]
- 51.Brooks TA, Kennedy DR, Gruol DJ, Ojima I, Baer MR, Bernacki RJ. Structure–activity analysis of taxane-based broad-spectrum multidrug resistance modulators. Anticancer Res. 2004;24(2A):409–415. [PubMed] [Google Scholar]
- 52.Brooks TA, Minderman H, O’Loughlin KL, et al. Taxane-based reversal agents modulate drug resistance mediated by P-glycoprotein, multidrug resistance protein, and breast cancer resistance protein. Mol Cancer Ther. 2003;2(11):1195–1205. [PubMed] [Google Scholar]
- 53.Minderman H, Brooks TA, O’Loughlin KL, Ojima I, Bernacki RJ, Baer MR. Broad-spectrum modulation of ATP-binding cassette transport proteins by the taxane derivatives ortataxel (IDN-5109, BAY 59–8862) and tRA96023. Cancer Chemother Pharmacol. 2004;53(5):363–369. doi: 10.1007/s00280-003-0745-2. [DOI] [PubMed] [Google Scholar]
- 54.Ojima I, Borella CP, Wu X, Bounaud P-Y, Oderda CF, Sturm M, et al. Design, synthesis and structure–activity relationships of novel taxane-based multidrug resistance reversal agents. J Med Chem. 2005;48(6):2218–2228. doi: 10.1021/jm049483y. [DOI] [PubMed] [Google Scholar]
- 55.Ojima I, Bounaud P-Y, Bernacki RJ. New weapons in the fight against cancer. Chemtech. 1998;28(6):31–36. [Google Scholar]
- 56.Ojima I, Bounaud P-Y, Bernacki RJ. Designing taxanes to treat multidrug-resistant tumors. Mod Drug Discov. 1999;2(3):45, 47–48, 51–52. [Google Scholar]
- 57.Ojima I, Bounaud P-Y, Oderda CF. Recent strategies for the treatment of multi-drug resistance in cancer cells. Expert Opin Ther Pat. 1998;8(12):1587–1598. [Google Scholar]
- 58.Ojima I, Bounaud P-Y, Takeuchi C, Pera P, Bernacki RJ. New taxanes as highly efficient reversal agents for multi-drug resistance in cancer cells. Bioorg Med Chem Lett. 1998;8(2):189–194. doi: 10.1016/s0960-894x(97)10218-9. [DOI] [PubMed] [Google Scholar]
- 59▪.Huang Q, Kirikae F, Kirikae T, et al. Targeting FtsZ for anti-tuberculosis drug discovery: noncytotoxic taxanes as novel anti-tuberculosis agents. J Med Chem. 2006;49(2):463–466. doi: 10.1021/jm050920y. Describes the first examples of noncytotoxic taxanes that show antitubercular activity and target M. tuberculosis FtsZ. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Appendino G, Danieli B, Jakupovic J, Belloro E, Scambia G, Bombardelli E. The chemistry and occurrence of taxane derivatives. XXX. Synthesis and evaluation of C-seco paclitaxel analogs. Tetrahedron Lett. 1997;38(24):4273–4276. [Google Scholar]
- 61.Taraboletti G, Micheletti G, Rieppi M, et al. Antiangiogenic and anti-tumor activity of IDN 5390, a new taxane derivative. Clin Cancer Res. 2002;8(4):1182–1188. [PubMed] [Google Scholar]
- 62.Kumar K, Lee S-Y, Zanardi I, Slayden RA, Ojima I. Synthesis and optimization of a library of novel benzimidazole leads for anti-tuberculosis drug discovery. Abstracts of Papers, 234th ACS National Meeting; Boston, MA, USA. 19–23 August; MEDI; 2007. p. 205. [Google Scholar]
- 63.Kumar K, Zanardi I, Ruzsicska B, Slayden RA, Walker SG, Ojima I. SAR study on novel benzimidazoles as potential broad-spectrum antibacterial agents targeting FtsZ. Abstracts of Papers, 236th ACS National Meeting; Philadelphia, PA, USA. 17–21 August; MEDI; 2008. p. 290. [Google Scholar]
- 64.Kumar K, Zanardi I, Berger WT, Ruzsicska B, Slayden RA, Ojima I. SAR study and optimization of novel benzimidazoles for anti-tuberculosis drug discovery, targeting FtsZ. Abstracts of Papers, 238th ACS National Meeting; Washington, DC, USA. 16–20 August; MEDI; 2009. p. 123. [Google Scholar]
- 65.Rai D, Singh JK, Roy N, Panda D. Curcumin inhibits FtsZ assembly: an attractive mechanism for its antibacterial activity. Biochem J. 2008;410(1):147–155. doi: 10.1042/BJ20070891. [DOI] [PubMed] [Google Scholar]
- 66.Lowe J, Amos LA. Tubulin-like protofilaments in Ca2+-induced FtsZ sheets. Embo J. 1999;18(9):2364–2371. doi: 10.1093/emboj/18.9.2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Scheffers D-J, de Wit JG, den Blaauwen T, Driessen AJM. GTP hydrolysis of cell division protein FtsZ: evidence that the active site is formed by the association of monomers. Biochemistry. 2002;41(2):521–529. doi: 10.1021/bi011370i. [DOI] [PubMed] [Google Scholar]
- 68.Wang J, Galgoci A, Kodali S, et al. Discovery of a small molecule that inhibits cell division by blocking FtsZ, a novel therapeutic target of antibiotics. J Biol Chem. 2003;278(45):44424–44428. doi: 10.1074/jbc.M307625200. [DOI] [PubMed] [Google Scholar]
- 69.Laeppchen T, Hartog AF, Pinas VA, Koomen G-J, Den Blaauwen T. GTP analogue inhibits polymerization and GTPase activity of the bacterial protein FtsZ without affecting its eukaryotic homologue tubulin. Biochemistry. 2005;44(21):7879–7884. doi: 10.1021/bi047297o. [DOI] [PubMed] [Google Scholar]
- 70.Laeppchen T, Pinas VA, Hartog AF, et al. Probing FtsZ and tubulin with C8-substituted GTP analogs reveals differences in their nucleotide binding sites. Chem Biol. 2008;15(2):189–199. doi: 10.1016/j.chembiol.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 71.Paradis-Bleau C, Beaumont M, Sanschagrin F, Voyer N, Levesque RC. Parallel solid synthesis of inhibitors of the essential cell division FtsZ enzyme as a new potential class of antibacterials. Bioorg Med Chem. 2007;15(3):1330–1340. doi: 10.1016/j.bmc.2006.11.015. [DOI] [PubMed] [Google Scholar]
- 72.Godowski KC. Antimicrobial action of sanguinarine. J Clin Dent. 1989;1(4):96–101. [PubMed] [Google Scholar]
- 73.Beuria TK, Santra MK, Panda D. Sanguinarine Blocks cytokinesis in bacteria by inhibiting FtsZ assembly and bundling. Biochemistry. 2005;44(50):16584–16593. doi: 10.1021/bi050767+. [DOI] [PubMed] [Google Scholar]
- 74.Mukherjee S, Robinson CA, Howe AG, et al. N-benzyl-3-sulfonamidopyrrolidines as novel inhibitors of cell division in E. coli. Bioorg Med Chem Lett. 2007;17(23):6651–6655. doi: 10.1016/j.bmcl.2007.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Anam EM. 2′-Hydroxy-3′-benzyluvarinol,2′-hydroxy-5′-benzylisouvarinol-A and 2′-hydroxy-5′-benzylisouvarinol-B: three novel tetra-C-benzylated flavanones from the root extract of Xylopia africana (Benth) Oliver (Annonaceae) Indian J Chem, Sect B Org Chem Incl Med Chem. 1994;33B(10):1009–1011. [Google Scholar]
- 76.Hufford CD, Lasswell WL., Jr Antimicrobial activities of constituents of Uvaria chamae. Lloydia. 1978;41(2):156–160. [PubMed] [Google Scholar]
- 77.Leegaard TM, Caugant DA, Froholm LO, Hoiby EA. Apparent differences in antimicrobial susceptibility as a consequence of national guidelines. Clin Microbiol Infect. 2000;6(6):290–293. doi: 10.1046/j.1469-0691.2000.00086.x. [DOI] [PubMed] [Google Scholar]
- 78.Urgaonkar S, La Pierre HS, Meir I, Lund H, Ray Chaudhuri D, Shaw JT. Synthesis of antimicrobial natural products targeting FtsZ: (+-)-dichamanetin and (+-)-2′-hydroxy-5′-benzylisouvarinol-B. Org Lett. 2005;7(25):5609–5612. doi: 10.1021/ol052269z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Domadia P, Swarup S, Bhunia A, Sivaraman J, Dasgupta D. Inhibition of bacterial cell division protein FtsZ by cinnamaldehyde. Biochem Pharmacol. 2007;74(6):831–840. doi: 10.1016/j.bcp.2007.06.029. [DOI] [PubMed] [Google Scholar]
- 80.Beuria TK, Singh P, Surolia A, Panda D. Promoting assembly and bundling of FtsZ as a strategy to inhibit bacterial cell division: a new approach for developing novel antibacterial drugs. Biochem J. 2009;423(1):61–69. doi: 10.1042/BJ20090817. [DOI] [PubMed] [Google Scholar]
- 81.Hwang BY, Roberts SK, Chadwick LR, Wu CD, Kinghorn AD. Antimicrobial constituents from goldenseal (the rhizomes of Hydrastis canadensis) against selected oral pathogens. Planta Med. 2003;69(7):623–627. doi: 10.1055/s-2003-41115. [DOI] [PubMed] [Google Scholar]
- 82.Villinski JR, Dumas ER, Chai H-B, Pezzuto JM, Angerhofer CK, Gafner S. Antibacterial activity and alkaloid content of Berberis thunbergii, Berberis vulgaris and Hydrastis canadensis. Pharm Biol. 2003;41(8):551–557. [Google Scholar]
- 83.Yu H-H, Kim K-J, Cha J-D, et al. Antimicrobial activity of berberine Alone and in combination with ampicillin or oxacillin against methicillin-resistant Staphylococcus aureus. J Med Food. 2005;8(4):454–461. doi: 10.1089/jmf.2005.8.454. [DOI] [PubMed] [Google Scholar]
- 84.Domadia PN, Bhunia A, Sivaraman J, Swarup S, Dasgupta D. Berberine targets assembly of Escherichia coli cell division protein FtsZ. Biochemistry. 2008;47(10):3225–3234. doi: 10.1021/bi7018546. [DOI] [PubMed] [Google Scholar]
- 85.Clement M-J, Jourdain I, Lachkar S, Savarin P, Gigant B, Knossow M, et al. N-terminal stathmin-like peptides bind tubulin and impede microtubule assembly. Biochemistry. 2005;44(44):14616–14625. doi: 10.1021/bi0512492. [DOI] [PubMed] [Google Scholar]
- 86.Clement M-J, Kuoch B-t, Ha-Duong T, et al. The stathmin-derived I19L peptide interacts with FtsZ and alters its bundling. Biochemistry. 2009;48(41):9734–9744. doi: 10.1021/bi900556a. [DOI] [PubMed] [Google Scholar]
- 87.Arbildua JJ, Brunet JE, Jameson DM, et al. Fluorescence resonance energy transfer and molecular modeling studies on 4′,6-diamidino-2-phenylindole (DAPI) complexes with tubulin. Protein Sci. 2006;15(3):410–419. doi: 10.1110/ps.051862206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bonne D, Heusele C, Simon C, Pantaloni D. 4′,6-diamidino-2-phenylindole, a fluorescent probe for tubulin and microtubules. J Biol Chem. 1985;260(5):2819–2825. [PubMed] [Google Scholar]
- 89.Ortiz M, Lagos R, Monasterio O. Interaction between the C-terminal peptides of tubulin and tubulin S detected with the fluorescent probe 4′,6-diamidino-2-phenylindole. Arch Biochem Biophys. 1993;303(1):159–164. doi: 10.1006/abbi.1993.1267. [DOI] [PubMed] [Google Scholar]
- 90.Nova E, Montecinos F, Brunet JE, Lagos R, Monasterio O. 4′,6-diamidino-2-phenylindole (DAPI) induces bundling of Escherichia coli FtsZ polymers inhibiting the GTPase activity. Arch Biochem Biophys. 2007;465(2):315–319. doi: 10.1016/j.abb.2007.06.032. [DOI] [PubMed] [Google Scholar]
- 91.Stokes NR, Sievers J, Barker S, et al. Novel Inhibitors of bacterial cytokinesis identified by a cell-based antibiotic screening assay. J Biol Chem. 2005;280(48):39709–39715. doi: 10.1074/jbc.M506741200. [DOI] [PubMed] [Google Scholar]
- 92.Ohashi Y, Chijiiwa Y, Suzuki K, et al. The lethal effect of a benzamide derivative, 3-methoxybenzamide, can be suppressed by mutations within a cell division gene, ftsZ, in Bacillus subtilis. J Bacteriol. 1999;181(4):1348–1351. doi: 10.1128/jb.181.4.1348-1351.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Haydon DJ, Stokes NR, Ure R, et al. An inhibitor of FtsZ with potent and selective anti-staphylococcal activity. Science. 2008;321(5896):1673–1675. doi: 10.1126/science.1159961. [DOI] [PubMed] [Google Scholar]
- 94.Andreu JM, Schaffner-Barbero C, Huecas S, et al. The antibacterial cell division inhibitor PC190723 is an FtsZ polymer-stabilizing agent that induces filament assembly and condensation. J Biol Chem. 2010;285(19):14239–14246. doi: 10.1074/jbc.M109.094722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Haney SA, Glasfeld E, Hale C, Keeney D, He Z, De Boer P. Genetic analysis of the Escherichia coli FtsZ. ZipA interaction in the yeast two-hybrid system Characterization of FtsZ residues essential for the interactions with ZipA and with FtsA. J Biol Chem. 2001;276(15):11980–11987. doi: 10.1074/jbc.M009810200. [DOI] [PubMed] [Google Scholar]
- 96.Singh JK, Makde RD, Kumar V, Panda D. A membrane protein, EzrA, regulates assembly dynamics of FtsZ by interacting with the C-Terminal tail of FtsZ. Biochemistry. 2007;46(38):11013–11022. doi: 10.1021/bi700710j. [DOI] [PubMed] [Google Scholar]
- 97.Singh JK, Makde RD, Kumar V, Panda D. SepF increases the assembly and bundling of FtsZ Polymers and stabilizes FtsZ protofilaments by binding along its length. J Biol Chem. 2008;283(45):31116–31124. doi: 10.1074/jbc.M805910200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98▪.Mosyak L, Zhang Y, Glasfeld E, et al. The bacterial cell-division protein ZipA and its interaction with an FtsZ fragment revealed by x-ray crystallography. EMBO J. 2000;19(13):3179–3191. doi: 10.1093/emboj/19.13.3179. Provides an attractive insight into FtsZ–ZipA crystal structure and its interactions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Moy FJ, Glasfeld E, Mosyak L, Powers R. Solution structure of ZipA, a crucial component of Escherichia coli cell division. Biochemistry. 2000;39(31):9146–9156. doi: 10.1021/bi0009690. [DOI] [PubMed] [Google Scholar]
- 100.Kenny CH, Ding W, Kelleher K, et al. Development of a fluorescence polarization assay to screen for inhibitors of the FtsZ/ZipA interaction. Anal Biochem. 2003;323(2):224–233. doi: 10.1016/j.ab.2003.08.033. [DOI] [PubMed] [Google Scholar]
- 101.Sutherland AG, Alvarez J, Ding W, et al. Structure-based design of carboxybiphenylindole inhibitors of the ZipA–FtsZ interaction. Org Biomol Chem. 2003;1(23):4138–4140. doi: 10.1039/b312016c. [DOI] [PubMed] [Google Scholar]
Patent
- 201.Ojima I, Lee S-Y. Benzimidazoles and pharmaceutical compositions thereof. 2008130669. International Patent Application WO. 2008
Websites
- 301.World Health Organization. Tuberculosis: data and country profiles. www.who.int/tb/country/en/
- 302.National Institute of Allergy and Infectious Disease (NIAID) website. www3.niaid.nih.gov/topics/tuberculosis/Understanding/WhatIsTB/ScientificIllustrations/multidrugResistantIllustration.htm.