Abstract
The phylogenetic relationships of numerous branches within the core Y-chromosome haplogroup R-M207 support a West Asian origin of haplogroup R1b, its initial differentiation there followed by a rapid spread of one of its sub-clades carrying the M269 mutation to Europe. Here, we present phylogeographically resolved data for 2043 M269-derived Y-chromosomes from 118 West Asian and European populations assessed for the M412 SNP that largely separates the majority of Central and West European R1b lineages from those observed in Eastern Europe, the Circum-Uralic region, the Near East, the Caucasus and Pakistan. Within the M412 dichotomy, the major S116 sub-clade shows a frequency peak in the upper Danube basin and Paris area with declining frequency toward Italy, Iberia, Southern France and British Isles. Although this frequency pattern closely approximates the spread of the Linearbandkeramik (LBK), Neolithic culture, an advent leading to a number of pre-historic cultural developments during the past ≤10 thousand years, more complex pre-Neolithic scenarios remain possible for the L23(xM412) components in Southeast Europe and elsewhere.
Keywords: Y-chromosome, haplogroup R1b, human evolution, population genetics
INTRODUCTION
The complex pattern of European Y-chromosome diversity has been ascribed to anatomically modern human dispersals, incorporating the combined heritage of initial upper Paleolithic colonization, secondary post-glacial mesolithic re-expansions and the Neolithic era demic diffusion of agriculturalists from the Near East.1 Regardless of possibly a minor autosomal contribution, as yet, there is no Y-chromosome evidence of hybridization between Neanderthals and modern human beings.2, 3
Although haploid genealogies capture only a narrow ancestry spectrum of the history of a population's gene pool, they afford a relatively uncomplicated and unique approach to disentangle and investigate complexities created by the superimposition of later gene flow patterns onto preexisting substrates, revealing population formation and affinities as well as insights into gender-related levels of reproductive success.2, 4 Using the conventional Y-chromosome haplogroup nomenclature, the majority of lineages observed in contemporary European populations fall into the following main haplogroups: E, G, I, J, N and R. Typically, >50% of men in Europe are affiliated with haplogroup R.5, 6 Members of haplogroup R are also widespread in Western,7, 8 Central9 and Southern Asia10, 11 as well as in some parts of the Sahel region of Africa.12, 13 In Europe, essentially all R associates belong to its sub-clade R1 defined by M173. Two R1 sub-clades show distinctive geographic distributions where Germany represents a major differentiation zone. R1a-M420 varieties are most frequent in the East and the R1b-M343 sub-clade is more common in the West.14 A further sub-clade of R1b, defined by the mutation M269, is the most common Y-chromosome haplogroup throughout Western Europe.15, 16 Notably, it also occurs at informative >5% frequencies in Northeast Italy and the Balkans,17 Turkey,7 Iran,8 the Caucasus (this study), Iraq and Lebanon,18 Pakistan10 and Algeria.19
Haplogroup R1b frequency in Europe is clinal with increasing frequencies observed in Northwest Europe, a pattern that has been ascribed to the persistance of Palaeolithic Y chromosomes in Europe after a Neolithic demic diffusion from the Near East.20, 21 Interestingly, attempts to date the Y-STR-based diversity of R1b-M269 chromosomes in populations from Europe and Turkey have yielded Holocene expansion times in both regions.7, 22, 23 These findings have led to the reappraisal that R1b-M269 in Europe is young and likely associated with a Neolithic demic expansion from the Near East through Anatolia.22, 23
A more precise understanding of the affinities among haplogroup R1b-M269 members has been concealed by a lack of phylogeographic knowledge concerning sub-haplogroup differentiation within the seemingly homogeneous R1b-M269 landscape. It is only recently that progress regarding the fractionation of additional informative (>5% in more than one population) phylogenetic sub-haplogroups has occurred.24, 25 This progress has been made possible mainly because of large-scale re-sequencing efforts and the deployment of parallel genotyping array technology for hundreds of Y-chromosome-specific SNPs.26, 27 Here, we evaluate the geographic distribution of prevalent sub-clades of Y-chromosome haplogroup R1b in Europe, the Circum-Uralic region and the Caucasus.
MATERIALS AND METHODS
All samples studied were obtained using locally approved informed consent protocols. A total of 2193 samples within the R-M343 component were genotyped in a hierarchical manner for the following SNP markers: M412, M415, M478, M520, M529, L11, L23 and S116 (Supplementary Table S1). In addition, markers M42013 and V8828 were genotyped according to the previously published protocols, as well as the following seven previously published markers in haplogroup R1b: P297, M73, M269, U106, U198, U152 and M222.29 The M479 SNP (specifications given in Supplementary Table S1) was typed in R-M207(xM173) samples.
Although not genotyped in the population data, representative M18-, M65-, M126-, M153-, M160-, M167 [AKA Sry2627]-, M415- and M520-derived samples were evaluated to determine the phylogenetic relationships of these markers. Following PCR amplification, SNP genotyping was accomplished by either denaturing high performance liquid chromatography (DHPLC), RFLP analysis, Taqman (Life Technologies Corp., Carlsbad, CA, USA) assay or direct-sequencing methodology. Although the phylogeny is under constant revision, the International Society of Genetic Genealogy Haplotype 2010 Tree provides a catalog of current refinements (http://www.isogg.org/tree/) that were followed in this study.
The ages of various haplogroups in populations were estimated using the methodology described by Zhivotovsky et al,30 modified according to Sengupta et al,10 using the evolutionary effective mutation rate of 6.9 × 10−4 per 25 years. The accuracy and appropriateness of this mutation rate has been independently confirmed in several deep-rooted pedigrees of the Hutterites.31 Important caveats to consider include the fact that coalescent times (Td) is sensitive to authentic rare outlier alleles and that multiple founders during population formation will inflate the age estimate of the event.
Coalescent times based on DYS19, DYS388, DYS389I, DYS389II, DYS390, DYS391, DYS392, DYS393, DYS439, DYS461 (TAGA counts) and median haplotypes of specific R1b sub-haplogroups by population are given in Supplementary Table S2. STR haplotype data are given in Supplementary Table S3. Although the DYS390 STR locus was typed in representative members of all haplogroup R chromosomes studied, the 19 repeat allele was observed only in a subset of R1b-M73-derived chromosomes. A 10-locus network analysis of 27 M73-derived chromosomes (results not shown) revealed a distinctive sub-cluster suggestive of recent common ancestry. As outlier alleles inflate Td estimates, we present separate Td estimates (Supplementary Table S2) that pivot about the 19 repeat allele. Freeware POPSTR program (http://harpending.humanevo.utah.edu/popstr/) was used to conduct principal component analyses based on haplogroup frequencies. The Moran's I coefficient for the spatial autocorrelation analysis was calculated using the PASSAGE software v.1.1 with binary weight matrix, nine distance classes and random distribution assumption. All of the four correlograms (S116, M529, U106, U152) were significant (P<0.05), and the Moran's I values for each distance class were denoted according to significance/non-significance. Correlation analysis of coalescent times (Td) with great circle distances in radians was conducted using the SPSS PC package to compare the geographic spread of coalescent times for the S116-related haplogroup component from Paris and Munich and the U106 sub-clade from Warsaw.
RESULTS
The phylogenetic relationships of rs9786140 [M412], rs9786194 [M415], rs17842518, and rs17250114 SNPs26 were initially investigated using DHPLC in a geographically diverse set of 30 haplogroup R samples. During this process, two new markers [M478, M479] were detected in the flanking regions of rs17842518 and rs17250114, respectively, and confirmed by direct sequencing. In addition, markers L11, L23, S116, M520 and M529 were evaluated in our data set based on unsolicited insights from the genetic genealogy community. The phylogenetic relationships of Y-chromosome haplogroup R components studied are presented in Figure 1a. It is important to recognize that ascertainment bias is likely responsible for the currently observed disproportionate branching patterns.5 All branches are shown with common marker names and labeled according to standard YCC nomenclature guidelines.29 Henceforth, for shortness and clarity, the haplogroups are referred to in the text by the defining mutations rather than the cumbersome YCC labels. Y-chromosome genotype data available for 10355 DNA samples were used to evaluate the diversification of 2193 haplogroup R-M343 samples, the majority (n=2043) of which were derived for the M269 mutation. All of the haplogroup R-M207 chromosomes studied were derived for either the R1-M173 or R2-M479 markers, ie no R-M207* chromosomes were detected in our sample (the star * symbol here refers to the unresolved status in the phylogeny beyond the given marker). However, we cannot rule out the possible existence of such lineages, as our study lacks coverage in Central Asia and India. It should be noted that some previous studies on India have reported the presence of R-M207*, ranging from ∼1–3%.11, 32 The frequencies of basal haplogroup R1a-M420* and various haplogroups associated with R2-M479 and R1b-M343 elements surveyed in populations from the West, North, East, Central and Southeast regions of Europe, the Circum-Uralic regions, the Caucasus and Near/Middle East, Turkey and Pakistan are presented in Supplementary Table S4.
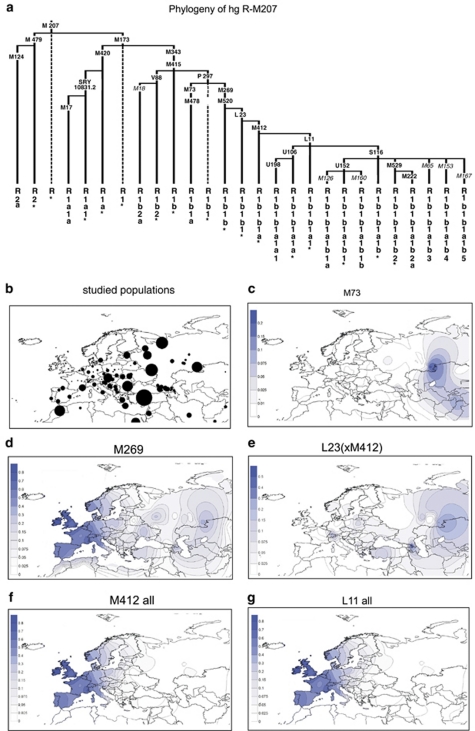
Figure 1.
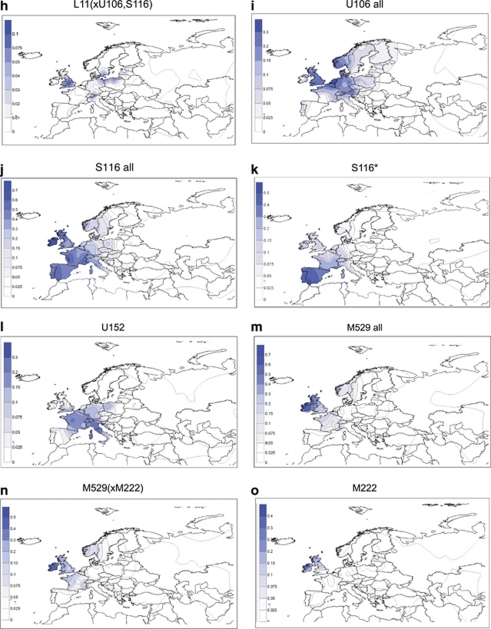
(a) Phylogenetic relationships of haplogroup R binary polymorphisms studied. The names of six polymorphisms whose phylogenetic positions were determined in representative-derived samples, but not surveyed in the entire sample collection are indicated in italics. Dashed lines indicate basal haplogroup branches that were not observed. The YCC nomenclature labels reflect the exclusion of the P25 SNP from the phylogeny given its innate instability.44 The asterisk (*) refers to the unresolved status of the phylogenetic haplogroups beyond the specified marker. (b) Approximate locations of the 118 studied populations appear as circles on the map that are proportional to sample sizes, the smallest n=9 and the largest n=522. (c–o). Spatial-frequency distributions of haplogroup-frequency data. Each map was obtained by applying the frequencies from Supplementary Table S4 for 10355 individuals distributed in 118 population samples that are either new or updated to the present phylogenetic-resolution level from literature (references listed in the Supplementary Table 4) plus R-M269 data from Cruciani et al13 for North African points. Data concerning the strong U152 founder effect signal with identical haplotypes in Northern Bashkirs is excluded from the plots. The frequency data were converted to spatial-frequency maps using Surfer software (version 7, Golden software Inc., Cold Spring Harbor, NY, USA), following the Kriging procedure.
Although the frequency of R1 lineages is currently the highest in Europe, the phylogeographic argument for their origin outside Europe, likely somewhere in West Asia, arises from the geographic distribution of the primary splits in the R1 phylogeny: at least three basic R-M207-derived haplogroups – R1a-M420*, R1b-M343* and R2 – occur mostly outside Europe. Figure 1b shows approximate locations of the 118 populations studied and proportional sample sizes. As the intensity of sampling is thin relative to the expanse of West Asia, the spatial-frequency surfaces for this region should be viewed as preliminary. Of the total of 193 R1b-M73 chromosomes detected, all except two Russians occurred outside Europe, either in the Caucasus, Turkey, the Circum-Uralic and North Pakistan regions (Figure 1c), in contrast to its considerably more widespread companion R1b-M269 clade (Figure 1d). With the exception of rare incidences of R1b-V88 in Corsica, Sardinia13 and Southern France (Supplementary Table S4), there is nearly mutually exclusive patterning of V88 across trans-Saharan Africa vs the prominence of P297-related varieties widespread across the Caucasus, Circum-Uralic regions, Anatolia and Europe. The detection of V88 in Iran, Palestine and especially the Dead Sea, Jordan (Supplementary Table S4) provides an insight into the back to Africa migration route.
The frequency data for 13 major R1b1-P297 components with minimum frequency ≥10% were used to create spatial distribution maps (Figure 1c–o), whereas the phylogenetic relationships of the haplogroups are shown in Figure 1a. Besides the obvious differences in the geographic spreads of the M73, M269 and V88 branches that stem out of the R1b-M343 node as noted above, there are apparent geographic patterns also in the downstream branches, between markers M412 and M222 (Figure 1f–o). Although it is likely that additional sub-haplogroups within the more numerous L23*(xM412) assemblage currently remain hidden, it is instructive that these chromosomes often exceed 10% frequency in the Caucasus, Turkey and some SE Europe and Circum-Uralic populations (Supplementary Table S4; Figure 1e), whereas conversely they typically display frequencies ≤5% in Western Europe (except for an instance of 27% in Switzerland's Upper Rhone Valley) in contrast to the prominent spread of derived M412 varieties in West Europe (Figure 1f).
Major R1b Founder Effect in West Europe
R1b-M412 appears to be the most common Y-chromosome haplogroup in Western Europe (>70%), while being virtually absent in the Near East, the Caucasus and West Asia (Figure 1f). Recent founder effects could explain why the M412-L11 assemblage of chromosomes is abundant and restricted to Western parts of Europe (Figure 1f and g).
Examples of additional founder effects and subsequent demographic expansions are evident among the more prominent L11-related, S116 (Figure 1i) and U106 (Figure 1k) components that generally distribute West and East of the Rhine river basin, respectively. Within the three major sub-haplogroups of the S116 assemblage further geographic localization is evident. Specifically, S116*(xU152, M529) occurrence is maximal in Iberia (Figure 1j), whereas the U152 branch is most frequent (20–44%) in Switzerland, Italy, France and Western Poland, with additional instances exceeding 15% in some regions of England and Germany (Figure 1l). Last, the M529 clade is highest (25–50%) in England and Ireland (Figure 1m and n), with the M222 sub-clade (Figure 1o) mainly restricted to Ireland.
Coalescent Times
As the methodology assumes one founder, the expansion times will be inflated if multiple founders or recurrent gene flows have occurred. Thus, these estimates should be viewed as the upper bounds of dispersal times. A total of 1029 chromosomes were included in the Y-STR-based coalescent analysis involving components of the R1b-M343-affiliated phylogeny. The coalescent estimate for the Y-STR network tree of 245 M269*+L23(xM412) chromosomes is 10 270±1680 years Before Present (BP). This estimate approximates the median TMRCA dates (8.5–12.5k years) of M269 clade across Europe based on alternative demographic inference methodology.33 Our estimate of 8870±1708 years BP, based on 757 M412 chromosomes, suggests that the M412 lineage evolved in Europe soon after the arrival of a L23* ancestor. The coalescent times for 11 sub-haplogroups averaged across populations in which the sample size was 5 are presented in Supplementary Table S2. Notable are the equivalent expansion times for all S116 (n=481), Td=8630±1529 years BP and U106 (n=239), Td 8742±1551 years BP-related lineages.
Archeologically, there are two attested phases regarding the geographic spread of the Linearbandkeramik (LBK). The first phase extended to the upper Danube river near Munich. The second phase extended further to the Paris basin.34 Furthermore, there is evidence of several post-LBK Neolithic expansions, ca 6000 years BP from the Paris basin region toward Northern Italy, Southern France and Iberia, characterized by the Chasseen horizon,35, 36 as well as to England.37 We examined the geographic regional patterns of S116, U106, U152 and M529 haplogroups more quantitatively within particular distance classes by spatial autocorrelation analysis. All these four sub-clades displayed clinal distributions of frequency variation (Supplementary Figure 1).
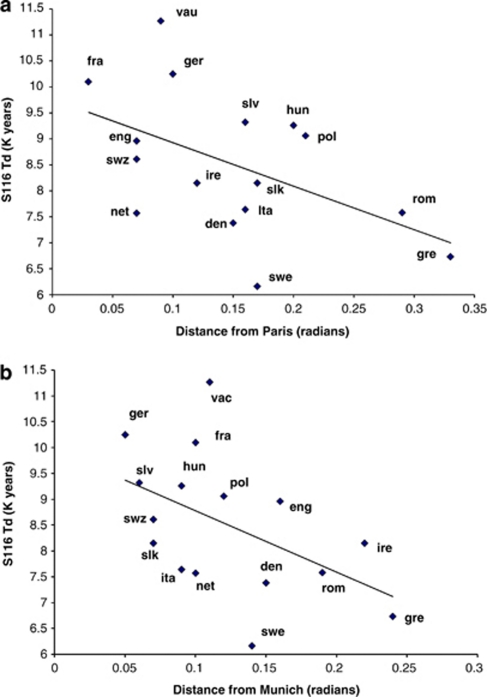
We investigated the association of Td for the S116 assemblage with the great circle distance from both Paris and Munich and U106 from Warsaw both as representations of the transition to agriculture in the North-Central European plain. Figure 2a shows that S116 Td decreases with distance from Paris (r=−0.51, P<0.025, n=16 one-tailed Pearson's) and Figure 2b with distance from Munich (r=−0.49, P<0.05, n=16 one-tailed Pearson's). There was no significant correlation of U106 Td with distance from Warsaw (r=−0.40, ns). As Td estimates are sensitive to outliers, we also calculated the correlations between the mean Y-STR variance at distances from Paris and Munich. Both correlations remained significant. It is important to recognize that we used regression analyses to identify the approximate geographic source of S116 diversity as it spreads outward and not to chronologically date the spreading events as multiple S116 lineages were likely involved.
Figure 2.
Regression plots of coalescent times for S116 lineages vs distance from (a) Paris and (b) Munich. Population codes: France (fra); Germany (ger); England (eng); Switzerland (swz); Netherlands (net), Ireland (ire); Denmark (den); Italy (ita); Slovakia (slk); Sweden (swe); Greece (gre); Romania (rom); Poland (pol); Hungary (hun); Slovenia (slo).
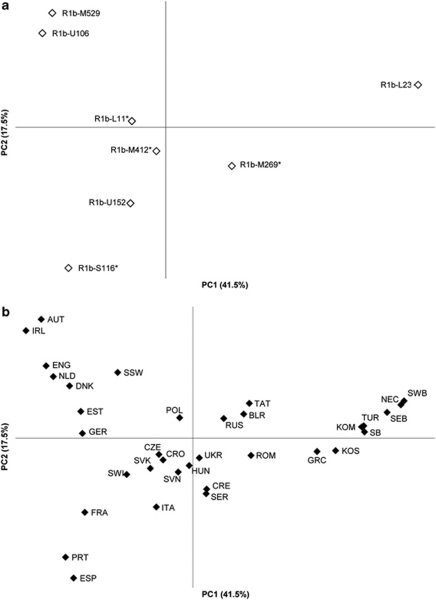
We conducted principal components analysis to investigate affinities of haplogroup R1b fractions among different populations based on the frequency distributions of M269*, L23, M412*, L11*, U106, S116*, U152 and M529 with respect to total M269. Figure 3a shows the contributions of the sub-haplogroups to the first two principal components. The first principal component separates L23 from M412 and its sub-clades, whereas the second differentiates the sub-haplogroups within M412. Figure 3b shows Western Europeans clustering in an approximately congruent manner with geography (according to the frequencies of M412 sub-clades) on the left, with Central and Eastern Europeans in the middle and a group of populations from the Balkans, Turkey, the Caucasus and the Circum-Uralic region on the right, separated by a high frequency of L23.
Figure 3.
Principal component analysis by haplogroup R1b sub-clades: (a) M269*, L23, M412*, L11*, U106, S116*, U152 and M529 sub-haplogroups with respect to total M269, and (b) by collapsing the 118 populations into 34 regionally defined populations. We excluded populations when the total R1b frequency was <5% or the count was less than n=5. Population codes: Austria (AUT); Belarus (BLR); Crete (CRE); Croatia (CRO); Czech Republic (CZE); Denmark (DNK); England (ENG); Estonia (EST); France (FRA); Germany (GER); Greece (GRC); Hungary (HUN); Ireland (IRL); Italy (ITA); Komis from Perm Oblast, Russia (KOM); Kosovo (KOS); Northeast Caucasus (NEC); Netherlands (NLD); Poland (POL); Portugal (PRT); Romania (ROM); Russians from Russia (RUS); Serbia (SER); Southern Bashkirs from Bashkortostan, Russia (SB); Southeastern Bashkirs from Bashkortostan, Russia (SEB); Southwestern Bashkirs from Bashkortostan, Russia (SWB); Slovakia (SVK); Slovenia (SVN); South Sweden (SSW); Spain (ESP); Switzerland (SWI); Tatars from Russia (TAT); Turkey (TUR); Ukraine (UKR).
DISCUSSION
The initial arrival of farmers from Southwest Asia to the present-day Greece occurred ca 9000 years BP.38 Outside of Southeast Europe, two episodes of early farming are attested archeologically.39 The first involved a maritime colonization of Crete ca 9000 years BP and Southern Italy ca 8000 years BP and subsequently spread to coastal Mediterranean France and Spain, as exemplified by impressed/cardial pottery. The second involved a migration to Central Europe, from Hungary to France, characterized by LBK (ca 7500 years BP). Within a 3k-year period, the agricultural economy spread across Europe, terminating in Britain and Scandinavia ∼6000 years BP.39
This study has evaluated the spatial and temporal distributions of sub-clades of Y-chromosome haplogroup R1b-M269 in Europe, the Near East, the Circum-Uralic region and the Caucasus, revealing the major M412-defined phylogenetic dichotomy between the Central/Western Europe and more easterly distributed representatives (Figure 1e and f, respectively). In addition, several additional sub-haplogroup varieties, especially those in Central and Western Europe, display patterns with geographic locality (Figure 1g–o) and clinality (Supplementary Figure 1). The enhanced resolution of M412-related lineages permits a finer-grained view of the proposal that R1b-M269 coincides with the arrival and spread of farming into Europe. A recent analysis of 9 Y-STR loci associated with 840 R1b chromosomes resolved just to the level of M269 concluded that all such chromosomes in Europe reflect a recent genetic heritage that was uniformly introduced by exogenous farmers migrating from Western Anatolia.23 Our high-resolution SNP genotype results show that the majority of Central and Western European haplogroups relate to common M412 founders whose sub-clades display phylogeographic and temporal patterns consistent with allele surfing at the periphery of expansions.40, 41 Opportunities for the establishment of new varieties on a regional basis would be enhanced if preexisting population densities were not excessive. Estimates of population densities in the early Neolithic suggest that they were low, 0.6 per square kilometer.42 Such low population densities would have helped to promote founder effects such as those seen for the more prominent L11-related, S116 (Figure 1j) and U106 (Figure 1i) components and their respective sub-haplogroups. This is shown in part as the inversely related decreasing expansion times of S116-related haplogroups with increased distance from high-diversity areas coincident with the establishment of the early Danubian Neolithic LBK horizon in Europe (Figure 2a and b).
Our results implicate complexity in the post-glacial formation and expansion of populations in Europe during the past ca 10 000 years. The narrow temporal window between potential expansions by Mesolithic foragers at the onset of the Holocene (10k years ago) and pioneer farmers from the Near East during the early Neolithic into Central Europe (7.5k years ago) is exceedingly difficult to discern with genetic tools.22 Thus, invoking the pronounced transformation of the pre-Neolithic European gene pool by intrusive pioneer farmers from the Near East must be viewed cautiously especially when such an argument is based on just a single incompletely resolved haplogroup. Although the transition to agriculture was a pivotal event in human history, the spread of specific haplogroups can occur in more than one migration event. Evidence of trade networks based on the exchange of commodities (eg salt, amber) along northwest to south and southeast directions, eg the Iron Age Hallstatt Culture,43 provided opportunities for potential gene dispersion. However, the magnitude of such putative commodity-driven gene flows remains uncertain until direct evidence from ancient DNA is provided in combination with potentially even more high-resolution and informative sub-haplogroup fractions relevant to particular trade routes or cultural horizons are detected and used to test hypotheses concerning post-Neolithic histories.
Acknowledgments
We thank all the men who donated DNA samples used in this study. We thank Vincent Vizachero for alerting us to the potential of M520, M529, S116, L11, L23 SNPs and for insight regarding the DYS390, 19 repeat allele and M73. Professor Joachim Hallmayer provided laboratory facilities for PAU and AAL. This research was supported by the European Union European Regional Development Fund through the Centre of Excellence in Genomics, Estonian Biocentre and Tartu University, Estonian Science Foundation Grant no. 7445 (to SR) and Estonian Basic Research Grant SF 0270177s08 (to RV) and by the Artur Lind scholarship from the Estonian Genome Foundation (to MJ) and the Croatian Ministry of Science, Education and Sports Grant 196-1962766-2751 (to PR) and the grants of the Russian foundation for basic research 10-06-00451 and 10-07-00515 to OB and AP. We thank Scott R Woodward and the Sorenson Molecular Genealogy Foundation for providing support for AAL and PAU. RJK was supported by a grant from the Stanford-France Interdisciplinary Program. JC was supported by a grant from the ANR Project AFGHAPOP, no. BLAN07-3_222301, CSD 9.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Barbujani G, Bertorelle G. Genetics and the population history of Europe. Proc Natl Acad Sci USA. 2001;98:22–25. doi: 10.1073/pnas.98.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underhill PA, Kivisild T. Use of Y chromosome and mitochondrial DNA population structure in tracing human migrations. Annu Rev Genet. 2007;41:539–564. doi: 10.1146/annurev.genet.41.110306.130407. [DOI] [PubMed] [Google Scholar]
- Green RE, Krause J, Briggs AW, et al. A draft sequence of the Neandertal genome. Science. 2010;328:710–722. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novelletto A. Y chromosome variation in Europe: continental and local processes in the formation of the extant gene pool. Ann Hum Biol. 2007;34:139–172. doi: 10.1080/03014460701206843. [DOI] [PubMed] [Google Scholar]
- Jobling MA, Tyler-Smith C. The human Y chromosome: an evolutionary marker comes of age. Nat Rev Genet. 2003;4:598–612. doi: 10.1038/nrg1124. [DOI] [PubMed] [Google Scholar]
- Chiaroni J, Underhill PA, Cavalli-Sforza LL. Y chromosome diversity, human expansion, drift, and cultural evolution. Proc Natl Acad Sci USA. 2009;106:20174–20179. doi: 10.1073/pnas.0910803106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cinnioğlu C, King R, Kivisild T, et al. Excavating Y-chromosome haplotype strata in Anatolia. Hum Genet. 2004;114:127–148. doi: 10.1007/s00439-003-1031-4. [DOI] [PubMed] [Google Scholar]
- Regueiro M, Cadenas AM, Gayden T, Underhill PA, Herrera RJ. Iran: tricontinental nexus for Y-chromosome driven migration. Hum Hered. 2006;61:132–143. doi: 10.1159/000093774. [DOI] [PubMed] [Google Scholar]
- Wells RS, Yuldasheva N, Ruzibakiev R, et al. The Eurasian heartland: a continental perspective on Y-chromosome diversity. Proc Natl Acad Sci USA. 2001;98:10244–10249. doi: 10.1073/pnas.171305098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta S, Zhivotovsky LA, King R, et al. Polarity and temporality of high-resolution Y-chromosome distributions in India identify both indigenous and exogenous expansions and reveal minor genetic influence of Central Asian pastoralists. Am J Hum Genet. 2006;78:202–221. doi: 10.1086/499411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahoo S, Singh A, Himabindu G, et al. A prehistory of Indian Y chromosomes: evaluating demic diffusion scenarios. Proc Natl Acad Sci USA. 2006;103:843–848. doi: 10.1073/pnas.0507714103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bereir RE, Hassan HY, Salih NA, et al. Co-introgression of Y-chromosome haplogroups and the sickle cell gene across Africa's Sahel. Eur J Hum Genet. 2007;15:1183–1185. doi: 10.1038/sj.ejhg.5201892. [DOI] [PubMed] [Google Scholar]
- Cruciani F, Trombetta B, Sellitto D, et al. Human Y chromosome haplogroup R-V88: a paternal genetic record of early mid Holocene trans-Saharan connections and the spread of Chadic languages. Eur J Hum Genet. 2010;18:800–807. doi: 10.1038/ejhg.2009.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayser M, Lao O, Anslinger K, et al. Significant genetic differentiation between Poland and Germany follows present-day political borders, as revealed by Y-chromosome analysis. Hum Genet. 2005;117:428–443. doi: 10.1007/s00439-005-1333-9. [DOI] [PubMed] [Google Scholar]
- Underhill PA.Inferring Human History: Clues from Y-Chromosome Haplotypes Cold Spring Harbor Symposia on Quantitative Biology Cold Spring Harbor Laboratory Press: NY, USA; 2003. Vol LXVIII, pp487–493. [DOI] [PubMed] [Google Scholar]
- Alonso S, Flores C, Cabrera V, et al. The place of the Basques in the European Y-chromosome diversity landscape. Eur J Hum Genet. 2005;13:1293–1302. doi: 10.1038/sj.ejhg.5201482. [DOI] [PubMed] [Google Scholar]
- Battaglia V, Fornarino S, Al-Zahery N, et al. Y-chromosomal evidence of the cultural diffusion of agriculture in Southeast Europe. Eur J Hum Genet. 2009;17:820–830. doi: 10.1038/ejhg.2008.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Zahery N, Semino O, Benuzzi G, et al. Y-chromosome and mtDNA polymorphisms in Iraq, a crossroad of the early human dispersal and of post-Neolithic migrations. Mol Phylogenet Evol. 2003;28:458–472. doi: 10.1016/s1055-7903(03)00039-3. [DOI] [PubMed] [Google Scholar]
- Robino C, Crobu F, Di Gaetano C, et al. Analysis of Y-chromosomal SNP haplogroups and STR haplotypes in an Algerian population sample. Int J Legal Med. 2008;122:251–255. doi: 10.1007/s00414-007-0203-5. [DOI] [PubMed] [Google Scholar]
- Rosser ZH, Zerjal T, Hurles ME, et al. Y-chromosomal diversity in Europe is clinal and influenced primarily by geography, rather than by language. Am J Hum Genet. 2000;67:1526–1543. doi: 10.1086/316890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semino O, Passarino G, Oefner PJ, et al. The genetic legacy of Paleolithic Homo sapiens sapiens in extant Europeans: a Y chromosome perspective. Science. 2000;290:1155–1159. doi: 10.1126/science.290.5494.1155. [DOI] [PubMed] [Google Scholar]
- Arredi B, Poloni ES, Tyler-Smith C. Anthropological Genetics: Theory, Methods and Applications. Cambridge University Press: Cambridge; 2010. The Peopling of Europe. [Google Scholar]
- Balaresque P, Bowden GR, Adams SM, et al. A predominantly neolithic origin for European paternal lineages. PLoS Biol. 2010;8:e1000285. doi: 10.1371/journal.pbio.1000285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims LM, Garvey D, Ballantyne J. Sub-populations within the major European and African derived haplogroups R1b3 and E3a are differentiated by previously phylogenetically undefined Y-SNPs. Hum Mutat. 2007;28:97. doi: 10.1002/humu.9469. [DOI] [PubMed] [Google Scholar]
- Myres NM, Ekins JE, Lin AA, Cavalli-Sforza LL, Woodward SR, Underhill PA. Y-chromosome short tandem repeat DYS458.2 non-consensus alleles occur independently in both binary haplogroups J1-M267 and R1b3-M405. Croat Med J. 2007;48:450–459. [PMC free article] [PubMed] [Google Scholar]
- Hinds DA, Stuve LL, Nilsen GB, et al. Whole-genome patterns of common DNA variation in three human populations. Science. 2005;307:1072–1079. doi: 10.1126/science.1105436. [DOI] [PubMed] [Google Scholar]
- Repping S, van Daalen SK, Brown LG, et al. High mutation rates have driven extensive structural polymorphism among human Y chromosomes. Nat Genet. 2006;38:463–467. doi: 10.1038/ng1754. [DOI] [PubMed] [Google Scholar]
- Underhill PA, Myres NM, Rootsi S, et al. Separating the post-Glacial coancestry of European and Asian Y chromosomes within haplogroup R1a. Eur J Hum Genet. 2009;18:479–484. doi: 10.1038/ejhg.2009.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karafet TM, Mendez FL, Meilerman MB, Underhill PA, Zegura SL, Hammer MF. New binary polymorphisms reshape and increase resolution of the human Y chromosomal haplogroup tree. Genome Res. 2008;18:830–838. doi: 10.1101/gr.7172008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhivotovsky LA, Underhill PA, Cinnioğlu C, et al. The effective mutation rate at Y chromosome short tandem repeats, with application to human population-divergence time. Am J Hum Genet. 2004;74:50–61. doi: 10.1086/380911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichler I, Fuchsberger C, Platzer C, et al. Drawing the history of the Hutterite population on a genetic landscape: inference from Y-chromosome and mtDNA genotypes. Eur J Hum Genet. 2010;18:463–470. doi: 10.1038/ejhg.2009.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivedi R, Sahoo S, Singh A, et al. Genetic imprints of Pleistocene origin of Indian populations: a comprehensive Phylogeographic sketch of Indian Y-chromosomes. Int J Hum Genet. 2008;8:97–118. [Google Scholar]
- Shi W, Ayub Q, Vermeulen M, et al. A worldwide survey of human male demographic history based on Y-SNP and Y-STR data from the HGDP-CEPH populations. Mol Biol Evol. 2010;27:385–393. doi: 10.1093/molbev/msp243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittle A.The first farmersIn: Cunliffe B (ed): Prehistoric Europe: An Illustrated History Oxford University Press: Oxford; 1998136–166. [Google Scholar]
- Burkill M.The Middle Neolithic of the Paris Basin and North-Eastern FranceIn: Scarre C (ed).: Ancient France, Neolithic Societies and Their Landscapes 6000-2000 BC The Edinburgh University Press: Edinburgh; 1983. p394 [Google Scholar]
- Sherratt A.The transformation of early agrarian Europe: the later Neolithic and Copper Ages 4500-2500 BCIn: Cunliffe B (ed): Prehistoric Europe: An Illustrated History Oxford University Press: Oxford; 1998167–201. [Google Scholar]
- Collard M, Edinborough K, Shennan S, Thomas M. Radiocarbon evidence indicates that migrants introduced farming to Britain. J Arch Sci. 2009;37:866–870. [Google Scholar]
- Perlès C.The Early Neolithic in Greece: The First Farming Communities in Europe Cambridge: University of Cambridge Press; 2001. p395 [Google Scholar]
- Price TD. Europe's First Farmers: An Introduction: Europe's First Farmers. Cambridge University Press: Cambridge; 2000. pp. 1–18. [Google Scholar]
- Edmonds CA, Lillie AS, Cavalli-Sforza LL. Mutations arising in the wave front of an expanding population. Proc Natl Acad Sci USA. 2004;101:975–979. doi: 10.1073/pnas.0308064100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klopfstein S, Currat M, Excoffier L. The fate of mutations surfing on the wave of a range expansion. Mol Biol Evol. 2006;23:482–490. doi: 10.1093/molbev/msj057. [DOI] [PubMed] [Google Scholar]
- Zimmermann A, Hilpert J, Wendt KP. Estimations of population density for selected periods between the Neolithic and AD 1800. Hum Biol. 2009;81:357–380. doi: 10.3378/027.081.0313. [DOI] [PubMed] [Google Scholar]
- Cuniliffe B.Iron Age societies in Western Europe and beyond, 900-140 BCIn: Cunliffe B (ed): Prehistoric Europe: An Illustrated History Oxford University Press: Oxford; 1998336–372. [Google Scholar]
- Adams SM, King TE, Bosch E, Jobling MA. The case of the unreliable SNP: recurrent back-mutation of Y-chromosomal marker P25 through gene conversion. Forensic Sci Int. 2006;159:14–20. doi: 10.1016/j.forsciint.2005.06.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.