Abstract
Background
Plasmids containing hylEfm (pHylEfm) were previously shown to increase gastrointestinal colonization and lethality of Enterococcus faecium in experimental peritonitis. The hylEfm gene, predicting a glycosyl hydrolase, has been considered as a virulence determinant of hospital-associated E. faecium, although its direct contribution to virulence has not been investigated. Here, we constructed mutants of the hylEfm-region and we evaluated their effect on virulence using a murine peritonitis model.
Results
Five mutants of the hylEfm-region of pHylEfmTX16 from the sequenced endocarditis strain (TX16 [DO]) were obtained using an adaptation of the PheS* system and were evaluated in a commensal strain TX1330RF to which pHylEfmTX16 was transferred by mating; these include i) deletion of hylEfm only; ii) deletion of the gene downstream of hylEfm (down) of unknown function; iii) deletion of hylEfm plus down; iv) deletion of hylEfm-down and two adjacent genes; and v) a 7,534 bp deletion including these four genes plus partial deletion of two others, with replacement by cat. The 7,534 bp deletion did not affect virulence of TX16 in peritonitis but, when pHylEfmTX16Δ7,534 was transferred to the TX1330RF background, the transconjugant was affected in in vitro growth versus TX1330RF(pHylEfmTX16) and was attenuated in virulence; however, neither hylEfm nor hylEfm-down restored wild type function. We did not observe any in vivo effect on virulence of the other deletions of the hylEfm-region
Conclusions
The four genes of the hylEfm region (including hylEfm) do not mediate the increased virulence conferred by pHylEfmTX16 in murine peritonitis. The use of the markerless counterselection system PheS* should facilitate the genetic manipulation of E. faecium in the future.
Background
Enterococcus faecium is a common enterococcal species increasingly isolated from hospital-associated infections in the USA [1]. Compelling evidence suggests that this substantial increase in E. faecium nosocomial infections is due to the worldwide occurrence of a genetic subcluster (designated clonal cluster 17, CC17) which encompasses clones that appear to have evolved independently [2-4]. Several genes have been associated with CC17 E. faecium including i) espEfm, encoding a surface protein which has been associated with increased biofilm formation and urinary tract infection (UTI) [4-6]; ii) some fms genes (two of which are also designated pilA and pilB), encoding putative microbial surface components recognizing adhesive matrix molecules (MSCRAMMs) or components of enterococcal pili (including the pilus operon ebpABCfm, which appear to play a role in biofilm formation and experimental UTI) [2,7-10]; iii) an intact acm gene encoding a collagen adhesin which was shown to be important in the pathogenesis of endocarditis [8] and, iv) plasmids carrying the hylEfm gene [11-14].
It has been previously shown that hylEfm is carried by large transferable megaplasmids of different sizes (145 to 375 kb) in hospital-associated E. faecium which are widely distributed worldwide [11-13,15] These plasmids also can harbour antibiotic resistance determinants and some pilus-encoding genes of E. faecium which are present with hylEfm in the same plasmid [15,16]. The acquisition of the hylEfm-plasmid by an E. faecium laboratory strain (D344SRF) from a US clinical isolate (C68) increased the colonization of the gastrointestinal tract of mice, an effect that was independent of the presence of antibiotic resistance determinants [17]. Moreover, the acquisition of the hylEfm-plasmid from another US clinical strain (TX16) increased the virulence of a commensal strain E. faecium TX1330RF in experimental peritonitis [11].
The HylEfm protein was initially predicted to have homology with hyaluronidases which have been associated with virulence in other gram-positive pathogens [18,19], although hyaluronidase activity has not been detected in E. faecium isolates carrying this gene [15]. The most recent annotation and sequence comparisons indicate that this protein is likely to encode a family 84 glycosyl hydrolase [12,13]. In fact, the homolog of hylEfm in Streptococcus pyogenes (spy1600) encoded in a genetic locus with a similar organization to that of the hylEfm-region and sharing 42% identity at the amino acid level (61% similarity), was recently shown not to have any detectable hyaluronidase activity. Spy1600 was characterized as a family 84 glycosyl hydrolase with β-N-acetyl-glucosaminidase specificity after purification and substrate analysis [20] and expression of spy1600 in S. pyogenes was found to be up-regulated during phagocytosis [21]. For this reason, and because of the almost exclusive occurrence of hylEfm in isolates from clinical origin in different surveillance studies [14,22-24], this gene has been postulated as an important pathogenic determinant of hospital-associated E. faecium. However, its exact role in virulence has not been established. In this work, we assess the role of the hylEfm-region in E. faecium pathogenesis of experimental peritonitis.
Methods
Bacterial strains and plasmids
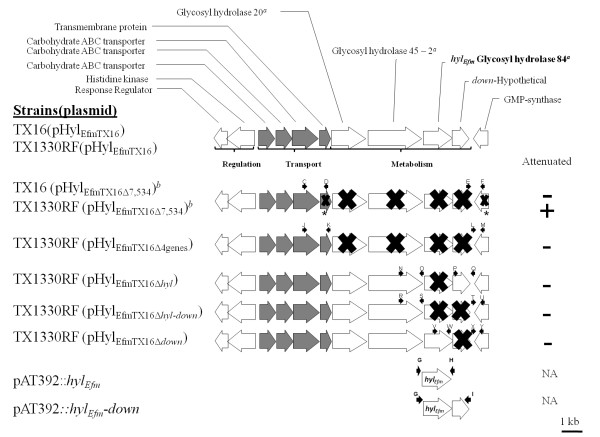
Table 1 and Figure 1 show the strains and plasmids used in this work and depict the genetic organization of the hylEfm-region in E. faecium strains and mutants.
Table 1.
E. faecium strains and plasmids used in this work
| Strains/Plasmids | Relevant Characteristics | Reference |
|---|---|---|
| Strains | ||
| E. faecium | ||
| TX16 (DO) | Sequenced endocarditis clinical isolate, Emr, Smr. ST-16a http://www.hgsc.bcm.tmc.edu | [35] |
| TX1330RF | Fsr and Rfr derivative of TX1330, a faecal colonizing strain from a healthy human volunteer | [11] |
| TX1330RF (pHylEfmTX16) | Derivative of TX1330RF to which the hylEfm-containing plasmid (pHylEfmTX16) was transferred by conjugation from TX16 (DO) (~250 kb) | [11] |
| TX1330RF (pHylEfmTX16Δ7,534) | Mutant with deletion of part or all of 6 genes of the hylEfm region of TX1330RF(pHylEfmTX16) | This work |
| TX1330RF (pHylEfmTX16Δ4genes) | Non-polar deletion of 4 genes of the hylEfm region of TX1330RF(pHylEfmTX16) | This work |
| TX1330RF (pHylEfmTX16Δhyl) | Non-polar deletion mutant of hylEfm of TX1330RF(pHylEfmTX16) | This work |
| TX1330RF (pHylEfmTX16Δhyl-down) | Non-polar deletion of hylEfm plus its downstream gene of TX1330RF(pHylEfmTX16) | This work |
| TX1330RF (pHylEfmTX16Δdown) | Non-polar deletion of the gene downstream of hylEfm of TX1330RF(pHylEfmTX16) | This work |
| E. faecalis | ||
| CK111 | OG1Sp upp4::P23 repA4 | [25] |
| Plasmids | ||
| pHylEfmTX16 | Conjugative and transferable megaplasmid (ca. 250 kb) of TX16 (DO) containing hylEfm | [11] |
| pCJK47 | Conjugative donor plasmid for markerless mutagenesis; oriTpCF10 and pheS* pORI280 derivative; confers Emr | [25] |
| pHOU1 | Derivative of pCJK47 in which the erm(C) gene was replaced by aph-2'-ID; confers Gmr | This work |
| pHOU2 | Derivative of pCJK47 in which the erm(C) gene was replaced by aph-2'-ID and cat was incorporated in the cloning site for allelic replacements; confers Gmr. | This work |
| pTEX5501ts | E. coli-enterococcal shuttle plasmid for mutagenesis using a temperature-sensitive replicon | [27] |
| pAT392 | oriRpAMβ1, oriRpUC oriTRK2 spc lacZα P2 aac(6')-aph(2") | [30] |
| pAT392::hylEfm | Derivative of pAT392 containing hylEfm (cloned with SacI and SmaI) under the control of the P2 promoter | This work |
| pAT392::hylEfm-down | Derivative of pAT392 containing both the hylEfm plus downstream genes (cloned with SacI and SmaI) under the control of the P2 promoter | This work |
Emr, erythromycin resistance; Fsr, fusidic acid resistance; Gmr, gentamicin resistance, Rfr, rifampin resistance; Smr, high-level resistance to streptomycin. aST refers to sequence type after multi-locus sequence typing. ST16 is part of CC17
Figure 1.
Physical map of the hylEfm-region in pHylEfmTX16. The annotated predicted function of the corresponding genes is shown above the genes. The genes were divided into three groups (metabolism, transport [in gray] and regulation based on putative functions). Strain nomenclature follows that specified in Table 1. Black arrows above the genes indicate the position of the primers used to obtain DNA fragments for mutagenesis and follow the nomenclature of Table 2. The crosses depict the genes that were deleted. The asterisks indicate only partial deletion of the gene was obtained. aThe number refers to the glycosyl hydrolase family with hylEfm depicted in bold; ballelic replacement with the chloramphenicol acetyl transferase gene (cat) was performed. NA, not applicable.
Construction of a deletion mutant of the hylEfm-region using the pheS* counter-selection system in TX16(pHylEfmTX16) and its transfer to TX1330RF
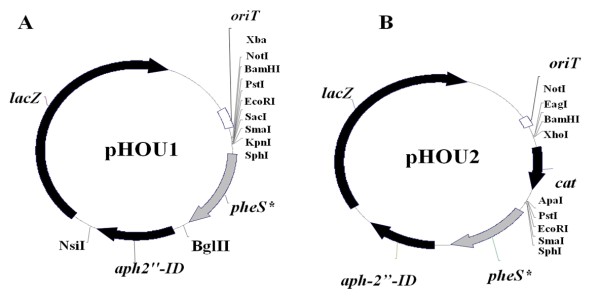
The pheS* system (previously used in Enterococcus faecalis) [25] is based on the acquired sensitivity of bacteria to p-chloro-phenylalanine (p-Cl-Phe) if they carry a pheS* allele encoding a phenylalanine tRNA synthetase with altered substrate specificity [25,26]. In order to apply this approach to E. faecium strains, which are commonly macrolide resistant, we constructed a derivative of the pheS* vector pCJK47 by replacing its erm(C) gene with aph2"-ID, which confers resistance to gentamicin. The full aph-2"-ID gene (including promoter and terminator regions) was amplified by PCR using plasmid pTEX5501ts [27] as the template with primers A and B (Table 2). The amplified fragment (1,089 bp) was digested with NsiI and BglII and ligated with pCJK47 digested with the same enzymes resulting in pHOU1 (Figure 2A). Subsequently, pHOU1 was digested with BamHI and PstI and ligated with a 992 bp fragment released from pTEX5501ts after digestion with the same enzymes and containing the chloramphenicol acetyl-transferase gene (cat), obtaining a 7,906 bp vector designated pHOU2 (Figure 2B).
Table 2.
Primers used in this work
| Primer | Sequence (5'-3') | Relevant Characteristics |
|---|---|---|
| A | gaagatctgagataggttatgcaagat | Forward, BglII site (underlined), used amplification of aph-2"-ID |
| B | ccaatgcatgattccggattctaaaaaagg | Reverse, NsiI site (underlined), used amplification of aph-2"-ID |
| C | cgggatccgtttaaaaccagctggaaaag | Forward, BamHI site (underlined), located 1,251 nucleotides upstream of the start codon of the gene encoding a putative glycosyl hydrolase family 20 (Figure 1.) |
| D | ccgctcgagcaattcaacattgcaaagac | Reverse, XhoI site (underlined), located 294 nucleotides upstream of the start codon of the gene encoding a putative glycosyl hydrolase family 20 (Figure 1.) |
| E | cgagggcccgtgaagtattgccagatgt | Forward, ApaI site (underlined); located 592 nucleotides downstream of the down gene (hypothetical, Figure 1.) |
| F | ccggaattcaaaagcagaattggaaatca | Reverse, EcoRI site, 1,571 nucleotides downstream of the down gene (hypothetical, Figure 1.) |
| G | gcgagctcgattactttcaaaggaga | Forward, SacI site (underlined), ribosomal binding site of hylEfm (italics) (Figure 1.) |
| H | tcccccgggctaacttttgataatttgctc | Reverse, SmaI site, (underlined) and stop codon of hylEfm (Figure 1.) |
| I | tcccccgggttagcgattgatcgagc | Reverse, SmaI site (underlined), stop codon of down (Figure 1.) |
| J | cgggatcccaatcaagaagtagcggatt | Forward, BamH site (underlined) 438 nucleotides upstream of the stop codon of carbohydrate ABC transporter gene (Figure 1.) |
| K | gcggccgctcgagggcccttagtgcgattgtatctgac | Reverse, stop codon of the gene that encodes to transmembrane protein (Figure 1.) |
| L | gggcccctcgaggcggccgcaaaattaaataaaaaatgg | Forward, ApaI, XhoI, NotI site, stop codon down (Figure 1.) |
| M | catgcatgaatcaggaactgaaactgc | Reverse, NsiI site, 1,091 nucleotides upstream of stop codon of GMP synthase (opposite orientation) (Figure 1.) |
| N | ccggaattccagtaaaaggcacagagc | Forward, EcoRI site (underlined), located 2,138 nucleotides down-stream of glycosyl hidrolase family 45-2 start codon (Figure 1.) |
| O | tcatctattttctcctttgaaagtaatcactatattcc | Reverse, stop codon of glycosyl hydrolase family 45-2 (Figure 1.) |
| P | tcaaaggagaaaatagatgaatatcttaaaaaataaaaagc | Forward, located 40 nucleotides upstream of down gene start codon (Figure 1.) |
| Q | ataagaatgcggccgcttagcgattgatcgagcg | Reverse, NotI site (underlined), stop codon of down (Figure 1.) |
| R | ataagaatgcggccgccagtaaaaggcacagagc | Forward, NotI site (underlined), located 2,138 nucleotides down-stream of glycosyl hydrolase family 45-2 start codon (Figure 1.) |
| S | tcatctattttctcctttgaaagtaatcactatattcc | Reverse, stop codon of glycosyl hydrolase family 45-2 (Figure 1.) |
| T | tcaaaggagaaaatagatgacaaaattaaataaaaaatgg | Forward, 1,973 nucleotides upstream of stop codon of GMP synthase (Figure 1.) |
| U | cggaattcgaatttgtatatgtcttcg | Reverse, EcoRI site (underlined), 994 nucleotides upstream of start codon of GMP synthase (opposite direction) (Figure 1.) |
| V | aaggaaaaaagcggccgccagaatatgataatcgtcatgg | Forward, NotI site (underlined), 902 nucleotides downstream of hylEfm start codon (Figure 1.) |
| W | tttgttctcctttttcttgctttttattttttaag | Reverse, stop codon of of hylEfm (Figure 1.) |
| X | gcaagaaaaaggagaacaaacaaaattaaataaaaaatgg | Forward, 1,973 nucleotides upstream of stop codon of GMP synthase (opposite direction) (Figure 1.) |
| Y | ccggaattcgaatcaggaactgaaactgccc | Reverse, EcoRI site (underlined), 1,094 nucleotides upstream of stop codon of GMP synthase (opposite direction) (Figure 1.) |
| A1 | cgcgtcgtattaaaaatcat | Forward, 143 nucleotides upstream of stop codon of GH20 (Figure 3.) |
| A2 | gatcgataaactggctcgt | Reverse, 139 nucleotides upstream of start codon of GH42 (Figure 3.) |
| B1 | acgcgtcgacagcagctggatatgctga | Forward, SalI site (underlined), 2,316 nucleotides downstream of start codon of GH42 (Figure 3.) |
| B2 | ggaagatctccggtttccagacttctt | Reverse, BglII site (underlined), 159 nucleotides downstream of start codon of hylEfm (Figure 3.) |
| C1 | gttagaagaagtctggaaaccg | Forward, 138 nucleotides downstream of start codon of hylEfm (Figure 3.) |
| C2 | tgctaagatattcctctactcg | Reverse, 798 nucleotides upstream of stop codon of hylEfm (Figure 3.) |
| D1 | acatgcatgcagaattggagccttggtt | Forward, SphI site (underlined), 169 nucleotides upstream of stop codon of hylEfm (Figure 3.) |
| D2 | cggaattctgcttccgcataagaaa | Reverse, EcoRI site (underlined), 319 nucleotides upstream of stop codon of down gene (Figure 3.) |
| E1 | gcaaggcttcttagaga | Forward, ddl E. faecium [32,33] |
| E2 | catcgtgtaagctaacttc | Reverse, ddl E. faecium [32,33] |
Figure 2.
Physical map of the plasmids pHOU1 and pHOU2 for targeted mutagenesis of E. faecium. A, plasmid used for construction of TX1330RF (pHylEfmTX16Δ4genes), TX1330RF(pHylEfmTX16Δhyl), TX1330RF(pHylEfmTX16Δhyl-down) and TX1330RF (pHylEfmTX16Δdown) deletion mutants (Figure 1); B, plasmid used for construction of the TX1330RF(pHylEfmTX16Δ7,534) deletion mutant (Figure 1)
In order to create a deletion mutant of the hylEfm-region (which contains genes predicted to be involved in carbohydrate metabolism and transport; Figure 1), fragments upstream (977 bp) and downstream (999 bp) of this region were amplified by PCR (with primers C-D and E-F, respectively; Table 2) and cloned upstream and downstream of the cat gene in pHOU2, respectively, using BamHI and XhoI for the upstream fragment and ApaI and EcoRI for the downstream fragment; the correct insert was confirmed by sequencing in both directions. This recombinant plasmid was introduced into E. faecalis CK111 by electroporation as described previously [25,28] and blue colonies were recovered on brain heart infusion (BHI) agar plates containing gentamicin (125 μg/ml) and X-Gal (200 μg/ml). Subsequently, the pHOU2 derivatives were introduced into strain TX16 by filter mating [29] with E. faecalis CK111 as the donor. Single cross-over integrants were selected on gentamicin (170 μg/ml) and erythromycin (200 mg/ml) and purified colonies were then resuspended in 50 μl of normal saline and plated on MM9YEG media (salts and yeast extract) supplemented with 7 mM of p-Cl-Phe [25] and incubated for 48 h at 37°C. To confirm that colonies which grew on MM9YEG media supplemented with p-Cl-Phe were excisants, the corresponding colonies were grown simultaneously on BHI agar in the presence and absence of gentamicin. Colonies that were susceptible to gentamicin were further screened by PCR, pulsed field gel electrophoresis (PFGE) and hybridizations with hylEfm and cat probes as described before [11]. The mutated region was also sequenced in order to confirm deletion of the corresponding genes. Subsequently, the mutated hylEfm-containing plasmid (pHylEfmTX16Δ7,534) was transferred from E. faecium TX16 to TX1330RF (a fusidic and rifampin resistant derivative of the commensal strain TX1330, Table 1) by filter mating as described previously [11] to obtain the strain TX1330RF(pHylEfmTX16Δ7,534). Acquisition of the mutated plasmid by TX1330RF was also confirmed by PFGE, PCR, hybridizations and sequencing. S1 nuclease digestion and PFGE was performed with the mutant to confirm that no other plasmid had transferred during the conjugation event as previously described [11].
Complementation of the hylEfm-region mutant TX1330RF(pHylEfmTX16Δ7,534)
The hylEfm gene was PCR amplified with primers G and H (including the ribosomal binding site and the stop codon of hylEfm) (Table 2) using total DNA from TX16 as template, and the DNA fragment (1,685 bp) cloned into the shuttle plasmid pAT392 [30] under the control of the P2 promoter (which allows constitutive expression of the cloned genes) and upstream of the aac(6')-aph(2") gene (which is co-transcribed from the same promoter) using SacI and SmaI sites (plasmid pAT392::hylEfm). In order to evaluate if the deletion of hylEfm had an effect in the downstream gene (encoding a hypothetical protein of 331 amino acids of unknown function), the hylEfm and down genes (Figure 1) were also cloned together into pAT392 following a similar strategy and using primers G and I (pAT392::hylEfm-down). Recombinant pAT392-derivatives were purified from E. coli grown on Luria-Bertani agar containing gentamicin (25 μg/ml) and all their DNA inserts sequenced. Subsequently, they were introduced into E. faecium TX1330RF, and the TX1330RF(pHylEfmTX16Δ7,534) mutant by electroporation. Stability of the plasmid constructs was tested by isolating ca. 100 colonies from overnight cultures (using BHI broth) and from the spleens of dead animals (in different experiments) after intraperitoneal inoculation of the corresponding strain (see below) and plating them simultaneously on BHI and BHI-gentamicin (125 μg/ml).
Construction of additional mutants of the hylEfm-region in E. faecium TX1330RF(pHylEfmTX16)
To investigate the specific role of the hylEfm locus in E. faecium pathogenesis, complete in-frame deletions of four genes of the hylEfm-region, hylEfm alone, hylEfm plus its downstream gene and the gene downstream of hylEfm were generated using TX1330RF(pHylEfmTX16). Fragments upstream and downstream of each region were amplified by PCR with the corresponding primers (Figure 1 and Table 2). These fragments, with overlapping ends, were subsequently amplified by crossover PCR and cloned into pHOU1 using EcoRI and NotI (for hylEfm, hylEfm plus its downstream gene and the downstream gene of hylEfm mutants); and BamHI and PstI (for the four gene mutant). The inserts were sequenced in both directions to confirm that no mutations had been introduced during the cloning process. The recombinant plasmids were electroporated or transferred by conjugation (using E. faecalis CK111) into TX1330RF(pHylEfmTX16). Single crossover events and deletions of targeted regions (Figure 1) were obtained by plating in BHI with gentamicin and p-Cl-Phe containing medium, respectively, as previously described [25]. Confirmation of the deletion was performed by PCR, PFGE, hybridizations and DNA sequencing.
RT-PCR
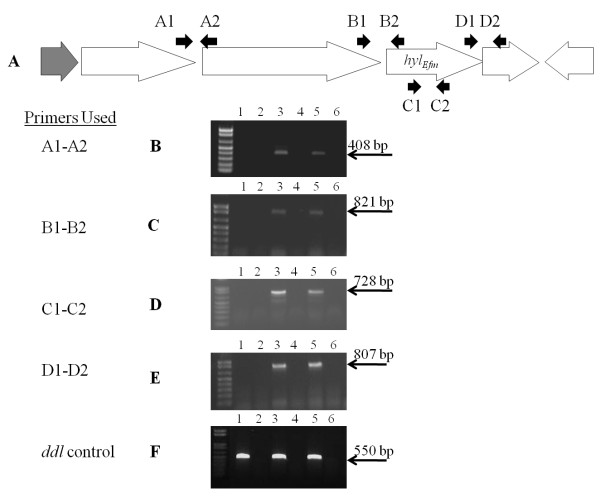
RNA was extracted from bacterial cells (TX16, TX1330RF(pHylEfmTX16), TX1330RF and strains containing pAT392 derivatives) grown in BHI broth at 37°C with mild agitation (logarithmic phase of growth, A600 0.8) as described before [31], and using the RNA isolation kit RNAwiz (Ambion, Austin, TX). RNA was treated twice with DNase (DNase-Free solution, Ambion) and synthesis of cDNA was performed using the commercial kit SuperScript One-Step reverse transcription-PCR (RT-PCR) with Platinum Taq (Invitrogen), according to the manufacturer's instructions. The mixture contained 0.2 μM of each primer, designed to detect overlapping transcripts of the four putative metabolic genes (Figure 3) and an internal transcript of hylEfm (Table 2). A primer pair directed to detect a 550-bp transcript of the housekeeping gene ddlE. faecium was used as an internal control for RT-PCR experiments [32,33].
Figure 3.
Transcriptional analysis of genes in the hylEfm region using reverse transcriptase (RT)-PCR. A, physical map of the hylEfm region and primers used for RT-PCR experiments. Black arrows above the genes indicate the position of the primers used to amplify DNA sequences from the cDNA obtained after reverse transcription. B, RT-PCR using primers A1-A2; C, RT-PCR using primes B1-B2; D, RT-PCR using primers C1-C2; E, RT-PCR using primers D1-D2; F, RT-PCR with ddl as the target gene using primers E1-E2 (Table 2) [32,33]. Lanes 1 and 2, TX1330RF (RT-PCR reaction and control without RT enzyme, respectively); lanes 3 and 4, TX1330RF(pHylEfm16) (RT-PCR reaction and control without RT enzyme, respectively); lanes 5 and 6 TX16(pHylEfm16) (RT-PCR reaction and control without RT enzyme respectively). The molecular weight of the bands is indicated to the right.
Mouse peritonitis model
Female (4 to 6 week old), outbred ICR mice (Harlan Sprague Dawley, Houston) were used as previously described [34]. Groups of 10 mice per inoculum (ranging from 2.3 × 108 to 3.1 × 109 CFU/ml) were included in each experiment. Inocula for each peritonitis experiment were prepared by growing bacteria initially on BHI agar plates. Subsequently, one colony was grown in BHI broth for 24 h at 37°C and the cells were concentrated in saline (0.9%) to an A600 of ca. 1.2. Strains containing pAT392 and derivatives were handled similarly before the intraperitoneal inoculation, except that the BHI agar and broth contained gentamicin (125 μg/ml). Comparison of the survival curves at similar inocula was performed using a log-rank test with Prism for Windows®. A P < 0.05 was considered significant. All experiments were approved by the Animal Welfare committee, University of Texas Health Science Center at Houston.
Results and Discussion
Deletion of 6 genes in the E. faecium hylEfm-region altered in vitro growth and attenuated virulence of TX1330RF(pHylEfmTX16) but not TX16(pHylEfmTX16) in murine peritonitis
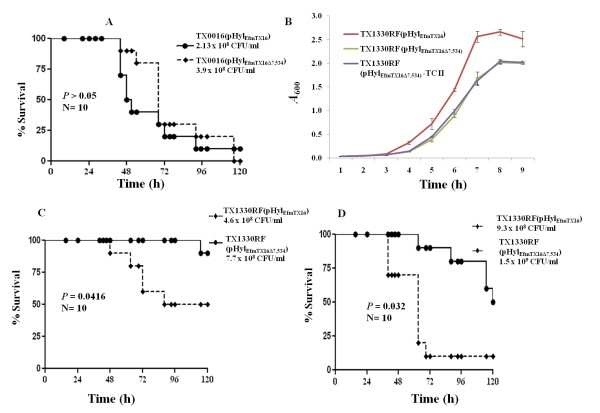
Since acquisition of the transferable pHylEfmTX16 by TX1330RF conferred increased virulence in experimental peritonitis [11], we explored the possibility that the hylEfm region was an important mediator of this effect. Using RT-PCR assays, we were able to detect in vitro expression of hylEfm during the exponential phase of growth in both TX16 and TX1330RF (pHylEfmTX16) (Figure 3). RT-PCR with primers located at the 3' and 5' ends of contiguous genes yielded products of the expected size in each case, suggesting that these genes are likely to be co-transcribed (Figure 3). Then, we adapted the pheS* counter-selection system [25] developed for E. faecalis to obtain several deletions of the hylEfm-region. The hylEfm gene in E. faecium TX16 (http://www.ncbi.nlm.nih.gov/genomeprj/30627, Genbank accession number ACIY00000000) is located in a cluster of genes whose putative function appears to involve the transport and breakdown of carbohydrates (Figure 1) [13]. As an initial step to test the mutagenesis system, a relatively large deletion (7,534 bp) from pHylEfmTX16 was obtained. The deletion involved three genes predicted to encode glycosyl hydrolases (including hylEfm) and a gene downstream of hylEfm whose function is unknown (Figure 1). Part (226 nucleotides) of a gene encoding a hypothetical transmembrane protein and located upstream of the putative family 20 glycosyl hydrolase gene and part (202 nucleotides) of a gene located 1,332 nt downstream of hylEfm encoding a putative GMP-synthase and likely transcribed in the opposite direction from the hylEfm cluster (Figure 1) were also deleted. As it is shown in Figure 4A, the deletion of 7,534 bp in the hylEfm-region did not affect the virulence of TX16 (DO) in murine peritonitis.
Figure 4.
Growth and survival curves in the mouse peritonitis model of E. faecium TX0016(pHylEfmTX16) and TX1330RF(pHylEfmTX16), carrying an intact hylEfm-region, and pHylEfmTX16Δ7,534 (6 gene mutant of the hylEfm-region). A, Survival curve of representative inoculum (5 inocula per experiment in two independent experiments) of TX0016(pHylEfmTX16) vs TX0016(pHylEfmTX16Δ7,534) in mouse peritonitis; B, growth curves of TX1330RF(pHylEfmTX16) vs TX1330RF(pHylEfmTX16Δ7,534) and a second transconjugant [TX1330RF(pHylEfmTX16Δ7,534)-TCII] obtained from the same mating experiment between TX16(pHylEfmTX16Δ7,534) and TX1330RF, expressed as optical density (A600) in brain heart infusion (BHI) broth (results of at least three experiments per strain). C and D, survival curves of TX1330RF(pHylEfmTX16) vs TX1330RF(pHylEfmTX16Δ7,534) obtained in the peritonitis model at different inocula in independent experiments performed at different days.
Next, we considered the possibility that an in vivo effect might be more clearly dissected if studies were performed in the background of a non-clinical strain. We hypothesized that an in vivo effect of a virulence determinant might more likely be seen in strains which are less successful clinically; that is, that a commensal strain such as TX1330RF [11] is likely to have decreased fitness or ability to produce disease compared to TX16 [35] and, thus, acquisition plus subsequent loss of a virulence determinant that alters such fitness would be easier to identify [11]. Thus, the mutated plasmid from strain TX16(pHylEfmTX16Δ7,534) was transferred to TX1330RF by conjugation and the in vivo effect of acquiring the intact plasmid [11] vs the plasmid carrying the deletion was evaluated. The two strains [TX1330RF(pHylEfmTX16) and TX1330RF(pHylEfmTX16Δ7,534)] appeared to differ only in the size of the hylEfm plasmid by PFGE and S1 nuclease assays [11] (not shown). Figure 4B shows that deletion of 7,534 bp in the hylEfm region of TX1330RF(pHylEfmTX16) caused an in vitro growth defect. The alteration of growth was also seen in a second transconjugant from the same mating experiment between TX16(pHylEfmTX16Δ7,534) and TX1330RF (TC-II in Figure 4B). The mutant strain TX1330RF(pHylEfmTX16Δ7,534) was attenuated in the mouse model of peritonitis (even when an increased intraperitoneal inoculum for the mutant were used) (Figure 4C and 4D) (P < 0.05). Due to the alterations produced in the growth of TX1330RF(pHylEfmTX16Δ7,534), these results suggest that the attenuation in virulence may have also been due to factors other than those specifically related to virulence.
Complementation of the hylEfm-region mutant with hylEfm and a combination of hylEfm and the downstream gene did not restore the virulence of TX1330RF(pHylEfmTX16Δ7,534)
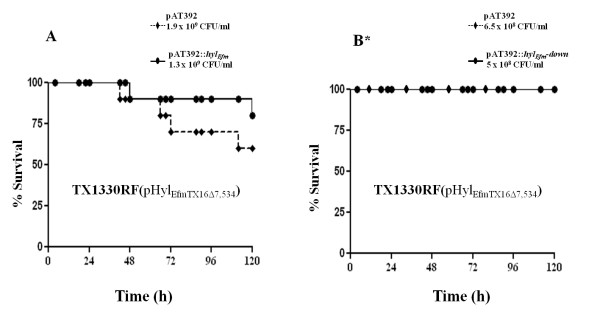
In order to further evaluate if the attenuation observed in TX1330RF(pHylEfmTX16Δ7,534) (as described above) was mediated by a direct effect of hylEfm in the peritonitis model, we explored complementation of this mutant in trans with the full hylEfm gene and a combination of hylEfm and the downstream gene using the shuttle vector pAT392 [30]. The cloning strategy placed these genes upstream of the aac(6')-aph(2") gene (which confers resistance to gentamicin) resulting in all open reading frames under the control of the constitutive P2 promoter. Up to 80% loss was observed with all strains in the absence of gentamicin; however, in the presence of the antibiotic during inoculum preparation, the TX1330RF(pHylEfmTX16Δ7,534)-derivatives containing the pAT392 constructs were stable both in vitro and in vivo (5% maximum percentage of plasmid loss). Introduction of hylEfm or a combination of hylEfm plus its downstream gene (cloned into pAT392) did not restore the virulence of the mutant strain TX1330RF(pHylEfmTX16Δ7,534), compared to pAT392 alone in the presence of gentamicin (Figure 5A and 5B). The results indicate that constitutive expression of hylEfm alone or in combination with its downstream gene (which was confirmed by RT-PCR, not shown) was not able to restore the phenotypic differences observed in the mutant strain TX1330RF(pHylEfmTX16Δ7,534), supporting the fact that hylEfm may not be directly responsible of the attenuation observed in the mutant.
Figure 5.
Survival curves in the mouse peritonitis model of E. faecium TX1330RF and derivatives. A and B show survival curves of the TX1330RF(pHylEfmTX16Δ7,534) (6 gene mutant in the hylEfm region) complemented with pAT392-derivatives (which include pAT392::hylEfm and pAT392::hylEfm-down) obtained in the peritonitis model at different inocula in independent experiments performed at different days. The asterisk indicates that the lines are superimposed since values are identical.
Under our experimental conditions, we cannot completely rule out that the in vivo attenuation observed with pHylEfmTX16Δ7,534 in the TX1330RF background may have been caused by the partial deletion of the hypothetical transmembrane protein or the putative GMP-synthase located upstream and downstream of the hylEfm-cluster, respectively. Indeed, a deletion of 76 amino acids in the C-terminus of the hypothetical membrane protein occurred in this plasmid, resulting in the deletion of three predicted transmembrane helices. Similarly, 68 amino acids in the C-terminus of the putative GMP-synthase were deleted; the removal of these amino acids is likely to disturb the dimerization domain of this protein [36] affecting its function in nucleotide metabolism. Moreover, a second TX1330RF(pHylEfmTX16Δ7,534) mutant also exhibited an almost identical growth defect (Figure 4B). Thus, it is tempting to speculate that changes in these two genes may have affected the "metabolic" fitness of the TX1330RF(pHylEfmTX16Δ7,534) strain. However, since no evident change in fitness or virulence was observed with the mutated plasmid in the TX16 background, another possibility is that an extraneous change elsewhere in the plasmid (or chromosome) occurred during the conjugation process that influenced the in vitro growth of the TX1330RF(pHylEfmTX16Δ7,534) mutant(s) and its virulence.
Additional deletions of genes in the hylEfm-region did not alter the virulence of TX1330RF(pHylEfmTX16) in the mouse peritonitis model
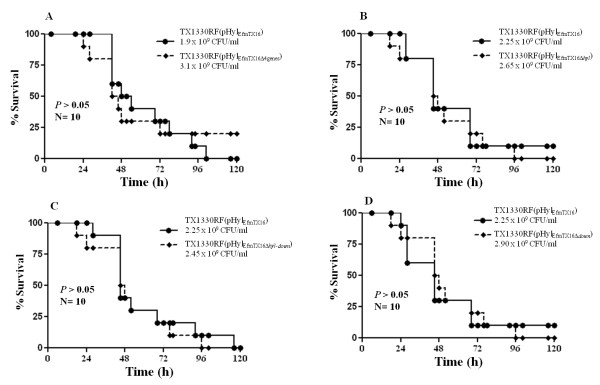
In order to dissect further the in vivo role of hylEfm and the adjacent genes, we produced several in-frame deletions of these genes (Figure 1) including: i) a four gene mutant of the hylEfm-region (including hylEfm) [TX1330RF(pHylEfmTX16Δ4genes)], ii) a deletion of hylEfm alone [TX1330RF (pHylEfmTX16Δhyl)], iii) a deletion of hylEfm plus its downstream gene mutant [TX1330RF (pHylEfmTX16Δhyl-down)] and, iv) a single deletion of the gene located downstream from hylEfm [TX1330RF (pHylEfmTX16Δdown)]. The mutagenesis strategy removed the open reading frame from the start codon of the first gene to the stop codon of the last gene (in case of multiple genes). In case of single gene deletion, the complete ORF (start to stop codon) was removed, leaving the surrounding DNA intact as in the wild type plasmid. None of the four mutants of the hylEfm-region showed a deleterious effect in the growth kinetics compared to TX1330RF (pHylEfmTX16) (harbouring an intact plasmid, Additional file 1). Moreover, we were unable to observe any attenuation of virulence in the mouse peritonitis model compared to the parental strain with the intact plasmid (Figure 6A-D), which further supports the fact that the four genes of the hylEfm region do not appear to be directly involved in increasing the pathogenic potential of pHylEfmTX16 in strain TX1330RF(pHylEfmTX16).
Figure 6.
Survival curves in the mouse peritonitis model of E. faecium TX1330RF(pHylEfmTX16) and deletion mutants (Figure 1 and Table 1) showing representative inocula (5 inocula per each experiment). A, TX1330RF(pHylEfmTX16) vs TX1330RF(pHylEfmTX16Δ4genes); B, TX1330RF(pHylEfmTX16) vs TX1330RF (pHylEfmTX16Δhyl); C, TX1330RF(pHylEfmTX16) vs TX1330RF(pHylEfmTX16Δhyl-down); D, TX1330RF(pHylEfmTX16) vs TX1330RF(pHylEfmTX16Δdown)
Megaplasmids (>145 kb, with or without hylEfm) have been recently found to be widespread among clinical isolates of E. faecium worldwide [12,13,15]. The proportion of these plasmids carrying hylEfm appears to vary according to geographical location (ca. 11 to 36%) [12,13]. Our findings indicate that the four genes of the hylEfm-cluster studied here, including hylEfm are not the main mediators of the virulence effect conferred by the plasmid carrying them in experimental peritonitis. Since the pHylEfm plasmids are large, it is presumed that other genes (i.e., upstream or downstream of the glycoside hydrolase-encoding genes) are more relevant in mediating this effect. Additionally, we cannot exclude that the hylEfm cluster studied in this work may play a role in other infections such as endocarditis or urinary tract infections (a subject of our ongoing studies). As a final remark, the adaptation of the pheS* counter-selection system for targeted mutagenesis in plasmid and chromosomal genes of E. faecium will facilitate the understanding of the role of other specific plasmid genes in the pathogenesis of E. faecium infections in the near future.
Conclusions
We provided evidence that four genes of the hylEfm-region (including hylEfm) do not mediate the virulence effect of the E. faecium plasmid pHylEfm in experimental peritonitis. The adaptation of the PheS* counter-selection system for targeted mutagenesis of E. faecium should facilitate the study of the role of other pHylEfm genes in the pathogenesis of murine peritonitis.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
DP carried out molecular genetics studies, animal experiments and participated in editing the manuscript. MCM, SR and MFM performed molecular genetics experiments. KVS carried out part of the animal work. BEM and LBR participated in editing the manuscript and data analysis. CAA is the principal investigator, conceived the study, designed the experiments, performed data analysis and wrote the manuscript. All authors read and approved the final version of the manuscript.
Supplementary Material
Growth curves of E. faecium and mutants. The strains were incubated in BHI broth and the A600 were measured every hour.
Contributor Information
Diana Panesso, Email: diana.panesso@uth.tmc.edu.
Maria C Montealegre, Email: mmontealegre@cideim.org.co.
Sandra Rincón, Email: sandralrn30@yahoo.com.
Maria F Mojica, Email: mmojica@cideim.org.co.
Louis B Rice, Email: LRice@lifespan.org.
Kavindra V Singh, Email: kavindra.singh@uth.tmc.edu.
Barbara E Murray, Email: barbara.e.murray@uth.tmc.edu.
Cesar A Arias, Email: cesar.arias@uth.tmc.edu.
Acknowledgements
CAA is supported by NIH Pathway to independence award R00 AI72961 from the National Institute of Allergy and Infectious Diseases (NIAID). This work was also supported in part by NIH grant R56 AI042399 and R01 AI067861 (to BEM) and R01 grant AI045626 (to LBR) from the NIAID. DP was partially funded by a graduate scholarship from The Instituto Colombiano para el Desarrollo de la Ciencia y Tecnología, "Francisco José de Caldas", COLCIENCIAS. SR was supported by an ASM-PAHO Infectious Disease Epidemiology and Surveillance Fellowship. We are grateful to Patrice Courvalin and Gary Dunny for providing plasmids pAT392 and pCJK47, respectively, and Pontificia Universidad Javeriana, (Bogotá, Colombia) for logistic support. We are grateful to Shreedhar Nallapareddy for useful discussions and experimental advice.
References
- Hidron AI, Edwards JR, Patel J, Horan TC, Sievert DM, Pollock DA, Fridkin SK. NHSN annual update: antimicrobial-resistant pathogens associated with healthcare-associated infections: annual summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2006-2007. Infect Control Hosp Epidemiol. 2008;29(11):996–1011. doi: 10.1086/591861. [DOI] [PubMed] [Google Scholar]
- Willems RJ, van Schaik W. Transition of Enterococcus faecium from commensal organism to nosocomial pathogen. Future Microbiol. 2009;4:1125–1135. doi: 10.2217/fmb.09.82. [DOI] [PubMed] [Google Scholar]
- van Schaik W, Top J, Riley DR, Boekhorst J, Vrijenhoek JE, Schapendonk CM, Hendrickx AP, Nijman IJ, Bonten MJ, Tettelin H. et al. Pyrosequencing-based comparative genome analysis of the nosocomial pathogen Enterococcus faecium and identification of a large transferable pathogenicity island. BMC Genomics. 2010;11:239. doi: 10.1186/1471-2164-11-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems RJ, Top J, van Santen M, Robinson DA, Coque TM, Baquero F, Grundmann H, Bonten MJ. Global spread of vancomycin-resistant Enterococcus faecium from distinct nosocomial genetic complex. Emerg Infect Dis. 2005;11(6):821–828. doi: 10.3201/eid1106.041204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heikens E, Bonten MJ, Willems RJ. Enterococcal surface protein Esp is important for biofilm formation of Enterococcus faecium E1162. J Bacteriol. 2007;189(22):8233–8240. doi: 10.1128/JB.01205-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leendertse M, Heikens E, Wijnands LM, van Luit-Asbroek M, Teske GJ, Roelofs JJ, Bonten MJ, van der Poll T, Willems RJ. Enterococcal surface protein transiently aggravates Enterococcus faecium-induced urinary tract infection in mice. J Infect Dis. 2009;200(7):1162–1165. doi: 10.1086/605609. [DOI] [PubMed] [Google Scholar]
- Hendrickx AP, Bonten MJ, van Luit-Asbroek M, Schapendonk CM, Kragten AH, Willems RJ. Expression of two distinct types of pili by a hospital-acquired Enterococcus faecium isolate. Microbiology. 2008;154(Pt 10):3212–3223. doi: 10.1099/mic.0.2008/020891-0. [DOI] [PubMed] [Google Scholar]
- Nallapareddy SR, Singh KV, Murray BE. Contribution of the collagen adhesin Acm to pathogenesis of Enterococcus faecium in experimental endocarditis. Infect Immun. 2008;76(9):4120–4128. doi: 10.1128/IAI.00376-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sillanpaa J, Prakash VP, Nallapareddy SR, Murray BE. Distribution of genes encoding MSCRAMMs and Pili in clinical and natural populations of Enterococcus faecium. J Clin Microbiol. 2009;47(4):896–901. doi: 10.1128/JCM.02283-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sillanpaa J, Nallapareddy SR, Singh KV, Prakash VP, Fothergill T, Ton-That H, Murray BE. Characterization of the ebp pilus-encoding operon of Enterococcus faecium and its role in biofilm formation and virulence in a murine model of urinary tract infection. Virulence. 2010;1(4):236–246. doi: 10.4161/viru.1.4.11966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias CA, Panesso D, Singh KV, Rice LB, Murray BE. Cotransfer of antibiotic resistance genes and a hylEfm-containing virulence plasmid in Enterococcus faecium. Antimicrob Agents Chemother. 2009;53(10):4240–4246. doi: 10.1128/AAC.00242-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panesso D, Reyes J, Rincon S, Diaz L, Galloway-Pena J, Zurita J, Carrillo C, Merentes A, Guzman M, Adachi JA. et al. Molecular epidemiology of vancomycin-resistant Enterococcus faecium: a prospective, multicenter study in South American hospitals. J Clin Microbiol. 2010;48(5):1562–1569. doi: 10.1128/JCM.02526-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitas AR, Tedim AP, Novais C, Ruiz-Garbajosa P, Werner G, Laverde-Gomez JA, Canton R, Peixe L, Baquero F, Coque TM. Global spread of the hyl(Efm) colonization-virulence gene in megaplasmids of the Enterococcus faecium CC17 polyclonal subcluster. Antimicrob Agents Chemother. 2010;54(6):2660–2665. doi: 10.1128/AAC.00134-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice LB, Carias L, Rudin S, Vael C, Goossens H, Konstabel C, Klare I, Nallapareddy SR, Huang W, Murray BE. A potential virulence gene, hylEfm, predominates in Enterococcus faecium of clinical origin. J Infect Dis. 2003;187(3):508–512. doi: 10.1086/367711. [DOI] [PubMed] [Google Scholar]
- Laverde Gomez JA, van Schaik W, Freitas AR, Coque TM, Weaver KE, Francia MV, Witte W, Werner G. A multiresistance megaplasmid pLG1 bearing a hyl(Efm) genomic island in hospital Enterococcus faecium isolates. Int J Med Microbiol. 2011;301(2):165–175. doi: 10.1016/j.ijmm.2010.08.015. [DOI] [PubMed] [Google Scholar]
- Kim DS, Singh KV, Nallapareddy SR, Qin X, Panesso D, Arias CA, Murray BE. The fms21 (pilA)-fms20 locus encoding one of four distinct pili of Enterococcus faecium is harboured on a large transferable plasmid associated with gut colonization and virulence. J Med Microbiol. 2010;59(Pt 4):505–507. doi: 10.1099/jmm.0.016238-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice LB, Lakticova V, Carias LL, Rudin S, Hutton R, Marshall SH. Transferable capacity for gastrointestinal colonization in Enterococcus faecium in a mouse model. J Infect Dis. 2009;199(3):342–349. doi: 10.1086/595986. [DOI] [PubMed] [Google Scholar]
- Ferretti JJ, McShan WM, Ajdic D, Savic DJ, Savic G, Lyon K, Primeaux C, Sezate S, Suvorov AN, Kenton S. et al. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc Natl Acad Sci USA. 2001;98(8):4658–4663. doi: 10.1073/pnas.071559398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu T, Ohtani K, Hirakawa H, Ohshima K, Yamashita A, Shiba T, Ogasawara N, Hattori M, Kuhara S, Hayashi H. Complete genome sequence of Clostridium perfringens, an anaerobic flesh-eater. Proc Natl Acad Sci USA. 2002;99(2):996–1001. doi: 10.1073/pnas.022493799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon WL, Macauley MS, Taylor EJ, Robinson CE, Charnock SJ, Davies GJ, Vocadlo DJ, Black GW. Functional analysis of a group A streptococcal glycoside hydrolase Spy1600 from family 84 reveals it is a beta-N-acetylglucosaminidase and not a hyaluronidase. Biochem J. 2006;399(2):241–247. doi: 10.1042/BJ20060307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voyich JM, Sturdevant DE, Braughton KR, Kobayashi SD, Lei B, Virtaneva K, Dorward DW, Musser JM, DeLeo FR. Genome-wide protective response used by group A Streptococcus to evade destruction by human polymorphonuclear leukocytes. Proc Natl Acad Sci USA. 2003;100(4):1996–2001. doi: 10.1073/pnas.0337370100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galloway-Pena JR, Nallapareddy SR, Arias CA, Eliopoulos GM, Murray BE. Analysis of clonality and antibiotic resistance among early clinical isolates of Enterococcus faecium in the United States. J Infect Dis. 2009;200(10):1566–1573. doi: 10.1086/644790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vankerckhoven V, Van Autgaerden T, Vael C, Lammens C, Chapelle S, Rossi R, Jabes D, Goossens H. Development of a multiplex PCR for the detection of asa1, gelE, cylA, esp, and hyl genes in enterococci and survey for virulence determinants among European hospital isolates of Enterococcus faecium. J Clin Microbiol. 2004;42(10):4473–4479. doi: 10.1128/JCM.42.10.4473-4479.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner G, Klare I, Fleige C, Witte W. Increasing rates of vancomycin resistance among Enterococcus faecium isolated from German hospitals between 2004 and 2006 are due to wide clonal dissemination of vancomycin-resistant enterococci and horizontal spread of vanA clusters. Int J Med Microbiol. 2008;298(5-6):515–527. doi: 10.1016/j.ijmm.2007.05.008. [DOI] [PubMed] [Google Scholar]
- Kristich CJ, Chandler JR, Dunny GM. Development of a host-genotype-independent counterselectable marker and a high-frequency conjugative delivery system and their use in genetic analysis of Enterococcus faecalis. Plasmid. 2007;57(2):131–144. doi: 10.1016/j.plasmid.2006.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kast P, Wehrli C, Hennecke H. Impaired affinity for phenylalanine in Escherichia coli phenylalanyl-tRNA synthetase mutant caused by Gly-to-Asp exchange in motif 2 of class II tRNA synthetases. FEBS Lett. 1991;293(1-2):160–163. doi: 10.1016/0014-5793(91)81176-9. [DOI] [PubMed] [Google Scholar]
- Nallapareddy SR, Singh KV, Murray BE. Construction of improved temperature-sensitive and mobilizable vectors and their use for constructing mutations in the adhesin-encoding acm gene of poorly transformable clinical Enterococcus faecium strains. Appl Environ Microbiol. 2006;72(1):334–345. doi: 10.1128/AEM.72.1.334-345.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth R, An FY, Clewell DB. Highly efficient protoplast transformation system for Streptococcus faecalis and a new Escherichia coli-S. faecalis shuttle vector. J Bacteriol. 1986;165(3):831–836. doi: 10.1128/jb.165.3.831-836.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita H, Pierson C, Lim SK, Clewell DB, Ike Y. Possible connection between a widely disseminated conjugative gentamicin resistance (pMG1-like) plasmid and the emergence of vancomycin resistance in Enterococcus faecium. J Clin Microbiol. 2002;40(9):3326–3333. doi: 10.1128/JCM.40.9.3326-3333.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur M, Depardieu F, Snaith HA, Reynolds PE, Courvalin P. Contribution of VanY D,D-carboxypeptidase to glycopeptide resistance in Enterococcus faecalis by hydrolysis of peptidoglycan precursors. Antimicrob Agents Chemother. 1994;38(9):1899–1903. doi: 10.1128/aac.38.9.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgogne A, Hilsenbeck SG, Dunny GM, Murray BE. Comparison of OG1RF and an isogenic fsrB deletion mutant by transcriptional analysis: the Fsr system of Enterococcus faecalis is more than the activator of gelatinase and serine protease. J Bacteriol. 2006;188(8):2875–2884. doi: 10.1128/JB.188.8.2875-2884.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutka-Malen S, Evers S, Courvalin P. Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J Clin Microbiol. 1995;33(1):24–27. doi: 10.1128/jcm.33.1.24-27.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutka-Malen S, Evers S, Courvalin P. Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J Clin Microbiol. 1995;33(5):1434. doi: 10.1128/jcm.33.5.1434-1434.1995. Erratum. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh KV, Qin X, Weinstock GM, Murray BE. Generation and testing of mutants of Enterococcus faecalis in a mouse peritonitis model. J Infect Dis. 1998;178(5):1416–1420. doi: 10.1086/314453. [DOI] [PubMed] [Google Scholar]
- Nallapareddy SR, Weinstock GM, Murray BE. Clinical isolates of Enterococcus faecium exhibit strain-specific collagen binding mediated by Acm, a new member of the MSCRAMM family. Mol Microbiol. 2003;47(6):1733–1747. doi: 10.1046/j.1365-2958.2003.03417.x. [DOI] [PubMed] [Google Scholar]
- Bork P, Koonin EV. A P-loop-like motif in a widespread ATP pyrophosphatase domain: implications for the evolution of sequence motifs and enzyme activity. Proteins. 1994;20(4):347–355. doi: 10.1002/prot.340200407. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Growth curves of E. faecium and mutants. The strains were incubated in BHI broth and the A600 were measured every hour.