Abstract
The public sharing of primary research datasets potentially benefits the research community but is not yet common practice. In this pilot study, we analyzed whether data sharing frequency was associated with funder and publisher requirements, journal impact factor, or investigator experience and impact. Across 397 recent biomedical microarray studies, we found investigators were more likely to publicly share their raw dataset when their study was published in a high-impact journal and when the first or last authors had high levels of career experience and impact. We estimate the USA’s National Institutes of Health (NIH) data sharing policy applied to 19% of the studies in our cohort; being subject to the NIH data sharing plan requirement was not found to correlate with increased data sharing behavior in multivariate logistic regression analysis. Studies published in journals that required a database submission accession number as a condition of publication were more likely to share their data, but this trend was not statistically significant. These early results will inform our ongoing larger analysis, and hopefully contribute to the development of more effective data sharing initiatives.
Keywords: bibliometrics, bioinformatics, policy evaluation, data sharing
1. Introduction
Sharing and reusing primary research datasets has the potential to increase research efficiency and quality. Raw data can be used to explore related or new hypotheses, particularly when combined with other available datasets. Real data is indispensable for developing and validating study methods, analysis techniques, and software implementations. The larger scientific community also benefits: sharing data encourages multiple perspectives, helps to identify errors, discourages fraud, is useful for training new researchers, and increases efficient use of funding and population resources by avoiding duplicate data collection.
Eager to realize these benefits, funders, publishers, societies, and individual research groups have developed tools, resources, and policies to encourage investigators to make their data publicly available. For example, some journals require the submission of detailed biomedical datasets to publicly available databases as a condition of publication (McCain, 1995; Piwowar & Chapman, 2008). Many funders require data sharing plans as a condition of funding: since 2003, the National Institutes of Health (NIH) in the USA has required a data sharing plan for all large funding grants(NIH, 2003) and has more recently introduced stronger requirements for genome-wide association studies (NIH, 2007). Several government whitepapers(Cech, 2003; Fienberg, Martin, & Straf, 1985) and high-profile editorials(Data's shameful neglect, 2009; Got data?, 2007; Time for leadership, 2007) call for responsible data sharing and reuse. Large-scale collaborative science is increasing the need to share datasets (Kakazu, Cheung, & Lynne, 2004; The GAIN Collaborative Research Group, 2007), and many guidelines, tools, standards, and databases are being developed and maintained to facilitate data sharing and reuse (Schofield et al., 2009; Barrett et al., 2007; Brazma et al., 2001).
Despite these investments of time and money, we do not yet understand the impact of these initiatives. There is a well-known adage: you cannot manage what you do not measure. For those with a goal of promoting responsible data sharing, it would be helpful to evaluate the effectiveness of requirements, recommendations, and tools. When data sharing is voluntary, insights could be gained by learning which datasets are shared, on what topics, by whom, and in what locations. When policies make data sharing mandatory, monitoring is useful to understand compliance and unexpected consequences.
Dimensions of data sharing action and intension have been investigated by a variety of studies. Manual annotations and systematic data requests have been used to estimate the frequency of data sharing within biomedicine (Kyzas, Loizou, & Ioannidis, 2005; Noor, Zimmerman, & Teeter, 2006; Ochsner et al., 2008; Reidpath & Allotey, 2001), though few attempts were made to determine patterns of sharing and withholding within these samples. Blumenthal(2006), Campbell(2002), Hedstrom(2006) and others have used survey results to correlate self-reported instances of data sharing and withholding with self-reported attributes like industry involvement, perceived competitiveness, career productivity, and anticipated data sharing costs. Others have used surveys and interviews to analyze opinions about the effectiveness of mandates (Ventura, 2005) and the value of various incentives (Giordano, 2007; Hedstrom, 2006; Hedstrom & Niu, 2008; Niu, 2006). A few inventories list the data-sharing policies of funders (Lowrance, 2006; University of Nottingham) and journals (Brown, 2003; McCain, 1995), and some work has been done to correlate policy strength with outcome (McCullough, McGeary, & Harrison, 2008; Piwowar & Chapman, 2008). Surveys and case studies have been used to develop models of information behavior in related domains, including knowledge sharing within an organization(Constant, Kiesler, & Sproull, 1994; Matzler et al., 2008), physician knowledge sharing in hospitals (Ryu, Ho, & Han, 2003), participation in open source projects(Bitzer, Schrettl, & Schröder, 2007), academic contributions to institutional archives(Kim, 2007; Seonghee & Boryung, 2008), the choice to publish in open access journals(Warlick & Vaughan, 2007), sharing social science datasets (Hedstrom, 2006), and participation in large-scale biomedical research collaborations (Lee, Dourish, & Mark, 2006).
Although these studies provide valuable insights and their methods facilitate investigation into an author’s intentions and opinions, they have several limitations. First, associations between an investigator’s intention to share data do not directly translate to an association with actually sharing data (Kuo & Young, 2008). Second, associations that rely on self-reported data sharing and withholding likely suffer from underreporting and confounding, since people admit withholding data much less frequently than they report having experienced the data withholding of others (Blumenthal et al., 2006).
We suggest a supplemental approach for investigating research data sharing behavior. As part of an ongoing doctoral dissertation project, we are collecting and analyzing a large set of observed data sharing actions and associated policy, investigator, and environmental variables. In this report we provide preliminary findings on a small collection of studies and a few key questions: Are studies led by experienced and prolific primary investigators more likely to share their data than those led by junior investigators? Do funder and publisher requirements for data sharing increase the frequency with which data is shared? Are other funder and publisher characteristics associated with data sharing frequency?
We choose to study data sharing for one particular type of data: biological gene expression microarray intensity values. Microarray studies provide a useful environment for exploring data sharing policies and behaviors. Despite being a rich resource valuable for reuse (Rhodes et al., 2004), microarray data are often but not yet universally shared. Best-practice guidelines for sharing microarray data are fairly mature (Brazma et al., 2001; Hrynaszkiewicz & Altman, 2009). Two centralized databases have emerged as best-practice repositories: the Gene Expression Omnibus (GEO)(Barrett et al., 2007) and ArrayExpress(Parkinson et al., 2007). Finally, high-profile letters have called for strong journal data sharing policies(Ball et al., 2004), resulting in unusually strong data sharing requirements in some journals (Microarray standards at last, 2002).
2. Methods
We identified a set of studies in which the investigators had generated gene expression microarray datasets, and which of these had made their datasets publicly available on the internet. We analyzed variables related to the investigators, journals, and funding of these studies to determine which attributes were associated an increased frequency of data sharing.
2.1. Studies for analysis
Ochsner et al. (2008) manually reviewed articles in 20 journals to identify studies in which the authors generated microarray data. They found 397 such studies. Ochsner and co-investigators then searched online databases and web pages to determine which of these studies had made their gene expression profile datasets publicly available on the internet.
We use the Ochsner dataset as the article cohort, and their identification of data sharing as our dependent variable. In addition, we collected additional covariates to use as independent variables, as described below.
2.2. Author impact and experience
For each study, we collected variables related to the number of authors and the address of the corresponding author from PubMed® metadata.
We also characterized “author experience” of each first and last author (customarily the most hands-on and senior investigators, respectively, of a biomedical study) through a combination of their h-index (Hirsch, 2005), a-index (Jin, 2006), and number of years since publishing their first paper. We chose h-index and a-index since recent research suggests they represent different aspects of an investigator’s research productivity and impact (Bornmann, Mutz, & Daniel, 2008). We estimated these indices using data from PubMed, PubMed Central®, and the Author-ity name disambiguation engine (Torvik & Smalheiser, 2009; Torvik et al., 2005) to derive what we call an author’s “pubmedi” h-index and a-index.
Briefly, for each first and last author, we submitted the author’s name and known publication PubMed ID to the 2008 Author-ity web service (http://128.248.65.210/arrowsmith_uic/author2.html). Author-ity returned a list of publications estimated to be authored by the given investigator. For each of the PubMed publications in the Author-ity results list, we queried PubMed Central for the list of PubMed Central articles that cite the given PubMed publication, using the eUtils web service interface at http://www.ncbi.nlm.nih.gov/entrez/query/static/eutils_help.html. Finally, we used the list of {PubMed ID, Number of times the PubMed ID was cited by an article in PubMed Central} pairs to compute the pubmedi h-index and a-index surrogates for the author.
We also estimated the number of years since each author published his or her first paper (as indexed by PubMed) as a proxy for author age and/or career experience. We calculated this duration by extracting the earliest publication year from each author’s Author-ity publication list.
To conserve degrees of freedom in this pilot study, we clustered the pubmedi h-index, a-index and career length into one variable through principal component analysis. We call the first principal component coefficient of these variables (the log transform of h-index and a-index, and career length as number of years between the publication of first paper and 2008) the “author experience” in subsequent analysis. The author experience metric for first authors and last authors were computed separately. Each has a mean of 0.0 and a 25th and 75th percentile at approximately −1.0 and +1.0, respectively.
2.3. Grant funding and mandates
For this pilot study, we focused on funding by the National Institutes of Health (NIH), the major government biomedical funding body in the United States. For studies funded by the NIH, we collected the history of applicable grants, the total amount of funding per fiscal year, and we attempted to estimate whether the grant was subject to the NIH’s data sharing plan requirements. A data sharing plan is a required component of all proposals submitted to the NIH after 2003 that request more than $500 000 (United States dollars) in direct funding per year. Total grant funding from the NIH usually consists of direct funding (that which is specifically applicable to the proposal, such as investigator salaries and research supplies) and indirect funding (general facilities and administrative overhead, such as building depreciation and maintenance costs). The relative amounts of direct and indirect funding in a grant vary per institution.
We determined which studies were funded by the NIH using PubMed metadata. For studies with NIH grants, we looked up each of the NIH grant numbers in the NIH grant database (http://report.nih.gov/award/state/state.cfm). From this information we tabulated the amount of total funding received for each of the fiscal years 2003–2007. We assumed that those with more than $750 000 in total funding in any given year had received at least $500 000 in direct funding. We also estimated the date of renewal by identifying the most recent year in which the grant number was prefixed by a “1” or “2” -- indication that the grant is “new” or “renewed.” Finally, we assumed that any grant with more than $750 000 in total funds in a single year and with a new or renewed grant since 2003 was subject to the NIH data sharing plan requirement.
For studies identified in PubMed as funded by the NIH but without a complete NIH grant number listed in the PubMed metadata, we imputed the sum of the maximum award amount and whether the NIH data sharing requirement would have applied based on all other available variables using a multivariate tree classification as performed by Harrell (2001).
2.4. Journal impact and mandates
For each study, we collected two variables related to the journal in which it was published: impact factor, and the strength of the journal’s policy on data sharing. We gathered the impact factor for each of the 20 journals from ISI Journal Citation Reports 2007. We identified the strength of a journal’s data sharing policy by inspecting its “Instruction to Author” statement. As in (Piwowar & Chapman, 2008), we grouped journals with no mention of data sharing applicable to microarrays as “no mention”, a request or unenforceable requirement as a “weak policy,” and those that required an accession number from submission to an online database as a “strong policy.”
2.5. Statistical Methods
We used multivariate logistic regression to evaluate the association between authorship, grant, and journal attributes of a study and the public availability of its microarray data. Statistical analysis was performed using the Hmisc and Design packages for regression and visualization (http://biostat.mc.vanderbilt.edu/wiki/Main/Hmisc) in R version 2.7 (http://www.R-project.org). P-values are two-tailed.
2.6. Data and code availability
Statistical scripts and the raw dataset are included as supplemental data and are also available at www.researchremix.org.
3. Results
We studied the data sharing patterns of 397 gene expression microarray studied published in 2007 within 20 journals, as identified in a systematic review by Ochsner et al(Ochsner et al., 2008). Almost half of the studies made their raw datasets available (47%).
We found that 41 of the articles acknowledged NIH funding but did not reveal specific grant numbers; these studies appear to be randomly distributed throughout the sample, so we estimated their levels of NIH funding from other attributes, as described in the methods section.
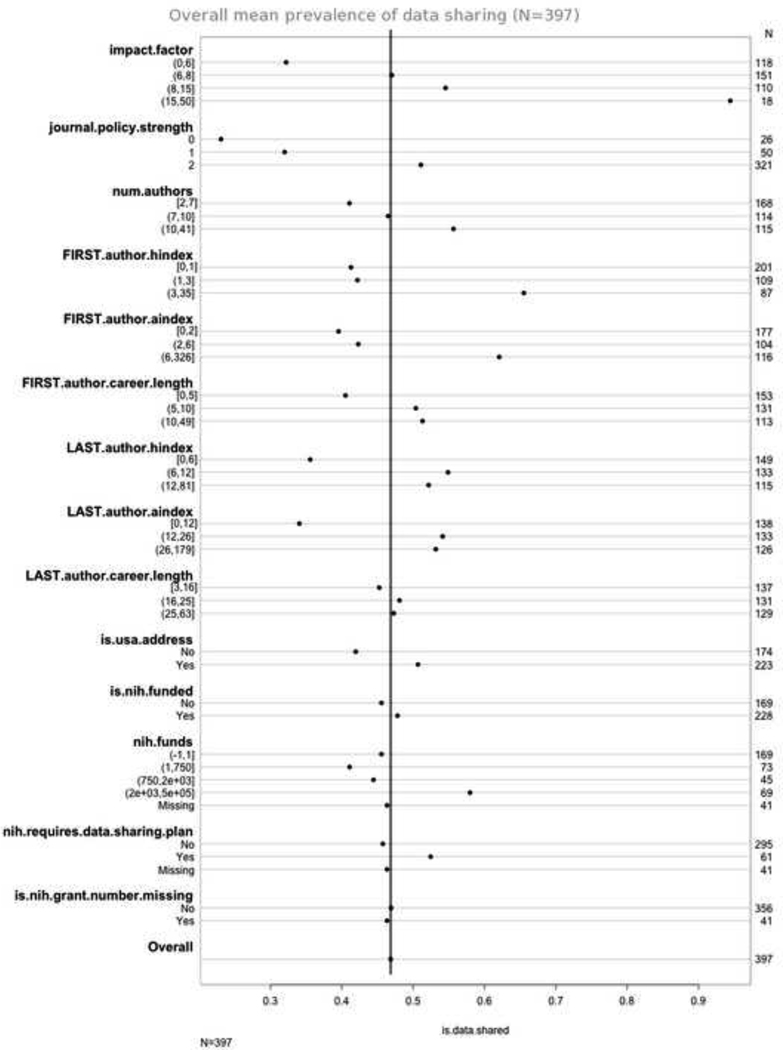
As seen in Figure 1, in univariate analysis many variables were found to vary with prevalence of data sharing: the impact factor of the journal, the strength of the journal’s data sharing policy, the number of authors, the h-index and a-index of the first and last author, the career length of the first author, whether the corresponding author had a US-based address, and whether the study was funded by more than $2 million dollars of NIH grants.
Figure 1.
Proportion of studies with publicly shared datasets, in univariate analysis. Overall mean prevalence of data sharing was 47%.
We estimated that the NIH data sharing policy applied to 61 of the studies (the study was submitted to the NIH after 2003 and received more than $500 000 in direct NIH funding per year): these 61 studies had a slightly higher frequency of data sharing in univariate analysis (52% vs. 46%). Last author career length and whether or not they received any NIH funding appeared to be largely independent of data sharing frequency.
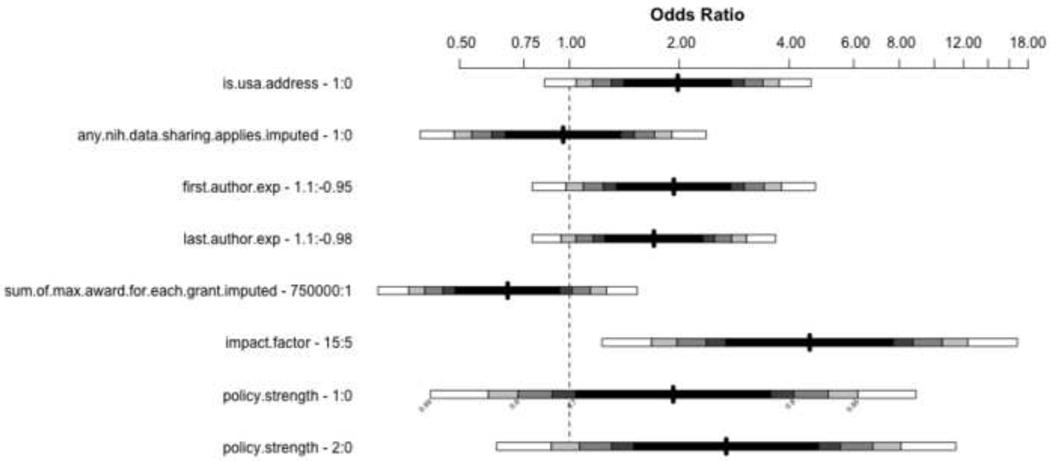
The results of multivariate analysis are shown in Table 1. The impact factor of the journal and the experience component for the first and the last authors were significantly associated with an increase in data sharing prevalence, with corresponding author country and journal policy strength is trending towards but not reaching statistical significance.
Table 1.
Significance of covariates in multivariate logistic regression on demonstrated data sharing.
| Covariate | p-value |
|---|---|
| First author experience | .03 |
| Last author experience | .01 |
| Corresponding author has a USA address? | .09 |
| Impact factor of journal | .03 |
| Strength of journal data sharing policy | .20 |
| Is NIH funded? | .72 |
| Does NIH data sharing mandate apply? | .98 |
| Sum of max award for each grant | .97 |
| Interaction between USA address and NIH funding | .43 |
| TOTAL (17 degrees of freedom) | <.001 |
The size of these effects are illustrated in Figure 2: a study with a corresponding address in the USA was twice as likely to publicly share its microarray data; a study published in a journal with an impact factor of 15 was 4.5 times as likely to have shared data compared to a study published in a journal with an impact factor of 5 (assuming all other covariates are held constant), increased author experience suggests a increased odds of data sharing, and the odds of data sharing is higher in journals with a weak data sharing policy than in a journal with no data sharing policy, and higher yet in journals with a strong data sharing policy.
Figure 2.
Odds ratios in multivariate logistic regression for a given change in variable; for first and last author experience, the given experience values represent an increase of experience from the first to the third quartile of our “author experience” metric, from approximately −1.0 to +1.0.
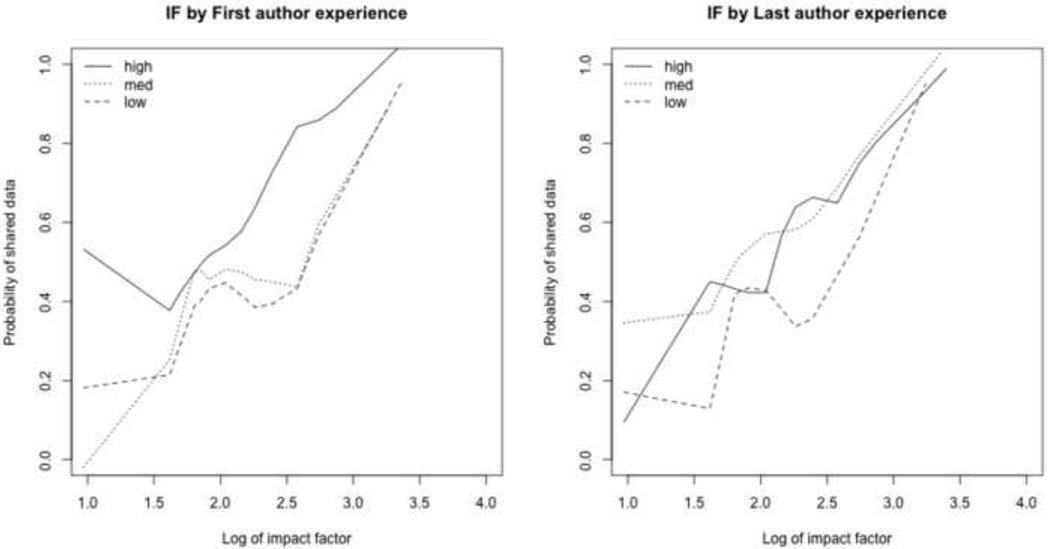
We display stratified relationships in Figures 3, 4, and 5 to explore interactions for future studies. For example, in Figure 3a we can see that first authors with a “high” level of experience are more likely to share data across the full range of impact factors, while Figure 3b suggests that last authors with a “low” level of experience are less likely to share data.
Figure 3.
Impact factor (IF) vs. probability of data sharing by author experience. Author experience calculated as the first principle component of the author’s h-index, a-index, and number of years since publishing first paper.
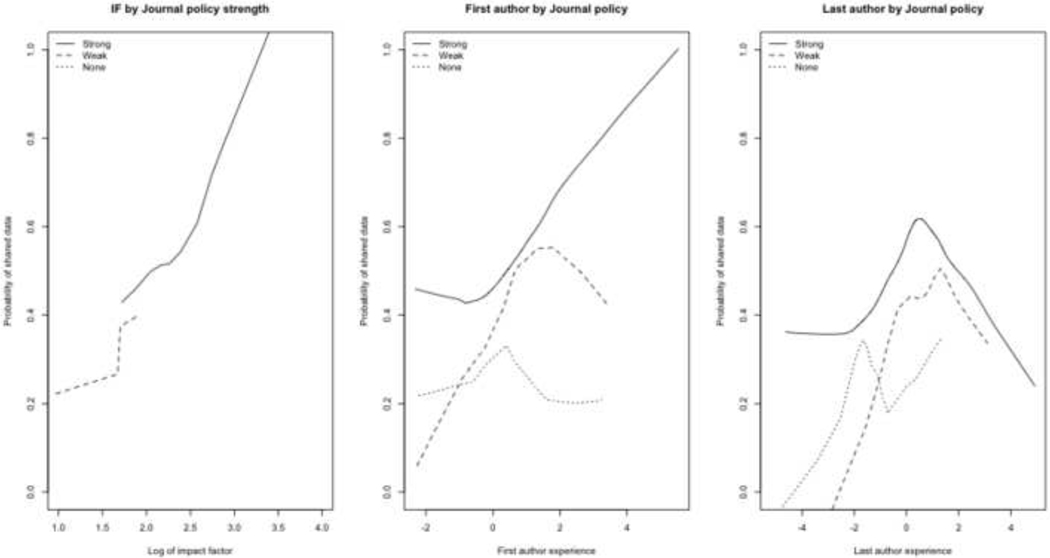
Figure 4.
Impact factor (IF) and author experience by journal policy strength. The first and last author experience values were calculated independently: each has a mean value at 0.0 and a first and third quartile at approximately −1.0 and +1.0, respectively.
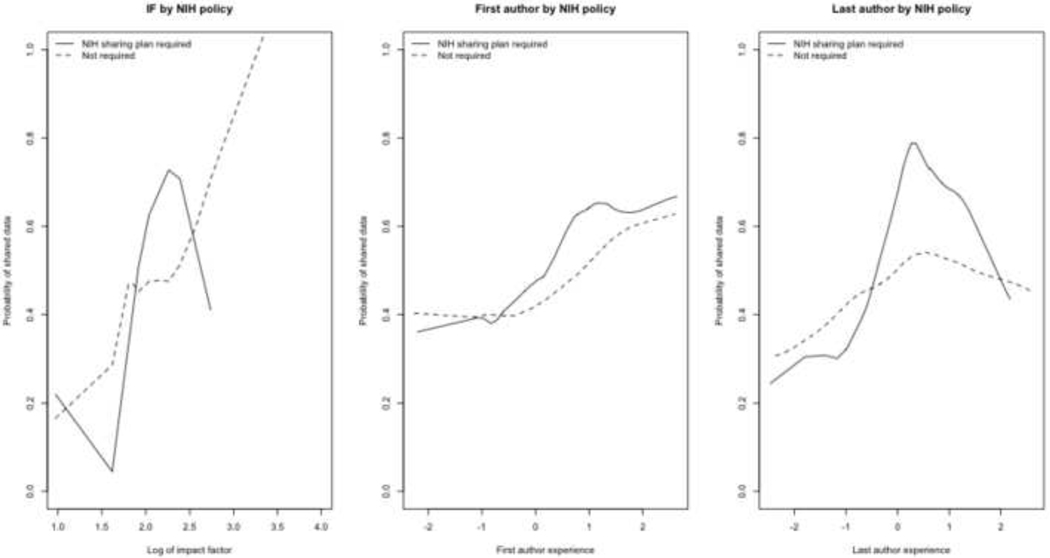
Figure 5.
Impact factor (IF) and author experience by applicability of the NIH’s data sharing policy. The first and last author experience values were calculated independently: each has a mean value at 0.0 and a first and third quartile at approximately −1.0 and +1.0, respectively.
Figure 4 illustrates interactions with journal policy strength. As seen in Figure 4a, impact factor is very strongly associated with journal policy strength. Figures 4b and 4c suggest that Strong journal policies are associated with an increased frequency of data sharing for all levels of first and last author experience, for a given journal impact factor.
Finally, Figure 5 shows the weak association between NIH policy and data sharing. There does not appear to be a systematic trend with impact factor, though there is faint evidence that authors are more likely to respond to an NIH data sharing policy as they become more “experienced” (as the first principal component of their h-index, a-index, and number of years since first publication increases).
4. Discussion
This study explored the association between policy variables, author experience, selected article attributes, and frequency of data sharing within 397 recent gene expression microarray studies. We found that data sharing was more prevalent for studies published in journals with a higher impact factor and by authors with more experience. Whether or not the study was funded by the NIH had little impact on data sharing rates. We estimate the NIH data sharing policy applied to only 19% of the studies, and was not correlated with an increase in data sharing in multivariate analysis. Articles published in journals with a strong data sharing requirement were more likely to share their data, but this trend was not statistically significant in this pilot study.
We note that the associations we have identified do not imply causation. It is possible, for example, that publishing in a high-impact journal and deciding to share data are not causally related but rather both a result of a high level of funding that facilitates impactful research and resources for sharing data.
The data collection and analysis methods used in this pilot study confer additional limitations. Several of our independent variables were highly correlated with one another: future work should cluster correlated variables together to improve robustness of the association estimates. Assumptions about proportions of direct and indirect funding costs may have been incorrect, leading to over- or underestimations of the applicability of NIH mandates. Our focus on measured variables, and particularly the narrow scope of the pilot study, suggest that the given analysis omits many important associations with data sharing. Consequently, our results may suffer from Simpson’s paradox (Simpson, 1951) in which the consideration of additional variables reveals contradictory association patterns.
The results of this pilot study may not be generally applicable. Because articles in this study were drawn from 20 relatively-high impact journals, the results may not accurately characterize data sharing behavior of studies published in low impact journals. It is not known to what degree the data sharing behavior we discovered would generalize to other data types. This pilot study does not consider directed sharing, such as peer-to-peer data exchange or sharing within a defined collaboration network, and thus underestimates the amount of data sharing in all its forms. We only looked at funder policies for the NIH: adherence to the policies of other funders may be different.
It was surprising how seldom the NIH data sharing plan applied to the studies in our cohort, by our estimation. The NIH data sharing requirement stipulates that all proposal submitted to the NIH after October 2003 and requesting more than $500 000 in direct costs per fiscal year must submit a data sharing plan. We did not have access to data that explicitly stated which grants required the submission of a data sharing plan, so we attempted to infer the information based on the grant numbers listed in the study articles and the NIH grants database. Most NIH grants listed in our cohort had been renewed since 2003 (141 of 187 studies for which we had NIH grant information), but only about half of these received more than $750 000 in total costs during any fiscal year (our approximation to $500k in direct costs). Our conclusions about the NIH data sharing policy should be considered preliminary, given this relatively small number of applicable studies included in our current dataset.
The strength of a journal’s data sharing policy was not significantly correlated with data sharing frequency. We believe the trend found by this pilot study would likely strengthen when we widen our dataset to include a larger selection of journals, as found by Piwowar & Chapman (2008). We note that for the most strict journal policies, those that require an accession number for publication, the data sharing frequency is only 51%. Clearly publishers do not enforce their policies very rigorously, especially in lower-impact journals.
Our “author experience” proxy variable was associated with data sharing frequency for both the first and last author (in biomedicine, customarily, the first and last authors make the largest contributions to a study and have the most power in publication decisions). We plan to investigate this finding in greater detail in future work, to isolate whether the correlation between data sharing behavior and “experience” has more to do with author age, tenure status, previous experience in the field, or previous experience sharing data. Although we used h-index and a-index citation metrics in this preliminary investigation, correlation with other citation index variants may be more appropriate for our model and will be explored in future research.
We intend to refine the preliminary work presented here through the collection and analysis of additional data points and variables. We hope that by using text analysis we can automatically identify many more studies that generate gene expression microarray data than the 397 in this sample, and thus investigate additional publication years and journals. Using this expanded set, we plan to analyze several additional variables. Previous work (Blumenthal et al., 1997) found that investigator gender was correlated with data withholding. Given that male scientists are more likely than women to have large NIH grants(Hosek et al., 2005), it is possible that gender is confounding our findings. We hope to estimate investigator gender using first names, perhaps using a web-context tool (http://www.gpeters.com/names/baby-names.php). Also, investigators who have submitted data before may be more likely to do so again. We will investigate this effect by including variables about whether the first or last authors have previously shared microarray datasets. We also hope to explore whether the authors have previously published in open access journals; the size, type, and research orientation of the corresponding author’s institution; and the disease and organism focus of the study.
Although the current work is too preliminary for actionable conclusions, it is instructive to speculate about what observations about policy impact could be drawn in the future, should these results be confirmed. First, it appears that the NIH data sharing policy only applies to minority of the NIH funded studies that generate gene expression microarray data. Second, studies that are required to submit a data sharing plan as a condition of NIH funding seem no more likely to share data than other similar studies. Third, policies that require database submission accession numbers are more effective than those that request or require submission in a non-enforceable manner.
These conclusions would have several policy implications for increasing the effectiveness of data sharing policies. The first suggestion is an expansion of the NIH data sharing policy inclusion criteria, either by lowering the funding amount to which the rules apply or making them applicable to all grants upon a certain milestone, such as publication. The second suggestion is to make policies more enforceable. Whenever there is a requirement for data sharing by a journal or a funder, require that the investigators cite the database submission accession number in their publication or future grant submission correspondence, in similar treatment to the current PubMed Central open-access accession number. Third, we suggest considering the approaches by which high-impact journals achieve such high rates of data sharing compliance: their combination of strict policies, active oversight, and visibility are likely related to the impressive 94% data sharing rate we found for studies published in journals with an impact factor above 15.
5. Conclusions
We believe our emphasis on observed variables facilitates measurement of important quantitative associations. Data sharing policies are controversial (Campbell, 1999; Cecil & Boruch, 1988; King, 1995), and thus deserve to be thoughtfully considered and evaluated. We hope the results from our analyses will contribute to a deeper understanding of information behavior for research data sharing, and eventually more effective data sharing initiatives so that the value of research related output can be most fully realized.
Supplementary Material
Acknowledgments
HAP was supported by NLM training grant 5T15-LM007059-19 and the Department of Biomedical Informatics at the University of Pittsburgh. WWC is funded through NLM grant 5R01-LM009427-0.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anonymous. Microarray standards at last. Nature. 2002;419(6905):323. doi: 10.1038/419323a. [DOI] [PubMed] [Google Scholar]
- Anonymous. Time for leadership. Nature Biotechnology. 2007;25(8):821–821. doi: 10.1038/nbt0807-821. [DOI] [PubMed] [Google Scholar]
- Anonymous. Got data? Nature Neuroscience. 2007;10(8):931–931. doi: 10.1038/nn0807-931. [DOI] [PubMed] [Google Scholar]
- Anonymous. Data’s shameful neglect. Nature. 2009;461(7261):145. doi: 10.1038/461145a. [DOI] [PubMed] [Google Scholar]
- Ball CA, et al. Submission of microarray data to public repositories. PLoS Biology. 2004;2(9):e317. doi: 10.1371/journal.pbio.0020317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett T, et al. NCBI GEO: mining tens of millions of expression profiles--database and tools update. Nucleic Acids Research. 2007;35(Database issue):D760–D765. doi: 10.1093/nar/gkl887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitzer J, Schrettl W, Schröder PJH. Intrinsic motivation in open source software development. Journal of Comparative Economics. 2007;35(1):160–169. [Google Scholar]
- Blumenthal D, et al. Withholding research results in academic life science. Evidence from a national survey of faculty. Journal of the American Medical Association. 1997;277(15):1224–1228. [PubMed] [Google Scholar]
- Blumenthal D, et al. Data withholding in genetics and the other life sciences: prevalences and predictors. Academic Medicine. 2006;81(2):137–145. doi: 10.1097/00001888-200602000-00006. [DOI] [PubMed] [Google Scholar]
- Bornmann L, Mutz R, Daniel H. Are there better indices for evaluation purposes than the h index? A comparison of nine different variants of theh index using data from biomedicine. Journal of the American Society for Information Science and Technology. 2008;59(5):830–837. [Google Scholar]
- Brazma A, et al. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nature Genetics. 2001;29(4):365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- Brown C. The changing face of scientific discourse: Analysis of genomic and proteomic database usage and acceptance. Journal of the American Society for Information Science and Technology. 2003;54(10):926–938. [Google Scholar]
- Campbell EG, et al. Data withholding in academic genetics: evidence from a national survey. Journal of the American Medical Association. 2002;287(4):473–480. doi: 10.1001/jama.287.4.473. [DOI] [PubMed] [Google Scholar]
- Campbell P. Controversial Proposal on Public Access to Research Data Draws 10,000 Comments. The Chronicle of Higher Education. 1999:A42. [Google Scholar]
- Cech T. Sharing Publication-Related Data and Materials: Responsibilities of Authorship in the Life Sciences. Washington: National Academies Press; 2003. [PubMed] [Google Scholar]
- Cecil JS, Boruch R. Compelled Disclosure of Research Data: An early warning and suggestions for psychologists. Law and Human Behavior. 1988;12(2):181–189. [Google Scholar]
- Constant D, Kiesler S, Sproull L. What’s mine is ours, or is it? A study of attitudes about information sharing. Information Systems Research. 1994;5(4):400–421. [Google Scholar]
- Fienberg SE, Martin ME, Straf ML. Sharing research data. Washington, D.C.: National Academy Press; 1985. [Google Scholar]
- Giordano R. The Scientist: Secretive, Selfish, or Reticent? A Social Network Analysis. [Accessed: 2009-11-17];E-Social Science 2007, from http://ess.si.umich.edu/papers/paper166.pdf. 2007 (Archived by WebCite® at http://www.webcitation.org/5lMqU0Ceq).
- Herrell FE. Regression Modeling Strategies: with applications to linear models, logistic regression, and survival analysis. New York: Springer; 2001. [Google Scholar]
- Hedstrom M. Producing Archive-Ready Datasets: Compliance, Incentives, and Motivation; IASSIST Conference 2006: Presentations; 2006. [Google Scholar]
- Hedstrom M, Niu J. Research Forum Presentation: Incentives to Create "Archive-Ready" Data: Implications for Archives and Records Management; Society of American Archivists Annual Meeting; 2008. [Google Scholar]
- Hirsch JE. An index to quantify an individual's scientific research output. Proceedings of the National Academy of Sciences. 2005;102(46):16569–16572. doi: 10.1073/pnas.0507655102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosek SD, et al. [Accessed: 2009-11-17];Gender Differences in Major Federal External Grant Programs, from http://www.rand.org/pubs/technical_reports/2005/RAND_TR307.sum.pdf. 2005 (Archived by WebCite® at http://www.webcitation.org/5lMtlPjAj)
- Hrynaszkiewicz I, Altman D. Towards agreement on best practice for publishing raw clinical trial data. Trials. 2009;10(1):17. doi: 10.1186/1745-6215-10-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin B. h-index: an evaluation indicator proposed by scientist. Science Focus. 2006;1:8–9. [Google Scholar]
- Kakazu KK, Cheung LW, Lynne W. The Cancer Biomedical Informatics Grid (caBIG): pioneering an expansive network of information and tools for collaborative cancer research. Hawaii Medical Journal. 2004;63(9):273–275. [PubMed] [Google Scholar]
- Kim J. Motivating and Impeding Factors Affecting Faculty Contribution to Institutional Repositories. Journal of Digital Information. 2007;8(2) [Google Scholar]
- King G. A Revised Proposal, Proposal. PS: Political Science and Politics. 1995;XXVIII(3):443–499. [Google Scholar]
- Kuo F, Young M. A study of the intention–action gap in knowledge sharing practices. Journal of the American Society for Information Science and Technology. 2008;59(8):1224–1237. [Google Scholar]
- Kyzas PA, Loizou KT, Ioannidis JP. Selective reporting biases in cancer prognostic factor studies. Journal of the National Cancer Institute. 2005;97(14):1043–1055. doi: 10.1093/jnci/dji184. [DOI] [PubMed] [Google Scholar]
- Lee CP, Dourish P, Mark G. The human infrastructure of cyberinfrastructure; Proceedings of the 2006 20th anniversary conference on Computer Supported Cooperative Work; 2006. [Google Scholar]
- Lowrance W. [Accessed: 2009-11-17];Access to Collections of Data and Materials for Heath Research: A report to the Medical Research Council and the Wellcome Trust, from http://www.wellcome.ac.uk/stellent/groups/corporatesite/@msh_grants/documents/web_document/wtx030842.pdf. 2006 (Archived by WebCite® at http://www.webcitation.org/5lMu93MZe)
- Matzler K, et al. Personality traits and knowledge sharing. Journal of Economic Psychology. 2008;29(3):301–313. [Google Scholar]
- McCain K. Mandating Sharing: Journal Policies in the Natural Sciences. Science Communication. 1995;16(4):403–431. [Google Scholar]
- McCullough BD, McGeary KA, Harrison TD. Do Economics Journal Archives Promote Replicable Research? Canadian Journal of Economics. 2008;41(4):1406–1420. [Google Scholar]
- NIH. NOT-OD-03-032: Final NIH Statement on Sharing Research Data. 2003
- NIH. NOT-OD-08-013: Implementation Guidance and Instructions for Applicants: Policy for Sharing of Data Obtained in NIH-Supported or Conducted Genome-Wide Association Studies (GWAS) 2007
- Niu J. [Accessed: 2009-11-17];Incentive study for research data sharing. A case study on NIJ grantees, from http://icd.si.umich.edu/twiki/pub/ICD/LabGroup/fieldpaper_6_25.pdf. 2006 (Archived by WebCite® at http://www.webcitation.org/5lMuGFvSy)
- Noor MA, Zimmerman KJ, Teeter KC. Data Sharing: How Much Doesn't Get Submitted to GenBank? PLoS Biology. 2006;4(7):e228. doi: 10.1371/journal.pbio.0040228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochsner SA, et al. Much room for improvement in deposition rates of expression microarray datasets. Nature Methods. 2008;5(12):991. doi: 10.1038/nmeth1208-991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson H, et al. ArrayExpress--a public database of microarray experiments and gene expression profiles. Nucleic Acids Research. 2007;35(Database issue):D747–D750. doi: 10.1093/nar/gkl995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piwowar HA, Chapman WW. A review of journal policies for sharing research data; International Conference on Electronic Publishing (ELPUB); 2008. [Google Scholar]
- Reidpath DD, Allotey PA. Data sharing in medical research: an empirical investigation. Bioethics. 2001;15(2):125–134. doi: 10.1111/1467-8519.00220. [DOI] [PubMed] [Google Scholar]
- Rhodes DR, et al. Large-scale meta-analysis of cancer microarray data identifies common transcriptional profiles of neoplastic transformation and progression. Proceedings of the National Academy of Sciences. 2004;101(25):9309–9314. doi: 10.1073/pnas.0401994101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu S, Ho SH, Han I. Knowledge sharing behavior of physicians in hospitals. Expert Systems With Applications. 2003;25(1):113–122. [Google Scholar]
- Seonghee K, Boryung J. An analysis of faculty perceptions: Attitudes toward knowledge sharing and collaboration in an academic institution. Library. 2008;30(4):282–290. [Google Scholar]
- Schofield PN, et al. Post-publication sharing of data and tools. Nature. 2009;461(7261):171. doi: 10.1038/461171a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson EH. The interpretation of interaction in contingency tables. Journal of the Royal Statistical Society. Series B (Methodological) 1951:238–241. [Google Scholar]
- The GAIN Collaborative Research Group. New models of collaboration in genome-wide association studies: the Genetic Association Information Network. Nature Genetics. 2007;39(9):1045–1051. doi: 10.1038/ng2127. [DOI] [PubMed] [Google Scholar]
- Torvik V, Smalheiser N. Author Name Disambiguation in MEDLINE. ACM Transactions on Knowledge Discovery from Data. 2009;3(3):11. doi: 10.1145/1552303.1552304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torvik V, et al. A probabilistic similarity metric for Medline records: A model for author name disambiguation. Journal of the American Society for Information Science and Technology. 2005;56(2):140–158. [Google Scholar]
- [Accessed: 2009-11-17];University of Nottingham JULIET: Research funders' open access policies, from http://www.sherpa.ac.uk/juliet/index.php. (Archived by WebCite® at http://www.webcitation.org/5lMuVgcOk)
- Ventura B. Mandatory submission of microarray data to public repositories: how is it working? Physiological Genomics. 2005;20(2):153–156. doi: 10.1152/physiolgenomics.00264.2004. [DOI] [PubMed] [Google Scholar]
- Warlick SE, Vaughan KT. Factors influencing publication choice: why faculty choose open access. Biomedical Digitital Libraries. 2007;4(1):1. doi: 10.1186/1742-5581-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Statistical scripts and the raw dataset are included as supplemental data and are also available at www.researchremix.org.