Abstract
In all, 80% of antenatal karyotypes are generated by Down's syndrome screening programmes (DSSP). After a positive screening, women are offered prenatal foetus karyotyping, the gold standard. Reliable molecular methods for rapid aneuploidy diagnosis (RAD: fluorescence in situ hybridization (FISH) and quantitative fluorescence PCR (QF-PCR)) can detect common aneuploidies, and are faster and less expensive than karyotyping.
In the UK, RAD is recommended as a standalone approach in DSSP, whereas the US guidelines recommend that RAD be followed up by karyotyping. A cost-effectiveness (CE) analysis of RAD in various DSSP is lacking. There is a debate over the significance of chromosome abnormalities (CA) detected with karyotyping but not using RAD. Our objectives were to compare the CE of RAD versus karyotyping, to evaluate the clinically significant missed CA and to determine the impact of detecting the missed CA. We performed computer simulations to compare six screening options followed by FISH, PCR or karyotyping using a population of 110 948 pregnancies. Among the safer screening strategies, the most cost-effective strategy was contingent screening with QF-PCR (CE ratio of $24 084 per Down's syndrome (DS) detected). Using karyotyping, the CE ratio increased to $27 898. QF-PCR missed only six clinically significant CA of which only one was expected to confer a high risk of an abnormal outcome. The incremental CE ratio (ICER) to find the CA missed by RAD was $66 608 per CA. These costs are much higher than those involved for detecting DS cases. As the DSSP are mainly designed for DS detection, it may be relevant to question the additional costs of karyotyping.
Keywords: Down's syndrome, prenatal diagnosis, chromosome abnormalities
Introduction
About 80% of antenatal cytogenetic referrals are in the context of a screening program for DS.1 In the last 15 years, major advancements have been made in prenatal screening for DS and prenatal diagnostic testing (amniocentesis or chorionic villus sampling) is offered only to pregnant women at high risk,2, 3 of whom about 7% reveal a chromosomal abnormality (CA).4
The screening methods that identify women at high risk of having a foetus with DS include biochemical or ultrasonography markers (or both) combined with age-related algorithms.1 Similar to worldwide-advised procedures,5, 6 six screening options consistent with US and Canadian guidelines are available:7, 8 quadruple, combined, integrated and serum-only integrated tests, as well as stepwise sequential (sequential) and contingent sequential (contingent) screening approaches.
After a positive prenatal screening test, women are usually offered foetal karyotyping, which is considered as the gold standard to confirm the presence or absence of CA by counting the number of chromosomes and looking for structural changes.1 However, the main limitation of karyotyping remains the requirement for cell culture, resulting in a delay of 10–14 days for test results in many clinical genetic laboratories.9 Reliable10, 11, 12, 13 molecular methods that are faster (≤1–3 days)14 and less expensive than karyotyping15, 16, 17 have been developed to detect common aneuploidies, which account for >80% of the clinically relevant CA,14 although they do not provide a full photographic display of all chromosome pairs. Such methods include interphase FISH and QF-PCR, collectively referred to as RAD.18
There is still no consensus on the most cost-effective strategy that should be implemented to diagnose affected foetuses in DS screening programs. For some authors, the implementation of RAD services for all prenatal samples is considered to be a major step towards the optimisation of prenatal services.17 For the UK government19 and the UK National Screening Committee, new screening programs for DS need not include karyotyping and can offer diagnosis with RAD as a standalone approach.1
Nevertheless, a joint statement by the American College of Medical Genetics and the American Society for Human Genetics20 reaffirmed that all RAD test results must be followed up with karyotyping.1 Reports11, 16, 21, 22 indicate that 15–30% of CA detected by karyotyping would not be detected by RAD,1 but the number of these CA with a risk of adverse outcome above background levels are much lower,1 and the relevance of diagnosing them via DS screening programs is debated owing to their clinical significance1, 16, 17, 18
Actually, there is insufficient data available to resolve this question. Grimshaw et al15 noted that most investigations of these molecular tests focused on test errors rather than on CE analysis. So far, published reports addressing the CE of RAD versus full karyotyping have not estimated their use in relation with the various screening options that would comply with current guidelines for DS,15, 16 and concluded that RAD would be less cost-effective than karyotyping owing to the cost of clinically significant CA missed by RAD, although the screening programs are not designed to detect them.
Given the numerous screening options to compare, any single empirical or clinical study is unlikely to evaluate all available strategies. Computer simulations are an elegant alternative to identify which strategy is likely to be the most cost-effective.23, 24 Using simulations, we recently reported the impact of various first-trimester risk cutoffs for three different screening strategies combining first- and second-trimester analyses.25
In the current study, we performed simulations using data from the Serum, Urine and Ultrasound Screening Study (SURUSS)23, 26 for DS prenatal screening and from Caine et al1 for the expected CA missed by RAD with a potential level of adverse outcome, to: (i) compare the CE of three approaches to diagnose CA (karyotyping, FISH or QF-PCR) combined with six DS screening options, matched to worldwide-advised procedures5, 6, 7, 8 (quadruple, combined, integrated and serum integrated tests, sequential and contingent screening approaches) for the detection of DS through prenatal screening programs; (ii) estimate their performance in identifying clinically significant CA; (iii) determine the impact of detecting missed CA by RAD in DS screening programs by calculating the ICER if karyotyping is performed.
Methods
Design, data, screening options and end points
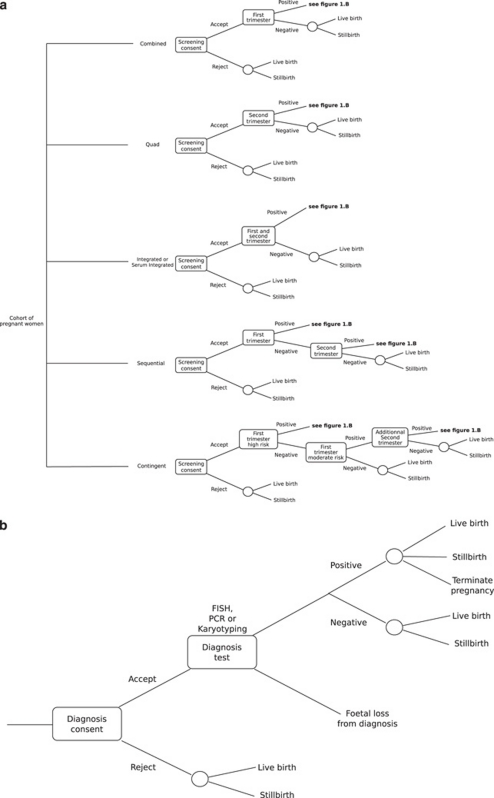
Using the modelling approach previously developed,25 we performed computer simulations to estimate the performance of six screening options recommended by guidelines in the US and Canada (quadruple, combined, integrated and serum integrated tests, sequential and contingent screening approaches).7, 8 We evaluated these six screening options combined with three diagnostic approaches (karyotyping, FISH or QF-PCR) for end points that cover the main outcomes in DS prenatal screening.23, 24 This yielded 18 potential screening algorithms. Figure 1a and b presents a simplified version of the decision model. The end points include: (1) the global costs; (2) the false-positive rate, which defines the number of scheduled prenatal diagnosis tests (amniocentesis or chorionic villus sampling); (3) the number of procedure-related euploid miscarriages; (4) the CE ratios (costs per DS detected); (5) the missed CA by RAD and (6) the ICER to detect them by karyotyping.
Figure 1.
Simplified version of the decision trees: (a) algorithms for screening options; (b) algorithms for the diagnosis procedure. Not shown in this simplified depiction, but included in our model, is the possibility that miscarriage occurs before testing or after releasing results.
The virtual population comprised 110 948 pregnancies corresponding to the number of pregnancies in 2001 in the province of Quebec.25 It allowed the generation of a virtual population with characteristics related to the prevalence and clinical course of non-DS or DS pregnancies, and a maternal age distribution identical to that found in the population of Quebec, and to simulate the costs and outcomes of all options under consideration.
To evaluate the impact of screening tests based on the gold standard of prenatal care, only amniocentesis was used for prenatal diagnostic testing following second-trimester screening results. However, for women who were tested positive in the first-trimester screening, a trans-abdominal chorionic villous sampling (CVS) karyotyping was considered.
The screening markers and procedures used are shown in Table 1. All input variables for simulations (screening procedures and expected CA) and their sources are presented in Tables 2, 3 and 4.
Table 1. Definitions of screening procedures.
| Combined test | First-trimester test based on combining nuchal translucency measurement (NT, an ultrasound measurement of the width of an area of translucency at the back of the foetal neck early in pregnancy) with free human chorionic gonadotropin (free β-hCG), pregnancy-associated plasma protein A (PAPP-A) and maternal age. |
| Quadruple test | Second-trimester test based on the measurement of AFP, uE3, free β-hCG (or total hCG) and inhibin-A together with maternal age. |
| Integrated test | The integration of measurements performed at different times of pregnancy into a single test result. Unless otherwise qualified, ‘integrated test' refers to the integration of NT and PAPP-A in the first trimester with the quadruple test markers in the second. The first-trimester screening marker results are not analyzed until the second-trimester markers are evaluated, at which point they are both assessed together. |
| Serum integrated test42 | A variant of the integrated test without NT (using PAPP-A in the first trimester and quadruple test markers in the second trimester). |
| Sequential screening23 | Screening in which a first-trimester test is performed (NT, free β-hCG and PAPP-A) and the result is interpreted immediately. If this is positive, a diagnostic test is offered (chorionic villous sampling, CVS), but if it is not positive, second-trimester serum markers are measured (quadruple test markers) and the first-trimester markers are reused to form an integrated test. |
| Contingent screening23 | Screening in which a first-trimester test (NT, free β-hCG and PAPP-A) is used to triage the population of women screened into three groups: one group (high-risk screen-positive) that is immediately offered a diagnostic test (CVS), a second group (screen-negative) that receives no further screening and a third intermediate (or low-risk screen-positive) group that has second-trimester markers measured (quadruple test markers) and the first-trimester measurements reused to form an integrated test. |
| Risk cutoff | The risk or likelihood of the condition being present in the foetus above which a prenatal diagnosis test is proposed. |
Table 2. Analysis input variables.
| Cost ($) | References | |
|---|---|---|
| Input costs of screening or diagnostic tests or events | ||
| Integrated test | 65 | 25 |
| Sequential screening | 105 | 25 |
| Contingent screening | 55 | 25 |
| Serum integrated test | 35 | 25 |
| Quadruple test | 25 | 25 |
| Combined test | 40 | 25 |
| Consulting with a genetic counselor | 73.90 | 25 |
| CVS + diagnostic procedure karyotyping | 876 | 25 |
| Amniocentesis + diagnostic procedure karyotyping | 500 | 25 |
| CVS + diagnostic procedure QF-PCR (Aneufast) | 198 | 25,43 |
| Amniocentesis + diagnostic procedure QF-PCR (Aneufast) | 198 | 25,43 |
| CVS + diagnostic procedure FISH | 422 | 25,43 |
| Amniocentesis + diagnostic procedure FISH | 422 | 25,43 |
| Termination of pregnancy | 1357.33 | 25 |
| Input variables of screening tests with a 90% detection rate | False-positive rate (%) | References |
| Integrated test (1/230 cutoff) | 2.11 | 30,35,44 |
| Sequential screening (1/9 cutoff, first trimester)a | 2.25 | 23,25 |
| Contingent screening (1/9 cutoff, first trimester)a | 2.42 | 23,25 |
| Serum integrated test (1/355 cutoff) | 5.30 | 26,30 |
| Quadruple test (1/545 cutoff) | 10.60 | 26,30 |
| Combined test (1/625 cutoff) | 8.40 | 30,35 |
| Different diagnostic tools | Sensitivity (%) | References |
| Karyotyping | 99.40 | 45 |
| QF-PCR | 99.30 | 14 |
| FISH | 98.70 | 14 |
Abbreviations: $, Canadian dollars; CVS, chorionic villous sampling.
Sequential and contingent screening tests consist of a sequence of analysis with many possible cutoff combinations, notably in the first trimester23. A risk cutoff is the risk or likelihood of the condition being present in the foetus. A woman was classified as positive if her risk estimate was equal to or greater than the corresponding specific cutoff level. Given the published data, for sequential and contingent screening tests, the first-trimester high-risk cutoff we applied was one in nine as previously advised25 and in the contingent screening approach, the lower risk cutoff used in the first test was 1 in 2000.23
Table 3. Demographic characteristics of the population and assumptionsa.
| Population simulated | Number | References |
|---|---|---|
| Total pregnant women | 110 948 | 25 |
| DS pregnancies at first trimester | 290 | 25,27,28 |
| DS pregnancies at second trimester | 190 | 25,27,28 |
| DS pregnancies at third trimester | 140 | 25,27,28 |
| DS babies at birth | 131 | 25,27,28 |
| Pregnant women's age distribution | ||
| <20 | 9008 | 25 |
| 20–24 | 24 987 | 25 |
| 25–29 | 33 421 | 25 |
| 30–34 | 27 320 | 25 |
| 35–39 | 13 135 | 25 |
| 40–44 | 2925 | 25 |
| 45≥ | 152 | 25 |
| Events before or after screening and diagnostic intervention | Probability (%) | References |
| Consent to participate in prenatal screening | 70 | 25 |
| Consent for amniocentesis or CVS with screening positive | 90 | 25 |
| Foetal loss from amniocentesis | 0.5 | 25,37,46–49 |
| Foetal loss from CVS | 1.6 | 25,37,46–49 |
| Proportion who terminated pregnancy with foetal DS | 90 | 25 |
Abbreviations: DS, Down's syndrome; CVS, chorionic villous sampling.
Data simulations were performed on a virtual population of 110 948 pregnancies with demographic (maternal age distribution), genetic and phenotypic (regarding DS) characteristics of the Quebec population in the year 2001.25
Table 4. Classification of expected chromosome abnormalitiesa.
| Type of chromosome abnormalities | Detectability by karyotypingb | Detectability by RADb | Risk categories of clinical significancec |
|---|---|---|---|
| Down's syndrome T21 | 1 | 1 | 3 |
| Edwards' syndrome T18 | 1 | 1 | 3 |
| Patau's syndrome T13 | 1 | 1 | 3 |
| Triploidy | 1 | 1 | 3 |
| Tetraploidy | |||
| Balanced structural rearrangement (inherited) | 1 | 3 | 1 |
| Robertsonian translocation (de novo or of unknown origin) | 1 | 3 | 1 |
| Marker chromosome (inherited) | 1 | 3 | 1 |
| 45,X; 47,XXX; 47,XXY; 47,XYY | 1 | 1 | 2 |
| 45,X mosaic; 47,XXY mosaic or sex chromosome mosaic | 1 | 2 | 2 |
| 45,X structurally abnormal X or structurally abnormal X/Y chromosome | 1 | 2 | 2 |
| Balanced structural rearrangement (de novo or of unknown origin) | 1 | 3 | 2 |
| Marker chromosome (de novo or of unknown origin) | 1 | 3 | 2 |
| Other autosomal trisomy | 1 | 3 | 2 |
| Unbalanced structural rearrangement | 1 | 3 | 3 |
Abbreviation: RAD, rapid aneuploidy diagnosis.
Classification according to Caine et al1 depending on their type, their detectability by karyotyping or RAD, and their clinical significance.
Detectability: 1=detectable; 2=sometimes detectable; 3=undetectable.
Risk categories of clinical significance: 1=background; 2=low-to-high risk; 3=high risk.
The number of DS pregnancies shown in Table 3 represents all DS cases in pregnancies registered for the population at each pregnancy period in 2001. The rates of DS pregnancy losses seemed to be more important than in other series (43%27 and 30%28), because part of these DS pregnancies are comprised in the voluntary pregnancy terminations in the first and second trimesters observed in our population in 2001.25
CA missed by RAD
Because the clinical significance (ie, preventable morbidity and mortality) of detecting CA in perinatal and live born babies depends on the type of CA, we used results from Caine et al1 to simulate the number and type of expected CA in our population, to evaluate CA missed by RAD and to estimate their potential level of adverse outcome. The study by Caine et al1constitutes the largest published retrospective cytogenetic audit, assessing 142 605 prenatal diagnoses. In the simulations, the CA were classified as detectable, sometimes detectable or undetectable by RAD and for clinical significance, as background risk, low-to-high risk and high risk of adverse outcome (Table 4). Also, we estimated the additional costs of detecting clinically significant CA missed by RAD by calculating the ICER24 obtained if full karyotyping is performed.
Test performance
The distribution of SURUSS marker results in DS-affected and DS-unaffected pregnancies was used to determine the screening test performances.23, 26 All false-positive rates and risk cutoffs were standardised to the same gestational age (11-week gestation) for first-trimester measurements. To compare first-trimester versus second-trimester screening procedures on a common basis, we used a fixed detection rate, which is more applicable than a fixed false-positive rate because applying the latter to the first-trimester as compared with the second-trimester results in a different detection rate, notably from the spontaneous losses of DS foetuses between the first and second trimesters.29 We chose the 90% detection rate as it is that used in the literature for test performances and cutoffs specified for all the screening tests (integrated,26 sequential and contingent screening tests23) used in the first and second trimesters. Also, as of April 2010, the UK National Screening Committee targets a detection rate of more than 90%.6
Costs
In Canada, in accordance with the Canadian Health Care Act, all medical necessary services are provided under the public healthcare system and are free of charge. Costs from provincial technical units were used for laboratory and imaging tests as previously detailed.25 Costs reported in Table 2 for screening tests do not reflect the cost of any single procedure but the mean cost for all medical necessary services provided for each screening option. Items considered for costing included screening costs as well as healthcare and medical services related to the following outcomes: birth, spontaneous miscarriage, elective abortion or procedure-related euploid miscarriages. Costs are expressed in Canadian dollars (CAD). The average exchange rate in 2007 was: 1.0748 CAD=1.00 USD=0.73 EUR.
CE analysis and confidence intervals
All measured costs occurred within 1 year; therefore, there was no need to discount costs and effects over time.24 Univariate sensitivity analyses25 were performed on the rate of consent to participate in prenatal screening (65 and 80%), the rates of foetal loss from CVS (0.5, 1 and 2%) and the rates of foetal loss from amniocentesis (1 and 1.5%). Moreover, the sensitivities and false-positive rates of DS screening strategies varied over the ranges achieved in the SURUSS trial.23, 26 To generate 95% confidence intervals (CI 95%) for global cost estimates, a bootstrap method was used as previously described25.
Results
The global cost analysis results, including the outcomes as a function of the screening strategy and diagnostic tool that were used, are summarised in Table 5. The CE ratio to detect DS cases (with the same screening strategy and QF-PCR for diagnosis), the number of CA missed by RAD according to the risk category of an abnormal outcome and the ICER for detecting them by karyotyping are reported in Table 6.
Table 5. Global costs, amount of diagnostic procedures induced and number of procedure-related euploid miscarriages.
| Global costsa | Number of diagnostic procedures | ||||||
|---|---|---|---|---|---|---|---|
| Screening strategies | QF-PCR | FISH | Karyotyping | Difference in karyotyping and QF-PCRb | Amniocentesis | CVS | Procedure-related euploid miscarriages |
| Integrated | 3.06(0.0016) | 3.28(0.0016) | 3.36(0.0020) | 0.294 | 908(903–913) | 4(3.4–4.3) | |
| Sequential | 3.38(0.0028) | 3.62(0.0033) | 3.74(0.0048) | 0.358 | 883(878–889) | 102(100–103) | 5(4.4–5.3) |
| Contingent | 2.50(0.0021) | 2.76(0.0029) | 2.90(0.0035) | 0.399 | 995(989–1001) | 111(109–112) | 6(5.3–6.9) |
| Serum integrated | 2.14(0.0024) | 2.69(0.0038) | 2.88(0.0043) | 0.739 | 2.293(2.285–2.300) | 15(14–16) | |
| Quadruple | 2.08(0.0033) | 3.18(0.0057) | 3.55(0.0072) | 1.464 | 4.537(4.524–4.550) | 22(21–23) | |
| Combined8.4% | 2.78(0.0032) | 3.62(0.0053) | 5.35(0.0112) | 2.568 | 3.760(3.746–3.772) | 70(68–72) | |
Confidence intervals are given in brackets.
Global costs are expressed in MCAD (million in Canadian dollars).
Difference of costs between karyotyping and QF-PCR are expressed in MCAD.
Table 6. CE ratios to detect DS, number of chromosome abnormalities missed by RAD, and their ICER.
| Number of missed CA by RAD | ||||
|---|---|---|---|---|
| Screening strategies | CE ratios to detect DS by QF-PCR | Low-to-high (2) and high risk (3) | High risk (3) | ICER to detect CA of groups 2 and 3 by karyotyping |
| Integrated | 34 293 | 5 | 1 | 59 377 |
| Sequential | 33 227 | 5 | 1 | 71 646 |
| Contingent | 24 084 | 6 | 1 | 66 608 |
| Serum integrated | 24 103 | 13 | 2 | 59 034 |
| Quadruple | 23 754 | 25 | 5 | 59 077 |
| Combined8.4% | 24 853 | 20 | 4 | 125 278 |
Abbreviations: CE, cost-effectiveness; DS, Down's syndrome; CA, chromosome abnormality; RAD, rapid aneuploidy diagnosis; ICER, incremental cost-effectiveness ratio.
Costs and main outcomes of DS screening options
With the same DS detection rate, all screening strategies cost less when using QF-PCR. Using the FISH test was associated with higher costs than using QF-PCR (Table 5). When considering only the safer screening strategies (ie, with a number of procedure-related euploid miscarriages induced <10 per 100 000 pregnancies), the most cost-effective strategy was the contingent screening associated with QF-PCR (global cost of $2 497 610 in Table 5) and CE ratio of $24 084 per DS detected (Table 6). The CE ratio of this screening strategy associated with full karyotyping would be $27 898 per DS detected. The global cost and CE ratio of the least cost-effective screening strategy (combined test using karyotyping) would be $5 347 554 (Table 5) and $47 358 per DS detected, respectively.
The difference in global cost between FISH and QF-PCR varied from $844 915 for the combined test to $220 404 for the integrated screening (Table 5). The largest difference in global cost for 100 000 pregnancies between QF-PCR and karyotyping was observed with the combined test, $2 567 981 ($5 347 554 with karyotyping and $2 779 573 with QF-PCR). The highest and lowest numbers of procedure-related euploid miscarriages occurred with the combined test (n=70) and the integrated test (n=4), respectively (Table 5).
Missed CA
Clinically significant CA were missed if the diagnosis was performed by RAD instead of karyotyping. However, when applied to 100 000 pregnancies, the contingent screening method associated with QF-PCR missed only six clinically significant CA (belonging to risk categories 2 or 3, ie, low-to-high and high risk), of which only one was expected to confer a high risk of an abnormal outcome (Table 6). The additional cost of finding these CA (ICER) by karyotyping instead of QF-PCR was much higher than the CE ratios to find DS cases regardless of the screening strategy considered (Table 6). Depending on which screening option is used, the ICER to find the CA missed by RAD ranged from $59 034 to $125 278 per CA (Table 6). Although FISH missed the same number of clinically significant CA as QF-PCR, it was associated with higher costs (Table 5).
Discussion
Limitations
Although our study is based on computer modelling rather than prospective data, our results are strengthened by the use of empirical data and true healthcare costs. It is rather unlikely that a vast prospective clinical study comparing the 18 screening strategies studied could realistically be performed. There is strong evidence that computer simulations are a powerful alternative to such expensive and hard-to-manage large-scale clinical studies.25, 30
All the reported CE ratios were computed in the context of the Quebec health care system and costs. However, with respect to cost comparisons, relative costs (ranking of different scenarios) usually lend themselves better to comparison. Also, given the robustness of our findings in the sensitivity analyses, the relative performance (or ranking) of various scenarios would likely be similar in other jurisdictions. Furthermore, the demographics of the simulated population are comparable to those found in other Western countries. Mean maternal age and the proportion of women over 35 years are comparable to the SURUSS26 and the First- and Second-Trimester Evaluation of Risk trial (FASTER trial)31 populations, which are representative of women in the UK and the USA, respectively.
RAD are the most cost-effective diagnostic tools
Our results demonstrate that RAD are the most cost-effective diagnostic tools and, although FISH misses the same number of clinically significant CA as QF-PCR, it is associated with higher costs. As DS screening programs are mainly designed for DS detection, it may be relevant to question the additional costs of full karyotyping to detect other anomalies. Quality of life and anxiety measurements show a significantly increased health status after diagnosis with RAD,15 and RAD allows earlier decision making in cases where the foetus has a detected CA.32 Also, the anxiety generated after being diagnosed with a not requested and not clinically significant CA by karyotyping can be prevented by using RAD.33
Furthermore, it could be more cost-effective to promote the use of QF-PCR instead of FISH, which is generally the first choice in genetic laboratories, and to favour contingent screening, which this study suggests as being the most cost-effective screening strategy for DS and associated with a more acceptable rate of procedure-related euploid miscarriages.
The combined test was associated with the highest number of procedure-related euploid miscarriages, which are the main adverse outcomes that should be reduced in DS prenatal screening.34 This is certainly owing to the fact that CVS is associated with the highest rate of procedure-related euploid miscarriages in women who had tested positive in the first trimester. Furthermore, a higher number of unnecessary terminations could contribute to the observed excess of procedure-related euploid miscarriages. Indeed, too early, a diagnosis may induce an excess in unnecessary terminations of the DS cases screened,23 as a spontaneous miscarriage may occur between the first and second trimesters.28 Some women who were screened as positive in the first trimester actually prefer to wait for an amniocentesis in the second trimester to avoid these disadvantages of CVS. The false-positive rate used for the combined test in our study was 8.4% for a detection rate of 90%, as revised by the SURUSS research group.35 This agrees with the false-positive rate reported by Malone et al31 in the FASTER trial for first-trimester screening, as at 11 weeks, the observed false-positive rate was 3.8% for a detection rate of 85%, and 18% for a detection rate of 95%. As other prospective trials evaluating first-trimester screening have reported a better efficiency for this screening strategy (eg, 5% false-positive rate for a 90% detection rate36), we simulated the CE and the number of procedure-related euploid miscarriages using the latter values and even with these assumptions, we confirmed the costs of the combined test: $4 034 243 (4 024 213–4 044 274) and 34 (33–35 for CI) for global cost (if used with karyotyping) and for the number of procedure-related euploid miscarriages, respectively.
RAD detect the majority of clinically significant CA
Our findings show that only a very small number of clinically significant CA are missed when RAD is used as a standalone approach and that savings can be substantial depending on the screening strategy used. For most of the screening scenarios evaluated, the incidence of procedure-related euploid miscarriages, which represent foetal losses of normal babies, was higher than the number of missed CA by RAD, especially when only high-risk CA were considered. The choice of the screening strategy, especially when based on the false-positive rate involved, could therefore have a more damaging impact than the use of RAD as a standalone diagnostic approach in DS screening programs.
ICERs to detect clinically significant CA missed by RAD are higher than the CE ratios to detect DS cases
We have shown that the additional costs to detect clinically significant CA missed by QF-PCR were much higher than the cost to detect cases of DS, ie, between 1.7-fold with the integrated test to 5-fold higher with the combined test. As the screening programs developed are only designed for DS screening or other common aneuploidies (trisomy 13 or 18) also detectable with RAD, and as women are not always aware of the fact that other CA can be detected with karyotyping,1, 33 taking on the additional costs involved for full karyotyping may be questionable.
Caine et al1 report that out of the 98 166 amniotic fluids referred from DS screening programs, 293 substantial-risk CA would be missed if RAD had been used. Taking a procedure-related miscarriage rate of 0.5%,25, 37 these 98 166 amniocentesis tests would have entailed 491 procedure-related miscarriages, ie, nearly twice the number of substantial risk CA missed by RAD. Also, it remains to be established whether withdrawal of karyotyping for pregnant women using prenatal screening programs for DS will have a substantial effect on the incidence of hitherto preventable morbidity and mortality in perinatal and live born babies, as previously stated by Caine et al.1 The incidence of birth defects are estimated at about 3.4% in the general population38 and, in 50% of cases, medical tests show normal results including karyotyping.39 Therefore, some recent studies support the use of novel diagnostic approaches for these patients with DNA arrays instead of conventional karyotyping.40 Thus, diagnostic confirmatory tools should perhaps focus on the original goal of DS prenatal screening programs: diagnosing DS cases.
Nevertheless, pregnant women who benefit from an amniocentesis also take the risk of a procedure-related miscarriage. Therefore, dimensions such as woman's values that are challenged by the additional information provided by karyotyping (including the information brought by a normal result), by the unexpected diagnosis of CA, and the anxiety generated by the birth of a child with clinically significant CA that could have been detected by karyotyping should all be considered.41
Acknowledgments
This work was supported by the Canadian Institutes for Health Research (Institutes of Genetics and Institutes of Health Services Research), the Canadian Agency for Drugs and Technologies in Health, and the Canadian Heart and Stroke Foundation through a grant to the CanGéneTest research consortium on genetic laboratory services.
The authors declare no conflict of interest.
References
- Caine A, Maltby AE, Parkin CA, Waters JJ, Crolla JA. Prenatal detection of Down's syndrome by rapid aneuploidy testing for chromosomes 13, 18, and 21 by FISH or PCR without a full karyotype: a cytogenetic risk assessment. Lancet. 2005;366:123–128. doi: 10.1016/S0140-6736(05)66790-6. [DOI] [PubMed] [Google Scholar]
- Wald NJ, Hackshaw AK, Watt H. Nuchal translucency and trisomy 18. Prenat Diagn. 1999;19:995–996. [PubMed] [Google Scholar]
- Wapner R, Thom E, Simpson JL, et al. First-trimester screening for trisomies 21 and 18. N Engl J Med. 2003;349:1405–1413. doi: 10.1056/NEJMoa025273. [DOI] [PubMed] [Google Scholar]
- NEQAS National External Quality Assessment Scheme in clinical cytogenetics. Annu Rep. 2000.
- Boyd PA, Devigan C, Khoshnood B, Loane M, Garne E, Dolk H. Survey of prenatal screening policies in Europe for structural malformations and chromosome anomalies, and their impact on detection and termination rates for neural tube defects and Down's syndrome. BJOG. 2008;115:689–696. doi: 10.1111/j.1471-0528.2008.01700.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NSC . Fetal Anomaly Screening Programme - Screening for Down's Syndrome: UK NSC Policy recommendations 2007–2010: Model of Best Practice. UK: Department of Health; 2008. [Google Scholar]
- ACOG Committee on Practice Bulletins ACOG Practice Bulletin No 77: screening for fetal chromosomal abnormalities. Obstet Gynecol. 2007;109:217–227. doi: 10.1097/00006250-200701000-00054. [DOI] [PubMed] [Google Scholar]
- Summers AM, Langlois S, Wyatt P, Wilson RD. Prenatal screening for fetal aneuploidy. J Obstet Gynaecol Can. 2007;29:146–179. doi: 10.1016/S1701-2163(16)32379-9. [DOI] [PubMed] [Google Scholar]
- Waters JJ, Waters KS. Trends in cytogenetic prenatal diagnosis in the UK: results from UKNEQAS external audit, 1987–1998. Prenat Diagn. 1999;19:1023–1026. [PubMed] [Google Scholar]
- Cirigliano V, Voglino G, Canadas MP, et al. Rapid prenatal diagnosis of common chromosome aneuploidies by QF-PCR. Assessment on 18,000 consecutive clinical samples. Mol Hum Reprod. 2004;10:839–846. doi: 10.1093/molehr/gah108. [DOI] [PubMed] [Google Scholar]
- Hulten MA, Dhanjal S, Pertl B. Rapid and simple prenatal diagnosis of common chromosome disorders: advantages and disadvantages of the molecular methods FISH and QF-PCR. Reproduction. 2003;126:279–297. doi: 10.1530/rep.0.1260279. [DOI] [PubMed] [Google Scholar]
- Mann K, Donaghue C, Fox SP, Docherty Z, Ogilvie CM. Strategies for the rapid prenatal diagnosis of chromosome aneuploidy. Eur J Hum Genet. 2004;12:907–915. doi: 10.1038/sj.ejhg.5201224. [DOI] [PubMed] [Google Scholar]
- Verma L, Macdonald F, Leedham P, McConachie M, Dhanjal S, Hulten M. Rapid and simple prenatal DNA diagnosis of Down's syndrome. Lancet. 1998;352:9–12. doi: 10.1016/S0140-6736(97)11090-X. [DOI] [PubMed] [Google Scholar]
- Shaffer LG, Bui TH. Molecular cytogenetic and rapid aneuploidy detection methods in prenatal diagnosis. Am J Med Genet C Semin Med Genet. 2007;145C:87–98. doi: 10.1002/ajmg.c.30114. [DOI] [PubMed] [Google Scholar]
- Grimshaw GM, Szczepura A, Hulten M, et al. Evaluation of molecular tests for prenatal diagnosis of chromosome abnormalities. Health Technol Assess. 2003;7:1–146. doi: 10.3310/hta7100. [DOI] [PubMed] [Google Scholar]
- Evans MI, Henry GP, Miller WA, et al. International, collaborative assessment of 146,000 prenatal karyotypes: expected limitations if only chromosome-specific probes and fluorescent in-situ hybridization are used. Hum Reprod. 1999;14:1213–1216. doi: 10.1093/humrep/14.5.1213. [DOI] [PubMed] [Google Scholar]
- Mann K, Fox SP, Abbs SJ, et al. Development and implementation of a new rapid aneuploidy diagnostic service within the UK National Health Service and implications for the future of prenatal diagnosis. Lancet. 2001;358:1057–1061. doi: 10.1016/S0140-6736(01)06183-9. [DOI] [PubMed] [Google Scholar]
- Boormans EM, Birnie E, Bilardo CM, Oepkes D, Bonsel GJ, van Lith JM. Karyotyping or rapid aneuploidy detection in prenatal diagnosis? The different views of users and providers of prenatal care. BJOG. 2009;116:1396–1399. doi: 10.1111/j.1471-0528.2009.02229.x. [DOI] [PubMed] [Google Scholar]
- UK Department of Health . Our Inheritance, Our Future: Realising the Potential of Genetics in the NHS. London: Stationery Office; 2003. [Google Scholar]
- Test and Technology Transfer Committee Technical and clinical assessment of fluorescence in situ hybridization: an ACMG/ASHG position statement. I. Technical considerations. Genet Med. 2000;2:356–361. doi: 10.1097/00125817-200011000-00011. [DOI] [PubMed] [Google Scholar]
- Thein AT, Abdel-Fattah SA, Kyle PM, Soothill PW. An assessment of the use of interphase FISH with chromosome specific probes as an alternative to cytogenetics in prenatal diagnosis. Prenat Diagn. 2000;20:275–280. doi: 10.1002/(sici)1097-0223(200004)20:4<275::aid-pd799>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- Thilaganathan B, Sairam S, Ballard T, Peterson C, Meredith R. Effectiveness of prenatal chromosomal analysis using multicolor fluorescent in situ hybridisation. BJOG. 2000;107:262–266. doi: 10.1111/j.1471-0528.2000.tb11698.x. [DOI] [PubMed] [Google Scholar]
- Wald NJ, Rudnicka AR, Bestwick JP. Sequential and contingent prenatal screening for Down syndrome. Prenat Diagn. 2006;26:769–777. doi: 10.1002/pd.1498. [DOI] [PubMed] [Google Scholar]
- Caughey AB. Cost-effectiveness analysis of prenatal diagnosis: methodological issues and concerns. Gynecol Obstet Invest. 2005;60:11–18. doi: 10.1159/000083480. [DOI] [PubMed] [Google Scholar]
- Gekas J, Gagne G, Bujold E, et al. Comparison of different strategies in prenatal screening for Down's syndrome: cost effectiveness analysis of computer simulation. BMJ. 2009;338:b138. doi: 10.1136/bmj.b138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wald NJ, Rodeck C, Hackshaw AK, Walters J, Chitty L, Mackinson AM. First and second trimester antenatal screening for Down's syndrome: the results of the Serum, Urine and Ultrasound Screening Study (SURUSS) J Med Screen. 2003;10:56–104. doi: 10.1258/096914103321824133. [DOI] [PubMed] [Google Scholar]
- Morris JK, Wald NJ, Watt HC. Fetal loss in Down syndrome pregnancies. Prenat Diagn. 1999;19:142–145. [PubMed] [Google Scholar]
- Snijders R. Fetal loss in Down syndrome pregnancies. Prenat Diagn. 1999;19:1180. [PubMed] [Google Scholar]
- Spencer K. What is the true fetal loss rate in pregnancies affected by trisomy 21 and how does this influence whether first trimester detection rates are superior to those in the second trimester. Prenat Diagn. 2001;21:788–789. doi: 10.1002/pd.134. [DOI] [PubMed] [Google Scholar]
- Wald N Rodeck C, Hackshaw AK, Walters J, Chitty L, Mackinson AM, Bestwick JP. Correction to SURUSS report. J Med Screen. 2006;13:51–52. [Google Scholar]
- Malone FD, Canick JA, Ball RH, et al. First-trimester or second-trimester screening, or both, for Down's syndrome. N Engl J Med. 2005;353:2001–2011. doi: 10.1056/NEJMoa043693. [DOI] [PubMed] [Google Scholar]
- Leung WC, Lam YH, Wong Y, Lau ET, Tang MH. The effect of fast reporting by amnio-PCR on anxiety levels in women with positive biochemical screening for Down syndrome—a randomized controlled trial. Prenat Diagn. 2002;22:256–259. doi: 10.1002/pd.314. [DOI] [PubMed] [Google Scholar]
- Chitty LS, Kagan KO, Molina FS, Waters JJ, Nicolaides KH. Fetal nuchal translucency scan and early prenatal diagnosis of chromosomal abnormalities by rapid aneuploidy screening: observational study. BMJ. 2006;332:452–455. doi: 10.1136/bmj.38730.655197.AE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley F, Buckley SJ. Costs of prenatal genetic screening. Lancet. 2008;372:1789–1791. doi: 10.1016/S0140-6736(08)61751-1. [DOI] [PubMed] [Google Scholar]
- Wald N, Rodeck C, Rudnicka A, Hackshaw A. Nuchal translucency and gestational age. Prenat Diagn. 2004;24:150–151. doi: 10.1002/pd.840. [DOI] [PubMed] [Google Scholar]
- Nicolaides KH, Spencer K, Avgidou K, Faiola S, Falcon O. Multicenter study of firsttrimester screening for trisomy 21 in 75 821 pregnancies: results and estimation of the potential impact of individual risk-orientated twostage first-trimester screening. Ultrasound Obstet Gynecol. 2005;25:221–226. doi: 10.1002/uog.1860. [DOI] [PubMed] [Google Scholar]
- Sundberg K, Bang J, Smidt-Jensen S, et al. Randomised study of risk of fetal loss related to early amniocentesis versus chorionic villus sampling. Lancet. 1997;350:697–703. doi: 10.1016/S0140-6736(97)02449-5. [DOI] [PubMed] [Google Scholar]
- Jacobs PA, Browne C, Gregson N, Joyce C, White H. Estimates of the frequency of chromosome abnormalities detectable in unselected newborns using moderate levels of banding. J Med Genet. 1992;29:103–108. doi: 10.1136/jmg.29.2.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight SJ, Regan R, Nicod A, et al. Subtle chromosomal rearrangements in children with unexplained mental retardation. Lancet. 1999;354:1676–1681. doi: 10.1016/S0140-6736(99)03070-6. [DOI] [PubMed] [Google Scholar]
- Miller DT, Adam MP, Aradhya S, et al. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet. 2010;86:749–764. doi: 10.1016/j.ajhg.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seror V. Fitting observed and theoretical choices - women's choices about prenatal diagnosis of Down syndrome. Health Econ. 2008;17:557–577. doi: 10.1002/hec.1276. [DOI] [PubMed] [Google Scholar]
- Wald NJ, Watt HC, Hackshaw AK. Integrated screening for Down's syndrome on the basis of tests performed during the first and second trimesters. N Engl J Med. 1999;341:461–467. doi: 10.1056/NEJM199908123410701. [DOI] [PubMed] [Google Scholar]
- Faucher E, Bujold E, Brassard N, Himaya E, Gekas J. FISH analysis for fetal karyotype : what are the factors associated with its failure. Prenat Diagn. 2008;28:S27. [Google Scholar]
- Bestwick JP, Wald N, Rudnicka AR.Cost of integrated, sequential and contingent prenatal screening for Down's syndrome BMJ 2009. e-pub ahead of print 2 April 2009.
- Midtrimester amniocentesis for prenatal diagnosis Safety and accuracy. JAMA. 1976;236:1471–1476. doi: 10.1001/jama.1976.03270140023016. [DOI] [PubMed] [Google Scholar]
- Alfirevic Z, Sundberg K, Brigham S. Amniocentesis and chorionic villus sampling for prenatal diagnosis. Cochrane Database Syst Rev. 2003. p. (3): CD003252. [DOI] [PMC free article] [PubMed]
- Mujezinovic F, Alfirevic Z. Procedure-related complications of amniocentesis and chorionic villous sampling: a systematic review. Obstet Gynecol. 2007;110:687–694. doi: 10.1097/01.AOG.0000278820.54029.e3. [DOI] [PubMed] [Google Scholar]
- Tabor A, Alfirevic Z. Update on procedure-related risks for prenatal diagnosis techniques. Fetal Diagn Ther. 2009;27:1–7. doi: 10.1159/000271995. [DOI] [PubMed] [Google Scholar]
- Tabor A, Vestergaard CH, Lidegaard O. Fetal loss rate after chorionic villus sampling and amniocentesis: an 11-year national registry study. Ultrasound Obstet Gynecol. 2009;34:19–24. doi: 10.1002/uog.6377. [DOI] [PubMed] [Google Scholar]