Abstract
Proline switches, controlled by cis–trans isomerization, have emerged as a particularly effective regulatory mechanism in a wide range of biological processes. Here we report the structures of both the cis and trans conformers of a proline switch in Crk signaling protein. Proline isomerization toggles Crk between two conformations: an autoinhibitory, stabilized by the intramolecular association of two tandem SH3 domains in the cis form, and an uninhibited, activated conformation promoted by the trans form. In addition to acting as a structural switch the heterogeneous proline recruits cyclophilin A, which accelerates the interconversion rate between the isomers thereby regulating the kinetics of Crk activation. The data provide atomic insight into the mechanisms that underpin the functionality of this binary switch and elucidate its remarkable efficiency. The results also reveal novel SH3 binding surfaces highlighting the binding versatility and expanding the non-canonical ligand repertoire of this important signaling domain.
INTRODUCTION
The Crk family of adaptor proteins are ubiquitously expressed in most tissues and mediate the timely formation of protein complexes elicited by a variety of extracellular stimuli, including various growth and differentiation factors1,2. Crk proteins are overexpressed in many human cancers3–5 and were found to stimulate the activity of Abl6, a kinase whose fusion to Bcr causes chronic myelogenous leukemia7. Cellular Crk (c-Crk-II) consists of a single SH2 domain, an N-terminal SH3 (SH3N) and a C-terminal SH3 domain (SH3C) (Fig. 1a). The SH3N and SH3C domains are tethered by a 50-residue long linker, which contains a Tyr residue (Tyr222 in chicken) that becomes phosphorylated by Abl8. Crk exerts its function by using its SH2 and SH3N domains, which selectively bind to pY-x-x-P and P-x-L-P-x-K motifs, respectively, to interact with a variety of proteins9,10. In contrast, recent structural work on chicken11 and human Crk12 has shown that SH3C plays an autoregulatory role, consistent with earlier biological results13–16.
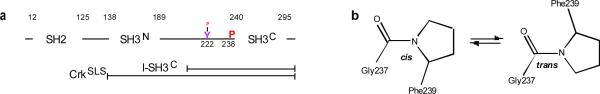
Figure 1. Domain organization of Crk.
(a) Schematic diagram of the domain organization of Crk. Pro238, which undergoes cis–trans isomerization, and the tyrosine residue (Tyr222) that becomes phosphorylated by Abl are indicated. (b) Schematic of cis–trans isomerization about the prolyl Gly237–Pro238 bond.
We have recently shown11 that the binding activity of chicken Crk is modulated by cis–trans isomerization at Pro238, a highly conserved proline residue (Supplementary Fig. 1)17,18. In contrast to covalent modifications afforded by post-translational processes, proline isomerization is an intrinsic conformational exchange process that has the potential to control protein activity19,20 without altering the covalent structure of the protein. Proline isomerization may exert its function through multiple, non-mutually-exclusive mechanisms, involving (i) significant conformational changes caused by the 180° rotation about the prolyl bond, (ii) slow kinetics of isomerization affording a molecular timer, (iii) and the recruitment of prolyl cis–trans isomerase enzymes (PPIases)21. For these reasons proline cis–trans isomerization is emerging as a critical component of an ever increasing number of important biological processes, including cell signaling11,22,23, neurodegeneration24, amyloidogenesis25, channel gating26, gene regulation27,28, phage and virus infection29,30, enzyme function31,32, and ligand recognition33. However, despite the importance of cis–trans isomerization, there is a surprising paucity of high-resolution structural data elucidating the effect that this process may elicit within a protein. Thus, how proline isomerization yields a functional binary switch remains poorly understood.
Here we present high-resolution structures of both the cis and trans conformers of Pro238 in Crk. The data showed that the linker interacts extensively with the SH3C domain and the interaction is strongly modulated by proline isomerization. The remarkable conformational rearrangement induced by cis–trans isomerization endows the conformers with distinct binding properties: in the cis form the two SH3 domains engage in an intramolecular fashion giving rise to an autoinhibitory conformation that prevents the binding of physiological ligands, such as the Abl kinase; in contrast, in the trans form the two SH3 domains do not interact and Crk exists in a uninhibited, ligand-binding-competent conformation. Interestingly, all of these interactions are mediated by novel SH3 binding surfaces. The results (i) provide unprecedented insight into the drastic, long-range conformational effect that proline isomerization may elicit constituting an elegant mechanism for protein activity regulation and (ii) highlight the versatile and unique binding properties of the SH3 modular domain.
RESULTS
Structure of the trans and cis conformers of Crk
We have previously shown11 that Pro238 in Crk undergoes cis–trans isomerization (Fig. 1) giving rise to two distinct, equally populated conformational states of the linker-SH3C (l-SH3C) polypeptide (Supplementary Fig. 2a). Because of the intrinsically high activation energy for rotation about the prolyl bond (~20 kcal mol−1)17, the two conformational states interconvert at a very slow rate. Moreover, the cis and trans conformers in Crk l-SH3C are equally populated (~50% each; Supplementary Fig. 2a). These favorable conditions enabled the complete NMR characterization and structure determination of both conformers (Fig. 2 and Supplementary Fig. 3 and Supplementary Methods).
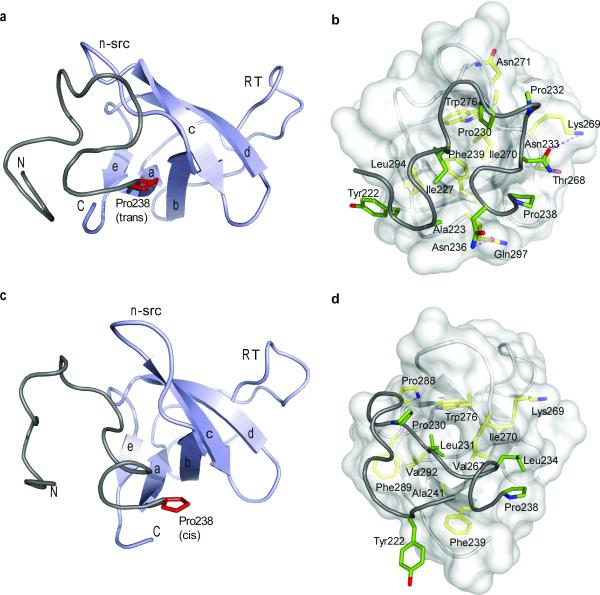
Figure 2. Structural characterization of the trans and cis Crk l-SH3C conformers.
(a,b) Structure of the Crk l-SH3C polypeptide in the trans conformation and (c,d) in the cis conformation. In (a and c) the SH3C domain is colored light blue and the linker is colored grey. Pro238 is shown in red. N and C denote the N- and C-terminus. In (b and d) the SH3C domain is displayed as a semi-transparent surface and residues involved in the linker–SH3C interactions are highlighted. Linker and SH3C residues are colored green and yellow, respectively. Dotted lines denote hydrogen bonding.
Pro isomerization modulates intramolecular interactions
The lowest-energy structure of the trans and cis conformers of l-SH3C are displayed in Figure 2a,b. In both conformers the SH3C domain adopts a typical SH3 fold; however, because of the lack of aromatic residues at its canonical binding site, SH3C does not bind polyproline II (PPII)-type sequences13,34. Superposition of the structures of the cis and trans conformers showed that isomerization about the Gly237–Pro238 prolyl bond results in notably distinct conformations of the linker with respect to SH3C (Fig. 3a). In the trans form the SH3C n-src loop adopts a typical conformation seen in the majority of the SH3 domains. However, in the cis form the n-src loop rearranges significantly to accommodate the linker, which, as a result of the cis conformation adopted by the Gly237–Pro238 prolyl bond, is forced further closer to SH3C (Fig. 3a,b). Interestingly, SH3C uses the same surface to interact with the linker in both the cis and trans conformers but distinct sets of interactions stabilize them.
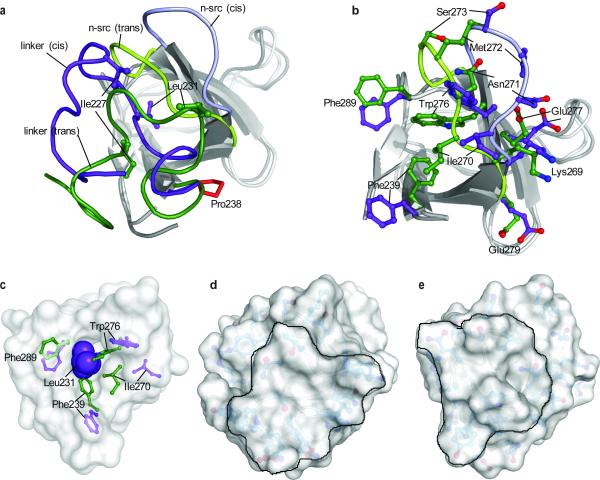
Figure 3. Structural comparison of the trans and cis isomers.
(a) Superposition of the trans and cis conformers of l-SH3C on the SH3C domain (residues 239–295). The SH3C domain in the trans and cis conformation is in light and dark grey, respectively. The linker adopts a very different structure in the trans (green) and the cis (purple) conformation. Representative residues at the linker are indicated. The conformation of the n-src loop is distinct in the trans (green) and the cis (purple) conformer. (b) Effect of cis–trans isomerization on the structure of SH3C. The side-chain conformation of select residues is shown. The side chain of Phe239 is solvent exposed in the cis isomer (purple) whereas it is buried in the trans isomer (green). Superposition of the structures and color code are as in (a). (c) Leu231 is completely buried into a hydrophobic pocket in SH3C in the cis conformation. The side-chain conformation of residues lining the hydrophobic pocket is shown in the cis (purple) and trans (green) isomers.(d,e) Solvent-accessible surface presentation of the trans (d) and cis (e) conformations. The models are rotated ~60° about the z axis relative to the models in (a) with the linker facing the reader. The surface presented by the linker is delineated.
The interaction of the linker with SH3C in the trans conformer is mediated primarily by hydrophobic contacts. More specifically, Ala223, Ile227, Pro230 and Pro232 at the linker interact with Phe239, Lys269, Ile270, Asn271, Trp276, and Leu294 at SH3C (Fig. 2b). The linker–SH3C interaction is further enhanced by hydrogen bonds formed between Asn233 and Lys269 and between Asn236 and Gln297. Overall, the linker–SH3C interaction buries a remarkable ~1,150 Å2 of surface (Supplementary Fig. 4a). The last 10 C-terminal residues of the linker (Ile227 to Asn236) interact intimately with the SH3C domain. In contrast, residues preceding Ile227, other than Ala223, do not appear to form strong and persistent contacts with SH3C, an observation that is further corroborated by NMR relaxation analysis showing that indeed the N-terminal linker residues are very flexible on the fast (pico-to-nanosecond, ps-ns) time scale (see below).
In contrast to the trans conformer, the linker–SH3C interaction in the cis conformer is mediated exclusively by hydrophobic contacts (Fig. 2d). In fact, only three residues from the linker (Pro230, Leu231, and Leu234) interact with SH3C (Fig. 2d and Supplementary Fig. 4b). Nevertheless, the linker–SH3C interaction in the cis conformer buries ~1,140 Å2 of surface, which equals the amount of surface buried in the trans conformer. The reason being that in the cis conformer the N-terminal part of the linker (residues Pro221 to Ile227) folds over and packs against the C-terminal part of the linker (residues Pro230 to Asn236) (Supplementary Fig. 5). Whereas in the trans conformer Pro230 and Pro232 are facing and interact with SH3C (Fig. 2b), in the cis conformer these two Pro residues form a hydrophobic pocket facing the linker with Pro225 and Ile227 fitting snugly into it (Supplementary Fig. 5). Interestingly, in the cis conformer the side chain of Leu231 is completely buried into a hydrophobic pocket in SH3C lined by residues Ala241, Val267, Trp276, Phe289, and Val292 (Fig. 3c). The strong linker–SH3C hydrophobic contacts result in significant stabilization of the interface and thus of the cis conformer, which is equally populated to the trans conformer.
Malleability of the SH3C surface
Prolyl cis–trans isomerization at Pro238 results in a large conformational rearrangement of the linker in the two conformers (Fig. 3a). As a consequence, the linker presents an entirely different surface to SH3C for binding. Notably, the SH3C surface that interacts with the linker adjusts drastically between the cis and trans conformers in order to optimize binding contacts with the alternative linker conformations (Fig. 3b). The SH3C n-src loop undergoes the most significant conformational change between the cis and trans conformers with a maximum translation of ~8 Å (Fig. 3b). As a result, many of the n-src and neighboring side chains rearrange, with notable examples being residues Phe239, Trp276, Phe289 and Ile270. In the trans conformer, the linker residues Ile227, Pro230 and Pro232 mediate all of the hydrophobic contacts with SH3C. In the cis conformer, Ile227 and Pro232 swing out of the linker–SH3C interface (Fig. 2b,c and 3a) in order to form, together with Pro230 and Pro225, a hydrophobic core within the linker (Supplementary Fig. 5). To compensate for the loss of these linker–SH3C hydrophobic contacts, as well as for the complete lack of polar interactions, the linker in the cis conformer uses Leu231 to interact firmly with SH3C (Fig. 3c). Ile270, Trp276 and Phe289 of SH3C adjust their side chain conformation to form a deep hydrophobic pocket wherein Leu231 side chain enters into and completely buries itself (Fig. 3c and Supplementary Movie). Interestingly, the conformation adopted by Phe239 in the trans conformer is not compatible with Leu231 binding to the deep pocket and as result of the trans-to-cis isomerization Phe239 is expelled from the interior of the protein (trans) to become completely solvent exposed (cis) (Fig. 3b,c). The drastically different SH3C and linker conformations induced by cis–trans isomerization give rise to distinct surfaces of the l-SH3C, potentially conferring the two conformers with distinct binding and physicochemical properties (Fig. 3d,e).
Dynamic changes caused by cis–trans isomerization
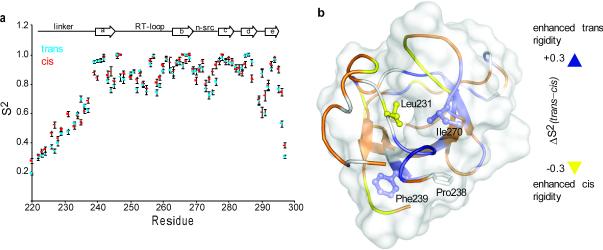
Cis–trans isomerization causes a drastic structural change in both the linker and the SH3C domain. Does the isomerization process also bring about changes in the dynamics of the protein? To address this question we measured backbone relaxation rates35,36 to obtain site-specific information about the N-H bond order parameters (Supplementary Fig. 6). The order parameter, S2, is a measure of the amplitude of internal motions on the pico-to-nanosecond (ps-ns) timescale and may vary from S2=1, for a bond vector having no internal motion, to S2=0, for a bond vector rapidly sampling multiple orientations36.
Cis–trans isomerization at Pro238 results in significant changes in S2 throughout Crk l-SH3C (Fig. 4). In the region around Pro238 there is significant enhancement in the flexibility on going from the trans to the cis isomer presumably as a result of the increase in solvent exposure of Phe239 (Fig. 4). A similar trend was also seen for the region around Ile270 for the same reason. In contrast, trans-to-cis isomerization causes significant rigidification in the linker, especially nearby Leu231, the residue that is completely buried into a hydrophobic pocket in the SH3C domain in the cis isomer (Fig. 3c). Moreover, there is significant rigidification in most of the n-src loop as a result of the tighter packing between this loop and the linker. In addition, most of the linker residues appear to be slightly more rigid in the cis conformer than in the trans one primarily because of the improved packing within the linker in the cis conformer (Supplementary Fig. 5). Thus, the data indicate that cis–trans isomerization causes not only significant structural changes (Fig. 2,3) but also significant changes in the motions of the protein (Fig. 4).
Figure 4. Dynamic characterization of the trans and cis isomers.
(a) N-H bond order parameters, S2, plotted as a function of the primary sequence, for the trans (blue) and cis (red) conformers of l-SH3C. (b) Changes in order parameters, ΔS2, between the trans and the cis conformers mapped using a gradient color code on the structure of cis l-SH3C. ΔS2 is given as S2 (trans) – S2 (cis), so positive ΔS2 values denote enhanced rigidity of the protein backbone in the trans over the cis conformation.
Structural basis for the SH3N–SH3C interaction
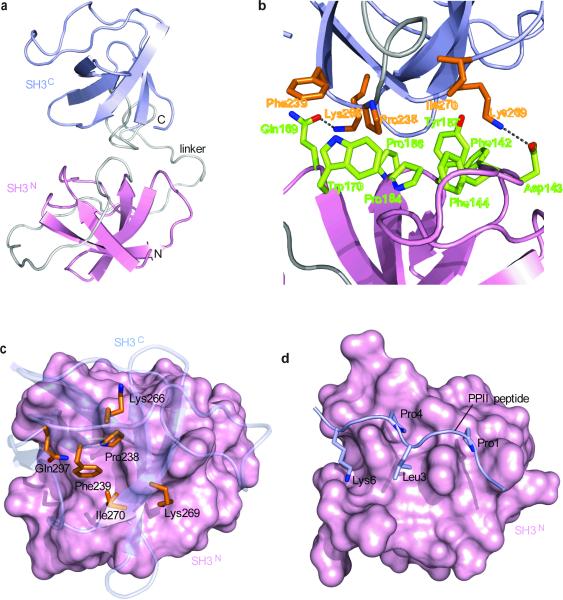
Combined chemical shift and relaxation rate analyses have indicated11 that Crk SH3N-linker-SH3C (CrkSLS) (Fig. 1a) exists in two conformations in solution: a major one (90%), wherein Gly237–Pro238 adopts the cis conformation and the two SH3 domains (SH3N and SH3C) interact in an intramolecular fashion (closed conformation), and a minor one (10%), wherein Gly237–Pro238 adopts the trans conformation and the two SH3 domains do not interact (open conformation) (Supplementary Fig. 7). NMR analysis shows that the full-length chicken Crk adopts a similar closed conformation. We now provide atomic insight into this very interesting binding process by having determined the structure of the closed conformation of CrkSLS (Fig. 5a and Supplementary Fig. 3c).
Figure 5. Structural characterization of CrkSLS in the closed, autoinhibited conformation.
(a) Lowest-energy structure of CrkSLS shown as cartoon with SH3N in pink, SH3C in light blue, and linker in grey. (b) Close-up view of the SH3N–SH3C interface. SH3N and SH3C residues are colored green and orange, respectively. (c) View from above SH3C, which is shown as transparent blue cartoon. SH3N is displayed as pink solvent-accessible surface. SH3C residues interacting with SH3N are shown as orange sticks. (d) Structure of SH3N in complex with a consensus PPII peptide (P-x-P-L-x-K)10. The orientation of SH3N in (c) and (d) is the same. Comparison of (c) and (d) show that the PPII-binding site in CrkSLS is not accessible.
The structural data showed, in agreement with the chemical shift analysis data, that the two SH3 domains interact extensively in the closed CrkSLS (Fig. 5a). The SH3N domain uses its canonical binding site, typically interacting with PPII ligands, to interact with SH3C. Specifically, a cluster of SH3N aromatic residues (Phe142, Phe144, Trp170 and Tyr187) along with Pro184 and Pro186 form an elongated hydrophobic solvent-exposed surface comprising its canonical ligand-binding site (Fig. 5b). This site is well suited to interact with hydrophobic sequences, such as proline-rich sequences. In Crk, SH3C uses the hydrophobic residues Pro238, Phe239 and Ile270 to interact with the canonical binding-site of SH3N (Fig. 5b,c). The interaction is further enhanced by the formation of two polar contacts: a hydrogen bond between Gln169 and Lys266 and a salt bridge between Asp143 and Lys269. Overall, the SH3N–SH3C interaction buries a total surface of ~800 Å2.
The Crk SH3N domain binds selectively to PPII ligands carrying the P-x-L-P-x-K sequence motif37 (Fig. 5d), such as the Abl kinase38. Pro1, Leu3 and Pro4 residues of the PPII ligand are closely packed against the hydrophobic residues in SH3N, whereas Lys6 forms salt bridges with conserved acidic residues10 (Fig. 5d). SH3C uses a similar set of interactions to bind specifically to SH3N: Pro238 and Phe239 of SH3C occupy similar positions, relative to SH3N, to Pro4 and Leu3, respectively, of the PPII peptide. Gln297, which forms a hydrogen-bond to SH3N, is found in a similar position to Lys6 of the PPII peptide, whereas Lys269 forms a salt-bridge to Asp143 (Fig. 5b), a highly conserved residue that does not participate though in the interaction between SH3N and the PPII peptide (Fig. 5d). Interestingly, the juxtaposition of SH3N with the PPII ligand appears to be more optimal than with SH3C since complex formation buries ~1,120 Å2 in the first case but only ~800 Å2 in the latter. The rather small interface between SH3N and SH3C is consistent with the low free energy (ΔG) of SH3N–SH3C interaction, which is only ~1.4 kcal mol−1 (Supplementary Fig. 7b).
The SH3N–SH3C interaction is mediated by the cis conformer
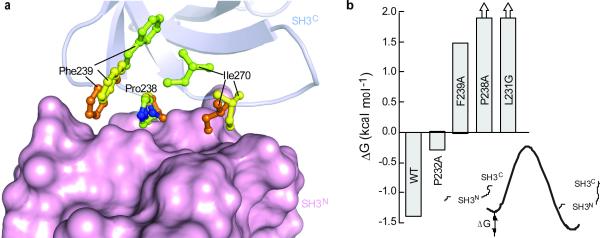
Analysis of the closed CrkSLS structure showed that Pro238 adopts uniquely the cis conformation. Interestingly, Pro238 adopts the trans conformation in the minor, open conformation of CrkSLS. Thus, in agreement with our previous chemical shift analysis (Supplementary Fig. 7a)11, the structural data demonstrated that the intramolecular interaction between the two SH3 domains is mediated exclusively by the cis conformer. The present data provide now the structural basis for this observation. Figure 6a displays the overlay of the structures of CrkSLS and the cis and trans conformers of l-SH3C superimposed on the SH3C domain. The SH3N–SH3C interaction is primarily mediated by SH3C residues Pro238, Phe239, and Ile270. The structural data clearly show that all these three residues are well poised to interact with the SH3N domain in the cis conformer. In contrast, in the trans conformer Phe239 is located in the interior of the SH3C domain and Ile270 swings towards the center of the SH3C domain as a result of the drastic rearrangement that n-src loop undergoes triggered by cis–trans isomerization (Fig. 3b and 6a). Thus, neither Phe239 nor Ile270 are poised to interact with the SH3N domain in the trans conformation (Supplementary Movie).
Figure 6. Structural basis for conformer-specific SH3N–SH3C interaction in CrkSLS.
(a) Superposition of the cis and trans l-SH3C and CrkSLS on the SH3C domain. The side chains of Pro238, Phe239, and Ile270, residues primarily mediating the SH3N–SH3C interaction, are shown in green for trans and yellow for cis l-SH3C and orange for CrkSLS. SH3N is shown as solvent-accessible surface. (b) Effect of single amino acid substitution on the stability of the closed conformation of CrkSLS as assessed by measuring the population of the closed and open conformations of CrkSLS by NMR. ΔG for P238A and L231G is a lower-bound limit since populations less than ~5% are beyond the NMR detection limit.
Phe239 undergoes the most dramatic conformational change upon cis–trans isomerization (Fig. 3b and 6a) and seems to play a key role in mediating SH3N–SH3C binding. Indeed, substitution of Phe239 by Ala abolishes the intramolecular interaction and CrkSLS-F239A adopts predominantly the open conformation (Fig. 6b and Supplementary Fig. 8). Similarly, the P238A substitution also abrogates the closed conformation (Fig. 6b). To test the effect of destabilizing the cis conformation on the integrity of the closed conformation we prepared and characterized substitutions of Pro232 and Leu231, two residues that are important for stabilizing the cis conformer through hydrophobic interactions (Supplementary Fig. 5). Indeed, the P232A substitution decreases the stability of the closed conformation of CrkSLS by ~1.1 kcal mol−1, whereas the L231G substitution appears to completely abrogate the closed conformation (Fig. 6b and Supplementary Fig. 8). Interestingly, Pro238 is compatible with SH3N binding in both the cis and trans conformations (Fig. 6a). Therefore, prolyl isomerization does not regulate binding specificity directly by means of the actual conformation of the proline itself, but rather indirectly by “sculpturing” the SH3C surface responsible for binding to SH3N.
The SH3N–SH3C interaction results in Crk autoinhibition
Previous experiments have shown that the binding of PPII peptides to the SH3N domain in Crk is much weaker compared to the binding of the PPII ligand to the isolated SH3N domain11. The present data provide the structural basis for the inhibition. SH3C binds to SH3N and masks the canonical PPII-binding site thereby providing an autoregulatory mechanism. Indeed, Abl kinase, which interacts with Crk SH3N through a P-x-L-P-x-K sequence motif, binds to the closed, autoinhibited form of CrkSLS with a 10-fold lower affinity than to the open form of CrkSLS, induced by the F239A substitution (Supplementary Fig. 9). Thus, ligand binding to Crk is inhibited in the cis form but it is promoted in the trans form.
Novel SH3 binding surfaces
The linker–SH3C and SH3N–SH3C interactions are mediated by a novel set of SH3 surfaces and contacts not seen previously in SH3 domains. Specifically, the linker interacts extensively with a surface of the SH3C domain delineated by β-strands a and e and the n-src loop (Supplementary Fig. 10). Notably, the linker interacts with this SH3C surface in both the cis and trans conformers, although a distinct set of contacts stabilize the two conformations. The interaction is quite extensive as it buries ~1,150 Å2 of surface. Moreover, SH3C uses a novel binding surface to interact with the other SH3 domain in Crk, SH3N (Supplementary Fig. 10). The two surfaces are distinct and not mutually exclusive since in the closed form of CrkSLS SH3C is bound to both the linker and SH3N simultaneously. Thus, the present data further highlight the binding versatility of SH3 domains and expands the list of the non-canonical ligands that may interact with this small signaling domain.
DISCUSSION
Proline cis–trans isomerization is an emerging regulatory mechanism involved in the activity regulation of a wide range of biomolecular processes. Understanding the underlying mechanisms of such molecular switches necessitates the availability of structural data on both the cis and trans conformers. The limited structural information19,20,39 on both conformers available so far has been suggestive of a rather local effect caused by proline isomerization. Here we provide atomic details of the structural and dynamic changes elicited by proline cis–trans isomerization in the Crk signaling protein. The data demonstrate that proline isomerization causes a remarkable reorganization of the interface between the linker and the SH3C domain, to the extent that the two resultant conformers present drastically different surfaces (Fig. 3d,e). Notably, the two conformers are conferred with distinct binding properties, with the cis one promoting the intramolecular engagement of the two SH3 domains to assemble an autoregulatory conformation and the trans conformer promoting a dumbbell-like, uninhibited conformation (Supplementary Movie).
How can a 180° rotation about a single prolyl bond cause such a strong effect on the global structure of the protein? Pro238 in Crk is located at a strategic position where the linker connects to the SH3C domain. In the trans conformer the linker interacts extensively with the SH3C domain. Trans-to-cis isomerization at Pro238 forces a new conformation to the linker thereby disrupting all contacts between the linker and the SH3C present in the trans form. Notably, both the linker and the SH3C domain readjust their structure and a new set of contacts is formed to stabilize the cis conformer. As a result, the SH3C domain exposes several residues that form a binding surface capable of mediating the specific interaction between SH3C and SH3N (Fig. 6a and Supplementary Movie). In the trans form this set of residues is buried and thus the SH3N-SH3C interaction is not possible resulting in a dumbbell-like conformation. Thus, the structural basis for the function of Pro238 as a molecular switch is the surface “sculpturing” afforded by the actual isomerization process.
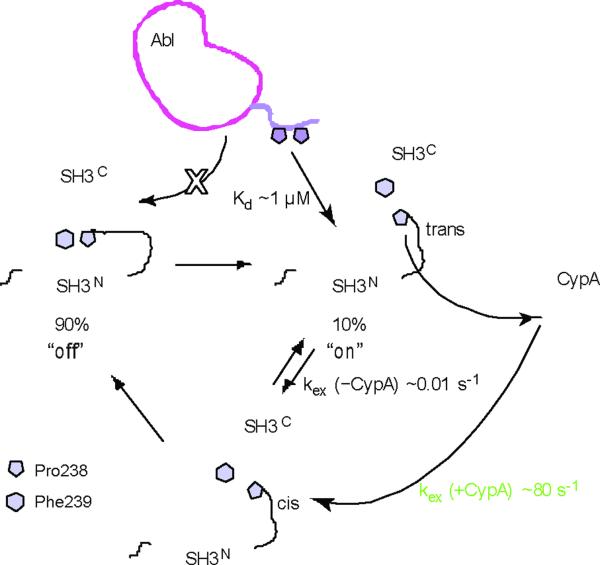
The proline switch in Crk plays a very important physiological role as it toggles the protein between a closed and an open conformation (Fig. 7). In fact, the open conformation, which is lowly populated (~10%), constitutes the activated form of Crk wherein the PPII-binding site is accessible to physiological ligands. Indeed, Abl binds preferentially to the open form of Crk (Supplementary Fig. 9)11. The energetics of the equilibrium between the auto-inhibited and the activated form is regulated by the proline switch since only the cis isomer stabilizes the closed form. However, proline isomerization is intrinsically a very slow process (of the order of minutes) rendering the kinetics of the molecular switch particularly slow (Fig. 7). Interestingly, cyclophilin A (CypA) accelerates dramatically the rate of the interconversion11, to the millisecond regime at physiological temperatures, a more meaningful timescale for modulating the kinetics of biological processes. Thus, proline isomerization in Crk functions as a molecular timer whereby the energetics and rate of signaling complexes formation can be regulated.
Figure 7. Mechanistic basis for the the regulation of Crk activity.
Crk adopts predominantly (~90%) the closed, auto-inhibited conformation but a minor population (~10%) adopts the open, uninhibited conformation wherein the PPII-binding site on SH3N is accessible for binding. The P-x-L-P-x-K motif of Abl binds to SH3N with a relatively strong affinity (Kd ~1 μM). The open conformation exists as an equilibrium between the cis and the trans isomer, but only the cis one forms the closed conformation. The rates of the cis–trans interconversion are regulated by the action of CypA, which accelerates the interconversion by four orders of magnitude11.
To what extent this regulatory mechanism is shared by other Crk family members remains to be addressed. Pro238 is strictly conserved among all Crk proteins (Supplementary Fig. 1) raising the possibility that proline isomerization may be a conserved feature. Interestingly, our data also indicate that the identity of the residue following the proline determines the population of the cis form. For example, the F239A substitution decreases the cis population to ~8%, from 50% in the wild type protein. This position is variable among Crk proteins (Supplementary Fig. 1). Thus, it is possible that proline isomerization is present in other Crk proteins although the relative population of the two conformers may not be as high as in chicken Crk. Interestingly, tyrosine (Tyr222) phosphorylation40 and the presence of an Abl SH3 binding site within the Crk SH2 domain41 provide additional regulatory mechanisms of Crk activity. It is intriguing that multiple layers of regulation are present in an adaptor protein, and it will be important to determine the relative contribution of each one of these mechanisms in modulating the response of Crk to different stimuli.
The present results, in agreement with previous observations11,12, highlight the importance of the SH3C domain in establishing and stabilizing the auto-inhibitory conformation in Crk. Point mutations or deletions in the SH3C domain were shown to stimulate the transforming activity of Crk6,13,16 and increase its expression levels in malignant brain and lung tumors3. For example, v-Crk, the viral oncogenic form of Crk that was originally isolated as an avian sarcoma virus in chicken tumors42, and c-Crk-I, an alternative splicing cellular form, lack the SH3C domain and the linker and they are associated with increased transforming activity12 that cannot be controlled. Even single point mutations in the SH3C domain (e.g. W276K) known to disrupt the folding of the domain are sufficient to significantly increase Crk activity43. Interestingly, structural studies on human Crk12 and chicken Crk (this work) show that in both cases the SH3N domain is autoinhibited but different structural elements are responsible for blocking the PPII-ligand binding site (Supplementary Fig. 11).
A particularly striking result in the present work is related to the versatility of the SH3 domain as a binding partner44,45. The interaction between SH3N and SH3C in Crk provides the first example of an SH3–SH3 complex mediated by direct contacts between the two domains. Moreover, the surfaces used by SH3C to interact with the linker and SH3N are novel binding surfaces for SH3 domains. It is remarkable that almost the entire surface (Supplementary Fig. 10) of such a small domain can be molded to accommodate a wide range of ligands with no apparent sequence or structure similarity.
METHODS
Protein preparation
The following Crk (chicken) constructs were used: full-length Crk (residues 1–305), CrkSLS (residues 139–305), and l-SH3C (residues 218–305). The various Crk constructs were amplified by PCR, using as a template the plasmid that encodes the full-length Crk protein, inserted into the pGEX6P-1 vector engineered with a PreScission protease cleavage site and transformed into BL21(DE3) cells. NMR samples were prepared as described in Supplementary Information.
NMR spectroscopy
NMR experiments were performed on Varian 800- and 600-MHz and Bruker 700- and 600-MHz spectrometers. Complete backbone and side-chain assignment for the Crk proteins studied was achieved using standard triple-resonance pulse sequences. NOEs were measured using 2D and 3D NOESY (13C and 15N edited) experiments using mixing times of 80 and 100 ms. All NMR samples were prepared in 50 mM KPi (pH 5.5), 140 mM NaCl, 1 mM DTT and 1 g l−1 NaN3. Pro238 isomerization as well as the relative population of the isomers are not sensitive to pH within the range 5.5–7.5 (Supplementary Fig. 12). Concentration of proteins was 0.6–0.8 mM. Spectra were recorded at 22 °C. RDC data46 were collected and analyzed47 as detailed in Supplementary Information.
Structure calculation and refinement
Structure calculations were performed with Xplor-NIH48. The 13Cα, 13Cβ, 13C', Hα, 15N and NH chemical shifts served as input for the TALOS program49 to extract dihedral (φ and ψ) angles. Distance restraints derived from the NOESY spectra were categorized in 3 bins with upper-bound distances of 2.8, 3.5, and 5.0 Å. RDC restraints were included in the final stages of the calculation. Ramachandran statistics for cis l-SH3C, trans l-SH3C, and CrkSLS, respectively, are as follows: residues in most favored regions: 71, 66, and 65; residues in additionally allowed regions: 26, 32, and 29; residues in generously allowed regions: 3, 2, and 6; residues in disallowed regions: 0, 0, and 0. The summary of NMR restraints and structure refinement statistics is presented in Supplementary Table 1.
Relaxation
Three relaxation parameters were measured for all backbone amides of Crk proteins: the 1H-15N NOE, the longitudinal relaxation rate R1 and the transverse relaxation rate R250. Details are provided in Supplementary Information.
ITC experiments
Calorimetric titrations were performed on an iTC200 microcalorimeter (Microcal) as described in detail in Supplementary Information.
Accession codes
Protein Data Bank: Coordinates for the cis and trans conformers of l-SH3C and CrkSLS have been deposited with accession codes 2L3P, 2L3Q, and 2L3S, respectively.
Supplementary Material
Acknowledgements
This work was supported by the U.S. National Institute of Health (GM80308 to C.G.K.).
Footnotes
Competing financial interests The authors declare no competing financial interests.
References
- 1.Birge RB, Kalodimos C, Inagaki F, Tanaka S. Crk and CrkL adaptor proteins: networks for physiological and pathological signaling. Cell Commun Signal. 2009;7:13. doi: 10.1186/1478-811X-7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feller SM. Crk family adaptors-signalling complex formation and biological roles. Oncogene. 2001;20:6348–71. doi: 10.1038/sj.onc.1204779. [DOI] [PubMed] [Google Scholar]
- 3.Miller CT, et al. Increased C-CRK proto-oncogene expression is associated with an aggressive phenotype in lung adenocarcinomas. Oncogene. 2003;22:7950–7. doi: 10.1038/sj.onc.1206529. [DOI] [PubMed] [Google Scholar]
- 4.Nishihara H, et al. Molecular and immunohistochemical analysis of signaling adaptor protein Crk in human cancers. Cancer Lett. 2002;180:55–61. doi: 10.1016/s0304-3835(01)00763-7. [DOI] [PubMed] [Google Scholar]
- 5.Linghu H, et al. Involvement of adaptor protein Crk in malignant feature of human ovarian cancer cell line MCAS. Oncogene. 2006;25:3547–3556. doi: 10.1038/sj.onc.1209398. [DOI] [PubMed] [Google Scholar]
- 6.Shishido T, et al. Crk family adaptor proteins trans-activate c-Abl kinase. Genes Cells. 2001;6:431–40. doi: 10.1046/j.1365-2443.2001.00431.x. [DOI] [PubMed] [Google Scholar]
- 7.Sirvent A, Benistant C, Roche S. Cytoplasmic signalling by the c-Abl tyrosine kinase in normal and cancer cells. Biol Cell. 2008;100:617–31. doi: 10.1042/BC20080020. [DOI] [PubMed] [Google Scholar]
- 8.Feller SM, Knudsen B, Hanafusa H. c-Abl kinase regulates the protein binding activity of c-Crk. EMBO J. 1994;13:2341–51. doi: 10.1002/j.1460-2075.1994.tb06518.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Songyang Z, et al. SH2 domains recognize specific phosphopeptide sequences. Cell. 1993;72:767–78. doi: 10.1016/0092-8674(93)90404-e. [DOI] [PubMed] [Google Scholar]
- 10.Wu X, et al. Structural basis for the specific interaction of lysine-containing proline-rich peptides with the N-terminal SH3 domain of c-Crk. Structure. 1995;3:215–26. doi: 10.1016/s0969-2126(01)00151-4. [DOI] [PubMed] [Google Scholar]
- 11.Sarkar P, Reichman C, Saleh T, Birge RB, Kalodimos CG. Proline cis-trans isomerization controls autoinhibition of a signaling protein. Mol Cell. 2007;25:413–26. doi: 10.1016/j.molcel.2007.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kobashigawa Y, et al. Structural basis for the transforming activity of human cancer-related signaling adaptor protein CRK. Nat Struct Mol Biol. 2007;14:503–10. doi: 10.1038/nsmb1241. [DOI] [PubMed] [Google Scholar]
- 13.Reichman C, et al. Transactivation of Abl by the Crk II adapter protein requires a PNAY sequence in the Crk C-terminal SH3 domain. Oncogene. 2005;24:8187–99. doi: 10.1038/sj.onc.1208988. [DOI] [PubMed] [Google Scholar]
- 14.Akakura S, et al. C-terminal SH3 domain of CrkII regulates the assembly and function of the DOCK180/ELMO Rac-GEF. J Cell Physiol. 2005;204:344–51. doi: 10.1002/jcp.20288. [DOI] [PubMed] [Google Scholar]
- 15.Kizaka-Kondoh S, Matsuda M, Okayama H. CrkII signals from epidermal growth factor receptor to Ras. Proc Natl Acad Sci USA. 1996;93:12177–82. doi: 10.1073/pnas.93.22.12177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ogawa S, et al. The C-terminal SH3 domain of the mouse c-Crk protein negatively regulates tyrosine-phosphorylation of Crk associated p130 in rat 3Y1 cells. Oncogene. 1994;9:1669–78. [PubMed] [Google Scholar]
- 17.Nicholson LK, Lu KP. Prolyl cis-trans Isomerization as a molecular timer in Crk signaling. Mol Cell. 2007;25:483–5. doi: 10.1016/j.molcel.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 18.Isakov N. A new twist to adaptor proteins contributes to regulation of lymphocyte cell signaling. Trends Immunol. 2008;29:388–96. doi: 10.1016/j.it.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 19.Lu KP, Finn G, Lee TH, Nicholson LK. Prolyl cis-trans isomerization as a molecular timer. Nat Chem Biol. 2007;3:619–29. doi: 10.1038/nchembio.2007.35. [DOI] [PubMed] [Google Scholar]
- 20.Andreotti AH. Native state proline isomerization: an intrinsic molecular switch. Biochemistry. 2003;42:9515–24. doi: 10.1021/bi0350710. [DOI] [PubMed] [Google Scholar]
- 21.Gothel SF, Marahiel MA. Peptidyl-prolyl cis-trans isomerases, a superfamily of ubiquitous folding catalysts. Cell Mol Life Sci. 1999;55:423–36. doi: 10.1007/s000180050299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brazin KN, Mallis RJ, Fulton DB, Andreotti AH. Regulation of the tyrosine kinase Itk by the peptidyl-prolyl isomerase cyclophilin A. Proc Natl Acad Sci USA. 2002;99:1899–904. doi: 10.1073/pnas.042529199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wulf G, Finn G, Suizu F, Lu KP. Phosphorylation-specific prolyl isomerization: is there an underlying theme? Nat Cell Biol. 2005;7:435–41. doi: 10.1038/ncb0505-435. [DOI] [PubMed] [Google Scholar]
- 24.Pastorino L, et al. The prolyl isomerase Pin1 regulates amyloid precursor protein processing and amyloid-beta production. Nature. 2006;440:528–34. doi: 10.1038/nature04543. [DOI] [PubMed] [Google Scholar]
- 25.Calabrese MF, Eakin CM, Wang JM, Miranker AD. A regulatable switch mediates self-association in an immunoglobulin fold. Nat Struct Mol Biol. 2008;15:965–71. doi: 10.1038/nsmb.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lummis SC, et al. Cis-trans isomerization at a proline opens the pore of a neurotransmitter-gated ion channel. Nature. 2005;438:248–52. doi: 10.1038/nature04130. [DOI] [PubMed] [Google Scholar]
- 27.Nelson CJ, Santos-Rosa H, Kouzarides T. Proline isomerization of histone h3 regulates lysine methylation and gene expression. Cell. 2006;126:905–16. doi: 10.1016/j.cell.2006.07.026. [DOI] [PubMed] [Google Scholar]
- 28.Wang Z, et al. Pro isomerization in MLL1 PHD3-bromo cassette connects H3K4me readout to CyP33 and HDAC-mediated repression. Cell. 2010;141:1183–94. doi: 10.1016/j.cell.2010.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eckert B, Martin A, Balbach J, Schmid FX. Prolyl isomerization as a molecular timer in phage infection. Nat Struct Mol Biol. 2005;12:619–23. doi: 10.1038/nsmb946. [DOI] [PubMed] [Google Scholar]
- 30.Franke EK, Yuan HE, Luban J. Specific incorporation of cyclophilin A into HIV-1 virions. Nature. 1994;372:359–62. doi: 10.1038/372359a0. [DOI] [PubMed] [Google Scholar]
- 31.OuYang B, Pochapsky SS, Dang M, Pochapsky TC. A functional proline switch in cytochrome P450cam. Structure. 2008;16:916–23. doi: 10.1016/j.str.2008.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vogel M, Bukau B, Mayer MP. Allosteric regulation of Hsp70 chaperones by a proline switch. Mol Cell. 2006;21:359–67. doi: 10.1016/j.molcel.2005.12.017. [DOI] [PubMed] [Google Scholar]
- 33.Severin A, Joseph RE, Boyken S, Fulton DB, Andreotti AH. Proline isomerization preorganizes the Itk SH2 domain for binding to the Itk SH3 domain. J Mol Biol. 2009;387:726–43. doi: 10.1016/j.jmb.2009.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muralidharan V, et al. Solution Structure and Folding Characteristics of the C-Terminal SH3 Domain of c-Crk-II. Biochemistry. 2006;45:8874–8884. doi: 10.1021/bi060590z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Palmer AG., 3rd NMR characterization of the dynamics of biomacromolecules. Chem Rev. 2004;104:3623–40. doi: 10.1021/cr030413t. [DOI] [PubMed] [Google Scholar]
- 36.Mittermaier A, Kay LE. New tools provide new insights in NMR studies of protein dynamics. Science. 2006;312:224–8. doi: 10.1126/science.1124964. [DOI] [PubMed] [Google Scholar]
- 37.Knudsen BS, et al. Affinity and specificity requirements for the first Src homology 3 domain of the Crk proteins. EMBO J. 1995;14:2191–8. doi: 10.1002/j.1460-2075.1995.tb07213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ren R, Ye ZS, Baltimore D. Abl protein-tyrosine kinase selects the Crk adapter as a substrate using SH3-binding sites. Genes Dev. 1994;8:783–95. doi: 10.1101/gad.8.7.783. [DOI] [PubMed] [Google Scholar]
- 39.Mallis RJ, Brazin KN, Fulton DB, Andreotti AH. Structural characterization of a proline-driven conformational switch within the Itk SH2 domain. Nat Struct Biol. 2002;9:900–5. doi: 10.1038/nsb864. [DOI] [PubMed] [Google Scholar]
- 40.Rosen MK, et al. Direct demonstration of an intramolecular SH2-phosphotyrosine interaction in the Crk protein. Nature. 1995;374:477–9. doi: 10.1038/374477a0. [DOI] [PubMed] [Google Scholar]
- 41.Donaldson LW, Gish G, Pawson T, Kay LE, Forman-Kay JD. Structure of a regulatory complex involving the Abl SH3 domain, the Crk SH2 domain, and a Crk-derived phosphopeptide. Proc Natl Acad Sci USA. 2002;99:14053–8. doi: 10.1073/pnas.212518799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mayer BJ, Hamaguchi M, Hanafusa H. A novel viral oncogene with structural similarity to phospholipase C. Nature. 1988;332:272–5. doi: 10.1038/332272a0. [DOI] [PubMed] [Google Scholar]
- 43.Zvara A, et al. Activation of the focal adhesion kinase signaling pathway by structural alterations in the carboxyl-terminal region of c-Crk II. Oncogene. 2001;20:951–61. doi: 10.1038/sj.onc.1204173. [DOI] [PubMed] [Google Scholar]
- 44.Li SS. Specificity and versatility of SH3 and other proline-recognition domains: structural basis and implications for cellular signal transduction. Biochem J. 2005;390:641–53. doi: 10.1042/BJ20050411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vaynberg J, et al. Structure of an ultraweak protein-protein complex and its crucial role in regulation of cell morphology and motility. Mol Cell. 2005;17:513–23. doi: 10.1016/j.molcel.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 46.Bax A, Grishaev A. Weak alignment NMR: a hawk-eyed view of biomolecular structure. Curr Opin Struct Biol. 2005;15:563–70. doi: 10.1016/j.sbi.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 47.Popovych N, Tzeng SR, Tonelli M, Ebright RH, Kalodimos CG. Structural basis for cAMP-mediated allosteric control of the catabolite activator protein. Proc Natl Acad Sci USA. 2009;106:6927–32. doi: 10.1073/pnas.0900595106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schwieters CD, Kuszewski JJ, Clore MG. Using Xplor-NIH for NMR molecular structure determination. Progress in Nuclear Magnetic Resonance Spectroscopy. 2006;48:47–62. [Google Scholar]
- 49.Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 50.Tzeng SR, Kalodimos CG. Dynamic activation of an allosteric regulatory protein. Nature. 2009;462:368–372. doi: 10.1038/nature08560. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.