Recently, Whibley et al1 reported on a family carrying a microdeletion at Xp11.3 involving only the MAOA and MAOB genes. The affected brothers in this family suffered from severe mental retardation, epilepsy, stereotypic hand movements, and lip smacking. Here we report on a male patient with mental retardation and intractable epilepsy carrying a duplication of Xp11.3 involving the MAOA, MAOB, and NDP genes.
The patient was the first child of unrelated parents. Pregnancy was complicated with oedemas, and benzodiazepine use because of anxiety. The patient was born 2 weeks post term (BL: 57 cm, BW: 4800 g, OFC: 36 cm). The neonatal period was complicated by opistotonus and by failure to thrive due to Hirschprung disease, for which he was successfully surgically treated at the age of 16 months. Early developmental milestones were delayed. The patient presented with febrile seizures at the age of 12 months. From 3 years of age, afebrile epileptic seizures in terms of generalised atonic, myoclonic, and tonic–clonic seizures were observed. Despite treatment, the epilepsy was intractable. Clinical examination at 32 years of age revealed moderate-to-severe mental retardation, normal stature (185 cm, 87 kg), osteoporosis, slight scoliosis, friendly mood, and a high-pitched voice. The patient had normal vision, normal hearing, and normal sexual maturation.
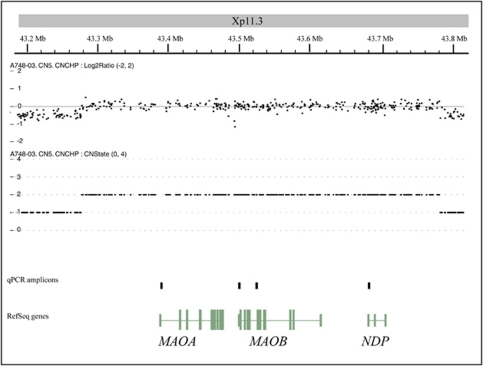
Genome-Wide Human SNP Array 6.0 (Affymetrix) performed on DNA from peripheral blood lymphocytes revealed a 0.5-Mb duplication at chromosome Xp11.3 (chrX: 43290667–43794205, Hg18) (Figure 1). The duplication included the MAOA, MAOB, and NDP genes, and was validated using real-time quantitative PCR (qPCR) and the 2(−Delta Delta C(T)) method. One control region outside and four exonic regions inside the duplicated region were amplified using GAPDH as an internal standard (Supplementary Figure 1).
Figure 1.
Duplication of Xp11.3 in a male patient with mental retardation and epilepsy. Three genes are located in the duplicated region: MAOA, MAOB, and NDP. The duplication was verified by qPCR with amplicons in all three genes.
In this patient, a total of 16 copy number variants (CNVs) of minimum 100 kb were detected (Table 1). Only four of these did not overlap completely with known CNVs reported in the Database of Genomic Variants, and of these only the Xp11.3 duplication contained RefSeq genes. The three other unknown CNVs did not overlap with reported non-genic regulatory landscapes2, 3, 4 around key developmental genes, making it less likely that they could be pathogenic, for example, involved in long-range position effects.
Table 1. Copy number variants (CNVs), larger than 100 kb, detected in a male patient with mental retardation and intractable epilepsy.
| Chr. | Cytoband | Size (kb) | Gain/loss | Start position (Hg 18) | End position (Hg 18) | % CNV overlap |
|---|---|---|---|---|---|---|
| 1 | q23.3–q23.3 | 142 | Gain | 159 763 524 | 159 905 125 | 100 |
| 4 | q13.2–q13.2 | 114 | Loss | 69 054 586 | 69 168 562 | 100 |
| 8 | p23.1–p23.1 | 610 | Gain | 7 237 778 | 7 847 304 | 100 |
| 8 | p11.23–p11.23 | 133 | Loss | 39 354 748 | 39 488 053 | 100 |
| 8 | q21.13–q21.13 | 212 | Loss | 83 306 931 | 83 519 284 | 28 |
| 9 | p11.2–p11.2 | 294 | Loss | 43 445 836 | 43 740 170 | 100 |
| 14 | q11.1–q11.2 | 632 | Loss | 18 860 343 | 19 492 423 | 100 |
| 15 | q14–q14 | 108 | Loss | 32 510 712 | 32 618 224 | 100 |
| 16 | p11.2–p11.2 | 497 | Loss | 32 066 096 | 32 563 012 | 100 |
| 16 | p11.2–p11.2 | 369 | Loss | 33 311 629 | 33 680 554 | 100 |
| 17 | q21.31–q21.32 | 404 | Gain | 41 703 504 | 42 107 467 | 100 |
| 19 | p12–p12 | 120 | Loss | 20 388 034 | 20 508 217 | 100 |
| 22 | q11.21–q11.21 | 238 | Gain | 19 937 072 | 20 175 282 | 100 |
| X | p11.3–p11.3 | 504 | Gain | 43 290 667 | 43 794 205 | 0 |
| X | q21.31–q21.31 | 230 | Gain | 88 352 639 | 88 583 010 | 0 |
| Y | q11.221–q11.221 | 157 | Gain | 16 835 918 | 16 992 884 | 0 |
In total, this patient had 16 CNVs, whereof one partially overlaps with a known CNV and three did not overlap any known CNV region. The 0.5-Mb duplication of Xp11.3 is shown in bold.
Reciprocal deletions and duplications of several genes/loci may result in either different or similar clinical features depending on the gene/locus involved.5 Deletions or point mutations of NDP cause Norrie disease (OMIM #310600), an X-linked disorder characterised by early childhood blindness and progressive sensorineural hearing loss. MAOA and MAOB oxidise biogenic amines in neuronal as well as non-neuronal tissue. Point mutations in MAOA have been associated with ‘antisocial behaviour following childhood maltreatment' (OMIM #309850). Male patients with contiguous deletions of NDP, MAOA, and MAOB present with atypical Norrie disease with a more severe cognitive outcome than the classical Norrie disease. These findings support the importance of MAOA and MAOB for cognitive function reported by Whibley et al.
Only one of the two previously reported duplications of Xp11.36, 7 has been molecularly characterised – Tzschach et al7 described a male patient with mental retardation and epilepsy carrying an inherited 9.3-Mb duplication including MAOA, MAOB, and NDP. Our patient with a much smaller duplication involving only NDP, MAOA, and MAOB also presented with mental retardation and epilepsy. Combined, these findings support that tight dosage regulation of one or more of the three genes is important for normal cognitive function and development of the central nervous system. In contrast to the two patients with Xp11.3 duplication described in the literature, our patient was not obese but only slightly overweight (BMI=25.4). Thus, MAOA is not the only gene determining obesity, consistent with a multifactorial aetiology of obesity.8
The identification of additional cases with Xp11.3 duplications should further contribute to delineation of the associated clinical spectrum and elucidate the specific candidate gene(s) for mental retardation and epilepsy.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Whibley A, Urquhart J, Dore J, et al. Deletion of MAOA and MAOB in a male patient causes severe developmental delay, intermittent hypotonia and stereotyped hand movements Eur J Hum Genet 2010. E-pub 19 May 2010. [DOI] [PMC free article] [PubMed]
- Sandelin A, Bailey P, Bruce S, et al. Arrays of ultraconserved non-coding regions span the loci of key developmental genes in vertebrate genomes. BMC Genomics. 2004;5:99. doi: 10.1186/1471-2164-5-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovcharenko I, Loots GG, Nobrega MA, Hardison RC, Miller W, Stubbs L. Evolution and functional classification of vertebrate gene deserts. Genome Res. 2005;15:137–145. doi: 10.1101/gr.3015505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolfe A, Goodson M, Goode DK, et al. Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol. 2005;3:e7. doi: 10.1371/journal.pbio.0030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Lupski JR. CNV and nervous system disease—What's new. Cytogenet Genome Res. 2008;123:54–64. doi: 10.1159/000184692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen KB, Langkjaer F. Inherited partial X chromosome duplication in a mentally retarded male. J Med Genet. 1982;19:222–224. doi: 10.1136/jmg.19.3.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzschach A, Chen W, Erdogan F, et al. Characterization of interstitial Xp duplications in two families by tiling path array CGH. Am J Med Genet. 2008;146A:197–203. doi: 10.1002/ajmg.a.32070. [DOI] [PubMed] [Google Scholar]
- Hinney A, Vogel CIG, Hebebrand J. From monogenic to polygenic obesity: recent advances. Eur Child Adolesc Psychiatry. 2010;19:297–310. doi: 10.1007/s00787-010-0096-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.