Abstract
Clarin 1 (CLRN1) is a four-transmembrane protein expressed in cochlear hair cells and neural retina, and when mutated it causes Usher syndrome type 3 (USH3). The main human splice variant of CLRN1 is composed of three exons that code for a 232-aa protein. In this study, we aimed to refine the structure of CLRN1 by an examination of transcript splice variants and promoter regions. Analysis of human retinal cDNA revealed 11 CLRN1 splice variants, of which 5 have not been previously reported. We studied the regulation of gene expression by several promoter domains using a luciferase assay, and identified 1000 nt upstream of the translation start site of the primary CLRN1 splice variant as the principal promoter region. Our results suggest that the CLRN1 gene is significantly more complex than previously described. The complexity of the CLRN1 gene and the identification of multiple splice variants may partially explain why mutations in CLRN1 result in substantial variation in clinical phenotype.
Keywords: Clarin 1, CLRN1, Usher syndrome, USH3, alternative splicing, promoter
Introduction
Usher syndrome (USH) is the most common cause of hereditary combined deaf-blindness with the prevalence of 3–6 per 100 000.1 USH is a clinically and genetically heterogeneous disease: the three subtypes (USH1, USH2, and USH3) are distinguished by the age of onset, severity, and type of the hearing loss. Usher syndromes are caused by mutations in at least 11 genetic loci within 9 identified genes.2 USH3 is characterized by progressive hearing loss, retinitis pigmentosa, and variable vestibular dysfunction.3 Worldwide, USH3 is the rarest subtype accounting for only 1–6% of all Usher syndrome cases; however, among Ashkenazi Jews and in the Finnish population, up to 40% of the USH cases have the USH3 subtype.3, 4, 5 The causative gene for the USH3 phenotype is Clarin 1 (CLRN1).6, 7, 8
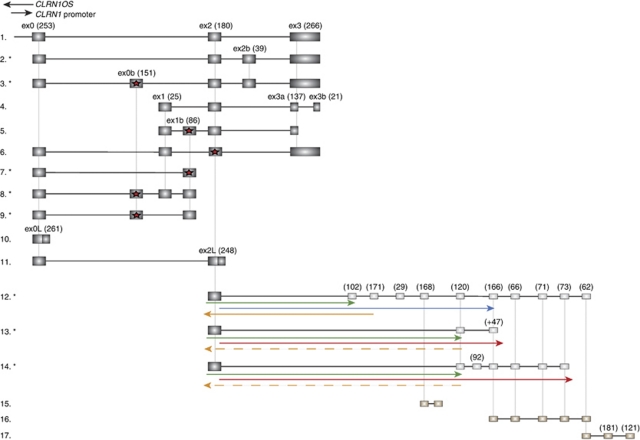
Initially, the gene was called USH3A and was reported to contain five exons and two splice variants: the main form contained exons 1, 2, 3 (exon 3a in this article), and 4 (exon 3b in this article) with a 120-aa open reading frame (ORF) (Figure 1, 4); the second variant also included exon 1b, between exons 1 and 2, and had an ORF of 30-aa (Figure 1, 5).6 Subsequent studies refined the gene structure and named the gene CLRN1. The initially identified four-exon form was determined to be a rare splice variant, with the main splice form containing exons 0 (named for its location upstream of exon 1), 2, and 3 (exon 3a continuing into the intron between exons 3a and 3b) with 232-aa ORF (Figure 1, 1).7, 8 To date, all of the known mutations causing USH3 are located within this three-exon main variant.7, 8, 9
Figure 1.
Known and novel splice variants of CLRN1. (1) NM_174878, the main CLRN1 variant comprises exons 0, 2, and 3. The 5′ UTR and 3′ UTRs are not counted in the exon nucleotide number (in brackets). Known human CLRN1 splice variants or EST sequences from gene databanks: (4) NM_052995, the original CLRN1 variant with exons 1, 2, 3a, and 3b reported in 2001; (5) AF388368, coding region only 30-aa long; (6) BM666773 found from retina, exons 1 and 2 are coded in an alternative reading frame compared with the other splice variants, and a stop codon appears in this frame in exon 2 (presence of a stop codon marked with a red star); (10) BX491536, exon 0 open reading frame (ORF) continues to the intron until stop codon, potentially codes for 87-aa protein; and (11) CV570593, exon 2 continues to the intron, codes for a potentially 167-aa long protein. Novel splice variants found in human retina cDNA library (marked with an asterisk): (2) HM626132, main variant with added exon 2b; (3) HM626133, main variant with added exons 0b and 2b; and (7–9) HM626134, HM626135 and HM626136, splice variants between exons 0 and 1b. CLRN1 exon 2 is also connected by splicing to three EST sequences downstream of CLRN1. Downstream EST sequences from databanks are (15) BE673203, (16) DV080481, and (17) DV080691. The 5′ UTR is unknown, but presumed in this figure to start from exon 2. Possible ORFs in these variants (12–14: HM626137, HM626138, and HM626139) are depicted as arrows. ORFs that continue the same ORF as in CLRN1 exon 2 are depicted as green arrows, reading frames in blue and red begin from exon 2, but are not in the same ORF as CLRN1. Two ORFs running in opposite direction than CLRN1 ORF are depicted as orange in either solid or dashed line. The exon and intron sizes are not drawn in scale.
The CLRN1 gene is rather conserved throughout evolution and the primary three-exon variant is ubiquitously expressed in many human tissues.6, 8 Computer analyses predict a four-transmembrane domain tetraspanin-like secondary/tertiary structure for the CLRN1 protein.7, 8 When transfected into and expressed in cultured baby hamster kidney (BHK) cells under CMV promoter, the wild-type full-length CLRN1 protein is trafficked to the plasma membrane,9, 10, 11 and endogenous CLRN1 protein in UB/OC-1 immortal auditory hair cell line is trafficked to the post-trans Golgi vesicles,12 whereas the mutated forms are retained in the endoplasmic reticulum (ER).9, 11 CLRN1 has also been reported to be involved with F-actin organization11 and synaptic maturation.12 The localization has been characterized in the mouse cochlea, where Clrn1 is expressed in hair cells and spiral ganglion cells,7, 12 and the absence of Clrn1 in null mice leads to disorganization of the hair cell stereocilia.10, 13 In the murine retina, Clrn1 expression was found in Müller glia,10 whereas immunohistochemical analyses suggest protein localization in the photoreceptor connecting cilia, inner segments, and ribbon synapses.12 The pathophysiology of USH3 remains unexplained. This study was conducted to elucidate the complete gene structure of CLRN1: its promoter regions, alternative splice variants, and the possible implications of these splice variants on CLRN1 function.
Methods
Amplification and sequencing of alternative splice variants
Alternative CLRN1 splice variants were amplified from Human retinal cDNA library (Clontech, Mountain View, CA, USA) with several combinations of primers (The representative primers used to obtain the previously known and the novel splice variants reported in this study are given in Supplementary Table 1; others are available upon request.), variable annealing temperatures, and different DNA polymerases (Advantage-GC 2 (Clontech); AmpliTaq Gold (Applied Biosystems, Foster City, CA, USA); FastStart Taq (Roche Diagnostics, Basel, Switzerland); and Titanium Taq (Clontech)). cDNA from human retinal pigment epithelial cell line (ARPE-19) was obtained from American Type Culture Collection (ATCC, Manassas, VA, USA) and isolated according to Roomets et al,14 human cochlear cDNA was a gift from Dr Frans Cremers, and additional tissue-specific cDNAs were from Human Multiple Tissue cDNA panels I and II (Clontech). These cDNAs were used to study the tissue-specific expression of alternative splice variants. The amplified products were electrophoresed through agarose gels and any amplified DNA sequences that differed in size from the main variant were collected using a Qiaquick Gel Extraction Kit (Qiagen GmbH, Hilden, Germany). The purified amplicons were cloned using TOPO TA cloning kit (Invitrogen, Carlsbad, CA, USA) and subsequently sequenced using BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems).
In silico studies of promoter regions
We searched for promoter region conservation between human and mouse with William Pearson's lalign program (http://www.ch.embnet.org/software/LALIGN_form.html)15 and ClustalW2 (http://www.ebi.ac.uk/Tools/clustalw2/index.html).16 Transcription factor binding sites were predicted with TESS (http://www.cbil.upenn.edu/tess)17 and EMBOSS CpG island detection prediction (http://emboss.ch.embnet.org/wEMBOSS/)18 software packages. Transmembrane regions of splice variants were predicted using TMHMM (http://www.cbs.dtu.dk/services/TMHMM/)19 and TMpred (http://www.ch.embnet.org/software/TMPRED_form.html)20 programs.
Promoter analysis
Candidate promoter regions that were identified in silico were further analyzed for promoter activity in vitro. The 1550 nt region upstream of the first exon of the primary transcript, exon 0, showed the most potential in silico and the rest of the in vitro studied promoters were set to the same size. We cloned the 1550 nt segments in front of exons 0, 1, 2, and 3a into a pGluc-Basic vector (New England BioLabs, Ipswich, MA, USA) and used pCMV-Gluc control plasmid (New England BioLabs) as a positive control. The vector constructs were then transfected in triplicate with Fugene 6 transfection reagent (Roche Diagnostics) into BHK-21 cells (CCL-10, ATCC). The promoter activities in transfected cells were studied using Gaussian Luciferase Assay kit (New England Biolabs) and Victor 2 Wallac 1420 multilabel counter (Perkin Elmer, Waltham, MA, USA) at 48 h after transfection.
Results
Alternative splice variants
We found 10 alternative CLRN1 splice variants in addition to the main variant containing exons 0, 2, and 3. Five of these variants were novel. Alternative splice variants either included new exons, excluded previously known ones, or were found to use alternative 3′ splice sites (Figure 1, 2–11). All of these variants included at least one of the three exons belonging to the main variant (Figure 1, 1). Each of the exons 0, 2, and 3 have alternative 3′ splice sites leading to elongated exon 0 (transcript 0 long, 0L; Figure 1, 10), elongated exon 2 (transcript 0-2 long, 2L; Figure 1, 11), and shortened exon 3 (3a in original 1-2-3a-3b transcript; Figure 1, 4), respectively. There are interrupting stop codons in exons 0b, 1b, and in exon 2 of the 0-1-2-3 splice variant (Figure 1, 3, 5–9). Splice variants, 0-2-3, 1-2-3a-3b, 1-1b-2-3, 0-1-2-3, 0L, and 0-2L, were also identified in existing gene sequence databases. The splice variants 0-2-2b-3 (HM626132), 0-0b-2-2b-3 (HM626133), 0-1b (HM626134), 0-0b-1-1b (HM626135), and 0-0b-1b (HM626136) described in this study are novel (Table 1). We sequenced the additional CLRN1 exons (except 0b because of early stop codon) for sequence changes from approximately 65 USH patients with a phenotype compatible with USH3 and no mutations in the exons of the main variant (0, 2, and 3), but we were unable to identify mutations in exons not included in the main variant. We also found splice variants in human retinal cDNA that contained CLRN1 exon 2, which was connected by splicing to exons from EST sequences BE673203, DV080481, and DV080691 located downstream from CLRN1. The 5′ ends of these transcripts remain unknown, and therefore it remains unclear as to whether these sequences are translated into protein. There are, however, ORFs in all these splice variants (Figure 1, 12–14). All the intron–exon splice sites in CLRN1 splice variants follow the GT-AG rule.21 Most of the splice variants within the CLRN1 gene contain predicted transmembrane regions (Supplementary Figure 1).
Table 1. Splice sites and sizes of alternative exons and introns.
| Exon/intron size | 5′ splice site | 3′ splice site | |
|---|---|---|---|
| Exon 0 | 253 nt (544 nt with 5′ UTR) | CGGTTCTCAT/gtaagtagca | |
| Exon 0L | 0L: 261 nt (598 nt) | ||
| Intron (0 to 0b] | 26 792 nt | ||
| Exon 0b | 151 nt | ttgtgagaag/ATAAAGGAAA | CTTATTTCAT/gtatgtacat |
| Intron (0b to 1) | 1537 nt | ||
| Exon 1 | 25 nt (172 nt as a cassette exon) (417 nt with 5′ UTR) | gttgatgcag/GCTATTTTCT | CAGCAACCAG/gtaggggtgc |
| Intron (1 to 1b) | 1182 nt | ||
| Exon 1b | 68 nt (86 nt as a cassette exon) | cccattaggt/TATAAGCTCT | CTGGCACATA/gtaggtgctc |
| Intron (1b to 2) | 810 nt | ||
| Exon 2 | 180 nt | ttatcttcag/TTTTTCCAGA | TTCATTTCAG/gtaagtacaa |
| Exon 2L | 2L: 248 nt (347 nt with 3′ UTR) | ||
| Intron (2 to 2b) | 1063 nt | ||
| Exon 2b | 39 nt | tctcctgaag/TTGCCCTTTG | CAGGCTCAAG/gtactttctt |
| Intron (2b to 3) | 12 279 nt | ||
| Exon 3 | 266 nt (1369 nt with main variant 3′ UTR) (141 nt with 1b variant 3′ UTR) | tctgttgcag/GCTCCTGTGG | |
| Exon 3a+3b | 3a: 137 nt intron: 1192 nt 3b: 21 nt (689 nt with 3′ UTR) | tgatctgcag/CTGACTAAAG | CTCATTCTGG/gtcattttct |
Uncoding UTRs are calculated in brackets.
All the splice variants were initially detected from human retinal cDNA. The main variant was expressed in several tissues (Supplementary Figure 2) including retinal pigment epithelial ARPE-19 cells (data not shown), retina, and cochlea. Other tissues such as heart, lung, skeletal muscle, spleen, thymus, and peripheral blood leukocytes had either very weak or unmeasurable expression (Supplementary Figure 2). Splice variant 0-2-2b-3, which most likely encodes a functional protein isoform as it only changes the primary product by 13-aa (Supplementary Figure 1b), was detected in the retina, cochlea, heart, brain, placenta, lung, skeletal muscle, pancreas, and ovary (Supplementary Figure 2).
Promoter region analysis
Apart from one CpG island that was detected −4400 nt 5′ of exon 1, which is not included in the main splice variant, no detectable CpG islands were found from the studied CLRN1 promoter regions (newcpgreport and newcpgseek; EMBOSS). In silico predictions (TESS) found several potential transcription factor binding sites, including C/EBP, GATA, H1, Sp1, TBP, YY1, and WT1-KTS upstream of exon 0; C/EBP, GATA, H1, TBP, and YY1 upstream of exon 2; CACCC, H1, Sp1, and YY1 upstream of exon 1; and CACCC, Sp1, and YY1 upstream of exon 3 (Figure 2).
Figure 2.
Potential sites of gene expression regulation in the proximal 1550 nt of each of CLRN1's primary exons.
Promoter constructs (5′ 1550 nt long sequences upstream of translated regions of exons 0, 1, 2, and 3a) were transfected in triplicate into BHK-21 cells and luciferase activity was measured from the culture media (Figure 3). The strongest activity was induced by the 1000 nt region upstream of exon 0 (the canonical CLRN1 promoter). There was a significant drop in activity when the region between 1000 and 1550 nt upstream of exon 0 was included. In this region, (CA)23 repeat was located 1107–1152 nt 5′ of translation start site. The second strongest activity level was induced by the region upstream of exon 2 whereas the region upstream of exon 1 induced expression only slightly above the negative control (media), and less signal than the baseline control (unmodified pGluc vector; Figure 3). The experiment was replicated three times to confirm the results.
Figure 3.
CLRN1 promoter region activity levels. Possible promoter regions were inserted in the pGLuc expression vector and transfected in triplicate into BHK cells. The luciferase activity was measured from the following constructs (upstream of translation start site or exon splice site): (1) exon 0: 1–500 nt, (2) exon 0: 1–1000 nt, (3) exon 0: 1–1550 nt, (4) exon 1: 1–1550 nt, (5) exon 2: 1–1550 nt, (6) exon 3: 1–1550 nt, and (7) unmodified pGluc vector. The relative activity levels were set to positive control pCMV-Gluc vector (New England BioLabs) as 100% and untransfected cell culture media signal level was subtracted from these values. Error bars reflect 1 SD. Promoter region ClustalW2 scores (sequence conservation between mouse and human) are displayed as an insert: (a) 500 nt upstream exon 0, (b) 500–1000 nt upstream exon 0, (c) 1000–1550 nt upstream exon 0, (d) 1–1550 nt upstream exon 0, (e) 1–1550 nt upstream exon 1, (f) 1–1550 nt upstream exon 2, and (g) 1–1550 nt upstream exon 3.
Discussion
This study demonstrates that the structure of CLRN1 gene is significantly more complex than previously indicated. The complexity is especially evident in the first intron (3′ of exon 0) the longest of the CLRN1 introns, which is likely re-spliced at the 0b, 1, and 1b exon junctions before the main intron–exon junction at exon 2.22 The function of these splice variants remains unknown: some may be errors in the splicing process and some may generate functional molecules. Among other USH genes, for example, harmonin is known to undergo alternative splicing, a characteristic that is known to be important for proper gene function. Alternative harmonin isoforms localize to separate compartments in photoreceptor cells and, moreover, have also been found to exhibit different tissue specificities.23
In CLRN1, some of the alternative splice variants have ORFs that could be translated into functional proteins. For example, the variant including exon 2b results in the addition of 13 extra amino acids onto the 232-aa main isoform, and thus very likely encodes a functional protein (Supplementary Figure 1b). Small, tissue-specific alternative exons that provide crucial functional modifications to the proteins are especially important in nervous system-specific isoforms.24 The 232-aa CLRN1 isoform forms dimers and multimers when expressed in cell cultures,9 and there is a possibility that the alternative isoforms are also included in these multimer structures. Many CLRN1 splice variants have translation stop codons before the final exon (Figure 1, 3, 5–9). They are most likely untranslated, but may have a regulatory function at the RNA level, influencing the expression of the primary transcript, similar to other known premature termination codons within alternative splice variants.25 Some of these untranslated variants may be degraded by nonsense-mediated decay (NMD) that is known to affect splice variants having a stop codon >50–55 nt upstream of the last spliced exon–exon boundary.26, 27 In light of these data, the splice variants including exon 0b are most likely affected by NMD as the stop codon is −101 nt upstream of its exon–exon junction. Variants with exon 1b (stop codon −21 nt upstream of exon splice site), and 0-1-2-3 with a stop codon in exon 2 (stop codon −50 nt upstream of exon–exon splice site) may also be affected by NMD if the UTR continues splicing as in isoform 1-1b-2-3. Alternatively, NMD may not influence expression if the UTR does not continue past exon 2. The complete structure of the 3′ UTR in all exon 1b splice variants is still unknown.
It is also plausible that the CLRN1 splice variants exhibit different tissue specificities. Tissue-specific splicing requires unique combinations of negative and positive influences by transcription factors and other regulatory elements. These signals are difficult to accurately determine using in silico examination.22
In the BHK cells that we studied, the main CLRN1 promoter region is 1000 nt 5′ to exon 0 and likely regulates the weak expression levels seen for the main CLRN1 splice variant in most tissue types.7, 8 We studied other possible promoter regions responsible for this activity and other possible promoter regions for the alternative splice variants in transfected cells to identify all regions required for the basal activity. Our studies showed that although promoter region activity levels varied among transfected cell culture sets according to cell culture age, stage, and other variables, the CMV promoter expression level (used as a positive control) also varied correspondingly and the relationship between weaker and stronger regulatory regions remained constant. Also, the promoter region conservation between human and mouse is in concordance with the observed higher activity level of the more conserved 5′ region upstream of exon 0, when compared with the weaker region upstream of exon 1 (Figure 3). It is, however, unlikely that the proximal promoter contains all the required information for correct transcriptional control of CLRN1 expression in all tissues and developmental stages. Additional elements such as enhancers and/or silencers may be located more distantly, downstream or upstream. In the retina and cochlea where USH3 manifests itself, the studied CLRN1 promoter sequences are probably augmented by cell type and developmental stage-specific signals that could not be recapitulated here (perhaps with the exception of in silico studies). For example, the potential promoter regions for the main CLRN1 transcript contain the H1 core sequence (TAATC) that is thought to be a binding site for the photoreceptor-specific homeodomain transcription factor Crx28 (Figure 2).
Our results suggest that the dominant promoter elements proximal to the CLRN1 gene are located within 1000 nt upstream of exon 0. When an additional 550 nt (5′ to the 1000 nt) were added to the expression construct, expression decreased significantly, suggesting the presence of negatively acting control elements in the region between −1000 and −1550 nt 5′ of the translation start site. The CA repeat region in this area has potential binding site for WT1-KTS that can function in transcriptional repression.29 Similar results in cell culture studies have been reported, with extended promoter sequences having an inhibitory effect on active promoter regions: examples include Pcdh15, the gene associated with USH1F,30 and RK (rhodopsin kinase), the gene associated with Oguchi disease.28, 31, 32 The situation is further complicated by the pseudogene CLRN1OS (AF388367) located on the opposite strand.6, 7 CLRN1OS and the main CLRN1 splice variant have overlapping first exon 5′ UTRs running in opposite directions (Figure 1). This pseudogene may have an important role in antisense transcriptional control of CLRN1 either by hybridizing to the CLRN1 coding DNA strand or interfering with transcription or mRNA stability similar to cases reported by Katayama et al.33
The main variant of CLRN1 seems to be expressed rather ubiquitously,7, 8 which would correlate with the presence of a CpG island that is often found in promoters of genes with no or little tissue specificity.34 The only clear CpG island near CLRN1 could be detected using in silico studies within −4400 nt upstream of exon 1. TATA boxes are usually associated with tissue specificity, but in CLRN1, the function of recognized TATA binding protein (TBP) binding sites may well be compensated by the presence of Sp1 and YY1 factor binding sites, which are either weakly or strongly, respectively, associated with less tissue-specific genes34 (Figure 2).
Knowing the structure and function of the CLRN1 gene is a prerequisite for understanding the pathophysiology of USH3, and for developing therapies for the disease. All the known mutations have been found from the exons included in the main splice variant (Figure 1, exons 0, 2, and 3), but most of the mutations occur in the exons 0 and 2,9 which are also included in some of the alternative CLRN1 protein isoforms (Supplementary Figure 1). Curiously, USH3 has highly variable progression and severity, even among siblings carrying the same mutations.3, 4, 5, 35, 36 Some of this phenotypic variety may be explained by the complex structure of CLRN1, the use of alternative promoters and the expression of alternatively spliced variants. As the CLRN1 protein is suggested to form multimers, the presence of alternative protein isoforms in these multimeric complexes may be crucial to the correct function of CLRN1 in human cochlea and retina.
Acknowledgments
We thank Drs Frans Cremers, Eva Roomets, and Tiina Tyni for providing us with cDNA samples. This study was supported by the Finnish Eye and Tissue Bank Foundation, the Finnish RP Society, the Foundation Fighting Blindness, the Medicinska understödsföreningen Liv och Hälsa, Otto A. Malmin lahjoitusrahasto, the Research Foundation of the University of Helsinki, Sokeain Ystävät ry, and Hope for Vision.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Spandau UH, Rohrschneider K. Prevalence and geographical distribution of Usher syndrome in Germany. Graefes Arch Clin Exp Ophthalmol. 2002;240:495–498. doi: 10.1007/s00417-002-0485-8. [DOI] [PubMed] [Google Scholar]
- Saihan Z, Webster AR, Luxon L, Bitner-Glindzicz M. Update on Usher syndrome. Curr Opin Neurol. 2009;22:19–27. doi: 10.1097/wco.0b013e3283218807. [DOI] [PubMed] [Google Scholar]
- Pakarinen L, Tuppurainen K, Laippala P, Mäntyjärvi M, Puhakka H. The ophthalmological course of Usher syndrome type III. Int Ophthalmol. 1995;19:307–311. doi: 10.1007/BF00130927. [DOI] [PubMed] [Google Scholar]
- Ness SL, Ben-Yosef T, Bar-Lev A, et al. Genetic homogeneity and phenotypic variability among Ashkenazi Jews with Usher syndrome type III. J Med Genet. 2003;40:767–772. doi: 10.1136/jmg.40.10.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen M, Bitner-Glindzicz M, Luxon L. The changing face of Usher syndrome: clinical implications. Int J Audiol. 2007;46:82–93. doi: 10.1080/14992020600975279. [DOI] [PubMed] [Google Scholar]
- Joensuu T, Hämäläinen R, Yuan B, et al. Mutations in a novel gene with transmembrane domains underlie Usher syndrome type 3. Am J Hum Genet. 2001;69:673–684. doi: 10.1086/323610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adato A, Vreugde S, Joensuu T, et al. USH3A transcripts encode clarin-1, a four-transmembrane-domain protein with a possible role in sensory synapses. Eur J Hum Genet. 2002;10:339–350. doi: 10.1038/sj.ejhg.5200831. [DOI] [PubMed] [Google Scholar]
- Fields RR, Zhou G, Huang D, et al. Usher Syndrome type III: revised genomic structure of the USH3 gene and identification of novel mutations. Am J Hum Genet. 2002;71:607–617. doi: 10.1086/342098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isosomppi J, Västinsalo H, Geller S, et al. Disease-causing mutations in the CLRN1 gene alter normal CLRN1 protein trafficking to the plasma membrane. Mol Vis. 2009;15:1806–1818. [PMC free article] [PubMed] [Google Scholar]
- Geller SF, Guerin KI, Visel M, et al. CLRN1 is nonessential in the mouse retina but is required for cochlear hair cell development. PLoS Genet. 2009;5:e1000607. doi: 10.1371/journal.pgen.1000607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian G, Zhou Y, Hajkova D, et al. Clarin-1, encoded by the Usher syndrome III causative gene, forms a membranous microdomain: possible role of Clarin-1 in organizing the actin cytoskeleton. J Biol Chem. 2009;284:18980–18993. doi: 10.1074/jbc.M109.003160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zallocchi M, Meehan DT, Delimont D, et al. Localization and expression of clarin-1, the Clrn1 gene product, in auditory hair cells and photoreceptors. Hear Res. 2009;255:109–120. doi: 10.1016/j.heares.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng R, Geller SF, Hayashi T, et al. Usher syndrome IIIA gene clarin-1 is essential for hair cell function and associated neural activation. Hum Mol Genet. 2009;18:2748–2760. doi: 10.1093/hmg/ddp210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roomets E, Kivelä T, Tyni T. Carnitine palmitoyltransferase I and Acyl-CoA dehydrogenase 9 in retina: insights of retinopathy in mitochondrial trifunctional protein defects. Invest Ophthalmol Vis Sci. 2008;49:1660–1664. doi: 10.1167/iovs.07-1094. [DOI] [PubMed] [Google Scholar]
- Huang X, Miller W. A time-efficient, linear-space local similarity algorithm. Adv Appl Math. 1991;12:337–357. [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Schug J. Using TESS to predict transcription factor binding sites in DNA sequence. Curr Protoc Bioinformatics. 2008;2:2.6. doi: 10.1002/0471250953.bi0206s21. [DOI] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. EMBOSS: the European molecular biology open software suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Hofmann K, Stoffel W. TMbase – A database of membrane spanning proteins segments. Biol Chem Hoppe-Seyler. 1993;374:166. [Google Scholar]
- Breathnach R, Benoist C, O'Hare K, Gannon F, Chambon P. Ovalbumin gene: evidence for a leader sequence in mRNA and DNA sequences at the exon-intron boundaries. Proc Natl Acad Sci USA. 1978;75:4853–4857. doi: 10.1073/pnas.75.10.4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- Reiners J, Reidel B, El-Amraoui A, et al. Differential distribution of harmonin isoforms and their possible role in Usher-1 protein complexes in mammalian photoreceptor cells. Invest Ophthalmol Vis Sci. 2003;44:5006–5015. doi: 10.1167/iovs.03-0483. [DOI] [PubMed] [Google Scholar]
- Stetefeld J, Ruegg MA. Structural and functional diversity generated by alternative mRNA splicing. Trends Biochem Sci. 2005;30:515–521. doi: 10.1016/j.tibs.2005.07.001. [DOI] [PubMed] [Google Scholar]
- Lamba JK, Adachi M, Sun D, et al. Nonsense mediated decay downregulates conserved alternatively spliced ABCC4 transcripts bearing nonsense codons. Hum Mol Genet. 2003;12:99–109. doi: 10.1093/hmg/ddg011. [DOI] [PubMed] [Google Scholar]
- Behm-Ansmant I, Kashima I, Rehwinkel J, Sauliere J, Wittkopp N, Izaurralde E. mRNA quality control: an ancient machinery recognizes and degrades mRNAs with nonsense codons. FEBS Lett. 2007;581:2845–2853. doi: 10.1016/j.febslet.2007.05.027. [DOI] [PubMed] [Google Scholar]
- Lejeune F, Maquat LE. Mechanistic links between nonsense-mediated mRNA decay and pre-mRNA splicing in mammalian cells. Curr Opin Cell Biol. 2005;17:309–315. doi: 10.1016/j.ceb.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Young JE, Vogt T, Gross KW, Khani SC. A short, highly active photoreceptor-specific enhancer/promoter region upstream of the human rhodopsin kinase gene. Invest Ophthalmol Vis Sci. 2003;44:4076–4085. doi: 10.1167/iovs.03-0197. [DOI] [PubMed] [Google Scholar]
- Hewitt SM, Fraizer GC, Wu YJ, Rauscher FJ, III, Saunders GF. Differential function of Wilms' tumor gene WT1 splice isoforms in transcriptional regulation. J Biol Chem. 1996;271:8588–8592. doi: 10.1074/jbc.271.15.8588. [DOI] [PubMed] [Google Scholar]
- Alagramam KN, Miller ND, Adappa ND, et al. Promoter, alternative splice forms, and genomic structure of protocadherin 15. Genomics. 2007;90:482–492. doi: 10.1016/j.ygeno.2007.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto S, Sippel KC, Berson EL, Dryja TP. Defects in the rhodopsin kinase gene in the Oguchi form of stationary night blindness. Nat Genet. 1997;15:175–178. doi: 10.1038/ng0297-175. [DOI] [PubMed] [Google Scholar]
- Hayashi T, Gekka T, Takeuchi T, Goto-Omoto S, Kitahara K. A novel homozygous GRK1 mutation (P391H) in 2 siblings with Oguchi disease with markedly reduced cone responses. Ophthalmology. 2007;114:134–141. doi: 10.1016/j.ophtha.2006.05.069. [DOI] [PubMed] [Google Scholar]
- Katayama S, Tomaru Y, Kasukawa T, et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- Schug J, Schuller WP, Kappen C, Salbaum JM, Bucan M, Stoeckert CJ. Promoter features related to tissue specificity as measured by Shannon entropy. Genome Biol. 2005;6:R33. doi: 10.1186/gb-2005-6-4-r33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera W, Aleman TS, Cideciyan AV, et al. Retinal disease in Usher syndrome III caused by mutations in the clarin-1 gene. Invest Ophthalmol Vis Sci. 2008;49:2651–2660. doi: 10.1167/iovs.07-1505. [DOI] [PubMed] [Google Scholar]
- Plantinga RF, Kleemola L, Huygen PL, et al. Serial audiometry and speech recognition findings in Finnish Usher syndrome type III patients. Audiol Neurootol. 2005;10:79–89. doi: 10.1159/000083363. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.