Abstract
It is now accepted that breast cancer is not a single disease, but instead it is composed of a spectrum of tumor subtypes with distinct cellular origins, somatic changes, and etiologies. Gene expression profiling using DNA microarrays has contributed significantly to our understanding of the molecular heterogeneity of breast tumor formation, progression, and recurrence. For example, at least two clinical diagnostic assays exist (i.e., OncotypeDX RS and Mammaprint®) that are able to predict outcome in patients using patterns of gene expression and predetermined mathematical algorithms. In addition, a new molecular taxonomy based upon the inherent, or “intrinsic,” biology of breast tumors has been developed; this taxonomy is called the “intrinsic subtypes of breast cancer,” which now identifies five distinct tumor types and a normal breast-like group. Importantly, the intrinsic subtypes of breast cancer predict patient relapse, overall survival, and response to endocrine and chemotherapy regimens. Thus, most of the clinical behavior of a breast tumor is already written in its subtype profile. Here, we describe the discovery and basic biology of the intrinsic subtypes of breast cancer, and detail how this interacts with underlying genetic alternations, response to therapy, and the metastatic process.
Mammary tumors have a variety of cellular origins and display significant heterogeneity. A new molecular taxonomy defines five tumor subtypes and can predict patient relapse, survival, and responses to therapy.
DISCOVERY OF BREAST CANCER INTRINSIC SUBTYPES
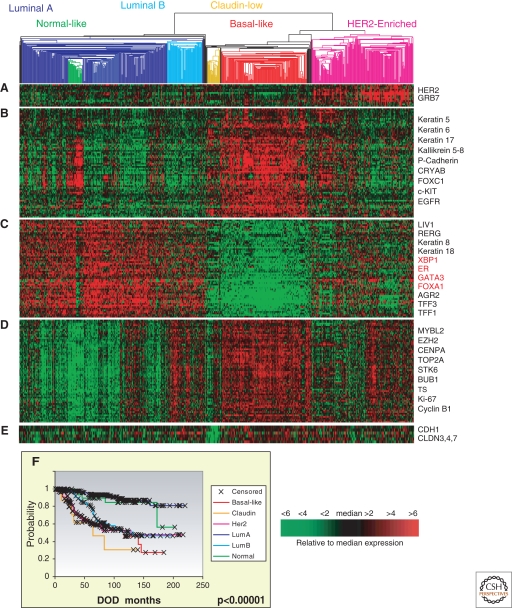
In 2000, a team led by Drs. David Botstein, Patrick Brown, and Anne-Lise Børresen-Dale used a semiunsupervised approach to identify what should be the naturally occurring breast cancer subtypes, using 40 patients with locally advanced breast cancers (Perou et al. 2000). They identified 496 genes, termed the “intrinsic gene set,” by searching for genes that showed little variance within repeated tumor samples (i.e., before and after neoadjuvant chemotherapy pairs), but high variance across different tumors, and then used this gene set for tumor subtype identification. Among these breast tumors, they found that the patterns of expression of these genes identified four distinct tumor subtypes and a normal breast-like group. These so called “intrinsic subtypes,” named because the gene list that defines them reflects the intrinsic properties of these breast cancers, have been consistently identified in independent data sets using different methods and multiple microarray platforms (Sorlie et al. 2001, 2003; Sotiriou et al. 2003; Abd El-Rehim et al. 2004; Carey et al. 2006; Hu et al. 2006; Parker et al. 2009). These subtypes are also conserved across ethnic groups (Yu et al. 2004), and are present even at the ductal carcinoma in situ (DCIS) stage (Livasy et al. 2007; Allred et al. 2008). Importantly, the intrinsic subtypes segregated tumors by expression of hormone receptors (both estrogen receptor [ER] and progesterone receptor [PR]) and the genes they regulate, supporting earlier epidemiologic and biomarker studies, suggesting that ER-positive and ER-negative breast cancers are distinct. At least two hormone-receptor-positive subtypes were identified that were called “luminal A” and “luminal B.” Conversely, there were several subtypes characterized by low expression of hormone receptors and their regulated genes, one of which was called the “HER2-enriched” subtype and another called the “basal-like” subtype (Fig. 1). The fifth subtype, the normal breast-like group, is a less clear subtype; it is acknowledged that the normal breast-like group is a heterogeneous group including those with a high stromal content, those with high lymphocyte infiltration, and those with true normal epithelial cell contamination of a low malignant cell content tumor. In Figure 1, the normal breast-like group is composed of many true normal breast samples from reduction mammoplasties and some tumors, which upon hematoxylin and eosin (H&E) examination show <50% tumor tissue. In this figure, the normal breast-like group is likely clustering with the luminal A subtype due to their common low proliferation rates and moderate expression of luminal epithelial genes. Alternatively, a normal breast-like cell line may exist including the PMC42 line, which has stem-cell-like properties (Git et al. 2008). Ongoing studies have recently identified a new and intriguing subtype called the “claudin-low” group (Herschkowitz et al. 2007), but for the time being, these four tumor subtypes and the normal breast-like group are the ones consistently identified and commonly accepted. Although the intrinsic subtypes were identified regardless of outcome (i.e., no knowledge of patient outcomes was used to select the intrinsic gene set), these subtypes have strong prognostic implications (Fig. 1F); in particular, patients with basal-like, HER2-enriched, and luminal B tumors show significantly poorer outcomes when compared to patients with luminal A tumors (Sorlie et al. 2001, 2003; Sotiriou et al. 2003; Yu et al. 2004; Carey et al. 2006; Hu et al. 2006; Langerod et al. 2007; Parker et al. 2009).
Figure 1.
677 breast tumors analyzed using hierarchical clustering and the Intrinsic/UNC 1300 gene list. A single data set of 340 samples from UNC and 337 from the Netherlands Cancer Institute were combined using Distance Weighted Discrimination (Benito et al. 2004), and then clustered together to yield a large and homogenous data set containing over 470 different tumors with RFS and OS data. The clustering analysis identified the 5 major intrinsic subtypes of luminal A, luminal B, normal-like, basal-like, and HER2-enriched, and also identified the newest subtype in the center called the “claudin-low” group. The gene sets most definitive of each subtype are shown and are (A) HER2-amplicon gene set, (B) basal epithelial gene set, (C) luminal epithelial gene set containing ER, and (D) proliferation gene set. (E) Claudin-low gene set including E-cadherin and claudin 3, 4, and 7. (F) Kaplan-Meier plot for survival based upon disease-specific survival (DOD) for the six groups described here. Scale bar showing the expression levels of each gene relative to the median expression.
A critical aspect of biomarker biology is validation, and the intrinsic subtypes have been validated on many independent data sets (Sorlie et al. 2003; Hu et al. 2006; Langerod et al. 2007; Naume et al. 2007; Parker et al. 2009). For example, Sorlie et al. (2003) showed that when using this classification method on multiple data sets, the subtypes were represented with similar distributions, despite differences in the populations (i.e., the original gene expression study was based upon high-risk and locally-advanced tumors treated with chemotherapy, whereas the NKI 295-patient data set included women under 55 with lymph-node-negative tumors that did not receive adjuvant systemic therapy (van 't Veer et al. 2002; van de Vijver et al. 2002), and the West et al. data set was a mixture of stages, nodal status, and hormone receptor status (West et al. 2001). Since the clustering methodology for identifying intrinsic subtypes is suboptimal for reproducible classifications, a promising alternative approach for reproducible subtype classifications has been developed based upon identifying the mean expression profiles, called centroids, for each subtype (Hu et al. 2006; Parker et al. 2009). Hu and colleagues (Hu et al. 2006) developed the Single Sample Predictor (SSP) tool to serve as an unchanging prognostic indicator for individual patient samples; the SSP compares the gene expression profile of an unknown sample to a prototypical profile of each intrinsic subtype and classifies the unknown sample according to the profile/centroid it most closely matches. Recently this approach has been expanded upon to include statistically robust and objective methods for selecting prototypical samples/tumors, and a robust method for gene selection, which has resulted in the PAM50 intrinsic subtype classifier. This algorithm uses 50 genes to identify the four major intrinsic subtypes (luminal A, luminal B, basal-like, HER2-enriched) and the normal breast-like group, and has the advantage that it can utilize RNA purified from fresh frozen tumors or Formalin-Fixed Paraffin Embedded (FFPE) materials, thus making it compatible with materials coming from a typical pathology archive (Parker et al. 2009).
TUMOR SUBTYPE BIOLOGY AND CLINICAL FEATURES
Luminal Subtypes
The most common breast cancers are ER-positive tumors, which, according to gene expression patterns, fall into the luminal subtypes, so-called because they have a gene expression pattern reminiscent of the luminal epithelial component of the breast (Perou et al. 2000). These tumors are characterized by expression of the ER, PR, and genes associated with ER activation such as LIV1, TFF1/pS2, and Cyclin D1 (Fig. 1C), as well as expression of luminal cytokeratins 8 and 18 (Perou et al. 2000; Sotiriou et al. 2003; Oh et al. 2006). Luminal tumors are often low-grade, and fewer than 20% have TP53 mutations (Sorlie et al. 2001; Sotiriou et al. 2003; Langerod et al. 2007; Naume et al. 2007). Within the broad and diverse luminal/ER+ group, there are at least two subtypes, luminal A and luminal B, and there are many relevant differences between these two groups, although it is not always easy to distinguish a luminal A from a luminal B, since the expression of the genes defining these groups are a continuum. For example, luminal A tumors generally have high expression of ER and ER-regulated genes, low expression of the HER2 cluster (which is variable in luminal B tumors), and low expression of proliferation-associated genes including Ki-67 (Sorlie et al. 2001, 2003; Hu et al. 2006). Conversely, luminal B tumors tend to be highly proliferative, tend to be TP53 mutant, and in general show lower expression of ER and ER-regulated genes.
Luminal tumors in general are defined by a quartet of transcription factors (Fig. 1C) that includes ER, GATA3, FOXA1, and XBP1. Since the initial description of this gene set, a great deal has become known about the role of these new players in breast luminal cell biology. For example, when GATA3 is deleted early in mammary development, ductal growth is greatly inhibited and very few ER+/luminal cells develop (Kouros-Mehr et al. 2006); additionally, if this deletion occurs during lactation using a WAP-Cre promoter, lobular-alveolar development is greatly impaired (Asselin-Labat et al. 2006), suggesting that GATA3 is a critical determinant of luminal cell formation. Carrol et al. (2005) went on to perform genome-wide chromatin immuno-precipitation (i.e., ChIP-chip) experiments using ER and showed that the majority of ER binding sites also had a close binding site for FOXA1, and that FOXA1 was required for the induction of most ER-regulated genes including XBP1. Usary et al. (2004) also showed that GATA3 is occasionally mutated in ER+/luminal tumors, and that these mutations abolish DNA-binding activity. They also showed, using ectopic expression of GATA3, that FOXA1 is a GATA3-regulated gene. When synthesized together, these data suggest that GATA3 is a critical and early determinant of the luminal lineage that may directly (or indirectly) turn on ER and FOXA1, which in turn act together to induce the expression of ER-regulated genes including XBP1 (thus explaining the mechanistic significance of this quartet of transcription factors). Interestingly, ER may also induce GATA3, and GATA3 also induces ER; thus, once this developmental program is turned on, it may be self sustaining (Eeckhoute et al. 2007).
In population-based studies, the luminal A subtype is the most common, representing approximately 40% of all breast tumors, while the luminal B subtype comprises approximately 10% (Table 1) (see Carey et al. 2006; Millikan et al. 2007; Morris et al. 2007). Although risk factors for all of the subtypes remain an area of intense research, it is clear that all traditional risk and/or protective factors (like protection of risk from pregnancy) are factors for luminal breast cancers. Importantly, many of the studies focused on the intrinsic subtypes in population-based studies also show that premenopausal women and African-American women tend to develop fewer of the good-prognosis luminal A tumors and more of the poor-prognosis basal-like tumors (described further below), which may contribute to the poorer outcomes associated with this ethnic group (Carey et al. 2006; Millikan et al. 2007; Yang et al. 2007).
Table 1.
Frequency of the intrinsic subtypes according to race and age. (Table is from Millikan et al. [2007] and reprinted with permission from Springer © 2007.) Distribution of breast cancer subtypes according to race and menopausal status using 1424 cases: invasive (1000) and in-situ (424) breast cancers.
| Breast cancer subtype | African-American premenopausal N (%) | African-American postmenopausal N (%) | White premenopausal N (%) | White postmenopausal N (%) |
|---|---|---|---|---|
| Luminal A N = 796 | 108 (41.4%) | 179 (56.3%) | 216 (57.4%) | 293 (66.5%) |
| Basal-like N = 225 | 70 (27.2%) | 52(16.0%) | 54 (14.5 %) | 49 (9.3%) |
| HER2+/ERN = 116 | 22 (8.4%) | 26 (7.7%) | 24 (5.6%) | 44 (6.0%) |
| Luminal B N = 137 | 19 (7.3%) | 26 (8.7%) | 46 (12.4%) | 46 (10.7%) |
| Unclassified N = 150 | 41 (15.7%) | 38 (11.3%) | 38 (10.1%) | 33 (7.5%) |
| Total: 1424 P < 0.0001 | 260 (100%) | 321 (100%) | 378 (100%) | 465 (100%) |
While a clinical assay to identify luminal A and luminal B by gene expression is not yet available, the OncotypeDX Recurrence Score™ (RS) assay includes many genes (HER2, GRB7, ER, SCUBE2, Bcl2, Ki-67, Survivin, MYBL2, Cyclin B1) that are also used to define luminal A versus luminal B tumors. To more directly compare the OncotypeDX Recurrence Score and the intrinsic subtypes of luminal A and luminal B, Fan and colleagues compared both classifiers on the NKI295 data set and showed that 50% of the luminal A tumors had low RS (i.e., good outcome), whereas only 2% of luminal B tumors had low RS (Fan et al. 2006). Moreover, other prognostic genomic predictors were also tested including a signature of activated fibroblasts (Chang et al. 2004) and the NKI-developed Mammaprint signature (van 't Veer et al. 2002). Nonetheless, all of these predictors showed a high degree of concordance when compared to each other, and in prognostication. These findings have a number of important implications, one of which is that despite having almost completely no overlapping genes, these four gene expression profiles showed a high degree of agreement when the actual patient clasifications were compared. Second are the therapeutic implications where a high OncotypeDX RS is associated with a higher risk of relapse despite tamoxifen (Paik et al. 2004), and a higher benefit of adjuvant chemotherapy (Paik et al. 2006), suggesting that similar predictions likely also hold true for the distinction of luminal A versus luminal B.
HER2-enriched Subtype
The hormone-receptor-negative tumors are largely comprised of the HER2-enriched, basal-like, and claudin-low subtypes. The HER2-enriched subtype is relatively infrequent, comprising only ∼10% of all breast cancers (Carey et al. 2006). Typically, this subtype shows elevated expression of HER2 and many other genes that reside near HER2 in the genome, including GRB7 (Fig. 1A), because of the known HER2 genomic loci DNA amplification (Slamon et al. 1987, 1989). We do note, however, that not all tumors of this subtype show HER2 amplification and/or overexpression, and thus, the term HER2-enriched is used to describe this group to signify this imperfect but high correlation with HER2 amplification status. Other expression features of this subtype include low expression of the luminal, hormone receptor-regulated gene cluster and low expression of basal-like genes. It is also critical to note that some, but not all, clinically defined HER2-positive breast cancers fall into this category; the clinically defined HER2-positive breast cancers that have high expression of the luminal cluster and are ER-positive fall into the luminal subtypes (typically luminal B); thus, there exists at least two types of HER2-amplified tumors.
Another important feature of tumors in the HER2-enriched subtype is high expression of the proliferation cluster (Fig. 1D). Befitting this expression pattern, 75% are high-grade tumors, and over 40% have p53 mutations (Carey et al. 2006). In the era before HER2-targeted therapies, the HER2-enriched subtype carried a poor prognosis (Sorlie et al. 2001, 2003; Hu et al. 2006; Parker et al. 2009). However, given the large benefit of anti-HER2-targeted therapies in HER2-positive patients (Mass 2004; Piccart-Gebhart et al. 2005), it is reasonable to presume that the HER2-enriched subtype has benefited from the HER2-targeting revolution, but formal identification of this relationship is yet to be demonstrated. There are no known specific risk factors for the HER2-enriched subtype, and there is no apparent interaction with race or age (Carey et al. 2006; Millikan et al. 2007). However, the risk factor profile of this subtype most closely mirrors the luminal tumors. These data suggest that although the majority of ER-negative tumors are either of the HER2-enriched or basal-like subtype, the risk factors of HER2-enriched versus basal-like are distinct, again suggesting that these are two different diseases, in terms of etiology, that should merit individual attention for prevention and treatment strategies.
Basal-like Subtype
Perhaps the greatest impact of the intrinsic subtype genomic taxonomy was the identification of the basal-like subtype. Before the use of DNA microarrays, ER-positive and HER2-positive tumors were clearly appreciated as distinct disease types, but what was not appreciated was that of the remaining tumors. A significant disease entity existed that showed an obvious and strong common biology. In clinical terms, this group has become known as “triple-negative” tumors (Schneider et al. 2008), due to their typical immunohistochemical (IHC) pattern of being negative for ER, PR, and HER2 (which are the three commonly scored for predictive markers in the breast cancer clinic), although this is not a definitive classification since ∼25% of basal-like tumors are not triple-negative. The basal-like subtype is characterized by low expression of the luminal genes, low expression of the HER2 gene cluster, high expression of the proliferation cluster, and high expression of a unique cluster of genes called the basal cluster (Fig. 1B). The basal gene cluster includes basal epithelial cytokeratins (CK) such as CK5, 6, 14, and 17; epidermal growth factor receptor (EGFR); c-Kit; Vimentin; P-Cadherin; Fascin; Caveolins 1 and 2; and αB-crystallin. Note that it was the expression of cytokeratins 5, 6, 14, and 17 that gave rise to the term “basal-like,” as these are typically cytokeratins that are expressed within basal epithelial cells of the skin and airways.
Several risk factors for developing basal-like tumors have been identified, with the most interesting being the link between the basal-like subtype and BRCA1 mutation carriers (Olopade and Grushko 2001; Foulkes et al. 2003, 2004; Sorlie et al. 2003). Specifically, this association is that in women who carry a deleterious mutation in BRCA1 and who develop breast cancer, over 80% of the time their cancer is of the basal-like subtype. However, while BRCA1 mutation carriers usually develop basal-like breast cancer, most basal-like breast cancers are sporadic, and the BRCA1 gene and protein appear intact in these tumors (Richardson et al. 2006). A commonly held, but not yet formally proven, hypothesis is that the broader BRCA1 pathway is aberrant in sporadic basal-like breast cancer, which, if true, has important therapeutic implications. For example, the BRCA1 (and BRCA2) are critical for properhomologous recombination-mediated DNA repair, which is a high-fidelity mechanism. When the homologous recombination pathway is lost or dysfunctional, DNA repair occurs by the more error-prone method that involves poly(ADP-ribose) polymerase (PARP), which can be inhibited by a novel class of drugs that are being tested in clinical trials. Exciting results have now shown that PARP inhibitors elicit measurable responses in known BRCA1 and 2 mutant breast tumors (Fong et al. 2009), and more importantly, also provided improvements in response rates and overall survival in metastatic triple-negative patients (O'Shaughnessy et al. 2009). This latter finding does suggest that sporadic basal-like patients have an impaired BRCA1/2-pathway, but the precise genetic lesion is not yet known. Other known molecular genetic defects present in basal-like tumors include a high TP53 mutation rate (>50% as determined by sequence analysis) (Sorlie et al. 2001; Carey et al. 2006), and loss of RB1 function (Gauthier et al. 2007; Herschkowitz et al. 2008) that likely results in the high proliferation rates that are manifested by the high expression of the so-called “proliferation signature” (Fig. 1D), which mostly contain E2F-regulated genes (Whitfield et al. 2006).
Loss of normal DNA repair is also implicated in sensitivity to chemotherapy, particularly to DNA-damaging agents such as platinum drugs (Kennedy et al. 2004), although recent studies suggest that basal-like breast cancers may have a general sensitivity to chemotherapy (Rouzier et al. 2005; Carey et al. 2007). Another notable association is between the basal-like subtype, race, and age (Table 1). Several independent population-based studies have suggested that the basal-like subtype is overrepresented in young women with breast cancer, in African-American women, and especially in young African-American women (Carey et al. 2006; Bauer et al. 2007; Millikan et al. 2007; Lund et al. 2008). In the Carolina Breast Cancer Study, basal-like breast cancer was the most common among premenopausal African-American women (27%), and least common among postmenopausal non African-American women (9%) (Millikan et al. 2007). Interestingly, this overrepresentation of basal-like cancers is even more prevalent in Africans (Nigerians) (Huo et al. 2009). These findings suggest that there may be genetic predisposition to basal-like tumors, which is supported by alleles of MYBL2, showing increased frequency in basal-like cases versus controls (Thorner et al. 2009). Millikan et al. have also shown that there are life history risk factors for developing basal-like cancers, including multiple pregnancies (which is a protective factor for luminal disease), no lactation, and having a high waist/hip ratio (which is a measure of obesity that was predisposing for all breast cancer subtypes) (Millikan et al. 2007).
Claudin-low Subtype
Recently, comparative mouse and human genomic analysis identified a unique subtype in both humans and mice, with the mouse tumors noted to display a “spindloid” morphology (Herschkowitz et al. 2007). The conserved claudin-low tumor subtype is characterized by the low expression of genes involved in tight junctions and cell–cell adhesion including claudin 3, 4, 7, Occludin, and E-cadherin (Fig. 1E), as well as by the high expression of many mesenchymal genes including Vimentin, Snail1, Snail 2, and Twist1. This lack of epithelial cell features and expression of mesenchymal traits is reminiscent of features associated with stem cells, which is precisely what was shown by Lim et al. (2009); specifically, Lim et al. purified and expression-profiled normal human mammary stem cells (CD49f+/EpCAM−) using fluorescence-activated cell sorting (FACS). Interestingly, the authors compared these profiles to the average profile of each intrinsic subtype and determined that it was the claudin-low subtype that was the most similar to the Mammary Stem Cell profile. In this study, Lim et al. also identified a luminal progenitor fraction (CD49f+/EpCAM+) that was not able to form ducts in vivo but that could give rise to luminal colonies in vitro, and it was this fraction that was the most similar to basal-like tumors. Additionally, in tissues from BRCA1 mutation carriers, there were an increased number of these luminal progenitor/basal-like cells, thus suggesting that BRCA1 loss causes an arrest at this stage of development.
Other studies on Tumor Initiating Cells (TIC) isolated from multiple human breast tumors has shown that the claudin-low tumors are also enriched for TIC features including high ALDH1, a high mRNA ratio of CD44+/CD24−, and a high mRNA ratio of CD29+/CD24− (Creighton et al. 2009). In addition, it was also shown that metaplastic carcinomas (a rare and aggressive breast tumor subtype) show claudin-low features, with both claudin-low tumors and metaplastic carcinomas exhibiting characteristics of epithelial-to-mesenchymal transition (Hennessy et al. 2009). In total, these results suggest that the intrinsic subtypes of breast cancer may reflect arrest of epithelial cells at different stages of mammary epithelial development, with claudin-low tumors representing arrest at the most primitive stem cell state, then followed by basal-like tumors that are arrested at the luminal progenitor state, followed by luminal A/B tumors that show the greatest amount of differentiation (Lim et al. 2009; Prat and Perou 2009).
IMPORTANCE OF TUMOR STROMA IN BREAST CANCER
The subtypes discussed above have been identified through gene expression profiling of primary tumors that contain multiple cell types including epithelial cells, fibroblasts, adipocytes, and immune cells, among others. The topic of regulation of tumor growth and progression via the stroma is covered in other articles on this topic; however, we do note here that it is important to highlight that numerous publications have now identified that tumor stromal cells (i.e., cells other than the malignant epithelial cells present within tumors) can contribute to cancer development and progression (reviewed in Joyce and Pollard 2009), and stromal cell profiles alone can predict clinical outcome in breast cancer (Finak et al. 2008). Importantly, it is also becoming more and more evident that immune cells are key modulators of tumor progression and metastasis (DeNardo et al. 2009).
GENOMIC ALTERATIONS ASSOCIATED WITH INTRINSIC SUBTYPES
Genomic instability was shown decades ago to be a hallmark of cancer (Nowell and Hungerford 1960). Since then, there have been numerous studies using different methods to identify breast cancer subclasses based on the genomic structure ranging from karyotype studies and Array-based Comparative Genomic Hybridization (aCGH) analyses, to large-scale SNP arrays. For example, karyotyping has shown that invasive lobular carcinomas (ILC) have a chromosome 16q loss in more than 60% of cases, correlating with loss of E-cadherin expression, a protein important for cell adhesion and motility (Vos et al. 1997). This loss is often combined with 1q gain, and a translocation resulting in a der(1,16) is considered as an early event in mammary carcinogenesis (Tsarouha et al. 1999). In a study by Nordgard et al. (2008), deletion of 16q was overrepresented in the good prognosis luminal A subgroup, but at the same time was a predictor of good prognosis also for the nonluminal subgroups.
Both conventional and array-based CGH have been applied on invasive breast carcinoma cohorts. Several groups have found genomic alterations by aCGH that seem to be more frequent in one or more of the intrinsic classes (Bergamaschi et al. 2006; Chin et al. 2006; Fridlyand et al. 2006). Bergamaschi et al. (2006) showed in an advance stage cohort, that the intrinsic subclasses harbored different genomic alterations. The basal-like tumors had higher numbers of gains and losses than luminal A and the luminal B, and HER2-enriched had more frequent high-level amplifications. Chin and Fridlyand compared their aCGH groups to the expression subtypes and found that luminal A tumors were dominating the 1q/16q group; luminal A and HER2 enriched the “mixed amplifier” group; and basal-like and luminal B tumors comprised the majority of the “complex” group. Another study (Chin et al. 2007) identified a group of tumors with low genomic instability, and found these tumors to be enriched for the basal-like subtype. Normal breast-like samples are often too few to be studied, and luminal B can be difficult to identify in some data sets (Langerod et al. 2007); thus, their copy number profiles are much less defined.
Four different patterns of alterations were identified by Hicks et al. (2006). The “Simplex” group had broad segments of duplications and deletions and often associated to luminal A tumors; deletion of 16q, 8p, and 22 as well as gain of 1q, 8q, and 16p was dominating. The “Complex I” had either a “saw tooth” appearance with narrow segments of deletions and duplications affecting more or less all chromosomes, with the basal-like tumors dominating this group of tumors. “Complex II” tumors resembled the “simplex,” but had at least one localized region of clustered peaks of amplifications called “firestorms,” and were typical for the HER2-enriched group. The fourth pattern was called “flat,” defining profiles with no clear gains or losses in terms of copy number.
Several studies have had quite divergent definitions on which genomic alterations characterize distinct subgroups of breast carcinomas, but a common finding is that the 1q and 16q alterations dominate in one type, and multiple alterations on several arms dominate others (Tirkkonen et al. 1998; Teixeira et al. 2002; Rennstam et al. 2003; Korsching et al. 2004; Baudis 2007; Climent et al. 2007; Andre et al. 2009). In a recent study of genomic copy number aberrations (Russnes et al. 2010), two platform-independent algorithms were developed to explore genomic architectural distortion using aCGH data to measure whole arm gains and losses (WAAI) and complex rearrangements (CAAI). By applying CAAI and WAAI to data from 595 breast cancer patients, the tumors could be separated into eight subgroups with different distributions of genomic distortion. Within each subgroup data, from expression analyses, sequencing, and ploidy indicated that progression occurs along separate paths into more complex genotypes. In particular, basal-like tumors were separated into genomic classes with distinct patterns of alterations, emphasizing genomic heterogeneity within this intrinsic subtype.
Genomic Profiles of Clinical Utility
Microarray-based analyses have resulted in an explosion of prognostic and predictive profiles, and for breast cancer alone, there are >100 publications purporting to have prognostic profiles. We apologize to the many authors who developed important signatures that were not discussed here; however, a small number of profiles/signatures have come into relatively common clinical use and thus, are discussed here. Of particular note are the OncotypeDX (Paik et al. 2004, 2006) and Mammaprint gene expression tests (van 't Veer et al. 2002; van de Vijver et al. 2002). These tests were both developed as outcome predictors for breast cancer patients, which are basically predictors of the likelihood to develop metastasis (which is discussed in greater detail below). Specifically for the OncotypeDX assay, this 16-gene assay was developed to predict the likelihood of recurrence for ER+/node-negative patients receiving tamoxifen (Paik et al. 2004). Building upon these results, Paik et al. (2004) also showed that this assay can be used to identify a subset of ER+ patients that gain a benefit from chemotherapy (CMF therapy specifically), which was a relatively small subset of patients (∼25% of the total tested). The great utility of such an assay is that typically all of these patients might be treated with chemotherapy; thus, by using this assay, only 25% of patients will receive chemotherapy, as they are the subset that benefit, thus sparing the remaining 75% the toxicity and cost associated with chemotherapy regimens. This assay is now being tested in a prospective trial called TAILORx (www.cancer.gov/clinicaltrials/digestpage/TAILORx).
Mammaprint was developed as a pure prognosis predictor, using time to distant metastasis formation as the supervising parameter in patients receiving no systemic adjuvant therapy (van 't Veer et al. 2002; van de Vijver et al. 2002). The utility of this assay is to identify those patients in whom the risk of relapse/metastasis formation is so low that they can be spared adjuvant chemotherapy because they would do very well if they received no therapy at all. This predictor is also being tested in a prospective clinical trial in Europe called MINDACT (www.eortc.be/services/unit/mindact/MINDACT_websiteii.asp). Interestingly, there was only one gene that overlapped between the OncotypeDX assay and the Mammaprint assay, and so this prompted us to perform a simple analysis where we took a single data set and applied four of the most well-known prognostic profiles, and we asked what level of patient classification concordance existed (Fan et al. 2006). These predictors included the aforementioned OncotypeDx assay, the Mammaprint assay, the intrinsic subtype classification, and a fibroblast-derived wound response signature from Howard Chang and colleagues (Chang et al. 2004). The result was that there was a large amount of agreement between these assays, with, for example, the OncotypeDx and Mammaprint assays agreeing in outcome predictions for ∼80% of the patients. These findings strongly suggest that individual gene identity is not a good means of determining similarity across profiles, and suggests that these apparently different gene sets may be recognizing a common biological background.
The Genomics of Metastasis
Metastases are the main cause of mortality for patients with breast cancer, and diagnosis of disseminated disease correlates with less than a 3% survival rate over 20 years (Greenberg et al. 1996). Genomic profiling of human tumors and model systems has provided important mechanistic information for metastasis biology. First, there are gene signatures in primary tumors that can be highly predictive of the development of future metastases (van 't Veer et al. 2002; van de Vijver et al. 2002; Ramaswamy et al. 2003; Paik et al. 2004; Wang et al. 2005). Second, primary tumors and metastases appear genomically very similar to one another, despite being separated in time or space (Fig. 2) (Perou et al. 2000; Weigelt et al. 2005; Hu et al. 2009). Third, human cell lines can be selected that have specific end-organ tropisms with distinct expression profiles in brain, bone, and lung, and specific sets of genes mediate these aggressive behaviors (Kang et al. 2003; Minn et al. 2005; Bos et al. 2009). Above, we discuss many of the “metastasis predictors,” which are basically the different outcome predictors for breast cancer patients, and below we discuss genomic predictors that have provided biological insights into metastasis biology.
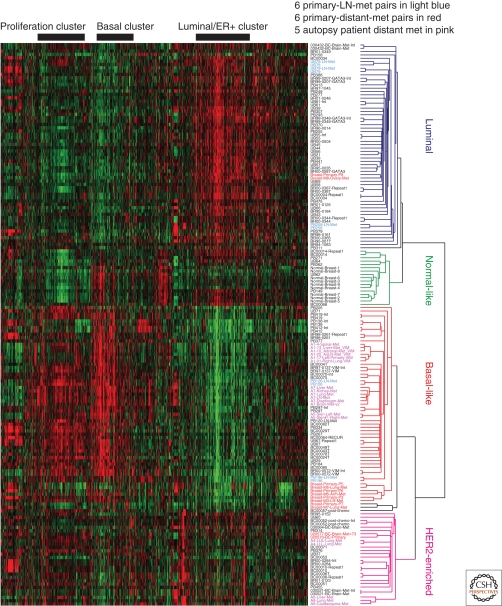
Figure 2.
Hierarchical clustering analysis of primary tumors and metastases taken from Weigelt et al. (2005) and reproduced with permission from AACR Publications © 2005. A hierarchical clustering analysis was performed using a 1300-gene intrinsic gene list from Hu et al. (2006). Genes were arranged in horizontal and samples in vertical. Sample names in red represent six primary tumor-distant metastasis pairs; those in light blue represent primary tumor-lymph node metastasis pairs; and those in pink represent local and distant metastasis samples from autopsy patients (which for patient A1 includes the primary).
Only a few years ago the predominant model of cancer progression stated that the capacity of a primary tumor to metastasize is acquired only rarely and late in tumorigenesis (Fidler and Kripke 1977). Belief in this model requires metastatic cells to be molecularly distinct from the majority of the tumor population they arose from. Global gene expression profiling is an ideal method to use to determine how different primary tumors are from their metastases. However, due mostly to the difficulties of harvesting metastatic tissues, there are only a few published studies that report on the gene expression similarity between primary tumors and their matched metastases. One study performed microarray analyses on eight pairs of tumors that were patient-matched primary and distant metastases (Weigelt et al. 2003), with the interval between the surgical removal of the primary tumors and metastases varying from 1.6 to 15 years. Unsupervised hierarchical clustering of these found that 6 of the 8 metastases were more similar to their primary tumor than to any other sample. On average, 92% of significantly expressed genes were similarly expressed between primary tumors and their metastases, and overall, no one gene was differently expressed in all metastases.
A second study with additional unmatched and matched primary tumor-metastasis samples confirmed these original results, finding that all primary breast tumors paired with their matching metastasis and maintained their intrinsic subtype (Weigelt et al. 2005). It is important to note that metastases occur in all subtypes (Fig. 2), although the rate and location of cancer progression are influenced by subtype (Smid et al. 2008). Interestingly, some of the markers for tumor aggressiveness were increased in six of the seven pairs of metastases, yielding a “poorer” 70-gene prognosis signature in the metastatic cells. These studies provide initial insights and suggest that metastases are much more similar to their primaries than was originally envisioned. It is this great similarity between primaries and their matched metastases, or between two pieces of any given tumor, that gave rise to the term “molecular portraits” because we learned that the genomic pattern of any individual tumor is as unique as a portrait of a given individual (Chung et al. 2002).
Site-specific Metastatic Signatures
A great deal of work has been done to determine if signatures of metastasis, particular signatures of metastasis to specific sites, exist. In 2003, Kang et al. described a genomic signature mediating breast cancer metastasis to bone (Kang et al. 2003). This study used an aggressive and metastatic human breast cancer cell line, MDA-MB-231, that can form osteolytic bone metastases 30% of the time after inoculation into the arterial circulation of mice. After recovering these bone metastases, and re-injecting them into mice serially, they developed cell lines with an increased propensity to grow in the bone. To determine the genes that contributed to increased bone metastases, microarray analyses were performed on the bone-tropic cells and the parental cells. Overall, they found few differences in the global gene expression profile, but they did find a small set of the genes that were more highly expressed in the bone tropic line that included cell membrane and secretory products that may function to affect the host environment to favor metastasis. Through in vitro single-cell cloning and microarray analyses, they found that the increase in aggressiveness observed in the in vivo isolated bone-seeking cells was due to specific cells within the parental cell line expressing five key metastasis genes. They concluded that overexpression of the small bone metastasis gene set was superimposed on a poor-prognosis gene expression signature already present in the parental breast cancer population, suggesting that metastasis requires a small set of functions beyond those already present within this aggressive cell line.
Further studies from the Massague laboratory used the same approach to identify genes that mediate breast cancer metastasis to the lung (Minn et al. 2005) and brain (Bos et al. 2009). Both studies identified relatively small gene sets that regulate lung and brain metastases with validation of their signatures using clinical outcomes. Interestingly, six of the 17 genes in the brain signature were also found in the lung signature, and this overlap suggests a partial sharing of mediators of metastasis to the brain and lungs, which is corroborated by observations from clinical practice. It should be stressed that these models require the initial injection of cancer cells directly into the bloodstream, hence the generation of these cells does not truly reflect metastatic events that occur from a primary tumor. Also, the MDA-MB-231 cells are reported to be a “basal B” cell line (Neve et al. 2006), or as more recently reported, a claudin-low cell line (Prat et al., unpubl.); therefore, the site-tropic signatures are superimposed on an already poor prognosis signature.
Using different approaches, other laboratories have also identified genes that are associated with distant metastases. In 2006, Smid et al. analyzed 107 microarrays of primary breast tumors/patients that had all experienced relapse (Smid et al. 2006). They found 69 genes that were significantly different between patients that experienced relapse to bone as compared to other distant sites, and the fibroblast growth factor receptor signaling pathway was identified as an important facilitator of bone colonization. They ultimately developed a 31-gene classifier that predicted bone relapse, and which could possibly be used to recommend bisphosphonate therapy to prevent osseous metastasis. In 2008, Landemaine et al. identified a six-gene signature that predicts lung metastasis (Landemaine et al. 2008). This study took a novel approach and compared the gene expression of five lung metastases against 18 metastases growing in other distant organs of patients. In 2009, Hu et al. (2009) identified a vascular endothelial growth factor (VEGF) signature that was associated with distant metastases and poor outcomes. This study compared gene expression profiles of 134 primary breast tumors, nine regional (lymph node) metastases, and 18 distant metastases in order to identify biological features associated with the distant metastases. Supervised analyses revealed very few differences between primary tumors and lymph node metastases; however, distant metastases had a distinct expression profile that was distinguished by the high expression of 13 genes, including VEGF, most of which were HIF1-alpha-regulated. The VEGF signature was able to predict survival in patients with breast cancer, lung cancer, and glioblastomas, suggesting that it may represent a widespread signature of hypoxia that is applicable to multiple tumors types.
Breast Cancer Stem Cells and Metastasis
Evidence for the existence of breast cancer stem cells is growing (Al-Hajj et al. 2003; Ginestier et al. 2007; Shipitsin et al. 2007; Wicha 2008), and is covered in other articles on this topic; however, mention of these unique cells is required here, as it is likely that they may be responsible for the formation of many metastases. Recent studies have shown that normal cells within the human and mouse mammary tissues have various amounts of differentiation and that certain rare cells, breast stem cells, can give rise to fully functional epithelial ductal structures (Raouf et al. 2008; Lim et al. 2009). Our understanding of the molecular mechanisms that give rise to breast tumors is incomplete, but it is likely that mutagenic agents target specific breast cell subpopulations (i.e., the cell of origin for the tumor), which is likely influenced by germline traits, to determine whether or not aberrant cell proliferation occurs. Thus, if highly differentiated cells are transformed, then ER+/luminal breast cancer may occur, while if more primitive stem, or committed, progenitor cells are targeted, a more stem-cell subtype (claudin-low) or luminal progenitor (basal-like) tumor may arise (Lim et al. 2009; Prat and Perou 2009). Interestingly, embryonic stem cell signatures have been identified and are genomically more related to poorly differentiated basal-like tumors than to luminal tumors (Ben-Porath et al. 2008). Furthermore, a recent study found that poorly differentiated tumors contain a higher content of prospectively isolated cancer stem cells compared to well-differentiated tumors (Pece et al. 2010). The incorporation of genome-wide expression studies described above has further validated the hypothesis that breast cancer cells within a tumor differ in their metastatic potential. Since breast cancer stem cells are relatively resistant to both chemotherapy and radiation (Phillips et al. 2006; Li et al. 2008), and because metastases nearly always recur after treatment, it is likely that stem cells are involved in metastatic progression.
Summary
Without a doubt, the past decade of breast cancer research has produced more advances in our understanding of the genetics of this disease than ever before. This knowledge has been significantly contributed to by the use of high-throughput DNA microarray analyses of primary tumors and metastases. The future of breast cancer genetic research looks encouraging, as new technologies are emerging that will couple DNA sequence and copy number changes with gene and protein expression. All of these advancements will further our understanding of this disease and will allow for improved targeted therapeutics to inhibit tumor growth and metastatic progression.
ACKNOWLEDGMENTS
We thank J. Chuck Harrell and Lisa Carey for assistance in writing and editing this article.
Footnotes
Editors: Mina J. Bissell, Kornelia Polyak, and Jeffrey Rosen
Additional Perspectives on The Mammary Gland as an Experimental Model available at www.cshperspectives.org
REFERENCES
- Abd El-Rehim DM, Pinder SE, Paish CE, Bell J, Blamey RW, Robertson JF, Nicholson RI, Ellis IO 2004. Expression of luminal and basal cytokeratins in human breast carcinoma. J Pathol 203: 661–671 [DOI] [PubMed] [Google Scholar]
- Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF 2003. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A 100: 3983–3988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allred DC, Wu Y, Mao S, Nagtegaal ID, Lee S, Perou CM, Mohsin SK, O'Connell P, Tsimelzon A, Medina D 2008. Ductal carcinoma in situ and the emergence of diversity during breast cancer evolution. Clin Cancer Res 14: 370–378 [DOI] [PubMed] [Google Scholar]
- Andre F, Job B, Dessen P, Tordai A, Michiels S, Liedtke C, Richon C, Yan K, Wang B, Vassal G, et al. 2009. Molecular characterization of breast cancer with high-resolution oligonucleotide comparative genomic hybridization array. Clin Cancer Res 15: 441–451 [DOI] [PubMed] [Google Scholar]
- Asselin-Labat ML, Sutherland KD, Barker H, Thomas R, Shackleton M, Forrest NC, Hartley L, Robb L, Grosveld FG, van der Wees J, et al. 2006. Gata-3 is an essential regulator of mammary-gland morphogenesis and luminal-cell differentiation. Nat Cell Biol 9: 201–209 [DOI] [PubMed] [Google Scholar]
- Baudis M 2007. Genomic imbalances in 5918 malignant epithelial tumors: An explorative meta-analysis of chromosomal CGH data. BMC Cancer 7: 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer KR, Brown M, Cress RD, Parise CA, Caggiano V 2007. Descriptive analysis of estrogen receptor (ER)-negative, progesterone receptor (PR)-negative, and HER2-negative invasive breast cancer, the so-called triple-negative phenotype: A population-based study from the California Cancer Registry. Cancer 109: 1721–1728 [DOI] [PubMed] [Google Scholar]
- Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, Weinberg RA 2008. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet 40: 499–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benito M, Parker J, Du Q, Wu J, Xiang D, Perou CM, Marron JS 2004. Adjustment of systematic microarray data biases. Bioinformatics 20: 105–114 [DOI] [PubMed] [Google Scholar]
- Bergamaschi A, Kim YH, Wang P, Sorlie T, Hernandez-Boussard T, Lonning PE, Tibshirani R, Børresen-Dale AL, Pollack JR 2006. Distinct patterns of DNA copy number alteration are associated with different clinicopathological features and gene-expression subtypes of breast cancer. Genes Chromosom Cancer 45: 1033–1040 [DOI] [PubMed] [Google Scholar]
- Bos PD, Zhang XH, Nadal C, Shu W, Gomis RR, Nguyen DX, Minn AJ, van de Vijver MJ, Gerald WL, Foekens JA, et al. 2009. Genes that mediate breast cancer metastasis to the brain. Nature 459: 1005–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey LA, Perou CM, Livasy CA, Dressler LG, Cowan D, Conway K, Karaca G, Troester MA, Tse CK, Edmiston S, et al. 2006. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA 295: 2492–2502 [DOI] [PubMed] [Google Scholar]
- Carey LA, Dees EC, Sawyer L, Gatti L, Moore DT, Collichio F, Ollila DW, Sartor CI, Graham ML, Perou CM 2007. The triple negative paradox: Primary tumor chemosensitivity of breast cancer subtypes. Clin Cancer Res 13: 2329–2334 [DOI] [PubMed] [Google Scholar]
- Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, Eeckhoute J, Shao W, Hestermann EV, Geistlinger TR, et al. 2005. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell 122: 33–43 [DOI] [PubMed] [Google Scholar]
- Chang HY, Sneddon JB, Alizadeh AA, Sood R, West RB, Montgomery K, Chi JT, van de Rijn M, Botstein D, Brown PO 2004. Gene expression signature of fibroblast serum response predicts human cancer progression: Similarities between tumors and wounds. PLoS Biol 2: E7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin K, DeVries S, Fridlyand J, Spellman PT, Roydasgupta R, Kuo WL, Lapuk A, Neve RM, Qian Z, Ryder T, et al. 2006. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell 10: 529–541 [DOI] [PubMed] [Google Scholar]
- Chin SF, Teschendorff AE, Marioni JC, Wang Y, Barbosa-Morais NL, Thorne NP, Costa JL, Pinder SE, van de Wiel MA, Green AR, et al. 2007. High-resolution aCGH and expression profiling identifies a novel genomic subtype of ER negative breast cancer. Genome Biol 8: R215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung CH, Bernard PS, Perou CM 2002. Molecular portraits and the family tree of cancer. Nat Genet 32: 533–540 [DOI] [PubMed] [Google Scholar]
- Climent J, Garcia JL, Mao JH, Arsuaga J, Perez-Losada J 2007. Characterization of breast cancer by array comparative genomic hybridization. Biochem Cell Biol 85: 497–508 [DOI] [PubMed] [Google Scholar]
- Creighton CJ, Li X, Landis M, Dixon JM, Neumeister VM, Sjolund A, Rimm DL, Wong H, Rodriguez A, Herschkowitz JI, et al. 2009. Residual breast cancers after conventional therapy display mesenchymal as well as tumor-initiating features. Proc Natl Acad Sci U S A 106: 13820–13825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeNardo DG, Barreto JB, Andreu P, Vasquez L, Tawfik D, Kolhatkar N, Coussens LM 2009. CD4(+) T cells regulate pulmonary metastasis of mammary carcinomas by enhancing protumor properties of macrophages. Cancer Cell 16: 91–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eeckhoute J, Keeton EK, Lupien M, Krum SA, Carroll JS, Brown M 2007. Positive cross-regulatory loop ties GATA-3 to estrogen receptor alpha expression in breast cancer. Cancer Res 67: 6477–6483 [DOI] [PubMed] [Google Scholar]
- Fan C, Oh DS, Wessels L, Weigelt B, Nuyten DS, Nobel AB, van't Veer LJ, Perou CM 2006. Concordance among gene-expression-based predictors for breast cancer. N Engl J Med 355: 560–569 [DOI] [PubMed] [Google Scholar]
- Fidler IJ, Kripke ML 1977. Metastasis results from preexisting variant cells within a malignant tumor. Science 197: 893–895 [DOI] [PubMed] [Google Scholar]
- Finak G, Bertos N, Pepin F, Sadekova S, Souleimanova M, Zhao H, Chen H, Omeroglu G, Meterissian S, Omeroglu A, et al. 2008. Stromal gene expression predicts clinical outcome in breast cancer. Nat Med 14: 518–527 [DOI] [PubMed] [Google Scholar]
- Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, Mortimer P, Swaisland H, Lau A, O'Connor MJ, et al. 2009. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med 361: 123–134 [DOI] [PubMed] [Google Scholar]
- Foulkes WD 2004. BRCA1 functions as a breast stem cell regulator. J Med Genet 41: 1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulkes WD, Stefansson IM, Chappuis PO, Begin LR, Goffin JR, Wong N, Trudel M, Akslen LA 2003. Germline BRCA1 mutations and a basal epithelial phenotype in breast cancer. J Natl Cancer Inst 95: 1482–1485 [DOI] [PubMed] [Google Scholar]
- Fridlyand J, Snijders AM, Ylstra B, Li H, Olshen A, Segraves R, Dairkee S, Tokuyasu T, Ljung BM, Jain AN, et al. 2006. Breast tumor copy number aberration phenotypes and genomic instability. BMC Cancer 6: 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier ML, Berman HK, Miller C, Kozakeiwicz K, Chew K, Moore D, Rabban J, Chen YY, Kerlikowske K, Tlsty TD 2007. Abrogated response to cellular stress identifies DCIS associated with subsequent tumor events and defines basal-like breast tumors. Cancer Cell 12: 479–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginestier C, Hur MH, Charafe-Jauffret E, Monville F, Dutcher J, Brown M, Jacquemier J, Viens P, Kleer CG, Liu S, et al. 2007. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell 1: 555–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Git A, Spiteri I, Blenkiron C, Dunning MJ, Pole JC, Chin SF, Wang Y, Smith J, Livesey FJ, Caldas C 2008. PMC42, a breast progenitor cancer cell line, has normal-like mRNA and microRNA transcriptomes. Breast Cancer Res 10: R54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg PA, Hortobagyi GN, Smith TL, Ziegler LD, Frye DK, Buzdar AU 1996. Long-term follow-up of patients with complete remission following combination chemotherapy for metastatic breast cancer. J Clin Oncol 14: 2197–2205 [DOI] [PubMed] [Google Scholar]
- Hennessy BT, Gonzalez-Angulo AM, Stemke-Hale K, Gilcrease MZ, Krishnamurthy S, Lee JS, Fridlyand J, Sahin A, Agarwal R, Joy C, et al. 2009. Characterization of a naturally occurring breast cancer subset enriched in epithelial-to-mesenchymal transition and stem cell characteristics. Cancer Res 69: 4116–4124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herschkowitz JI, Simin K, Weigman VJ, Mikaelian I, Usary J, Hu Z, Rasmussen KE, Jones LP, Assefnia S, Chandrasekharan S, et al. 2007. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol 8: R76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herschkowitz JI, He X, Fan C, Perou CM 2008. The functional loss of the retinoblastoma tumor suppressor is a common event in basal-like and luminal B breast carcinomas. Breast Cancer Res 10: R75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks J, Krasnitz A, Lakshmi B, Navin NE, Riggs M, Leibu E, Esposito D, Alexander J, Troge J, Grubor V, et al. 2006. Novel patterns of genome rearrangement and their association with survival in breast cancer. Genome Res 16: 1465–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Fan C, Oh DS, Marron JS, He X, Qaqish BF, Livasy C, Carey LA, Reynolds E, Dressler L, et al. 2006. The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genomics 7: 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Fan C, Livasy C, He X, Oh DS, Ewend MG, Carey LA, Subramanian S, West R, Ikpatt F, et al. 2009. A compact VEGF signature associated with distant metastases and poor outcomes. BMC Med 7: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo D, Ikpatt F, Khramtsov A, Dangou JM, Nanda R, Dignam J, Zhang B, Grushko T, Zhang C, Oluwasola O, et al. 2009. Population differences in breast cancer: Survey in indigenous African women reveals over-representation of triple-negative breast cancer. J Clin Oncol 27: 4515–4521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce JA, Pollard JW 2009. Microenvironmental regulation of metastasis. Nat Rev Cancer 9: 239–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y, Siegel PM, Shu W, Drobnjak M, Kakonen SM, Cordon-Cardo C, Guise TA, Massague J 2003. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell 3: 537–549 [DOI] [PubMed] [Google Scholar]
- Kennedy RD, Quinn JE, Mullan PB, Johnston PG, Harkin DP 2004. The role of BRCA1 in the cellular response to chemotherapy. J Natl Cancer Inst 96: 1659–1668 [DOI] [PubMed] [Google Scholar]
- Korsching E, Packeisen J, Helms MW, Kersting C, Voss R, van Diest PJ, Brandt B, van der Wall E, Boecker W, Burger H 2004. Deciphering a subgroup of breast carcinomas with putative progression of grade during carcinogenesis revealed by comparative genomic hybridisation (CGH) and immunohistochemistry. Br J Cancer 90: 1422–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouros-Mehr H, Slorach EM, Sternlicht MD, Werb Z 2006. GATA-3 maintains the differentiation of the luminal cell fate in the mammary gland. Cell 127: 1041–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landemaine T, Jackson A, Bellahcene A, Rucci N, Sin S, Abad BM, Sierra A, Boudinet A, Guinebretiere JM, Ricevuto E, et al. 2008. A six-gene signature predicting breast cancer lung metastasis. Cancer Res 68: 6092–6099 [DOI] [PubMed] [Google Scholar]
- Langerod A, Zhao H, Borgan O, Nesland JM, Bukholm IR, Ikdahl T, Karesen R, Børresen-Dale AL, Jeffrey SS 2007. TP53 mutation status and gene expression profiles are powerful prognostic markers of breast cancer. Breast Cancer Res 9: R30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Lewis MT, Huang J, Gutierrez C, Osborne CK, Wu MF, Hilsenbeck SG, Pavlick A, Zhang X, Chamness GC, et al. 2008. Intrinsic resistance of tumorigenic breast cancer cells to chemotherapy. J Natl Cancer Inst 100: 672–679 [DOI] [PubMed] [Google Scholar]
- Lim E, Vaillant F, Wu D, Forrest NC, Pal B, Hart AH, Asselin-Labat ML, Gyorki DE, Ward T, Partanen A, et al. 2009. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat Med 15: 907–913 [DOI] [PubMed] [Google Scholar]
- Livasy CA, Perou CM, Karaca G, Cowan DW, Maia D, Jackson S, Tse CK, Nyante S, Millikan RC 2007. Identification of a basal-like subtype of breast ductal carcinoma in situ. Hum Pathol 38: 197–204 [DOI] [PubMed] [Google Scholar]
- Lund MJ, Trivers KF, Porter PL, Coates RJ, Leyland-Jones B, Brawley OW, Flagg EW, O'Regan RM, Gabram SG, Eley JW 2008. Race and triple negative threats to breast cancer survival: A population-based study in Atlanta, GA. Breast Cancer Res Treat 113: 357–370 [DOI] [PubMed] [Google Scholar]
- Mass RD 2004. The HER receptor family: A rich target for therapeutic development. Int J Radiat Oncol Biol Phys 58: 932–940 [DOI] [PubMed] [Google Scholar]
- Millikan RC, Newman B, Tse CK, Moorman PG, Conway K, Smith LV, Labbok MH, Geradts J, Bensen JT, Jackson S, et al. 2007. Epidemiology of basal-like breast cancer. Breast Cancer Res Treat 109: 123–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, Viale A, Olshen AB, Gerald WL, Massague J 2005. Genes that mediate breast cancer metastasis to lung. Nature 436: 518–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris GJ, Naidu S, Topham AK, Guiles F, Xu Y, McCue P, Schwartz GF, Park PK, Rosenberg AL, Brill K, et al. 2007. Differences in breast carcinoma characteristics in newly diagnosed African-American and Caucasian patients: A single-institution compilation compared with the National Cancer Institute's Surveillance, Epidemiology, and End Results database. Cancer 110: 876–884 [DOI] [PubMed] [Google Scholar]
- Naume B, Zhao X, Synnestvedt M, Borgen E, Russnes HG, Lingjaerde OC, Stromberg M, Wiedswang G, Kvalheim G, Karesen R, et al. 2007. Presence of bone marrow micrometastasis is associated with different recurrence risk within molecular subtypes of breast cancer. Mol Oncol 1: 160–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, Clark L, Bayani N, Coppe JP, Tong F, et al. 2006. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell 10: 515–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordgard SH, Johansen FE, Alnaes GI, Bucher E, Syvanen AC, Naume B, Børresen-Dale AL, Kristensen VN 2008. Genome-wide analysis identifies 16q deletion associated with survival, molecular subtypes, mRNA expression, and germline haplotypes in breast cancer patients. Genes Chromosom Cancer 47: 680–696 [DOI] [PubMed] [Google Scholar]
- Nowell PC, Hungerford DA 1960. Chromosome studies on normal and leukemic human leukocytes. J Natl Cancer Inst 25: 85–109 [PubMed] [Google Scholar]
- O'Shaughnessy J, Osborne C, Pippen J, Patt D, Rocha C, Ossovshaya V, Sherman BM, Bradley CR 2009. Updated results of a randomized phase II study demonstrating efficacy and safety of BSI-201, a PARP inhibitor, in combination with gemcitabine/carboplatin in metastatic triple-negative breast cancer. San Antonio Breast Cancer Symposium, San Antonio, Texas [Google Scholar]
- Oh DS, Troester MA, Usary J, Hu Z, He X, Fan C, Wu J, Carey LA, Perou CM 2006. Estrogen-regulated genes predict survival in hormone receptor-positive breast cancers. J Clin Oncol 24: 1656–1664 [DOI] [PubMed] [Google Scholar]
- Olopade OI, Grushko T 2001. Gene-expression profiles in hereditary breast cancer. N Engl J Med 344: 2028–2029 [DOI] [PubMed] [Google Scholar]
- Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, Baehner FL, Walker MG, Watson D, Park T, et al. 2004. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med 351: 2817–2826 [DOI] [PubMed] [Google Scholar]
- Paik S, Tang G, Shak S, Kim C, Baker J, Kim W, Cronin M, Baehner FL, Watson D, Bryant J, et al. 2006. Gene expression and benefit of chemotherapy in women with node-negative, estrogen receptor-positive breast cancer. J Clin Oncol 24: 3726–3734 [DOI] [PubMed] [Google Scholar]
- Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T, Davies S, Fauron C, He X, Hu Z, et al. 2009. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol 27: 1160–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pece S, Tosoni D, Confalonieri S, Mazzarol G, Vecchi M, Ronzoni S, Bernard L, Viale G, Pelicci PG, Di Fiore PP 2010. Biological and molecular heterogeneity of breast cancers correlates with their cancer stem cell content. Cell 140: 62–73 [DOI] [PubMed] [Google Scholar]
- Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, et al. 2000. Molecular portraits of human breast tumours. Nature 406: 747–752 [DOI] [PubMed] [Google Scholar]
- Phillips TM, McBride WH, Pajonk F 2006. The response of CD24(-/low)/CD44+ breast cancer-initiating cells to radiation. J Natl Cancer Inst 98: 1777–1785 [DOI] [PubMed] [Google Scholar]
- Piccart-Gebhart MJ, Procter M, Leyland-Jones B, Goldhirsch A, Untch M, Smith I, Gianni L, Baselga J, Bell R, Jackisch C, et al. 2005. Trastuzumab after adjuvant chemotherapy in HER2-positive breast cancer. N Engl J Med 353: 1659–1672 [DOI] [PubMed] [Google Scholar]
- Prat A, Perou CM 2009. Mammary development meets cancer genomics. Nat Med 15: 842–844 [DOI] [PubMed] [Google Scholar]
- Prat A, Parker JS, Fan C, Karginova O, Livasy C, Herschkowitz J, He X, Perou CM Submitted Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswamy S, Ross KN, Lander ES, Golub TR 2003. A molecular signature of metastasis in primary solid tumors. Nat Genet 33: 49–54 [DOI] [PubMed] [Google Scholar]
- Raouf A, Zhao Y, To K, Stingl J, Delaney A, Barbara M, Iscove N, Jones S, McKinney S, Emerman J, et al. 2008. Transcriptome analysis of the normal human mammary cell commitment and differentiation process. Cell Stem Cell 3: 109–118 [DOI] [PubMed] [Google Scholar]
- Rennstam K, Ahlstedt-Soini M, Baldetorp B, Bendahl PO, Borg A, Karhu R, Tanner M, Tirkkonen M, Isola J 2003. Patterns of chromosomal imbalances defines subgroups of breast cancer with distinct clinical features and prognosis. A study of 305 tumors by comparative genomic hybridization. Cancer Res 63: 8861–8868 [PubMed] [Google Scholar]
- Richardson AL, Wang ZC, De Nicolo A, Lu X, Brown M, Miron A, Liao X, Iglehart JD, Livingston DM, Ganesan S 2006. X chromosomal abnormalities in basal-like human breast cancer. Cancer Cell 9: 121–132 [DOI] [PubMed] [Google Scholar]
- Rouzier R, Perou CM, Symmans WF, Ibrahim N, Cristofanilli M, Anderson K, Hess KR, Stec J, Ayers M, Wagner P, et al. 2005. Breast cancer molecular subtypes respond differently to preoperative chemotherapy. Clin Cancer Res 11: 5678–5685 [DOI] [PubMed] [Google Scholar]
- Russnes HG, Vollan HK, Lingjaerde OC, Krasnitz A, Lundin P, Naume B, Sørlie T, Borgen E, Rye IH, Langerød A, Chin SF, et al. 2010. Genomic architecture characterizes tumor progression paths and fate in breast cancer patients. Sci Transl Med 2: 38ra47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider BP, Winer EP, Foulkes WD, Garber J, Perou CM, Richardson A, Sledge GW, Carey LA 2008. Triple-negative breast cancer: Risk factors to potential targets. Clin Cancer Res 14: 8010–8018 [DOI] [PubMed] [Google Scholar]
- Shipitsin M, Campbell LL, Argani P, Weremowicz S, Bloushtain-Qimron N, Yao J, Nikolskaya T, Serebryiskaya T, Beroukhim R, Hu M, et al. 2007. Molecular definition of breast tumor heterogeneity. Cancer Cell 11: 259–273 [DOI] [PubMed] [Google Scholar]
- Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL 1987. Human breast cancer: Correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science 235: 177–182 [DOI] [PubMed] [Google Scholar]
- Slamon DJ, Godolphin W, Jones LA, Holt JA, Wong SG, Keith DE, Levin WJ, Stuart SG, Udove J, Ullrich A 1989. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science 244: 707–712 [DOI] [PubMed] [Google Scholar]
- Smid M, Wang Y, Klijn JG, Sieuwerts AM, Zhang Y, Atkins D, Martens JW, Foekens JA 2006. Genes associated with breast cancer metastatic to bone. J Clin Oncol 24: 2261–2267 [DOI] [PubMed] [Google Scholar]
- Smid M, Wang Y, Zhang Y, Sieuwerts AM, Yu J, Klijn JG, Foekens JA, Martens JW 2008. Subtypes of breast cancer show preferential site of relapse. Cancer Res 68: 3108–3114 [DOI] [PubMed] [Google Scholar]
- Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, et al. 2001. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A 98: 10869–10874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, Deng S, Johnsen H, Pesich R, Geisler S, et al. 2003. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci U S A 100: 8418–8423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotiriou C, Neo SY, McShane LM, Korn EL, Long PM, Jazaeri A, Martiat P, Fox SB, Harris AL, Liu ET 2003. Breast cancer classification and prognosis based on gene expression profiles from a population-based study. Proc Natl Acad Sci U S A 100: 10393–10398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira MR, Pandis N, Heim S 2002. Cytogenetic clues to breast carcinogenesis. Genes Chromosom Cancer 33: 1–16 [DOI] [PubMed] [Google Scholar]
- Thorner AR, Hoadley KA, Parker JS, Winkel S, Millikan RC, Perou CM 2009. In vitro and in vivo analysis of B-Myb in basal-like breast cancer. Oncogene 28: 742–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirkkonen M, Tanner M, Karhu R, Kallioniemi A, Isola J, Kallioniemi OP 1998. Molecular cytogenetics of primary breast cancer by CGH. Genes Chromosom Cancer 21: 177–184 [PubMed] [Google Scholar]
- Tsarouha H, Pandis N, Bardi G, Teixeira MR, Andersen JA, Heim S 1999. Karyotypic evolution in breast carcinomas with i(1)(q10) and der(1;16)(q10;p10) as the primary chromosome abnormality. Cancer Genet Cytogenet 113: 156–161 [DOI] [PubMed] [Google Scholar]
- Usary J, Llaca V, Karaca G, Presswala S, Karaca M, He X, Langerod A, Karesen R, Oh DS, Dressler LG, et al. 2004. Mutation of GATA3 in human breast tumors. Oncogene 23: 7669–7678 [DOI] [PubMed] [Google Scholar]
- van 't Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, et al. 2002. Gene expression profiling predicts clinical outcome of breast cancer. Nature 415: 530–536 [DOI] [PubMed] [Google Scholar]
- van de Vijver MJ, He YD, van't Veer LJ, Dai H, Hart AA, Voskuil DW, Schreiber GJ, Peterse JL, Roberts C, Marton MJ, et al. 2002. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med 347: 1999–2009 [DOI] [PubMed] [Google Scholar]
- Vos CB, Cleton-Jansen AM, Berx G, de Leeuw WJ, ter Haar NT, van Roy F, Cornelisse CJ, Peterse JL, van de Vijver MJ 1997. E-cadherin inactivation in lobular carcinoma in situ of the breast: An early event in tumorigenesis. Br J Cancer 76: 1131–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Klijn JG, Zhang Y, Sieuwerts AM, Look MP, Yang F, Talantov D, Timmermans M, Meijer-van Gelder ME, Yu J, et al. 2005. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet 365: 671–679 [DOI] [PubMed] [Google Scholar]
- Weigelt B, Glas AM, Wessels LF, Witteveen AT, Peterse JL, van't Veer LJ 2003. Gene expression profiles of primary breast tumors maintained in distant metastases. Proc Natl Acad Sci U S A 100: 15901–15905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigelt B, Hu Z, He X, Livasy C, Carey LA, Ewend MG, Glas AM, Perou CM, Van't Veer LJ 2005. Molecular portraits and 70-gene prognosis signature are preserved throughout the metastatic process of breast cancer. Cancer Res 65: 9155–9158 [DOI] [PubMed] [Google Scholar]
- West M, Blanchette C, Dressman H, Huang E, Ishida S, Spang R, Zuzan H, Olson JA Jr, Marks JR, Nevins JR 2001. Predicting the clinical status of human breast cancer by using gene expression profiles. Proc Natl Acad Sci U S A 98: 11462–11467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield ML, George LK, Grant GD, Perou CM 2006. Common markers of proliferation. Nat Rev Cancer 6: 99–106 [DOI] [PubMed] [Google Scholar]
- Wicha MS 2008. Cancer stem cell heterogeneity in hereditary breast cancer. Breast Cancer Res 10: 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XR, Sherman ME, Rimm DL, Lissowska J, Brinton LA, Peplonska B, Hewitt SM, Anderson WF, Szeszenia-Dabrowska N, Bardin-Mikolajczak A, et al. 2007. Differences in risk factors for breast cancer molecular subtypes in a population-based study. Cancer Epidemiol Biomarkers Prev 16: 439–443 [DOI] [PubMed] [Google Scholar]
- Yu K, Lee CH, Tan PH, Tan P 2004. Conservation of breast cancer molecular subtypes and transcriptional patterns of tumor progression across distinct ethnic populations. Clin Cancer Res 10: 5508–5517 [DOI] [PubMed] [Google Scholar]