Abstract
The Comoros Islands are situated off the coast of East Africa, at the northern entrance of the channel of Mozambique. Contemporary Comoros society displays linguistic, cultural and religious features that are indicators of interactions between African, Middle Eastern and Southeast Asian (SEA) populations. Influences came from the north, brought by the Arab and Persian traders whose maritime routes extended to Madagascar by 700–900 AD. Influences also came from the Far East, with the long-distance colonisation by Austronesian seafarers that reached Madagascar 1500 years ago. Indeed, strong genetic evidence for a SEA, but not a Middle Eastern, contribution has been found on Madagascar, but no genetic trace of either migration has been shown to exist in mainland Africa. Studying genetic diversity on the Comoros Islands could therefore provide new insights into human movement in the Indian Ocean. Here, we describe Y chromosomal and mitochondrial genetic variation in 577 Comorian islanders. We have defined 28 Y chromosomal and 9 mitochondrial lineages. We show the Comoros population to be a genetic mosaic, the result of tripartite gene flow from Africa, the Middle East and Southeast Asia. A distinctive profile of African haplogroups, shared with Madagascar, may be characteristic of coastal sub-Saharan East Africa. Finally, the absence of any maternal contribution from Western Eurasia strongly implicates male-dominated trade and religion as the drivers of gene flow from the North. The Comoros provides a first view of the genetic makeup of coastal East Africa.
Keywords: Y chromosome, mitochondrion, Indian Ocean, East Africa, Comoros
Introduction
The Indian Ocean can be considered as a closed sea, an afro-Asiatic Mediterranean,1, 2 around which populations have migrated and mixed. In contrast to the Atlantic Ocean, which was a formidable natural barrier to East–West migration, the Indian Ocean with its seasonal monsoon winds favoured such exchanges, and most of the early trade routes were maritime. The Comoros archipelago is situated in the western Indian Ocean, midway between the island of Madagascar and the coast of East Africa at the northern end of the Mozambique Channel. The archipelago is composed of four main islands: Grand Comore (Ngazidja), Anjouan (Ndzuani), Mohéli (Mwali) and Mayotte (Maore). The settlement of the four islands was an integral part of migration within the Indian Ocean, as they represent a potential maritime crossroads and juncture, between Bantu African, Middle Eastern and Southeast Asian (SEA) spheres of influence. The modern Comorian population is the result of a long-term process of biocultural admixture, mainly related to ancient trade and colonisation in the Indian Ocean.
The Comoros and Madagascar share obvious signs of SEA influence including the cultivation of rice (phased out during twentieth century), bananas and coconuts, and the use of outrigger canoes. Evidence from plant translocation suggests a migration from SEA 1500 Years Before Present (YBP).1, 3, 4 Clear genetic evidence for the SEA influence has been found on neighbouring Madagascar.5, 6, 7, 8 On the basis of Y chromosome and mitochondrial variation, ethnic groups with the strongest SEA biocultural features in Madagascar were estimated to have approximately 50% SEA ancestry.5, 8 In contrast to Madagascar where the language, Malagasy, is an Austronesian language with origins in SEA, the languages spoken on the Comoros are of Bantu origin. They are distinct from, but have close affinity to, Swahili, both branching from the precursor Sabaki language, 1000–2000 YBP.9
The cultural contributions of Middle Eastern civilisation are equally evident on the Islands. By 2000 YBP, a thriving commercial maritime network already existed, extending from the Middle East to India, and as far South as Tanzania on the East African coast. The name ‘Comoros' is from the Arabic Kmr, meaning ‘light in the sky'.3 From 1300 YBP, the Comoros archipelago served as a stepping stone, for Middle Eastern traders operating along the East African coast, and for SEA traders travelling to Madagascar and the East African coast.10, 11 By 1000 YBP, the Shirazi, traders with origins in the Persian city of Shiraz in present day Iran, had established themselves on the island of Kilwa. The Shirazi were responsible for the generalisation of Islam on the Swahili coast by 500 YBP. They had built mosques on Kilwa, Zanzibar and Anjouan by 800 YBP.12 Islam remains the religion of the Islands today.
An unambiguous genetic signal from the Middle East has not, however, been detected in East Africa further south than Ethiopia,13, 14 or in the ethnic groups sampled on Madagascar.5, 8 The Lemba people of South Africa, carrying a putative semitic Y chromosome, currently provide the only evidence for gene flow from the Middle East into southern Africa.15 The alleles of some autosomal genes found in the ex-patriot Comorian population living in Marseilles indicate a genetic contribution from Western Eurasia,16, 17, 18 but the populations living on the Comoros have until now not been studied.
In this context, the peopling of the Comoros is evidently integral to the movements of men and women across the entire Indian Ocean. To gain insights into this process, we therefore determined the Y chromosomal and mitochondrial genetic variation on the three Bantu-speaking islands of the Comoros Republic.
Materials and methods
Sample group
In February and March 2006, we obtained blood samples from 577 unrelated Comorian men (n=381) and women (n=196). We sampled the populations of three of the four islands of the Comoros archipelago (Grand Comore – 170 men, 67 women; Anjouan – 104 men, 69 women; and Moheli – 107 men, 60 women). In 2006, this represented approximately 0.1% of the total Comoros population of 690 000 people. Blood was collected in EDTA vacutainer tubes and DNA extracted using the salting-out method.19 Samples were collected from multiple towns and villages on each island (Supplementary Figure 1). Recruitment was achieved through contacts established by medical personnel who originated from each community sampled. Each donor included in the study had four grandparents who were born in the same island and were native speakers of the island's language (Shingazidja, Shindzuani or Shimwali). Informed consent was obtained from all participants.
Y-chromosome haplogroups and haplotypes
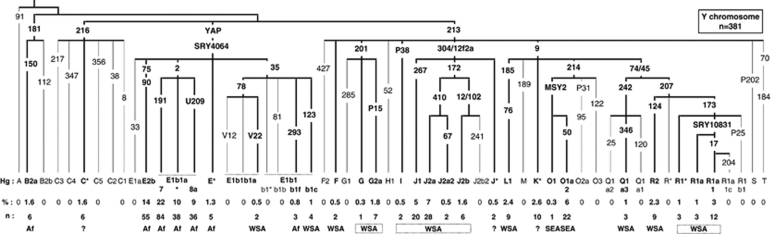
We typed 68 binary polymorphisms mainly by PCR-RFLP (Figure 1 and Supplementary Table 1). For 293 Y chromosomes, alleles of 17 short tandem repeat (STR) polymorphisms on the Y chromosome were amplified with the AmpFlSTR Yfiler PCR Amplification Kit (Applied Biosystems, Foster City, CA, USA). Y-STR haplotypes were determined for 15 of 38 E-M2(xM191,U209) and 19 of 84 E-M191 chromosomes, and for all other chromosomes (Supplementary Table 2).
Figure 1.
Frequencies (%) and numbers (n) of Y haplogroups in the Comoros population sample. Haplogroup names follow the 2008 nomenclature.20 Branches are labelled with the binary markers tested. Numbers without a letter represent ‘M' prefixed Y markers (eg, 50=M50).21 Putative geographic origin is indicated for each haplogroup: Af – sub-Saharan Africa, WSA – West and Southwest Asia, SEA – Southeast Asia and ? – uncertain. Frequencies of less than 5% have been rounded up or down to the nearest unit.
Mitochondrial haplogroups
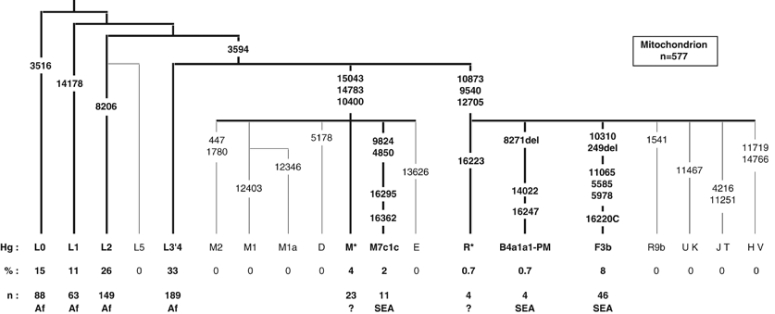
We typed 31 coding region polymorphisms in the mitochondrial genome mainly by PCR-RFLP (Figure 3, Supplementary Table 3). We also sequenced a 501-bp fragment, including HVS-I from all M, N(xR) and R samples (Supplementary Table 4 and GenBank: HM565257-HM565275). Choice of markers and branch designations was based on published data22 and trees presented at http://www.ianlogan.co.uk.
Data analysis
The genetic structure (haplogroup number, haplogroup diversity, population differentiation, Fst and Rst) of the study population sample was analysed using the ARLEQUIN package v. 3.01,23 and phylogenetic comparisons of Y-STR haplotypes and mitochondrial SNP haplogroups were examined by multidimensional scaling (MDS), based, respectively, on Rst or Fst distances,24, 25 using SPSS 10.00 software (Chicago, IL, USA). Admixture fractions were estimated using ADMIX 2.0.26 On the basis of our haplogroup and MDS analyses, and historical and linguistic data, we chose Borneo, Iran and East Africa (Y: Kenya and Tanzania and mitochondrial: Mozambique) as the most likely parent populations from published data.5, 13, 27, 28, 29, 30
Results
Y-chromosome diversity on the Comoros
We analysed 381 Y chromosomes from the Comoros and identified 28 distinct haplogroups belonging to 11 of the 20 major clades of the Y-chromosome tree as shown below (Figure 1).20 These fall into four groups, on the basis of the geographical distribution of haplogroups around the Indian Ocean: sub-Saharan African 59.6% Western and Southern Asia 29.7% Southeast Asia 6% and uncertain origin 4.7%. Four clades, E, J, O and R, have frequencies greater than 5% and represent 87.4% of the sample.
The paragroups C*(xC1-5), F*(xM282, M427), J* and K*(xLMNOPQRST) cannot be assigned an origin with certainty. Nevertheless, the high frequencies of C*-M216 (Borneo – 2.5–25%) and K* (2–30%),5, 31 in SEA, make an SEA origin probable. J* has been found in Bali (1.5%) but also on the island of Soqotra (71%) situated in the Gulf of Oman between Somalia and Yemen.32, 33 F*(xM282,M427) has been found mainly in the Indian subcontinent.34 A West or Southwest Asian origin is therefore more likely for the F* and J* chromosomes.
Y-STR analysis revealed a generally high variance (Table 1), which coupled with the large number of Y haplogroups, suggesting that genetic drift has not drastically reduced genetic diversity on the Comoros Islands.
Table 1. Variance of the principal Y haplogroups (n≥5) on the Comoros based on 15 Y-microsatellite loci.
| Haplogroup | Mutation | n | Variance |
|---|---|---|---|
| B2a | M150 | 6 | 1.01 |
| C* | M216 | 6 | 0.44 |
| E* | SRY4064 | 5 | 1.03 |
| E1b1a* | M2 | 15 | 0.46 |
| E1b1a7 | M191 | 19 | 1.13 |
| E1b1a8a | U209 | 36 | 0.66 |
| E2 | M75 | 14 | 0.75 |
| G2a | P15 | 7 | 0.10 |
| J1 | M267 | 20 | 0.54 |
| J2a | M410 | 28 | 0.60 |
| J2b | M12 | 6 | 0.57 |
| K* | M9 | 10 | 1.16 |
| L1 | M76 | 9 | 1.05 |
| O1a2 | M50 | 22 | 0.30 |
| R1a1 | M17 | 12 | 0.77 |
| R2 | M124 | 9 | 1.18 |
The markers DYS385a and b were not used in the calculation. The variance is calculated from STR data for all individuals of each haplogroup (n), except E1b1a7 (19 of 84 men) and E1b1a* (15 of 38 men).
Sub-Saharan African Y chromosomes
The most common Comorian haplogroups, E1b1-M2 (41%) and E2-M90 (14%), are those that are frequent in sub-Saharan Africa.13, 35, 36, 37, 38 They are present, respectively, at 56 and 6.4%, in Madagascar.8 Two haplogroups were identified under E1b1-M2, derived for markers M191 (22%) and U209 (9%). The haplogroup E1b1a-M191 has been found in east and west sub-Saharan Africa, 19% in Tanzania and 57% in Benin.13 The marker U209 was identified in Afro-Americans,39 and has not, until now, been tested for in African populations.
The low incidence of E-M293 (0.8%) and A-M91 (0%) on the Comoros contrasts strongly with the frequency of these haplogroups in East African populations. E-M293 is found mainly in East Africa, Kenya and Tanzania (18%).40 Furthermore, on the African mainland, M293 chromosomes carry either 10, or 13 and more repeats at the DYS389I STR locus,40 whereas on the Comoros, they have 12 repeats. Haplogroup A has a frequency of 14% in Kenyan Bantu and 7% in Tanzania.13
Other haplogroups of likely sub-Saharan African origin on the Comoros are E-SRY4064(xM2,M35,M75) (1.3%) and B2a (1.6%). B2a has a low frequency in southern Iran and Qatar,29, 41 but this is thought to be a consequence of the Arab slave trade. We therefore treat B2a as an African chromosome in this study.
Y chromosomes from around the Arabian Sea
The northern Y chromosomes on the Comoros, E-V22, E-M123, F*(xF2, GHIJK), G2a, I, J1, J2, L1, Q1a3, R1*, R1a*, R1a1 and R2 (29.7%), make up a diverse group. G2a, J1 and J2 (16.5%) are thought to have originated in the Middle East.14, 42 J1-M267 has mainly spread south and west into the Arabic Peninsula, and into North and Northeast Africa, whereas J2-M172 lineages have expanded north into Europe and east into Asia.13, 14, 41, 43, 44, 45 The M78 subclade, E-V22, and E-M123 are believed to have originated in Northeast Africa, with E-V22 spreading to the west of North Africa and to the Arabic peninsula by the Levantine corridor (United Arab Emirates (UAE) 6.7%),41, 46 whereas M123 spread mainly to the East (Yemen 8% Oman 12% Turkey 5.5% Iran 1%).13, 29, 41, 42 In contrast, the haplogroups L1, Q1a3, R1, R1a, R1a1 and R2 (10.5%) are thought to be of Central or Southern Asian origin and describe clines of decreasing frequency from India and Pakistan towards the Middle East.34
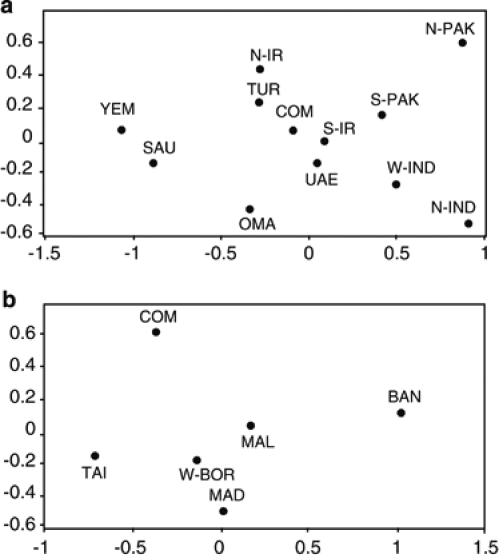
A comparison of the relative incidences of E-M78(V22), E-M123, G, J, L, Q and R on the Comoros with populations around the Arabian Sea shows greatest similarities with Southern Iran and, to a lesser extent, Turkey (Supplementary Figure 2).29, 42 The higher affinity to South Iran is also evident in the MDS analysis with the Comoros Y-STR data for the E-V22, E-M123, G, J, L Q and R haplogroups (Figure 2a). In the MDS, Comoros shows greatest affinity with UAE and South Iran. Southern Iran is the site of the first towns to develop in the Southern Middle East 2000–3000 years ago (Supplementary Figure 2).
Figure 2.
Multidimensional scaling (MDS) analysis plot of genetic distance (Rst) calculated from the incidence of alleles at eight Y-STR loci (DYS19, 389AB, 389CD, 390, 391, 392, 393, 439). The analysis was performed with subsets of the Comoros sample, which were created on the basis of putative haplogroup origin. (a) Middle East – haplogroups E-M123, E-V22, F, G, J, L, Q and R. (b) Southeast Asian – haplogroups O, C* and K*. The populations represented are the Comoros (COM), this study, Madagascar (MAD),8 Oman (OMA),13, 47 Turkey (TUR),42 North Pakistan (N-PAK), South Pakistan (S-PAK), North India (N-IND), South India (S-IND),34 Yemen (YEM), United Arab Emirates (UAE), Saudi Arabia (SAU),47 North Iran (N-IR),48 South Iran (S-IR),47, 48 Malaysia (MAL),49 Taiwan (Paiwan) (TAI),50 West Borneo (East Malaysia) (W-BOR)51 and Bangladesh (BAN).52
A possible source of the Northern Y chromosomes is therefore the Shirazi traders from Southern Iran who established trading posts on the Comoros by 800 YBP.12 It has previously been estimated that, at 9 Y-STR loci, 0–1 mutation will most likely separate the descendants of a single Y-chromosome haplotype after 40 generations (1000–1200 years).53, 54 Compatible with a Shirazi origin, we found that, at 9 Y-STR loci (DYS19, 389AB, 389CD, 390–393, 438 and 439), 42% of the Comoros Northern chromosomes differ by 0–1 mutation from chromosomes in Southern Iran.47, 48
SEA Y chromosomes
We found the O1 lineage (6%) in the Comoros sample, providing genetic evidence for an SEA influence. Haplogroup O has been found at highest frequencies in East Asia and Island Southeast Asia.55, 56 All but one of the Comorian O1 chromosomes are O1a-M50 (5.8%). The O1a-M50 Y chromosome has its highest incidence in SEA: Borneo (10–20%), Sulawesi (4%), Taiwanese aborigines (0–59%, mean 14%) and the Philippines (3–12%).5, 57, 58 It has not been detected in the Middle East or the Indian subcontinent.5, 29, 41, 34
We performed an MDS with our STR data for the Y haplogroups O, C* and K* together with available STR data from candidate SEA populations (Figure 2b). The Comoros show a low affinity to the populations selected, even when C* and K* are not included (not shown), suggesting that these populations are not the source of SEA chromosomes on the Comoros.
Mitochondrial diversity on the Comoros
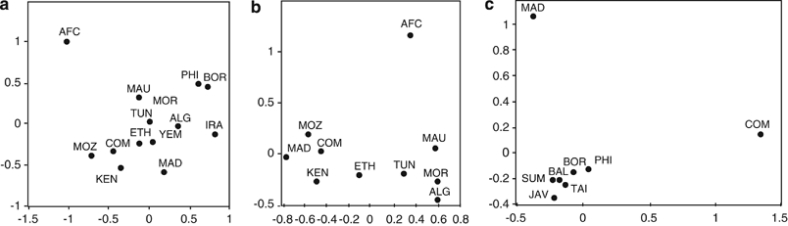
We have tested 577 Comorian samples for mitochondrial SNPs, and we define 9 distinct haplogroups (Figure 3). As for the Y chromosome, the majority of mitochondrial haplogroups on the Comoros are of African origin. The haplogroups L0, L1, L2 and L3′4(xMN) compose 84.7% of the mitochondria in the Comoros sample, and their relative proportions are most similar to profiles found in East and South East Africa.22, 59 The higher affinity with sub-Saharan East African populations is also evident in the MDS analysis (Figure 4a and b).
Figure 3.
Frequencies (%) and numbers (n) of mitochondrial haplogroups in the Comoros population sample. Numbers on branches refer to the position of polymorphisms in the CRS (Cambridge reference sequence). HVS-I sequence was not determined for L0, L1, L2 or L3′4(xMN). The HVS-I SNPs are shown for M and N haplogroups, only where they provide further definition than the coding SNPs. Putative geographic origin is indicated for each haplogroup: Af – sub-Saharan Africa, SEA – Southeast Asia and ? – uncertain.
Figure 4.
Multidimensional scaling (MDS) analysis plot of genetic distance (Fst) calculated from mitochondrial haplogroup frequencies. M* and R* were excluded from these analyses. (a) Africa, SEA and Iran – all Comoros haplogroups, except M* and R*. (b) and (c) MDS performed with subsets of the Comoros sample, defined on the basis of putative haplogroup origin. (b) Africa – Comoros haplogroups L. (c) SEA – Comoros and Madagascar haplogroups B4a, B4a1a1-PM, F3b, M7c1c and R9. The populations are Comoros (COM), this study, Madagascar (MAD),8 Central Africa (AFC),59 Iran (IRA), Mozambique (MOZ), Kenya (KEN),60 Ethopia (ETH),61 Tunisia (TUN), Algeria (ALG), Morocco (MOR), Mauritania (MAU),62 Taiwan (TAI), Philippines (PHI), Malaysia (MAL), Borneo (BOR), Sumatra (SUM), Bali (BAL) and Java (JAV).27
The remaining 15.3% of the Comoros sample is composed almost exclusively of haplogroups that can either be unambiguously identified as SEA (B4a1a1-PM, F3b and M7c1c – 10.6%),27 or fall into the paragroup M(xD, E, M1, M2, M7) (4%) (Figure 3). The latter haplogroups are probably also originally from Southeast Asia, but of the 12 different M* HVS-I sequences on the Comoros, only two match published sequences: two M(xM7) mitochondria found on Madagascar.8 We found no haplogroups that could be assigned to the Middle East.
SEA mitochondria
Of the SEA mitochondrial haplogroups present on the Comoros, F3b and M7c1c, similar to the Y-Hg O1-M50, each define an area of distribution that extends from Taiwan through the Philippines to Borneo27 (Supplementary Figure 3). The MDS analysis with SEA populations shows greatest affinity to the Philippines and Borneo, although affinity is relatively weak (Figure 4c). Linguistic studies indicate Southeast Borneo to be the probable origin of the migration from SEA to Madagascar.63 B4a1a1-PM (0.7%) is the major haplogroup throughout Polynesia (78%),31 and on Madagascar (25%), but, within island SEA, it has not been found further West than South Borneo (1%).5, 7, 8
Male-biased gene flow from the Middle East
There are no mitochondrial lineages on the Comoros that are frequent in the Middle East (Figure 3). We have tested for, but did not find, the R haplogroups, H, J, T, U and V, or N(xR) that represent 80% of the mitochondria in Iran.60 There is therefore striking evidence for male-biased gene flow from the Middle East to the Comoros, even if the unassigned mt-Hg M* and R* are designated as western Asian: 103/381 Y vs 27/577 mitochondria – Fisher's exact test, one-sided, P<10−22. This is entirely consistent with male-dominated trade and religious proselytisation being the forces that drove the Middle Eastern gene flow to the Comoros. For African and SEA contributions, if Y haplogroups C* and K* are counted as SEA, the under representation of the male lineages are similar (Y to mt ratio: Africa 0.69, SEA 0.66).
An opposite female gene flow from Africa to the Middle East is clearly evident in Yemen (34% mt-Hg L; 4% Y-Hg E-M2), Iraq and the Levant.64 However, no mt-Hg L has been found in Iran (n=712),60 despite the presence of Y-Hg E-M2 (1.7%),29 supporting the idea that the elevated mt-Hg L frequency in the western Middle East is not exclusively a consequence of the Arab slave trade, but also of geography.61
Discussion
We reveal the Comoros population to be a genetic mosaic, the result of tripartite gene flow from the North, the East and the West. Admixture analysis of the maternal and paternal contributions indicates the gene pool to be predominately African (72%), with significant contributions from Western Asia (17%) and Southeast Asia (11%). Our study therefore provides the first unequivocal evidence that the Middle Eastern trade routes that developed along the East African coast, during the last 2000 years, have left a genetic trace. Male and female SEA gene flow has already been described on Madagascar, in populations that speak Austronesian languages,5, 8 but here we show that this extends beyond Madagascar, into African populations speaking languages from the Bantu family. This raises the question of whether the demic migration from SEA reached the East African mainland.
The frequencies of Y-Hg E-V22, E-M123, G2a, J1, J2, R1a1 and R2 in the Comorian sample are compatible with gene flow from Iran.29, 34 This concords with historical data, which attests to the presence of traders from Shiraz in Iran on the Comoros, and also the Comorian's own oral traditions, which recount that Shirazi princes came in ships and established colonies on the islands. On the island of Anjouan the term ‘Shirazi' is used to refer to someone of Middle Eastern appearance. There is historical evidence that 1000 YBP Persian traders had an important role in trade along the East coast, and we therefore predict that an Iranian genetic signal will be detected among Swahili speakers at former Middle Eastern trading centres on the sub-Saharan East coast, such as the islands of Zanzibar and Kilwa off the coast of Tanzania.
Interestingly, there are a number of similarities between the genetic profile of the Comoros islanders and the Lemba of South Africa, a Bantu speaking people whose Semitic origins are evident at both the cultural and genetic level.15, 65 The Lemba have high frequencies of the Middle Eastern Y-chromosome HgJ-12f2a (25%), a potentially SEA Y, Hg-K(xPQR) (32%) and a Bantu Y, E-PN1 (30%) (similar to E-M2), raising the possibility that the Lemba and Comorian populations are consequences of similar demographic processes. The high-resolution genotyping of the Lemba Y chromosomes and mitochondria will elucidate this question.
The Comoros and Madagascar show similarities in the paternal and maternal contribution from SEA and Africa. The absence of a strong Middle Eastern signal on Madagascar could be due to sampling bias, as Arab or Persian traders are known to have established posts on the Northwest coast of Madagascar, whereas only populations from the centre and South of Madagascar have been studied to date.5, 8 The low frequencies of E-M293 and A-M91, on both the Comoros and Madagascar, contrasts with the high frequency found in inland populations from Tanzania and Kenya,13, 40 and could be characteristics of a genetic profile specific to sub-Saharan coastal East Africa.
The SEA haplogroups, shared with Madagascar, on the Comoros are O1a-M50 for the Y chromosome and M7c1c, F3b and B4a1a1-PM for the mitochondria. Consistent with their transit West across the Indian Ocean to the East African coast, O1a-M50, M7c1c and F3b are linked to maritime colonisation within island SEA. In contrast, B4a1a1-PM has not been found in island SEA further West than Southeast Borneo (1%) and has expanded mainly East into Polynesia, but also West to Madagascar where it predominates.27, 57 There are nevertheless several indicators that the Comoros' history of gene flow from SEA is distinct from Madagascar's: the absence of Y HgO2-M95 and the very low frequency of B4a1a1-PM, on the Comoros, the higher frequency of F3b (Comoros 8% Madagascar 3.7%), the dissimilarity of M* HVS-I sequences and the low affinity between the Comoros' O1a-M50 chromosomes and those of Madagascar. The fact that O1a-M50, M7c1c, F3b and B4a1a1-PM have not been found at sites around the Indian Ocean,28, 29, 41, 34, 60 outside SEA and now East Africa, is consistent with a colonising migration from SEA to East Africa directly across the Indian Ocean.
The Comoros population represents an exceptionally diverse genetic mosaic created by the complex process of human settlement around the Indian Ocean. The genealogy of our large sample is well documented and will provide a solid base from which to explore human diversity in Madagascar and coastal sub-Saharan East Africa.
Acknowledgments
We express our sincere gratitude to the people of the Comoros who participated in this study, and the medical personnel of the Comoros and the World Health Organization for sample collection, and in particular Dr Said Ahamada Fazul, Dr Islam Abdallah, Mr Ismail Msaidié, Mrs Wardat Abdoulkarim, Mme Oumrati Haribou, Mr A Djazza and Mr A.I Abdallah. We thank the EFS (French Blood Transfusion Service) Alpes-méditerranée for their support. We further thank Professor Nicolas Levy, Dr Catherine Badens, Caroline Lacoste, Julie di Cristofaro, Marie-Claude Bonino Fabien Ciné and Nathalie Eudes for help with genotyping. AD was supported by a MENRT post-graduate studentship from the French Government and a Fyssen post-doctoral fellowship. SM was supported by a studentship ‘Coopération France-Comores' given jointly from the French and the Comoros governments. This work was supported by the CNRS grant ‘OHLL – Origines de l'Homme, des langues et du language' and core funding from Inserm to MJM.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Beaujard P. The Indian Ocean in Eurasian and African World-Systems before the Sixteenth Century. J World Hist. 2005;16:411–465. [Google Scholar]
- Chaudhuri KN. Trade and Civilisation in the Indian Ocean: An Economic History from the Rise of Islam to 1750. Cambridge: Cambridge University Press; 1985. [Google Scholar]
- Allibert C. Le mot ‘ KOMR' dans l'océan indien. Etudes Océan Indien. 2001;31:13–33. [Google Scholar]
- Blench R.The ethnographic evidence for long-distance contacts between Oceania and East Africain Reade J (ed).: The Indian Ocean in Antiquity London, New York: Kegan Paul, British Museum; 1994461–470. [Google Scholar]
- Hurles ME, Sykes BC, Jobling MA, Forster P. The dual origin of the Malagasy in Island Southeast Asia and East Africa: evidence from maternal and paternal lineages. Am J Hum Genet. 2005;76:894–901. doi: 10.1086/430051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regueiro M, Mirabal S, Lacau H, Caeiro JL, Garcia-Bertrand RL, Herrera RJ. Austronesian genetic signature in East African Madagascar and Polynesia. J Hum Genet. 2008;53:106–120. doi: 10.1007/s10038-007-0224-4. [DOI] [PubMed] [Google Scholar]
- Soodyall H, Jenkins T, Stoneking M. ‘Polynesian' mtDNA in the Malagasy. Nat Genet. 1995;10:377–378. doi: 10.1038/ng0895-377. [DOI] [PubMed] [Google Scholar]
- Tofanelli S, Bertoncini S, Castri L, et al. On the origins and admixture of Malagasy: new evidence from high resolution analyses of paternal and maternal lineages. Mol Biol Evol. 2009;26:2109–2124. doi: 10.1093/molbev/msp120. [DOI] [PubMed] [Google Scholar]
- Nurse D, Hinnebusch TJ. Swahili and Sabaki: A Linguistic History. Berkeley and Los Angeles: University of California Press; 1993. [Google Scholar]
- Allibert C. Austronesian migration and the establishment of the Malagasy civilization: contrasted readings in linguistics, archaeology, genetics and cultural anthropology. Diogenes. 2008;55:7–16. [Google Scholar]
- Allibert C, Vérin P.The early pre-islamic history of the Comores islands: links with Madagascar and Africain Reade J (ed).: The Indian Ocean in Antiquity London, New York: Kegan Paul International, British Museum; 1996461–470. [Google Scholar]
- Allibert C.La chronique d'Anjouan par Said Ahmed Zakiin Allibert C (ed): Anjouan dans l'Histoire Paris: CEROI-INALCO; 2000. vol 29, pp9–92. [Google Scholar]
- Luis JR, Rowold DJ, Regueiro M, et al. The levant versus the horn of Africa: evidence for bidirectional corridors of human migrations. Am J Hum Genet. 2004;74:532–544. doi: 10.1086/382286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semino O, Magri C, Benuzzi G, et al. Origin, diffusion, and differentiation of Y-chromosome haplogroups E and J: inferences on the neolithization of Europe and later migratory events in the Mediterranean area. Am J Hum Genet. 2004;74:1023–1034. doi: 10.1086/386295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spurdle AB, Jenkins T. The origins of the Lemba ‘Black Jews' of southern Africa: evidence from p12F2 and other Y-chromosome markers. Am J Hum Genet. 1996;59:1126–1133. [PMC free article] [PubMed] [Google Scholar]
- Badens C, Martinez di Montemuros F, Thuret I, et al. Molecular basis of haemoglobinopathies and G6PD deficiency in the Comorian population. Hematol J. 2000;1:264–268. doi: 10.1038/sj.thj.6200042. [DOI] [PubMed] [Google Scholar]
- Chiaroni J, Touinssi M, Mazet M, De Micco P, Ferrera V. Adsorption of autoantibodies in the presence of LISS to detect alloantibodies underlying warm autoantibodies. Transfusion. 2003;43:651–655. doi: 10.1046/j.1537-2995.2003.00356.x. [DOI] [PubMed] [Google Scholar]
- Gibert M, Touinssi M, Reviron D, Mercier P, Boetsch G, Chiaroni J. HLA-DRB1 frequencies of the Comorian population and their genetic affinities with Sub-Saharan African and Indian Oceanian populations. Ann Hum Biol. 2006;33:265–278. doi: 10.1080/03014460600578599. [DOI] [PubMed] [Google Scholar]
- Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karafet TM, Mendez FL, Meilerman MB, Underhill PA, Zegura SL, Hammer MF. New binary polymorphisms reshape and increase resolution of the human Y chromosomal haplogroup tree. Genome Res. 2008;18:830–838. doi: 10.1101/gr.7172008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underhill PA, Passarino G, Lin AA, et al. The phylogeography of Y chromosome binary haplotypes and the origins of modern human populations. Ann Hum Genet. 2001;65:43–62. doi: 10.1046/j.1469-1809.2001.6510043.x. [DOI] [PubMed] [Google Scholar]
- Behar DM, Villems R, Soodyall H, et al. The dawn of human matrilineal diversity. Am J Hum Genet. 2008;82:1130–1140. doi: 10.1016/j.ajhg.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol Bioinform Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Reynolds J, Weir BS, Cockerham CC. Estimation of the coancestry coefficient: basis for a short-term genetic distance. Genetics. 1983;105:767–779. doi: 10.1093/genetics/105.3.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatkin M. A measure of population subdivision based on microsatellite allele frequencies. Genetics. 1995;139:457–462. doi: 10.1093/genetics/139.1.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupanloup I, Bertorelle G. Inferring admixture proportions from molecular data: extension to any number of parental populations. Mol Biol Evol. 2001;18:672–675. doi: 10.1093/oxfordjournals.molbev.a003847. [DOI] [PubMed] [Google Scholar]
- Hill C, Soares P, Mormina M, et al. A mitochondrial stratigraphy for island southeast Asia. Am J Hum Genet. 2007;80:29–43. doi: 10.1086/510412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metspalu M, Kivisild T, Metspalu E, et al. Most of the extant mtDNA boundaries in south and southwest Asia were likely shaped during the initial settlement of Eurasia by anatomically modern humans. BMC Genet. 2004;5:26. doi: 10.1186/1471-2156-5-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regueiro M, Cadenas AM, Gayden T, Underhill PA, Herrera RJ. Iran: tricontinental nexus for Y-chromosome driven migration. Hum Hered. 2006;61:132–143. doi: 10.1159/000093774. [DOI] [PubMed] [Google Scholar]
- Salas A, Richards M, De la Fe T, et al. The making of the African mtDNA landscape. Am J Hum Genet. 2002;71:1082–1111. doi: 10.1086/344348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayser M, Brauer S, Cordaux R, et al. Melanesian and Asian origins of Polynesians: mtDNA and Y chromosome gradients across the Pacific. Mol Biol Evol. 2006;23:2234–2244. doi: 10.1093/molbev/msl093. [DOI] [PubMed] [Google Scholar]
- Karafet TM, Lansing JS, Redd AJ, et al. Balinese Y-chromosome perspective on the peopling of Indonesia: genetic contributions from pre-neolithic hunter-gatherers, Austronesian farmers, and Indian traders. Hum Biol. 2005;77:93–114. doi: 10.1353/hub.2005.0030. [DOI] [PubMed] [Google Scholar]
- Cerny V, Pereira L, Kujanova M, et al. Out of Arabia-The settlement of Island Soqotra as revealed by mitochondrial and Y chromosome genetic diversity. Am J Phys Anthropol. 2008. [DOI] [PubMed]
- Sengupta S, Zhivotovsky LA, King R, et al. Polarity and temporality of high-resolution Y-chromosome distributions in India identify both indigenous and exogenous expansions and reveal minor genetic influence of Central Asian pastoralists. Am J Hum Genet. 2006;78:202–221. doi: 10.1086/499411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruciani F, Santolamazza P, Shen P, et al. A back migration from Asia to sub-Saharan Africa is supported by high-resolution analysis of human Y-chromosome haplotypes. Am J Hum Genet. 2002;70:1197–1214. doi: 10.1086/340257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passarino G, Semino O, Quintana-Murci L, Excoffier L, Hammer M, Santachiara-Benerecetti AS. Different genetic components in the Ethiopian population, identified by mtDNA and Y-chromosome polymorphisms. Am J Hum Genet. 1998;62:420–434. doi: 10.1086/301702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scozzari R, Cruciani F, Santolamazza P, et al. Combined use of biallelic and microsatellite Y-chromosome polymorphisms to infer affinities among African populations. Am J Hum Genet. 1999;65:829–846. doi: 10.1086/302538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underhill PA, Shen P, Lin AA, et al. Y chromosome sequence variation and the history of human populations. Nat Genet. 2000;26:358–361. doi: 10.1038/81685. [DOI] [PubMed] [Google Scholar]
- Sims LM, Garvey D, Ballantyne J. Sub-populations within the major European and African derived haplogroups R1b3 and E3a are differentiated by previously phylogenetically undefined Y-SNPs. Hum Mutat. 2007;28:97. doi: 10.1002/humu.9469. [DOI] [PubMed] [Google Scholar]
- Henn BM, Gignoux C, Lin AA, et al. Y-chromosomal evidence of a pastoralist migration through Tanzania to southern Africa. Proc Natl Acad Sci USA. 2008;105:10693–10698. doi: 10.1073/pnas.0801184105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadenas AM, Zhivotovsky LA, Cavalli-Sforza LL, Underhill PA, Herrera RJ. Y-chromosome diversity characterizes the Gulf of Oman. Eur J Hum Genet. 2008;16:374–386. doi: 10.1038/sj.ejhg.5201934. [DOI] [PubMed] [Google Scholar]
- Cinnioglu C, King R, Kivisild T, et al. Excavating Y-chromosome haplotype strata in Anatolia. Hum Genet. 2004;114:127–148. doi: 10.1007/s00439-003-1031-4. [DOI] [PubMed] [Google Scholar]
- Arredi B, Poloni ES, Paracchini S, et al. A predominantly neolithic origin for Y-chromosomal DNA variation in North Africa. Am J Hum Genet. 2004;75:338–345. doi: 10.1086/423147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan HY, Underhill PA, Cavalli-Sforza LL, Ibrahim ME. Y-chromosome variation among Sudanese: restricted gene flow, concordance with language, geography, and history. Am J Phys Anthropol. 2008;137:316–323. doi: 10.1002/ajpa.20876. [DOI] [PubMed] [Google Scholar]
- Tofanelli S, Ferri G, Bulayeva K, et al. J1-M267 Y lineage marks climate-driven pre-historical human displacements. Eur J Hum Genet. 2009. [DOI] [PMC free article] [PubMed]
- Cruciani F, La Fratta R, Trombetta B, et al. Tracing past human male movements in northern/eastern Africa and western Eurasia: new clues from Y-chromosomal haplogroups E-M78 and J-M12. Mol Biol Evol. 2007;24:1300–1311. doi: 10.1093/molbev/msm049. [DOI] [PubMed] [Google Scholar]
- Alshamali F, Pereira L, Budowle B, Poloni ES, Currat M. Local population structure in Arabian Peninsula revealed by Y-STR diversity. Hum Hered. 2009;68:45–54. doi: 10.1159/000210448. [DOI] [PubMed] [Google Scholar]
- Roewer L, Willuweit S, Stoneking M, Nasidze I. A Y-STR database of Iranian and Azerbaijanian minority populations. Forensic Sci Int Genet. 2009;4:e53–e55. doi: 10.1016/j.fsigen.2009.05.002. [DOI] [PubMed] [Google Scholar]
- Chang YM, Perumal R, Keat PY, Kuehn DL. Haplotype diversity of 16 Y-chromosomal STRs in three main ethnic populations (Malays, Chinese and Indians) in Malaysia. Forensic Sci Int. 2007;167:70–76. doi: 10.1016/j.forsciint.2006.01.002. [DOI] [PubMed] [Google Scholar]
- Wu FC, Ho CW, Pu CE, et al. Y-chromosomal STRs haplotypes in the Taiwanese Paiwan population Int J Legal Med 2010. DOI: 10.1007/s00414-009-0416-x. [DOI] [PMC free article] [PubMed]
- Chang YM, Swaran Y, Phoon YK, et al. Haplotype diversity of 17 Y-chromosomal STRs in three native Sarawak populations (Iban, Bidayuh and Melanau) in East Malaysia. Forensic Sci Int Genet. 2009;3:e77–e80. doi: 10.1016/j.fsigen.2008.07.007. [DOI] [PubMed] [Google Scholar]
- Alam S, Ali ME, Ferdous A, Hossain T, Hasan MM, Akhteruzzaman S. Haplotype diversity of 17 Y-chromosomal STR loci in the Bangladeshi population. Forensic Sci Int Genet. 2009;4:e59–e60. doi: 10.1016/j.fsigen.2009.05.005. [DOI] [PubMed] [Google Scholar]
- Capelli C, Onofri V, Brisighelli F, et al. Moors and Saracens in Europe: estimating the medieval North African male legacy in southern Europe. Eur J Hum Genet. 2009;17:848–852. doi: 10.1038/ejhg.2008.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh B. Estimating the time to the most recent common ancestor for the Y chromosome or mitochondrial DNA for a pair of individuals. Genetics. 2001;158:897–912. doi: 10.1093/genetics/158.2.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capelli C, Wilson JF, Richards M, et al. A predominantly indigenous paternal heritage for the Austronesian-speaking peoples of insular Southeast Asia and Oceania. Am J Hum Genet. 2001;68:432–443. doi: 10.1086/318205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayser M, Brauer S, Weiss G, Schiefenhovel W, Underhill PA, Stoneking M. Independent histories of human Y chromosomes from Melanesia and Australia. Am J Hum Genet. 2001;68:173–190. doi: 10.1086/316949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayser M, Choi Y, van Oven M, et al. The impact of the Austronesian expansion: evidence from mtDNA and Y chromosome diversity in the Admiralty Islands of Melanesia. Mol Biol Evol. 2008;25:1362–1374. doi: 10.1093/molbev/msn078. [DOI] [PubMed] [Google Scholar]
- Li H, Wen B, Chen SJ, et al. Paternal genetic affinity between Western Austronesians and Daic populations. BMC Evol Biol. 2008;8:146. doi: 10.1186/1471-2148-8-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonder MK, Mortensen HM, Reed FA, de Sousa A, Tishkoff SA. Whole-mtDNA genome sequence analysis of ancient African lineages. Mol Biol Evol. 2007;24:757–768. doi: 10.1093/molbev/msl209. [DOI] [PubMed] [Google Scholar]
- Abu-Amero KK, Gonzalez AM, Larruga JM, Bosley TM, Cabrera VM. Eurasian and African mitochondrial DNA influences in the Saudi Arabian population. BMC Evol Biol. 2007;7:32. doi: 10.1186/1471-2148-7-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kivisild T, Reidla M, Metspalu E, et al. Ethiopian mitochondrial DNA heritage: tracking gene flow across and around the gate of tears. Am J Hum Genet. 2004;75:752–770. doi: 10.1086/425161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turchi C, Buscemi L, Giacchino E, et al. Polymorphisms of mtDNA control region in Tunisian and Moroccan populations: an enrichment of forensic mtDNA databases with Northern Africa data. Forensic Sci Int Genet. 2009;3:166–172. doi: 10.1016/j.fsigen.2009.01.014. [DOI] [PubMed] [Google Scholar]
- Gray RD, Jordan FM. Language trees support the express-train sequence of Austronesian expansion. Nature. 2000;405:1052–1055. doi: 10.1038/35016575. [DOI] [PubMed] [Google Scholar]
- Richards M, Rengo C, Cruciani F, et al. Extensive female-mediated gene flow from sub-Saharan Africa into near eastern Arab populations. Am J Hum Genet. 2003;72:1058–1064. doi: 10.1086/374384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas MG, Parfitt T, Weiss DA, et al. Y chromosomes traveling south: the cohen modal haplotype and the origins of the Lemba – the ‘Black Jews of Southern Africa'. Am J Hum Genet. 2000;66:674–686. doi: 10.1086/302749. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.