Abstract
Purpose of review
We provide a long-term perspective of our experience with designing and managing a successful biorepository system. We include a brief history, a description of our current process, and lessons learned.
Recent findings
Biologic specimens, collected and stored as part of HIV-related research for years, are now being used for biomarker analyses that have important implications for both AIDS and non-AIDS events. If appropriately collected, documented and stored, biospecimens are a valuable resource that can help answer current and future scientific questions. International networks must be able to monitor and adhere to country-specific specimen use regulations. Specimens for human DNA research need increased levels of privacy protection. Issues to consider when designing a biorepository system include expertise, communication, data management, technology, standardized methods and procedures, shipping, and specimen use policies.
Summary
As biorepositories are an integral part of research their design should not be an afterthought. Good designs consider all stages of research, and the most critical components are expertise and planning. Successful biorepository systems must have a balance of flexibility and standardization. The need for adaptable data management systems, whether commercial products or systems developed specifically for the network, should not be underestimated. Investment in appropriate technology, including a barcoding system with high quality labels and printers, will pay off in the long term. To meet the needs of emerging technologies it is becoming increasingly important to document the conditions at the time of specimen collection and processing. Regular communication between all components of the biorepository system is critical.
Keywords: Biorepository, HIV, biomarkers, repository data management systems
Introduction
While biospecimens have been stored and used for retrospective studies as part of HIV-related research for many years an example illustrates their utility. A 2008 report from the Strategies for Management of Anti-Retroviral Therapy (SMART) study found that higher baseline levels of C-reactive protein (CRP), interleukin-6 and D-dimer measured on stored specimens from the baseline examination were associated with an increased risk of all-cause mortality[1], and that despite suppression of HIV RNA those markers remained elevated compared to the general population[2**]. Yet when the SMART study was designed in 2001 it was not yet known which biomarkers would eventually be analyzed. It may be many years after specimen collection and storage until promising biomarkers are identified. Enrollments in the Multiple Risk Factor Intervention Trial, a primary prevention trial of coronary heart disease (CHD), were completed in 1975 but it wasn’t until 20 years later that the relationship between CRP and CHD risk [3] was examined for the first time using stored specimens. In HIV research, a 1997 study using specimens that had been stored for more than 10 years found that plasma viral load was the single best predictor of clinical outcomes [4].
Biospecimens are being stored at a rapid rate. In the SMART study, with 3400 person years of follow-up for 5472 HIV-infected individuals, more than 325,000 biospecimen vials were stored. Of those over 40,000 vials have been assayed, resulting in 8 biomarker publications in print and many more in progress. If appropriately collected, documented and stored, biospecimens are a valuable resource that can help answer current and future scientific questions.
We review the 15-year process of designing and managing a successful biorepository system. The current system is managed by the International Network for Strategic Initiatives in Global HIV Trials (INSIGHT) network, a merger in 2006 of two HIV-research networks. As part of INSIGHT over 300 clinical sites in 38 different countries collect and ship specimens to biorepositories around the world. Because of its well-developed and international infrastructure INSIGHT quickly adapts to changing needs and is now also conducting two international observational studies on the pandemic influenza virus.
As a large global clinical research network the INSIGHT biorepository system has specific challenges, including working with clinical sites that are both small and large, in settings that are both community- and research-based; compliance with international shipping requirements; and collection of specimens for genomic research that require additional levels of identification protection [5–8]. Our considerations and lessons learned will be useful for many types of research settings.
A brief history of the INSIGHT biorepository system
The current INSIGHT repository system has its roots with the Community Programs for Clinical Research on AIDS (CPCRA), formed in 1989 to conduct research in U.S. primary care settings. In 1995 sites began shipping biospecimens, initially directly to a testing laboratory and beginning in 1998 to a central biorepository. Sites completed a specimen tracking form and submitted it to the Statistical and Data Management Center (SDMC), which communicated directly with the central biorepository to designate specimens for real-time testing or long term storage. From 1998–2002 the CPCRA sites did not use barcoded labels and the central biorepository used manual data entry for documenting the storage location and testing results.
In 2000 the Evaluation of Subcutaneous ProleukinR in a Randomized International Trial (ESPRIT) study [9**] opened and randomized 4150 participants from 25 countries within three years. The ESPRIT network used barcoded labels on specimens, virtually eliminating manual data entry. In 2002 the CPCRA network also became an international organization with the SMART study. Because the SMART study required immediate testing of various specimens at multiple locations CPCRA switched to using barcoded labels and the central repository incorporated real-time scanning and sorting of biospecimens.
The integration of CPCRA and ESPRIT into the INSIGHT network required extensive collaborations and communication, as hundreds of clinical sites and laboratories were involved. A Laboratory Procedures Group was an instrumental component, in which members addressed concerns in a structured manner. Throughout the process the INSIGHT SDMC, common to both CPCRA and ESPRIT, was the essential link between the collection sites, coordinating centers, testing laboratories, and biorepositories.
Current INSIGHT biorepository procedures
The INSIGHT network[10] includes four International Coordinating Centers (ICCs): the Copenhagen HIV Programme (CHIP), Copenhagen, Denmark; the Medical Research Council (MRC) Clinical Trials Unit, London, United Kingdom; the National Centre in HIV Epidemiology and Clinical Research (NCHECR), Sydney, Australia; and the Institute for Clinical Research at the Veterans Affairs Medical Center, Washington, D.C., USA. Each ICC is composed of multiple Site Coordinating Centers (SCCs) and individual clinical sites. The SDMC communicates directly with the ICCs, which in turn communicate with the SCCs and clinical sites.
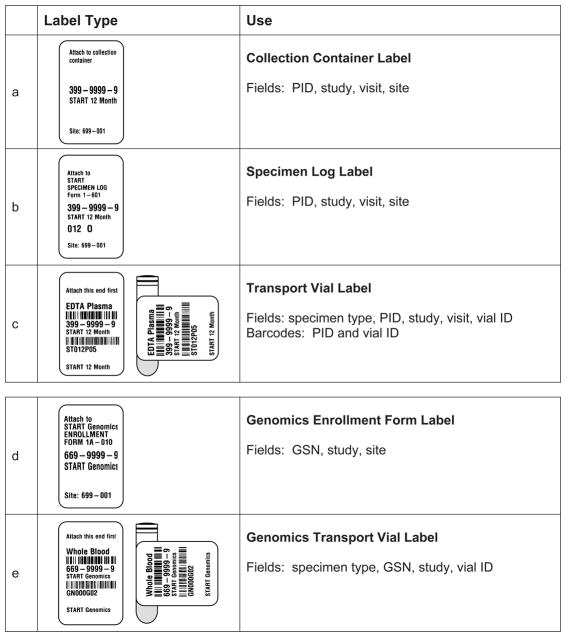
Each study participant is assigned a unique identification number (PID). The SDMC sends packets of pre-printed visit labels for the specimen collection containers, specimen logs, and transport vials. The labels for the containers and logs (Figure 1, panels a and b) are pre-printed with the PID, study, visit, and site code. Labels for the transport vials (Figure 1, panel c) have two barcodes: one with the PID and another with a code encrypted for study, visit, specimen type and vial number. All barcoded information is also printed in eye-readable form.
Figure 1.
Examples of labels used for specimen collection (panel a), shipping paperwork (panel b), transport vials (panels c and e), and an enrollment form for a genomic study (panel d).
Panels a–c are examples for the 12-month visit for the START study, and have a participant identification number (PID). Labels for genomic specimens (panels d and e) have a genomic sequence number (GSN) and not a PID, so that the biorepository can not associate a PID with a GSN. The barcoded vial identification (panels c and e) is code composed of two characters for the study, three digits for visit number, one character for specimen type, and two digits for vial number.
At the clinical site specimens are collected, processed and aliquoted according to study specifications and documented on a specimen log. Depending on the specific ICC structure the site either ships the specimens directly to the central biorepository or to an interim repository. Because of the unique barcoded identification specimens can be shipped together without sorting by study, PID, visit, or specimen type. The facility shipping to the central biorepository sends the specimen log to the SDMC. In addition, a shipping manifest is completed summarizing the type, volume and quantity of specimens, per the International Air Transport Association (IATA) regulations[11]. Copies of the manifest are included with the biospecimens and are electronically sent to the central biorepository and the SDMC.
The SDMC enters the data from the shipping log and manifest. If the study requires real-time assays the SDMC sends the central biorepository an electronic file specifying the PID, study, visit, and specimen type. The central biorepository loads that file into their Repository Data Management System (RDMS) before specimens are checked in. As barcoded specimens are scanned at the central repository the RDMS creates a database record and automatically alerts the technician to sort the vial into a long-term storage location or a temporary location prior to immediate testing or shipment to another laboratory or repository. The biorepository sends regular updates of the database to the SDMC.
When the use of stored specimens has been approved for a research project the SDMC sends the biorepository the authorizing paperwork and a list of specimens for withdrawal. The biorepository pulls the specimens, scans them out of the RDMS, and ships them to the testing laboratory. The left-over specimens, in the original transport vials, are rescanned into the RDMS when they are returned.
INSIGHT also collects biospecimens for genomic studies which involve a one-time specimen collection. The SDMC provides labels (Figure 1, panels d an e) with a Genomics Sequence Number (GSN). At the site level both a PID and a GSN are reported on the enrollment form, which is sent to the SDMC. As the biospecimen is barcoded with the GSN (Figure 1, panel e) the central biorepository never has the association between PID and GSN. When the genomic specimen is approved for use the SDMC sends the central biorepository a general identification number (GID) for each GSN. The GID label, designed to adhere to previously frozen specimens, is placed over the GSN label, effectively making the GSN illegible on the vial. The genomic specimen is then processed into multiple aliquots of DNA. The DNA cryovials are barcoded with only the GID and a vial identification code. One cryovial of DNA is shipped to the approved processing laboratory and the others are placed into long-term storage. The identification of the vials of DNA is now two levels removed from the PID. Only designated SDMC representatives can associate the PID, GSN, and GID.
Elements of a successful biorepository system (and lessons learned)
The considerations below are for research settings with the following goals: making specimen collection, documentation and shipping as easy as possible for clinical sites (which may not be at research institutions); obtaining high quality specimens with minimal on-site processing; and minimizing overall cost. Table 1 summarizes some of our challenges and solutions. Guidelines in the form of Best Practices for Repositories are also available from organizations such as the International Society for Biological and Environmental Repositories [12] and the National Cancer Institute [13] (NCI). Descriptions of the establishment and operation of biorepositories are also provided by Cortes et al [14*] for networks interested in shifting the specimen processing towards the physical specimen collection locations, and by Wich et al [15*] for a single-site multidisciplinary melanoma biospecimen bank linked to clinical information.
Table 1.
Challenges and solutions related to designing and managing a biorepository system for a multi-site network
| Challenge | Solution(s) |
|---|---|
| Designing a biorepository system that works for all stages of research | Expertise and buy-in at all levels (clinical, laboratory, biorepository, data management, statistical). Regular communication via a Laboratory Procedures Group to identify and solve problems and evolving issues in real time. A central statistical and data management center (SDMC) for specimen tracking and management, monitoring adherence to country- and site-specific regulatory requirements, and linking the clinical and laboratory data. |
| Flexibility to meet emerging research needs | Appropriate programming expertise at both the SDMC and the central biorepository. Both data management systems should accept multiple data formats and be quickly adaptable for including different data fields. |
| Obtaining high quality specimens from many types of clinical sites (from small clinics with little infrastructure or support to large research-based settings) | As much as possible standardize data collection and specimen collection, labeling and storage procedures. Minimize specimen processing at the clinical sites. Eliminate specimen-related data entry at the clinical sites. Use a bar-coding system for biospecimens with high quality labels and thermal label printers. Have pre-printed labels and pre-defined shipping schedules. Allow shipping boxes to include specimens of different types and from different studies. A well-designed and updated central website for information and training. |
| Preserving specimens collected specifically for future biomarker research (some assays are sensitive to collection and processing procedures) | Choose clinical sites that have the capability to collect and process specimens according to the study design. Document the conditions at the time of specimen collection (fasting status, time of day, quality of venipuncture, etc.) and during specimen processing (time from collection to processing, temperature during processing and storage, etc.) Documenting the conditions will allow future researchers to choose only specimens that were collected and processed appropriately for a particular assay. |
| Privacy protections for DNA specimens | Additional levels of privacy protection so that genetic information can not be linked back to a particular person, and appropriate security measures for the storage and use of sensitive data at the SDMC. |
| Logistics of specimen use at the biorepository (real-time testing, long-term storage, or batch testing) | An adaptable repository data management system (RDMS) that allows for real-time scanning and sorting of specimens. Using an electronic file provided by the SDMC before the specimens arrive, the RDMS alerts the technician when specimens should be used for immediate testing. |
| Protecting biospecimens as a valuable resource | A written specimen use procedure, including approval mechanisms for last vial use, using non-pristine (returned or leftover) vials when possible, and ensuring that specimens are returned to the biorepository. |
Expertise and communication
From design through management involve experts at all levels (clinical, laboratory, repository, data management and statistics) who are willing to communicate regularly and frequently. We have a Laboratory Procedures Group, with representation from ICCs, laboratories, clinicians, repositories and the SDMC. The group shares contacts and resources, meets with study teams and the executive committee, and can identify and resolve commonly encountered problems as well as unique situations.
Data management
The system(s) used for the biorepository and clinical data are crucial. Our biorepository system has several unique features. First, the system does not require specimen data entry at the site level, eliminating the need to have computers and software available and maintained at each site. There are other RDMS options available that require computer equipment at each site, including the Laboratory Data Management System[16*] and the Biological Specimen Inventory System[17*]. Second, the RDMS used at our central biorepository scans and sorts the specimens as they arrive, allowing for real-time testing or re-shipment with minimal handling and maximizing storage space in repository freezers. Other specimen management systems have a two-stage process of first scanning specimens and placing them into storage and then removing the specimen from storage for subsequent testing. Our RDMS requires programming expertise at the central repository but results in a product that is specific to our needs and quickly adaptable for emerging needs. Another strength of our data management system is a central statistical and data management center, where data in many formats can be imported and exported; new fields can be added to file structures as needed; country-specific specimen use regulations can be monitored and adhered to; and the clinical and specimen data can be linked. A central SDMC also facilitates the tracking and coordination of biospecimen availability and shipments, laboratory analysis results and statistical analyses for multiple collaborators.
Standardization and training
Simplify and minimize what the clinical sites need to do. Have clear instructions, materials, and frequently updated training programs readily available on a well-designed website. To increase efficiency, standardize the specimen and processing requirements between studies as much as possible. Consider the type and size of cryovials (internally versus externally threaded), the exact wording for specimen volume (e.g. ‘at least 1 mL’ versus ‘less than 1.5 mL’ for 2.0 mL cryovials), the size of cryogenic grid boxes, and temperature requirements. With earlier protocols that were not standardized across studies the sites needed to keep track of multiple requirements; specimens from studies that allowed larger transport vials required additional freeze/thaw cycles; and repositories needed additional freezer space to accommodate different sized grid boxes.
Data collection
Knowing the conditions and methods used at the time of specimen collection and processing is critical for some biomarker assays. Biomarker assays can be sensitive to many conditions, including diurnal rhythm, sample type, the time and temperatures from collection to storage, platelet contamination, and hemolysis [18–26]. Careful documentation of the methods used and the conditions during collection and processing will increase the future value of specimens.
Barcodes, labels, and printing
Avoid manual data entry by developing unique barcoded identifiers. Pre-print the labels with all barcoded information in eye-readable form (Figure 1, panels c and e). With our original studies the clinical sites manually wrote information on labels, resulting in transcription errors and discrepancies between labels and forms. We currently use Code 128, a high-density linear alphanumeric barcode symbology; 2-dimensional matrix barcodesare being considered for some projects. Design labels so that the barcode(s) will run parallel to the length of the vial (Figure 1, panels c and e). We use labels designed to withstand the temperature variation from room temperature collection to −80°C or vapor phase liquid nitrogen for storage [27, 28]. The labels are also designed so that when placed correctly on the vial a portion of the label overlaps on itself, increasing the label integrity and secure adherence on the vial. Additional technical and product-related information for labeling biospecimens is detailed in a White Paper [29].
Invest in a high-quality thermal barcode printer. We use high-volume, 600 dots per inch printers [27, 28]. We originally used laser and dot matrix printers, but some of those labels became so degraded during handling and storage that they were no longer readable and required relabeling, a very labor intensive process that can also introduce errors.
If labels are lost or need to be reprinted, new vial identification numbers must be used. We tried reprinting labels to exactly match the original set, but those ‘lost’ labels would occasionally be found and used, resulting in duplicate sets at the central biorepository.
We print all labels at the SDMC and ship to the ICCs for distribution, as the cost of central printing and shipping is lower than the fiscal and training costs of buying and maintaining expensive barcode printers at each site or ICC. Central printing also ensures that each vial can be uniquely identified.
Shipping
Appropriate regulations, including individual IATA record of training certifications[11], must be carefully followed. Clearly written procedures, including shipping frequency, must be readily available for all sites and repositories. For sites without regularly scheduled shipping days there should be a maximum storage time for specimens so that there is at least one shipment in every pre-defined interval, such as every 3 months. We learned that allowing long-term storage of specimens at clinical sites resulted in missing specimens, specimen integrity problems, and the inability to identify and resolve ongoing problems with specimen processing or labeling.
The choice of courier depends on country and study-specific requirements. Within the U.S. there are many shipping options, as cost is determined by container and overnight shipping is widely available if necessary. For international shipping it is most cost effective to ship multiple containers at the same time. We use a courier [30] that offers dry ice replenishment during transit as necessary.
The choice of packaging materials also depends on country. When available, it is advisable to use materials from companies [31, 32]that offer packaging components that have been tested as a system and meet IATA standards. Other countries may need to use courier-supplied materials.
International shipping also requires additional paperwork to be included with each shipment, including permits and labels from the Centers for Disease Control and Prevention[33*] and a Customs Invoice[34*, 35*]. A U.S. Department of Agriculture (USDA) permit is not required, but a separate USDA statement [36*] must accompany the shipment. We post the necessary forms, labels and documents for international shipping on the password-protected section of our website.
Specimen use procedures
Have a formal procedure to approve and track specimen use. The governing body for approval needs to be clearly specified and the mechanisms for verifying informed consent and tracking specimen shipment and subsequent return or destruction must be detailed. Because biospecimens are a limited and valuable resource our policy requires a series of written approvals before using the last specimen vial for a particular PID and study visit; having the testing laboratory re-freeze and return used specimens with documentation on the specimen volume used and freeze/thaw cycles; and preserving pristine vials by selecting a partially used specimen vial for use if appropriate. The SDMC tracks the status of the specimens at all times, including informed consent level, storage location, estimated volume, number of freeze/thaw cycles and final disposition.
Conclusions
Biorepositories are an integral part of research. Whether you are a small organization with a few thousand specimens or a large network with tens of thousands of specimens to collect and store, biorepository design should not be an afterthought. Good designs encompass and consider all stages of research, and the most critical components are expertise and planning at all levels. A successful biorepository system will be flexible enough to meet the needs of the smallest site but standardized as much as possible to meet the needs of the network. The need for adaptable data management systems, whether commercial products or systems developed specifically for the network, should not be underestimated. Investment in appropriate technology, including a barcoding system with high quality labels and printers, will pay off in the long term. International networks must have the capacity to monitor and adhere to country-specific specimen use regulations. Some specimens, such as those for human DNA research, need increased levels of privacy protection. To meet the needs of emerging and future technologies it is becoming increasingly important to document the conditions at the time of specimen collection and processing. Regular communication between all components of the biorepository system is critical.
Acknowledgments
We thank the following individuals for valuable assistance with this paper: Alain DuChene (Division of Biostatistics, University of Minnesota, Minneapolis, MN); Marie L. Hoover (Advanced BioMedical Laboratories, Cinnaminson, NJ); Julia A. Metcalf (National Institute of Allergy and Infectious Disease, Division of Clinical Research, Bethesda, MD); Terese Schultz (Division of Biostatistics, University of Minnesota, Minneapolis, MN); Russell P. Tracy (University of Vermont, Burlington, VT).
Disclosure of funding
This research was supported in part by the National Institute of Allergy and Infectious Disease, including grant number U01 AI068641.
This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Abbreviations
- GID
General Identification Number
- GSN
Genomics Sequence Number
- ICC
International Coordinating Center
- PID
Participant Identification Number
- RDMS
Repository Data Management System
- SDMC
Statistical and Data Management Center
References
- 1.Kuller LH, Tracy R, Belloso W, et al. Inflammatory and coagulation biomarkers and mortality in patients with HIV infection. PloS Medicine. 2008;5(10):1496–1508. doi: 10.1371/journal.pmed.0050203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2**.Neuhaus J, Jacobs DR, Baker JV, et al. Markers of inflammation, coagulation, and renal function in HIV-infected adults in SMART and in two large population-based cohorts. J Infect Dis. 2010;201(12):1788–1795. doi: 10.1086/652749. This study compares levels of biomarkers (hsCRP, IL-6, D-dimer, and cystatin C) in HIV-infected persons from the SMART study and HIV-uninfected persons from the MESA and CARDIA studies. Biomarker levels were higher among those with HIV infection and remained so even with virologic suppression and the use of ART. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kuller LH, Tracy RP, Shaten J, Meilahn EN. Relation of C-Reactive protein and coronary heart disease in the MRFIT nested case-control study. Am J Epidemiol. 1996;144(6):537–547. doi: 10.1093/oxfordjournals.aje.a008963. [DOI] [PubMed] [Google Scholar]
- 4.Mellors JW, Munoz A, Giorgi JV, et al. Plasma viral load and CD4+ lymphocytes as prognostic markers of HIV-1 infection. Annals of Internal Medicine. 1997;126(12):946–954. doi: 10.7326/0003-4819-126-12-199706150-00003. [DOI] [PubMed] [Google Scholar]
- 5.McGuire AL, Colgrove J, Whitney SN, et al. Ethical, legal, and social considerations in conducting the Human Microbiome Project. Genome Research. 2008;18:1861–1864. doi: 10.1101/gr.081653.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lowrance WW, Collins FS. Identifiability in genomic research. Science. 2007;317(5838):600–602. doi: 10.1126/science.1147699. [DOI] [PubMed] [Google Scholar]
- 7.Greenbaum D, Du J, Gerstein M. Genomic anonymity: have we already lost it? American Journal of Bioethics. 2008;8(10):71–81. doi: 10.1080/15265160802478560. [DOI] [PubMed] [Google Scholar]
- 8.Malin B, Sweeney L. How (not) to protect genomic data privacy in a distributed network: using trail re-identification to evaluate and design anonymity protection systems. Journal of Biomedical Informatics. 2004;37(3):179–192. doi: 10.1016/j.jbi.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 9**.The INSIGHT-ESPRIT Study Group and SILCAAT Scientific Committee. Interleukin-2 therapy in patients with HIV infection. N Engl J Med. 2009;361:1548–1559. doi: 10.1056/NEJMoa0903175. The results from 2 randomized studies (ESPRIT and SILCAAT) show that while the use of intermittent IL-2 with ART led to sustained CD4 cell count increases compared to the use of ART without IL-2, there were no clinical benefits (measured by opportunistic disease or death). The authors suggest two hypotheses to explain the results. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.International Network for Strategic Initiatives in Global HIV Trials (INSIGHT) http://insight-trials.org.
- 11.International Air Transport Association (IATA) [Online, accessed 24 May 2010];The IATA dangerous goods regulations. http://www.iata.org/whatwedo/cargo/dangerous_goods/Pages/infectious_substances.aspx.
- 12.International Society for Biological and Environmental Repositories. Best practices for repositories: Collection, storage, retrieval and distribution of biological materials for research. Cell Preservation Technology. 2008;6(1):1–58. http://www.isber.org/ibc.html.
- 13.National Cancer Institute Office of Biorepositories and Biospecimen Research. [Online, accessed 24 May 2010];Best Practices for Biospecimen Resources. http://biospecimens.cancer.gov/bestpractices.
- 14*.Cortes B, Schiffman M, Herrero R, et al. Establishment and operation of a biorepository for molecular epidemiologic studies in Costa Rica. Cancer Epidemiol Biomarkers Prev. 2010;19(4):916–922. doi: 10.1158/1055-9965.EPI-10-0066. For over 10 years this research group shipped biospecimens from Costa Rica to the U.S. on a monthly basis. This paper describes the planning and set-up of the technology transfer to a biorepository in Costa Rica, so that real-time testing for clinical management could be accomplished. [DOI] [PubMed] [Google Scholar]
- 15**.Wich LG, Hamilton HK, Shapiro RL, et al. Developing a multidisciplinary prospective melanoma biospecimen repository to advance translational research. Am J Transl Res. 2009;1(1):35–43. Although this biorepository was sent up by a single center the group thoughtfully describes the same issues and considerations that a large network would need to address when designing a biorepository, including the needs for adequate infrastructure, standardization of biospecimen collection, and adaptability. [PMC free article] [PubMed] [Google Scholar]
- 16*.Frontier Science and Technology Research Foundation [Amherst, NY USA] Laboratory Data Management System; http://www.fstrf.org/ldms. The LDMS system, developed specifically for AIDS research and used by several NIH-funded clinical trial networks, requires a computer, software and data entry at each processing site. Frontier Science does not support running the LDMS in a thin client environment. [Google Scholar]
- 17*.Information Management Services [Rockville, MD USA] Biological Specimen Inventory System; http://www.bsi-ii.com. BSI, a web-based program used by many NCI repositories, also requires a computer and data entry at each processing site. [Google Scholar]
- 18.Ambrosone CB. Sample collection, processing, and storage for large-scale studies: Biorepositories to support cancer research. Cancer Epidemiol Biomarkers Prev. 2006;15(9):1574. doi: 10.1158/1055-9965.EPI-06-0627. [DOI] [PubMed] [Google Scholar]
- 19.Tworoger SS, Hankinson SE. Collection, processing, and storage of biological samples in epidemiologic studies: sex hormones, carotenoids, inflammatory markers, and proteomics as examples. Cancer Epidemiol Biomarkers Prev. 2006;15(9):1578–1581. doi: 10.1158/1055-9965.EPI-06-0629. [DOI] [PubMed] [Google Scholar]
- 20.Vaught JB. Blood collection, shipment, processing, and storage. Cancer Epidemiol Biomarkers Prev. 2006;15(9):1582–1584. doi: 10.1158/1055-9965.EPI-06-0630. [DOI] [PubMed] [Google Scholar]
- 21.Weber M, Rabenau B, Stanisch M, et al. Influence of sample type and storage conditions on soluble CD40 ligand assessment. Clinical Chemistry. 2006;52(5):888–891. doi: 10.1373/clinchem.2005.062083. [DOI] [PubMed] [Google Scholar]
- 22.Macy E, Meilahn E, Declerck P, Tracy R. Sample preparation for plasma measurement of plasminogen activator inhibitor-1 antigen in large population studies. Arch Path Lab Med. 1993;117:67–70. [PubMed] [Google Scholar]
- 23.Refaai MA, Van Cott EM, Lukoszyk M, et al. Loss of Factor VIII and von Willebrand factor activities during cold storage of whole blood is reversed by warming. Laboratory Hematology. 2006;12:99–102. doi: 10.1532/LH96.05043. [DOI] [PubMed] [Google Scholar]
- 24.Stenner D, Tracy R, Riggs B, Mann K. Human platelets contain and secrete osteonectin, a major protein of mineralized bone. Proc Natl Acad Sci USA. 1986;83:6892–6896. doi: 10.1073/pnas.83.18.6892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henn V, Steinbach S, Buchner K, et al. The inflammatory action of CD40 ligand (CD154) expressed on activated human platelets is temporally limited by coexpressed CD40. Blood. 2001;98(4):1047–1054. doi: 10.1182/blood.v98.4.1047. [DOI] [PubMed] [Google Scholar]
- 26.Leyland-Jones BR, Ambrosone CB, Bartlett J, et al. Recommendations for collection and handling of specimens from group breast cancer clinical trials. Journal of Clinical Oncology. 2008;26(34):5638–5644. doi: 10.1200/JCO.2007.15.1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zebra Technologies [Lincolnshire, IL USA] http://www.zebra.com.
- 28.Brady Worldwide, Inc. [Milwaukee, WI USA] http://www/brady.com.
- 29.Cosentino M, Schoenborn J. Labeling of biological specimens. [Online, accessed 01 June 2010];White Paper. 2008 http://web.ncifcrf.gov/repository/cr/docs/WhitePaper.pdf.
- 30.World Courier Management, Inc. [Stamford, CT USA] http://www.worldcourier.com.
- 31.Saf-T-Pak Inc. [Edmonton, Alberta Canada] http://www.saftpak.com.
- 32.All-Pak, Inc. [Chicago, IL USA] http://www.all-pak.com.
- 33*.Centers for Disease Control and Prevention. [Online, accessed 24 May 2010];Etiologic agent import permit program. http://cdc.gov/od/eaipp/. This CDC website provides a clearly written summary of the Import Permit Program. It references specific federal regulations and includes valuable links for packaging guidelines.
- 34*.United States Bureau of Customs and Border Protection. [Online, accessed 01 June 2010];19CFR141.86. http://edocket.access.gpo.gov/cfr_2008/aprqtr/pdf/19cfr141.86.pdf Other terms for ‘customs invoice’ include ‘pro forma invoice’ and ‘commercial invoice’. This website provides the Code of Federal Regulations Section 141.86, including the contents of invoices and general requirements.
- 35*.United States Bureau of Customs and Border Protection. [Online, accessed 02 June 2010]; http://www.cbp.gov/linkhandler/cgov/newsroom/publications/trade/iius.ctt/iius.pdf This CBP guide includes suggestions to the exporter for faster clearance of goods at the U.S. border.
- 36*.United States Department of Agriculture Animal and Plant Health Inspection Service. [Online, accessed 24 May 2010];Guidelines for importation of human and non-human primate material. http://www.aphis.usda.gov/import_export/animals/animal_import/downloads/ihnhum.html. Specimens (such as peripheral blood mononuclear cells) that are processed using fetal calf serum or fetal bovine serum may be regulated by the USDA.