Abstract
Background
During ripening, climacteric fruits increase their ethylene level and subsequently undergo various physiological changes, such as softening, pigmentation and development of aroma and flavor. These changes occur simultaneously and are caused by the highly synchronized expression of numerous genes at the onset of ripening. In tomatoes, the MADS-box transcription factor RIN has been regarded as a key regulator responsible for the onset of ripening by acting upstream of both ethylene- and non-ethylene-mediated controls. However, except for LeACS2, direct targets of RIN have not been clarified, and little is known about the transcriptional cascade for ripening.
Results
Using immunoprecipitated (IPed) DNA fragments recovered by chromatin immunoprecipitation (ChIP) with anti-RIN antibody from ripening tomato fruit, we analyzed potential binding sites for RIN (CArG-box sites) in the promoters of representative ripening-induced genes by quantitative PCR. Results revealed nearly a 5- to 20-fold enrichment of CArG boxes in the promoters of LeACS2, LeACS4, PG, TBG4, LeEXP1, and LeMAN4 and of RIN itself, indicating direct interaction of RIN with their promoters in vivo. Moreover, sequence analysis and genome mapping of 51 cloned IPed DNAs revealed potential RIN binding sites. Quantitative PCR revealed that four of the potential binding sites were enriched 4- to 17-fold in the IPed DNA pools compared with the controls, indicating direct interaction of RIN with these sites in vivo. Near one of the four CArG boxes we found a gene encoding a protein similar to thioredoxin y1. An increase in the transcript level of this gene was observed with ripening in normal fruit but not in the rin mutant, suggesting that RIN possibly induces its expression.
Conclusions
The presented results suggest that RIN controls fruit softening and ethylene production by the direct transcriptional regulation of cell-wall-modifying genes and ethylene biosynthesis genes during ripening. Moreover, the binding of RIN to its own promoter suggests the presence of autoregulation for RIN expression. ChIP-based analyses identified a novel RIN-binding CArG-box site that harbors a gene associated with RIN expression in its flanking region. These findings clarify the crucial role of RIN in the transcriptional regulation of ripening initiation and progression.
Background
Ripening processes of many kinds of fruit involve various biochemical and physiological changes, such as softening, enrichment of pigments, organic acids and nutrients (e.g., vitamins and sugars), and development of aroma and flavor. These changes make fruits attractive for the human diet. For climacteric fruits, autocatalytic ethylene production and an increase in respiration occur during ripening, and ethylene has been well characterized as necessary for the coordination and completion of ripening [1]. At the onset of ripening, expression patterns of numerous genes involved in these ripening-associated phenomena are upregulated in a highly synchronized fashion, indicating that ripening is controlled by a highly systematic and sophisticated transcriptional mechanism. Therefore, much attention has been paid to how fruit ripening is regulated because ripening regulation is not only of agricultural importance but also of scientific interest in terms of the regulation of biological developmental processes. However, a substantial portion of the genetic regulatory mechanism controlling the process remains unclear.
The tomato (Solanum lycopersicum) is the most advantageous model plant for the study of fruit ripening due to its climacteric ripening nature, availability of the genome information and many suggestive mutations concerned in ripening [2,3]. Among the ripening mutations, ripening inhibitor (rin) is a well-characterized mutation that inhibits such characteristic phenomena observed during ripening as lycopene accumulation and softening, resulting in non-ripe fruit [4]. The rin mutation also inhibits autocatalytic ethylene production during ripening; thus, the wild-type gene on the rin locus has been regarded as a regulator responsible for the onset of ripening by acting upstream of both ethylene- and non-ethylene-mediated ripening control. The rin locus has been isolated and found to encode two MADS-box transcription factors, RIN and MC (Macrocalyx), and RIN is apparently responsible for the regulation of fruit ripening [5]. Molecular characterizations have revealed that RIN is expressed during ripening specifically, that the gene product exhibits transactivation activity and that RIN has the ability to bind to the specific DNA sequences known as C-(A/T)-rich-G (CArG) box, which is a typical binding sequence for MADS-box proteins [6].
To identify genes associated with ripening phenomena, the genes whose expressions are affected by the rin mutation have been extensively investigated. In ethylene biosynthesis and signaling, the transcription levels of the genes encoding 1-aminocyclopropane-1-carboxylic acid (ACC) synthase 2 (LeACS2), ACC synthase 4 (LeACS4), ACC oxidase 1 (LeACO1) and ethylene receptor protein 3 [ETR3; synonymous with NEVER RIPE (NR)] increase dramatically during ripening, but their transcriptions are inhibited by the rin mutation, indicating that these genes are responsible for the elevation of ethylene level and for ethylene signaling during ripening [7-9]. The rin mutation also inhibits upregulation of the genes involved in cell-wall modifications, such as Polygalacturonase (PG), β-Galactosidase 4 (TBG4), Endo-(1,4)-β-mannanase 4 (LeMAN4) and α-Expansin 1 (LeEXP1), all of which are assumed to be concerned with softening and the shelf life of fruit [10-19]. Ripening-associated upregulation of the gene for phytoene synthase 1 (PSY1), which is a rate-limiting enzyme for carotenoid production including lycopene in ripening tomatoes, and the gene for β-fructofuranosidase [also called invertase (INV)], which catalyzes hydrolysis of sucrose in ripening fruit, is also affected by the rin mutation [20-22]. In addition, a DNA microarray assay revealed that a large number of genes upregulated during ripening were suppressed by the rin mutation [23]. These results apparently indicate that the RIN protein is a transcriptional regulator triggering the onset of ripening by inducing the expression of these ripening-associated genes. Three pathways for the transcriptional regulation of ripening-associated genes by RIN are possible. The first pathway is that RIN binds to the promoter of target genes and directly regulates their expression. The second is that RIN induces ethylene production at the onset of ripening and ethylene-induced genes are consequently transcribed. The third is that RIN induces some transcription factors directly, and subsequently the transcription factors induce the ripening-specific gene expressions. It is expected that these three pathways act in parallel and regulate the expression of the large number of ripening-associated genes, although the elements of these transcriptional cascades remain largely unknown except that RIN binds to the promoter of LeACS2 [6].
To learn more about these pathways, we have developed ChIP-based analyses for fruit ripening. ChIP, a technique to collect target DNA sequences of a protein of interest as protein-DNA complexes (chromatin) with an antibody for the protein, is a powerful tool used to ascertain interactions of transcription factors with DNA in vivo [24]. Comprehensive ChIP-based approaches have been used for identifying potential target genes of a few floral homeotic MADS-box transcription factors in plants [25-27] but have not been applied for analysis of fruit ripening to date. Here, we report identification of the in vivo RIN-binding sites in the putative promoters of several ripening-induced genes through ChIP-based analyses using ripening tomato fruit. In addition, we screened novel RIN-binding sites containing CArG boxes and found a candidate gene possibly regulated by RIN. The results offer insights into the characteristics of RIN in the transcriptional regulation at the onset of ethylene production and cell-wall modifications, and in the autoregulation of RIN itself during ripening.
Results
In silico search of CArG boxes in the promoters of ripening-induced genes
The rin mutant lacks expression of LeACS2, LeACS4, TBG4, LeEXP1, LeMAN4 and PSY1, and shows decreased expression levels of LeACO1, ETR3, PG and INV, while these genes are highly upregulated in the wild-type fruit [8,13,28,29], indicating that all of these genes are regulated directly or indirectly by RIN. To detect potential RIN-binding sequences, a possible CArG-box motif [C(C/T)(A/T)6(A/G)G] [6] was searched against the promoters of these genes (~2 kb). The motif includes three groups of CArG-box sequences: SRF-like [canonical CArG-box, CC(A/T)6GG] [30], MEF2-like [CTA(A/T)4TAG] [31], and intermediate [CC(A/T)6AG] [6,32]. A motif search revealed that all the genes except LeACS4 and LeEXP1 have at least one typical CArG-box sequence in their promoters (Table 1 and Figure 1A). Instead, the promoter of LeEXP1 was found to contain no typical RIN target motif, but rather two atypical CArG-box sequences, CATTTATATG and CAATTTAAAG (underlines indicate atypical bases; Table 1 and Figure 1A). The promoter of LeACS4 was also found to carry no typical but three atypical sequences of CAAATATAAG, CAATTTTAAG and CTAGTTAAAG (underlines indicate atypical bases; Table 1 and Figure 1A). We also further analyzed these atypical CArG-box sequences in the LeEXP1 and LeACS4 promoters as well as analyzing the possible CArG boxes.
Table 1.
CArG-box sequences found in the promoters of ripening-induced genes.
| Site | CArG-box and its flanking sequences (5' to 3')1) | Motif 2) | CArG-box position (bps)3) |
|---|---|---|---|
| ACS2-a4) | AGCTATT-CTAAAAAAAG-TATCACATA5) | Possible | X59139 (1,365 - 1,374) (+) |
| ACS2-b | AAATGCAC-CCTAAATTAG-TCAAATAT5) | Possible | X59139 (2,654 - 2,663) (+) |
| ACS4-a | ATCAAACA-CAAATATAAG-TTTGGAAC5) | Atypical | M88487 (567 - 576) (+) |
| ACS4-b | ATTAAACA-CAATTTTAAG-AAACTTTT5) | Atypical | M88487 (1,201 - 1,210) (+) |
| ACS4-c | TGAAATAT-CTAGTTAAAG-ATATGTAC5) | Atypical | M88487 (1,802 - 1,811) (+) |
| ACO1 | GGTTGAAT-CTATAAAAAG-AAAAATAT | Possible | X58273 (1,285 - 1,294) (+) |
| ETR3-a | GGAGAAAT-CCTATAATAG-GGCAAACA | Intermediate | AY600437 (3,121 - 3,130) (+) |
| GAGAAATC-CTATAATAGG-GCAAACAC | Possible | AY600437 (3,122 - 3,131) (+) | |
| GGCAAACA-CCAAAAATAG-CTTGGAGT | Intermediate | AY600437 (3,139 - 3,148) (+) | |
| ETR3-b | AAATTTCA-CTTAATATGG-ACTAGAGA | Possible | AY600437 (3,745 - 3,754) (+) |
| PG-a | GCACCAAT-CTAATTTAGG-TTGAGCCG | Possible | scaffold01076 (1,534,540 - 1,534,549) (-) |
| PG-b | CTTAAAAT-CTATAAATAG-ACAAACCC | MEF2-like | scaffold01076 (1,533,632 - 1,533,641) (-) |
| TBG4-a | TATATGCT-CTATTTTTGG-ACGGCAGG5) | Possible | scaffold00061 (457,025 - 457,034) (+) |
| TBG4-b | TTTGGGCC-CCATTTAAGG-GATTGGGC5) | SRF-like | scaffold00061 (457,311 - 457,320) (+) |
| EXP1-a | TTATTTTA-CATTTATATG-TTATTATT | Atypical | scaffold00114 (3,118,161 - 3,118,170) (-) |
| EXP1-b | TGATGCTT-CAATTTAAAG-AAAATAAA5) | Atypical | scaffold00114 (3,117,729 - 3,117,738) (-) |
| MAN4 | TTTCTTTT-CCATTTATAG-AAAAACCA5) | Intermediate | scaffold01157 (9,316,653 - 9,316,662) (-) |
| PSY1 | TATGTGTA-CCAAAATTAG-AAAATCAG | Intermediate | scaffold00066 (364,464 - 364,473) (+) |
| CTTGTTGA-CTAAATATAG-AATGCATC | MEF2-like | scaffold00066 (364,504 - 364,513) (+) | |
| INV | TTATGATA-CTTAATATGG-TAATCTTT | Possible | Z12027 (1,634 - 1,643) (+) |
| TTCTCACT-CTATAAATAG-AGTTGTTC | MEF2-like | Z12027 (1,667 - 1,676) (+) | |
| RIN-a | GTTGCACT-CTAAAAAAAG-TTAAAAGG5) | Possible | scaffold00243 (210,835 - 210,844) (-) |
| RIN-b | ACAAAGAA-CCATTAAAAG-GTTAAAAA5) | Intermediate | scaffold00243 (210,262 - 210,271) (-) |
1) CArG-box sequences are underlined.
2) CArG-box sequences in the exhibited strand were grouped into motifs. Groups of the motif sequences are referenced in the text.
3) Scaffolds followed by Arabic numerals indicate that the sequences originate from WGS data. Symbols + and - in parentheses indicate that the CArG boxes displayed are presented in either the exhibited (+) or complementary (-) strands.
4) Site was previously described [6].
5) Sequences were used for in vitro gel retardation assays.
Figure 1.
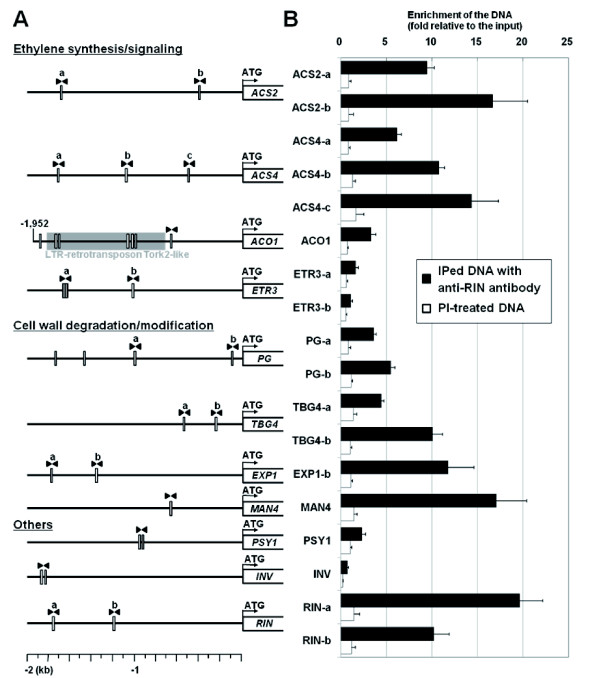
CArG-box sites in the ripening-associated gene promoters and their enrichment in ChIP-DNA. (A) Position of CArG-box sites (indicated by thin rectangles) found in the 2-kb potential promoter regions of the ripening-induced genes. A pair of primers specific to each site is indicated by pairs of filled arrowheads. When two or more sites exist in the same promoter, they are distinguished by the lower-case letters (a, b) above them. (B) Enrichment test of the CArG boxes. Bars represent the relative DNA amount of CArG boxes in IPed DNA recovered using either anti-RIN antibody or pre-immune serum (PI) to those in the total input chromatin DNA. (Note that the result of EXP1-a is omitted due to inadequate amplification efficiency in the real-time PCR analysis.) Data are the means from three independently prepared IPed DNAs. Error bars indicate standard error of each mean.
Binding of RIN to the CArG boxes in the ripening-induced gene promoters in vivo and in vitro
To distinguish the genes directly regulated by RIN, we applied the ChIP assay. Chromatin was prepared from ripening tomato fruits harvested at the pink coloring stage and was then immunoprecipitated with the anti-RIN antibody. The resulting immunoprecipitated DNAs (IPed DNAs) were assayed by quantitative-PCR analyses (qChIP-PCR) for the regions containing the putative RIN-binding sites described above, and then enrichment levels of the regions were evaluated. The qChIP-PCRs were carried out with a few considerations as follows: (1) when two or more CArG-box sequences were closely located (within ~150 bp region of each other), they were treated as one site and tested together with a pair of primers designed to include them both (Table 1, Figure 1A and Additional File 1); (2) in the case of PG, the two closest sites to the protein-coding region were tested (Figure 1A); (3) the five CArG-box sites found between the closest and the farthest sites in the promoter region (1,952 bp) of LeACO1 were excluded from the test because they were considered to be similar to an LTR-type retrotransposon (Tork2-like; Figure 1A) [33] whose replicates are dispersed throughout the genome, preventing site-specific amplification, and because the farthest site (at 1.8-kb upstream) from the protein-coding region of LeACO1 could not be examined by primer pairs of reasonable length.
The result of qChIP-PCR revealed significant enrichment of all the CArG boxes in the IPed DNA pools relative to the total chromatin DNA pools (the input DNA) (Figure 1B) except INV (0.7-fold) and EXP1-a (the enrichment of which failed to be monitored due to unstable PCR amplification). In particular, ACS2-a, ACS2-b, ACS4-b, ACS4-c, TBG4-b, EXP1-b, MAN4, RIN-a and RIN-b were extremely enriched to 9.5-, 16.7-, 10.8-, 14.4-, 10.1-, 11.8-, 17.1-, 19.6- and 10.2-fold, respectively (Figure 1B). Furthermore, ACS4-a, PG-b and TBG4-a were moderately enriched to 6.2-, 5.5- and 4.5-fold, respectively (Figure 1B). These enrichments were not observed in the immunoprecipitated DNA prepared with the pre-immune serum (PI-treated DNA pools) (0.2 to 1.7-fold; Figure 1B), indicating specific binding of RIN to these 12 CArG boxes in vivo. Other CArG boxes (ACO1, ETR3-a and -b, PG-a and PSY1) showed relatively lower enrichment (3.3-, 1.6-, 1.1-, 3.6-, 2.3-fold, respectively).
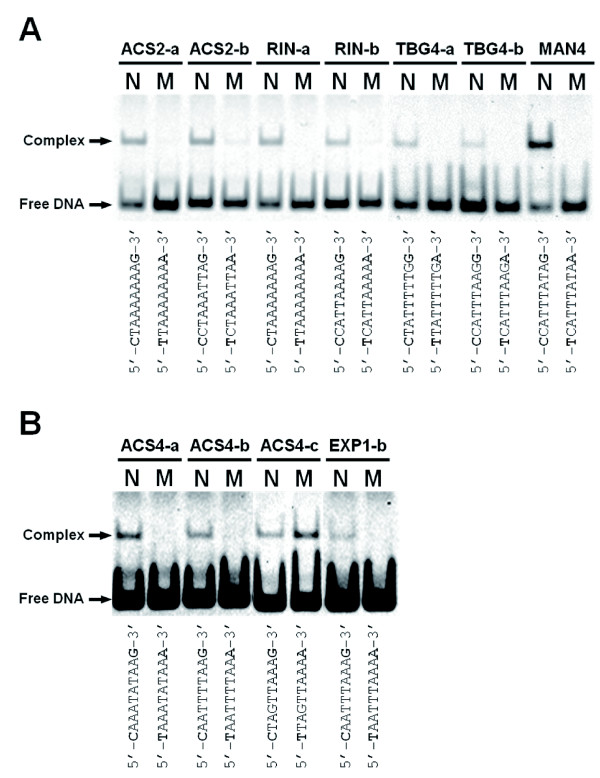
To identify the RIN-binding sequences within the enriched regions, the binding of RIN to the CArG-box sequences in the promoters of LeACS2, LeACS4, TBG4, LeEXP1, LeMAN4 and RIN were examined by in vitro gel retardation assay. Results showed that DNA fragments containing the CArG boxes were retarded in all the sites for these genes due to binding to the RIN protein (Figures 2A and 2B), although the signal intensity for atypical CArG-box sequences that include the three LeACS4 sites and EXP1-b appeared to be lower than that of the typical sequences, based on the ratio to the free DNA (Figures 2A and 2B). In contrast, by introducing mutations within the target sequences, the retardation was drastically inhibited in all target sequences except for ACS4-c (Figures 2A and 2B). These results indicate that RIN specifically recognizes the CArG-box sequences of the respective sites. In ACS4-c, the DNA fragment containing the mutated CArG box was retarded similarly to the normal sequence (Figure 2B), which is likely due to the unexpected generation of a sequence similar to the CArG box via base substitution (CTAAATATTT in the reverse strand; refer to the normal ACS4-c sequence in Table 1). This result indicates that RIN could bind to a wide spectrum of CArG-box sequences although in vitro RIN shows lower preference for the atypical motifs than for the typical CArG boxes.
Figure 2.
Gel retardation assay of CArG boxes in the ripening-induced gene promoters. Gel retardation assay of possible CArG boxes (A) and atypical CArG boxes (B) found in the promoters of the ripening-induced genes. DNA fragments of putative RIN-binding sites that contain a normal CArG-box sequence and flanking regions shown in Table 1 were reacted with the RIN-MIK protein in vitro and electrophoresed (lanes N). DNA fragments with mutations within the CArG box were also examined (lanes M). The normal CArG-box sequences (10 bp) and their mutant sequences are displayed below the gel images. Nucleotides substituted between the normal and mutated sequences are indicated by bold letters. The same amount of DNA fragments was applied to each lane in A and B. The image of B was adjusted to higher contrast than that of A due to the low intensities of the retarded signals for the sequences examined in B.
Genome mapping of DNA fragments recovered by ChIP and in silico detection of CArG-box sequences
To identify novel binding sites of RIN, we cloned the DNA fragments recovered by ChIP using the anti-RIN antibody. Of the IPed DNA clones, 51 were sequenced and mapped on independent genomic regions of the tomato (Table 2). The average length of the 51 cloned fragments was 380 bp (data not shown). To detect potential RIN-binding sequences, a possible CArG-box motif [C(C/T)(A/T)6(A/G)G] [6] was subsequently searched against the genomic regions. The search detected a total of 13 possible CArG boxes from 11 regions (Table 2). These boxes could be further categorized into four groups (Table 2). Because none of these regions has yet been reported to be bound by RIN, we considered them as novel potential RIN-binding sites and subjected them to further analyses.
Table 2.
CArG-box sequences found in regions for which IPed DNAs were mapped.
| CArG-box site | CArG-box and its flanking sequences (5' to 3')1) | Motif (strand)2) | CArG-box position (bps)3) |
|---|---|---|---|
| 009F | CCTAAATA-CTATTATAAG-AATGATCA | Possible (+, -) | scaffold01172 (3,039,503 - 3,039,512) |
| 016 | GTACAGCA-CCAAAATTGG-CGACCACA | SRF-like (+, -) | scaffold01172 (4,647,499 - 4,647,508) |
| 027R | AACTCTCC-CCTATATTGG-TGCTCAAT | SRF-like (+, -) | scaffold00008 (1,340,392 - 1,340,401) |
| 042F | GATAGATC-CTAATTTTGG-TAAGTGAC | Possible (+), Intermediate (-) | scaffold00077 (3,770,305 - 3,770,314) |
| 046F_1 | CTTTTGGG-CTTAATTTAG-GGATTTAC | Possible (+, -) | scaffold00162 (1,604,081 - 1,604,090) |
| 046F_2 | ACATTTTT-CCATATTTAG-TACTAGAT | Intermediate (+), Possible (-) | scaffold00162 (1,604,514 - 1,604,523) |
| 066F | ACTAGCAA-CTATTATAGG-GCCCTCCT | Possible (+), Intermediate (-) | scaffold00041 (5,908,092 - 5,908,101) |
| 073F | AAAGTCCC-CTTTTTTTGG-AAAAATAC | Possible (+, -) | scaffold00885 (1,175,686 - 1,175,695) |
| 090R | TATATTGT-CTATTATAGG-GGACGGTC | Possible (+), Intermediate (-) | scaffold00235 (679,848 - 679,857) |
| 100_1 | GCTGGATT-CTATTATAAG-GACATCAT | Possible (+, -) | scaffold00073 (9,132 - 9,141) |
| 100_2 | GCCAGATT-CCTATATTAG-CAGTATAG | Intermediate (+), Possible (-) | scaffold00073 (8,936 - 8,945) |
| 128R | CTTCATAC-CTTAATTAAG-CAACCTTA | Possible (+, -) | scaffold00183 (1,735,586 - 1,735,595) |
| 133R | AGAAAATG-CCATTTTTGG-AAGGAAGA | SRF-like (+, -) | scaffold00103 (1,409,234 - 1,409,243) |
1) CArG-box sequences are underlined. Sequences displayed are the same strand as the WGS data (version 1.03) released by SGN. They were used for in vitro gel retardation assay.
2) Symbols + and - in parentheses indicate that the CArG-box sequences in the exhibited (+) or complementary (-) strands could be grouped into the motifs. Groups of the motif sequences are referenced in the text.
3) Positions on the WGS are displayed.
Binding of RIN to the novel CArG-box sites in vivo and in vitro
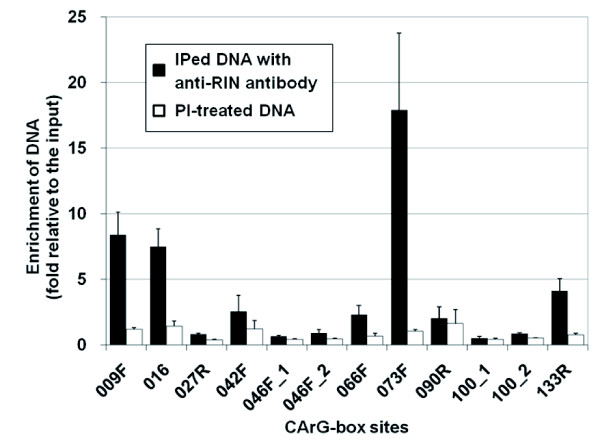
To examine the binding of RIN to the CArG-box sites within the cloned fragments, we monitored their enrichment levels in the IPed DNA pools by qChIP-PCR. Results showed that the DNA fragments of four sites, 009F, 016, 073F and 133R, were significantly enriched to 8.4-, 7.5-, 17.9- and 4.1-fold in the IPed DNA pools, respectively, compared with those in the input chromatin DNA. Such significant enrichment was not observed in the PI-treated DNA (only 0.4 to 1.4-fold; Figure 3), indicating that enrichment depends on the presence of the anti-RIN antibody, i.e., these sites are specifically bound by RIN in vivo. Compared with these four sites, the other eight sites examined here showed relatively lower enrichment (0.5 to 2.6-fold; Figure 3).
Figure 3.
Enrichment tests of the novel CArG boxes. Bars represent the relative DNA amount of CArG boxes in IPed DNA recovered either with anti-RIN antibody or pre-immune serum (PI) to those in the total input chromatin DNA. (Note that the result for 128R is omitted due to the inadequate amplification efficiency in the real-time PCR analysis.) Data are the means from three independently prepared samples by ChIP with the anti-RIN antibody or the pre-immune serum. Error bars indicate standard error of each mean.
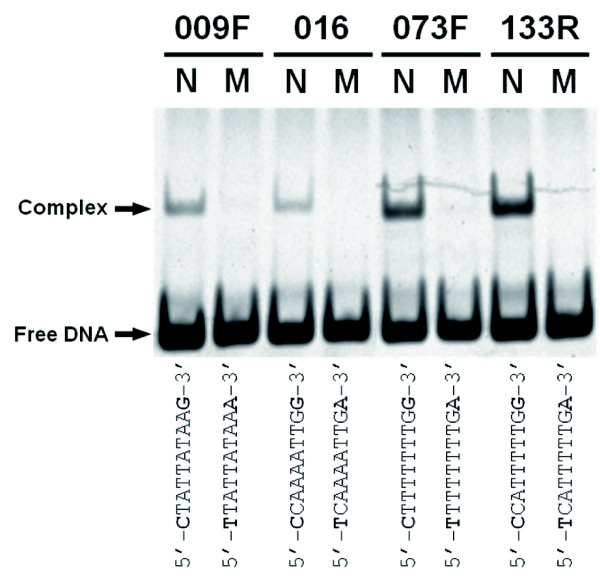
The binding of RIN to the four sites was also examined by in vitro gel retardation assay. Results showed that the mobility of DNA fragments containing the normal CArG-box sequences was delayed in all the sites by binding to the RIN protein (Figure 4). This retardation was not detected at any site when mutation-introduced target sequences were used (Figure 4), indicating that RIN binds specifically to these CArG boxes.
Figure 4.
Gel retardation assay of CArG boxes. DNA fragments of the sites containing the normal (lane N) and mutated (lane M) CArG boxes were reacted with the RIN-MIK protein in vitro and electrophoresed.
Genes in the flanking regions of the RIN-binding CArG boxes and their expression
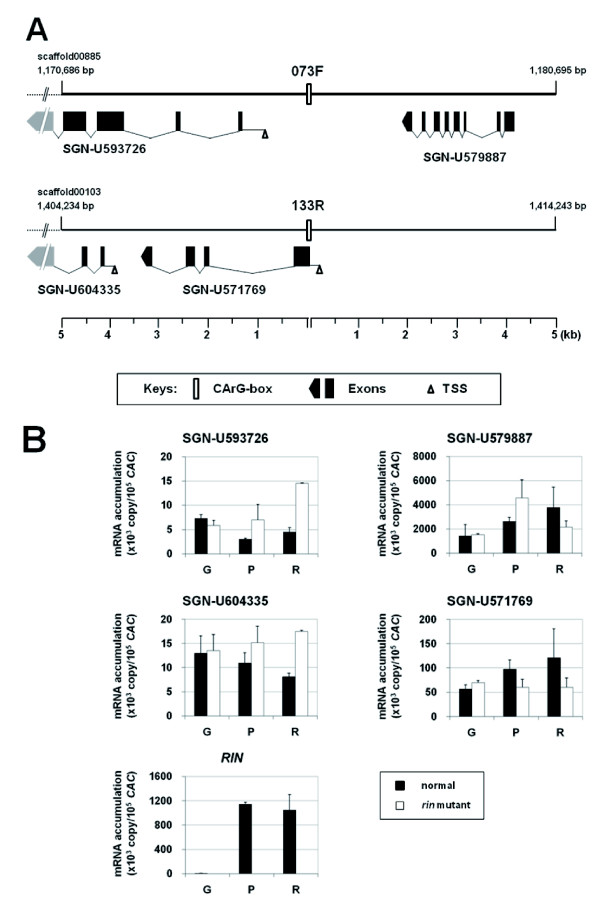
To detect potential target genes of RIN, we analyzed the 5-kb genomic regions flanking the four CArG boxes. A BLAST search of each flanking region of 073F and 133R using the SGN unigene set identified two potential genes corresponding to tomato ESTs, SGN-U579887 and SGN-U593726 for the former region, and SGN-U571769 and SGN-U604335 for the latter region (Figure 5A, Additional File 2). Gene predictions revealed that the CArG-box of 073F was located at the promoter region of the gene for SGN-U593726 [718 bp upstream of the predicted transcription start site (TSS)], while the CArG-box of 133R overlapped both the 5'-untranslated region (UTR) and the protein-coding sequence for SGN-U571769 (Figure 5A). By contrast, no EST was detected in the flanking 5-kb regions of 009F and 016.
Figure 5.
Position, structure and expression pattern of predicted genes on flanking genomic regions of CArG boxes. (A) Sequence positions of regions in the scaffolds from the WGS of tomato are indicated above the ends of the horizontal lines. Positions of the CArG boxes in the scaffolds are described in Table 1. All of the mapped ESTs were encoded on the complementary strand (in the right to left orientation). The exons mapped outside of the regions (broken lines) are shown in gray but are not to scale. Note that TSS for a gene corresponding to SGN-U579887 could not be predicted. The ruler below each diagram indicates the distance from the CArG boxes. (B) Expression analyses of the mapped ESTs. Lanes G, P and R represent the mature green, pink coloring and red ripe stages of tomato fruits, respectively. Data are the means from two independent experiments.
If these four genes are under the transcriptional regulation of RIN, their expression pattern should be associated with that of RIN. To examine this, we analyzed their expression in normal and rin mutant tomato fruits together with the expression of the RIN gene. Similar to previously reported observations [5,29], the RIN mRNA level was extensively increased in normal fruit at the ripening stages [pink coloring (P) and red ripe (R)] compared with the pre-ripening stage [mature green (G)], but not in the rin mutant at the corresponding stages (Figure 5B). Among the genes found around the RIN-binding sites, SGN-U571769 showed an increased mRNA level with ripening in normal fruit, but no such increase was observed in the mutant fruit. This expression pattern seemed to coincide with that of RIN although SGN-U571769 was expressed also in the G stage to some extent and its level was relatively lower (~1/10) than that of RIN at the ripening stages (Figure 5B). Unexpectedly, however, other genes examined did not exhibit the expression pattern associated with RIN (Figure 5B).
Discussion
Selectivity of target sequences of RIN for tomato genome
Based on our analyses to identify direct target genes of RIN, we successfully identified a number of sites to which RIN binds in the genome of ripening fruit cells by screening the promoters of the genes that are highly upregulated during ripening and also within the DNA fragments prepared by the ChIP with the anti-RIN antibody. Results for the RIN target sites provide insight into how RIN selects target genes within the tomato genome.
Our previous in vitro assays demonstrated that RIN can bind to sequences of C(C/T)(A/T)6(A/G)G, and that the preferential binding sequence is 'CCA(A/T)(A/t)(A/T)ATAG' [6]. Here, we demonstrated that a number of the ripening-associated genes contain these typical binding sites in their promoters and that RIN actually binds to these sites. In contrast, however, the promoter of LeACS4 contains no typical RIN-binding sequences but does contain three atypical CArG-box sequences, CAAATATAAG (ACS4-a), CAATTTTAAG (ACS4-b) and CTAGTTAAAG (ACS4-c) (underlines indicate atypical bases). Similarly, the promoter of LeEXP1 also contains one atypical CArG-box sequence, CAATTTAAAG. ChIP assays demonstrated that these four atypical sites were enriched within the IPed DNA at a high level comparable to the sites within the promoters of LeACS2 and LeMAN4 that contain the typical binding sequences. This result indicates that in vivo RIN binds to a wider spectrum of CArG-box sequences within the genome than previously expected based on in vitro assay.
This ability of RIN to bind with a wide range of CArG-box sequences in vivo suggests that other factors might be necessary for RIN to target only the ripening stage-specific genes. The binding site selectivity of RIN might be affected by chromatin structure (e.g., histone modification or DNA methylation) that controls the interaction of RIN with target DNAs in living cells. As another possibility, the selectivity of RIN might be increased by a tetramer- or higher-order multimer formation with other MADS-box proteins. For instance, in the 'quartet model' of floral organ specification, MADS-box proteins in floral organs form four combinations of tetramers that determine the identity of the different floral organs [34]. In this model, two dimers comprising the tetramer should recognize two different CArG-box sites, which confer higher selectivity of binding sites on transcription factor complexes [35]. In fact, Egea-Cortines et al. have proved that tetramer formation of MADS-box proteins dramatically increases the DNA-binding affinity [36]. In this study, we found that the cis-elements of LeACS2, LeACS4, TBG4 and RIN contain two CArG-box sites enriched at a significant level by ChIP using the anti-RIN antibody. These results suggest that RIN functions as a component of a tetramer that interacts with two binding sites of the target sites, just like the 'quartet model' transcription factor complexes. RIN possesses the ability to form a homodimer or heterodimers with other tomato MADS-box proteins belonging to the APETALA1/FRUITFULL subfamily (TM4, SLMBP7 and LeMADS1) and AG subfamily (TAGL1 and TAGL11) [6,37]. Because RIN belongs to the SEPALLATA (SEP) family among plant MADS-box group proteins [5,38], RIN might mediate interactions between other MADS-box proteins, similar to SEP proteins, which have been shown to mediate interactions between floral homeotic MADS-box proteins [39].
In this study, we identified novel RIN-binding sites from a screening of IPed DNA. At the regions flanking those sites, however, we found only one gene that is upregulated during ripening. Although we cannot exclude the possibility that the RINs binding to the other sites might regulate the genes located far from the sites (>5-kb), the bindings are likely not to activate any transcriptions. Similar observations were previously reported in the investigation of transcription factors of SEP3 through ChIP with Solexa sequencing (ChIP-SEQ), which showed that several percent of thousands of peaks are located far from protein-coding gene loci [27]. These facts mean that RIN-binding sites might not be restricted only to the promoter region of the RIN-inducible genes, but might include other genomic regions with CArG box-like sequences. If so, the binding of RIN at the cis-elements is necessary but not sufficient to induce the gene expressions required for fruit ripening, and certain factors are necessary for the specific transcriptional regulation by RIN. An assay using a yeast system revealed that RIN activates the transcriptional activity of an RNA polymerase that binds to the flanking genomic region [6], suggesting that the transactivation activity of RIN requires other transcriptional machinery at the RIN-binding sites. Higher-order multimer formation and other transcriptional machinery might allow RIN to induce specific gene expressions during ripening.
Autoregulation of RIN and the promising direct target genes of RIN
Our analysis revealed that RIN binds to the CArG-box sites in its own promoter in vivo, strongly suggesting that RIN autoregulates its own expression. The existence of this autoregulatory mechanism can explain how the rapid increase in the mRNA level of RIN at the onset of ripening is controlled [5,29] (Figure 5B), i.e., it is expected that autoregulation apparently controls the expression of RIN in a positive manner during ripening due to the transcriptional activation activity of RIN [6]. Autoregulation is frequently observed in plant floral homeotic MADS-box genes such as DEFICIENS (DEF) and GLOBOSA (GLO), Arabidopsis APETALA3 (AP3), PISTILLATA (PI), AG and embryogenetic AGL15 [25,36,40-42]. Positive or negative feedback loops in autoregulation maintain their expression in the development of floral organs or embryos after the initial induction. In tomatoes, TOMATO AGAMOUS-LIKE 1 (TAGL1) is possibly autoregulated [43]. The autoregulation of RIN might help to maintain its sufficiently high expression in ripening fruit, and consequently effectively regulate the expressions of direct target genes involved in the ripening process.
We also demonstrated here the direct interaction of RIN with the promoters of the genes involved in ethylene synthesis, LeACS2 and LeACS4, in vivo by ChIP. In the ethylene biosynthetic pathway, the conversion of S-adenosyl-L-methionine (SAM) to ACC by ACC synthase is a known rate-limiting step [44]. LeACS2 and LeACS4 regulate a massive increase in ethylene production of fruit associated with ripening, which is defined as system 2 ethylene production in contrast to system 1, which produces ethylene constitutively at a low level within pre-ripening fruits [7-9]. Recently, Yokotani et al. [9] demonstrated that ethylene production during ripening (system 2) consists of both autocatalytic and ethylene-independent systems and that transition from system 1 to system 2 occurs mainly via limited expression of LeACS2 and LeACS4, both of which are controlled by ethylene-independent developmental factors. Our finding strongly suggests that RIN directly regulates LeACS4 as well as LeACS2, and this finding is consistent with those of previous reports [6,8,9]. Meanwhile, direct regulation of LeACO1 and ETR3 expression by RIN cannot be confirmed at present due to relatively lower enrichment levels of the CArG boxes in their promoters in this study. In addition, a HD-Zip homeobox protein LeHB-1 directly regulates the expression of LeACO1 during fruit ripening [45], suggesting that RIN may not be involved in the direct regulation of LeACO1. However, we cannot exclude the possibility that RIN regulates the expression by binding to CArG boxes in the transposable element-like sequence in the LeACO1 promoter that could not be examined in this study. Regardless, our results certainly suggest that RIN contributes to the initiation of ethylene production at the onset of ripening through upregulation of LeACS2 and LeACS4.
We also demonstrated here the direct interaction of RIN with the promoters of ripening-induced genes, PG, TBG4, LeEXP1 and LeMAN4, in vivo using ChIP. This finding strongly suggests the direct transcriptional regulation of these genes by RIN, similar to its regulation of LeACS2 and LeACS4. Previous studies have shown that the expression of PG in tomato fruit is regulated by ethylene and that the promoter contains cis sites highly similar to the ethylene-responsive regions of the E8 and E4 genes [46-48]. However, Oeller et al. [49] previously revealed that PG expression during ripening is induced in an ethylene-independent manner, consistent with our view that RIN directly regulates the PG expression. These facts suggest that both pathways (ethylene and RIN) are effective in controlling the PG expression during ripening. Furthermore, previous analyses with suppression or overexpression of the four genes revealed that these genes are involved in cell wall modification but not enough to independently soften fruit [10-12,15,16,19,50,51]. These facts and our results therefore suggest that RIN induces expression of the genes involved in cell wall modification simultaneously during ripening, enabling the gene products to soften fruit in a cooperative manner.
ChIP revealed novel RIN-binding CArG-box sites and a possible RIN-regulating gene
We successfully identified four novel RIN-binding CArG-box sites from ripening tomato fruit by ChIP. This led to the detection of the genes near the CArG boxes. Among these genes, SGN-U571769 is possibly regulated by RIN during ripening due to the RIN-associated expression pattern. This gene encodes a protein similar to Trx y1, which serves as an oxidoreductase and is an activator of plastidal peroxiredoxin Q (Prx Q) [52]. Prx Q plays a role in peroxide detoxification, redox signaling and defense against pathogens [53,54]. When it decomposes peroxides, Trx y1 is used as an electron donor [55]. From these observations, we assume that RIN participates in these phenomena in ripening fruit via activation of Trx y1 expression. However, the involvement of Trx y1 in the ripening process needs further clarification because no functional evidence of such involvement or of the apparently irregular position of CArG-box 133R as a cis-acting element has been reported. More extensive ChIP-based analysis will lead to comprehensive identification of the target genes of RIN and will thus help clarify the transcriptional regulation of gene expression for fruit ripening.
Conclusions
Ripening of climacteric fruits is genetically controlled by intricate transcriptional cascades through ethylene and non-ethylene-mediated regulation. In this study, parts of the cascades involving RIN are revealed.
qChIP-PCR analysis of ripening tomato fruit and gel retardation assay demonstrated that RIN binds to the CArG boxes in the promoters of the genes involved in cell-wall modification (PG, LeEXP1, TBG4 and LeMAN4) and system-2 ethylene biosynthesis (LeACS2 and LeACS4) in vivo and in vitro. This suggests that RIN controls fruit softening and ethylene production by the direct transcriptional regulation of the cell-wall-modifying genes and ACS genes, respectively. The control of ethylene production by RIN during ripening means that RIN is also indirectly responsible for ethylene-inducible ripening processes together with an ethylene-mediated control. In addition, the presence of autoregulation of RIN itself is also suggested in this study, and thus explains how a rapid increase in the RIN transcription level occurs at the onset of fruit ripening.
Sequence analysis of the IPed DNAs from ripening tomato fruit, qChIP-PCR analysis and gel retardation assay revealed four new CArG-box sites bound by RIN. Of these four sites, the site 133R has a gene SGN-U571769 that encodes Trx y1 and shows a RIN-dependent expression pattern, suggesting that it is possibly regulated by RIN.
A series of binding analyses of RIN revealed that RIN can bind to a wider variety of CArG boxes in vivo than the consensus sequence previously determined in vitro. This implies the presence of other factors necessary for RIN to target fruit-ripening specific genes.
Methods
Chromatin immunoprecipitation
ChIP experiments were performed as previously described [6] using ripening tomato fruit at the pink coloring stage (4 days after breaker stage) where the expression of RIN is strongly induced. The immunoprecipitated (IPed) DNA fragments with anti-RIN antibody were recovered and purified. The resulting IPed DNA pool was used for qChIP-PCR as a template as described later.
The IPed DNA was also partially analyzed by sequencing and mapping as previously described [56]. In brief, both ends of the IPed DNAs were blunted with DNA polymerase I and phosphorylated with T4 kinase (Takara BKL kit; Takara Biotech, Otsu, Japan), followed by the addition of adenine (A) at the 3' end with Taq DNA polymerase (ExTaq, Takara Biotech) and by the ligation of an adaptor (prepared by annealing of two oligonucleotides, L: 5'-AGCACTCTCCAGCCTCTCACCGAGT-3' and S: 5'-CTCGGTGAGAGG-3'). The resulting DNA fragments were amplified by PCR with the L oligonucleotide as a primer. The PCR products were size-fractionated by agarose gel electrophoresis into 0.2-1 kb and cloned into a pBlueScriptII SK(-) vector that was digested with EcoRV and supplemented in advance with thymine (T) at the 3'end, followed by the introduction into Escherichia coli (JM109 strain) cells. The cloned DNA fragments were sequenced with a BigDye Terminator cycle sequencing kit v3.1 and analyzed by an ABI Prism 310 genetic analyzer (Applied Biosystems, Foster City, CA).
In silico motif search
Nucleotide sequences of the representative ripening-induced gene promoters were obtained from public databases: LeACS2 (accession no. X59139), LeACS4 (M88487), LeACO1 (X58273), ETR (NR, AY600437), PG (X14074), TBG4 (AF020390), LeEXP1 (U82123), LeMAN4 (AY046588), PSY1 (EF157835), INV (Z12027) and RIN (AF448522). Because the 5' region upstream of the protein-coding regions of PSY1, PG, TBG4, LeEXP1, LeMAN4 and RIN is less than 2 kb, their promoter sequences corresponding to 2 kb were complemented with Whole Genome Shotgun (WGS) data using the BLASTN program [57]. CArG boxes in the promoters were searched using the FUZZNUC program included in the EMBOSS package [58]. Sequence information for the promoters and motifs is summarized in Table 2.
The sequenced IPed DNA clones were mapped in silico on a draft genome sequence (WGS) of tomato released by the International Tomato Genome Sequencing Consortium (http://solgenomics.net/about/tomato_sequencing.pl) using BLASTN [57]. Using FUZZNUC [58], CArG-box motif sequences in the mapped regions were searched against the genomic sequences corresponding to the cloned IPed DNAs or the 1-kb genomic regions that mostly covered the clones when they were not fully sequenced.
Enrichment test for in vivo binding analysis of RIN
Enrichment levels of the CArG boxes by ChIP were monitored using quantitative real-time PCR (qChIP-PCR) as described below. The IPed DNA pool was used for qChIP-PCR as a template. The pre-immune serum (without anti-RIN antibody; PI)-treated chromatin DNA pools and the total input chromatin DNA (without ChIP treatment) pools were used as templates for the negative and standard controls, respectively. The nucleotide sequences of the oligonucleotide primers specific to the respective CArG-box sites used in this study are listed in Additional File 1.
Quantitative ChIP-PCR analyses were performed using Power SYBR green PCR master mix and a 7300 real-time PCR system (Applied Biosystems) according to the manufacturer's instructions. In a 50-μl reaction mixture, 2 μl of the IPed DNA, the PI-treated or the input DNA pool was applied as a template. The PCR conditions were as follows: 95°C for 10 min, 40 cycles of 95°C for 10 sec, and 60°C (or 57°C depending on the primers; see Additional File 1) for 1 min, followed by a dissociation step. The measurements [quantification cycle (Cq) values] for the CArG boxes were normalized with those for the Actin gene, which is non-bound by RIN and thereby used as a reference [6]. The enrichment levels were represented as fold changes relative to the input DNA. An outline/checklist for qChIP-PCR has been generated based on a template provided in [59] (Additional File 3).
Gel retardation assay
Gel retardation assays were performed as previously described [6] with a small modification. In brief, the DNA fragments including the normal or mutated CArG boxes (Table 1) were cloned. The mutated sequences were designed by replacing the first C and last G bases of the normal CArG boxes to T and A, respectively. The cloned DNAs were labeled by amplification with fluorescein isothiocyanate (FITC)-conjugated primers, purified and bound in vitro with RIN-MIK protein generated using a TnT SP6 quick-coupled transcription/translation system (Promega, Madison, WI) [6]. This protein, which lacks the C-terminal domain, was used because a clearer signal compared with that using the entire RIN protein could be generated [6]. The protein-DNA complexes were electrophoresed using polyacrylamide gel electrophoresis and detected using a typhoon 8600 (GE Healthcare Bio Science, Buckinghamshire, England) as previously reported [6].
Gene prediction
A sequence similarity search for the genomic ~5 kb flanking the CArG boxes enriched by ChIP was carried out against a tomato expressed sequence tag (EST) database provided by Solanaceae Genomics Network (SGN unigene set; available at ftp://ftp.sgn.cornell.edu/unigene_builds/) using the BLASTN program [57]. The ESTs with the highest sequence similarity (at least >100 bp alignment length and >90% identity) to the respective genomic sequences were adopted. A sequence similarity search for these ESTs was performed against the non-redundant (nr) protein database at the National Center for Biotechnology Information (NCBI) using the BLASTX program [57]. Gene structures were predicted using the sim4 [60] and the FGENESH 2.6 [61] programs with a parameter for the tomato genome (available at the Softberry site: http://linux1.softberry.com/berry.phtml)
Gene expression analysis
Expression levels of the genes were analyzed by quantitative real-time reverse transcription PCR (qRT-PCR) using oligonucleotide primers specific to each EST or RIN (Additional File 2). Using an RNeasy Plus Mini Kit (Qiagen, Hilden, Germany), total RNA was extracted and purified from tomato fruits of a normal (a genotype of RIN/RIN) plant at different stages (mature green, pink coloring and red ripe) and of a rin mutant (rin/rin) plant at periods corresponding to these stages, as previously described [29]. Complementary DNA was synthesized from total RNA using a PrimeScript II first cDNA strand synthesis kit (Takara Biotech) and then applied in real-time PCR as a template.
Quantitative RT-PCR analyses were performed in the same manner as for qChIP-PCR except that 2 μl of cDNA synthesis reaction mixture was applied as a template instead. Copy numbers of the objective transcripts were calculated from the measurements (Cq) using standard curves generated from a series of diluted PCR products for the respective genes. The data were normalized with that of a gene encoding clathrin adaptor complexes medium subunit (CAC; SGN-U314153) as a reference [62] (Additional File 2). An outline/checklist for qRT-PCR has been generated based on a template provided in [59] (Additional File 4).
Authors' contributions
MF and YI conceived the study and designed all the experiments. TN grew and prepared the tomato fruits. MF performed all the analyses. MF and YI interpreted the experimental data and participated in writing the manuscript. All the authors have read and approved the final manuscript.
Supplementary Material
Primer pairs specific to the CArG-box sites used for enrichment test. Primer pairs used in this study. The melting temperature (Tm) of each pair for qChIP-PCR is indicated in the right column.
Primer pairs specific to the tomato ESTs found in this study used for expression analysis and results of similarity searches for the ESTs. Primer pairs used in this study. Known proteins with the most significant similarity to proteins encoded by the ESTs are shown.
A checklist outlining the DNA to qChIP-PCR quality/methodology as described in [59].
A checklist outlining the RNA to qRT-PCR quality/methodology as described in [59].
Contributor Information
Masaki Fujisawa, Email: fmasaki@affrc.go.jp.
Toshitsugu Nakano, Email: tonakano@affrc.go.jp.
Yasuhiro Ito, Email: yasuito@affrc.go.jp.
Acknowledgements
The authors express their sincere thanks to Drs. M. Kitagawa and J. Kimbara, Research Institute, Kagome Co., Ltd (Tochigi, Japan), for providing tomato fruits. This work was supported in part by the Program for Promotion of Basic and Applied Researches for Innovations in the Bio-oriented Technology Research Advancement Institution (BRAIN) of Japan to Y. I.
References
- Prasanna V, Prabha TN, Tharanathan RN. Fruit ripening phenomena--an overview. Crit Rev Food Sci Nutr. 2007;47(1):1–19. doi: 10.1080/10408390600976841. [DOI] [PubMed] [Google Scholar]
- Giovannoni J. Molecular biology of fruit maturation and ripening. Annu Rev Plant Physiol Plant Mol Biol. 2001;52:725–749. doi: 10.1146/annurev.arplant.52.1.725. [DOI] [PubMed] [Google Scholar]
- Moore S, Vrebalov J, Payton P, Giovannoni J. Use of genomics tools to isolate key ripening genes and analyse fruit maturation in tomato. J Exp Bot. 2002;53(377):2023–2030. doi: 10.1093/jxb/erf057. [DOI] [PubMed] [Google Scholar]
- Tigchelaar E, McGlasson W, Buescher R. Genetic regulation of tomato fruit ripening. HortScience. 1978;13:508–513. [Google Scholar]
- Vrebalov J, Ruezinsky D, Padmanabhan V, White R, Medrano D, Drake R, Schuch W, Giovannoni J. A MADS-box gene necessary for fruit ripening at the tomato ripening-inhibitor (rin) locus. Science. 2002;296(5566):343–346. doi: 10.1126/science.1068181. [DOI] [PubMed] [Google Scholar]
- Ito Y, Kitagawa M, Ihashi N, Yabe K, Kimbara J, Yasuda J, Ito H, Inakuma T, Hiroi S, Kasumi T. DNA-binding specificity, transcriptional activation potential, and the rin mutation effect for the tomato fruit-ripening regulator RIN. Plant J. 2008;55(2):212–223. doi: 10.1111/j.1365-313X.2008.03491.x. [DOI] [PubMed] [Google Scholar]
- Nakatsuka A, Murachi S, Okunishi H, Shiomi S, Nakano R, Kubo Y, Inaba A. Differential expression and internal feedback regulation of 1-aminocyclopropane-1-carboxylate synthase, 1-aminocyclopropane-1-carboxylate oxidase, and ethylene receptor genes in tomato fruit during development and ripening. Plant Physiol. 1998;118(4):1295–1305. doi: 10.1104/pp.118.4.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry CS, Llop-Tous MI, Grierson D. The regulation of 1-aminocyclopropane-1-carboxylic acid synthase gene expression during the transition from system-1 to system-2 ethylene synthesis in tomato. Plant Physiol. 2000;123(3):979–986. doi: 10.1104/pp.123.3.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokotani N, Nakano R, Imanishi S, Nagata M, Inaba A, Kubo Y. Ripening-associated ethylene biosynthesis in tomato fruit is autocatalytically and developmentally regulated. J Exp Bot. 2009;60(12):3433–3442. doi: 10.1093/jxb/erp185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CJ, Watson CF, Morris PC, Bird CR, Seymour GB, Gray JE, Arnold C, Tucker GA, Schuch W, Harding S. et al. Inheritance and effect on ripening of antisense polygalacturonase genes in transgenic tomatoes. Plant Mol Biol. 1990;14(3):369–379. doi: 10.1007/BF00028773. [DOI] [PubMed] [Google Scholar]
- Smith CJS, Watson CF, Ray J, Bird CR, Morris PC, Schuch W, Grierson D. Antisense RNA inhibition of polygalacturonase gene-expression in transgenic tomatoes. Nature. 1988;334(6184):724–726. doi: 10.1038/334724a0. [DOI] [Google Scholar]
- Smith DL, Abbott JA, Gross KC. Down-regulation of tomato β-galactosidase 4 results in decreased fruit softening. Plant Physiol. 2002;129(4):1755–1762. doi: 10.1104/pp.011025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DL, Starrett DA, Gross KC. A gene coding for tomato fruit β-galactosidase II is expressed during fruit ripening. Cloning, characterization, and expression pattern. Plant Physiol. 1998;117(2):417–423. doi: 10.1104/pp.117.2.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington CMS, Vendrell M, Dominguez-Puigjaner E. Characterisation of an endo-(1,4)-β-mannanase (LeMAN4) expressed in ripening tomato fruit. Plant Sci. 2002;163(3):599–606. doi: 10.1016/S0168-9452(02)00167-X. [DOI] [Google Scholar]
- Wang A, Li J, Zhang B, Xu X, Bewley JD. Expression and location of endo-β-mannanase during the ripening of tomato fruit, and the relationship between its activity and softening. J Plant Physiol. 2009;166(15):1672–1684. doi: 10.1016/j.jplph.2009.04.008. [DOI] [PubMed] [Google Scholar]
- Brummell DA, Harpster MH, Civello PM, Palys JM, Bennett AB, Dunsmuir P. Modification of expansin protein abundance in tomato fruit alters softening and cell wall polymer metabolism during ripening. Plant Cell. 1999;11(11):2203–2216. doi: 10.1105/tpc.11.11.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummell DA, Harpster MH, Dunsmuir P. Differential expression of expansin gene family members during growth and ripening of tomato fruit. Plant Mol Biol. 1999;39(1):161–169. doi: 10.1023/A:1006130018931. [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ, Bedinger P, Durachko DM. Group I allergens of grass pollen as cell wall-loosening agents. Proc Natl Acad Sci USA. 1997;94(12):6559–6564. doi: 10.1073/pnas.94.12.6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose JK, Lee HH, Bennett AB. Expression of a divergent expansin gene is fruit-specific and ripening-regulated. Proc Natl Acad Sci USA. 1997;94(11):5955–5960. doi: 10.1073/pnas.94.11.5955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartley GE, Viitanen PV, Bacot KO, Scolnik PA. A tomato gene expressed during fruit ripening encodes an enzyme of the carotenoid biosynthesis pathway. J Biol Chem. 1992;267(8):5036–5039. [PubMed] [Google Scholar]
- Fray RG, Grierson D. Identification and genetic analysis of normal and mutant phytoene synthase genes of tomato by sequencing, complementation and co-suppression. Plant Mol Biol. 1993;22(4):589–602. doi: 10.1007/BF00047400. [DOI] [PubMed] [Google Scholar]
- Elliott KJ, Butler WO, Dickinson CD, Konno Y, Vedvick TS, Fitzmaurice L, Mirkov TE. Isolation and characterization of fruit vacuolar invertase genes from two tomato species and temporal differences in mRNA levels during fruit ripening. Plant Mol Biol. 1993;21(3):515–524. doi: 10.1007/BF00028808. [DOI] [PubMed] [Google Scholar]
- Kitagawa M, Moriyama T, Ito H, Ozasa S, Adachi A, Yasuda J, Ookura T, Inakuma T, Kasumi T, Ishiguro Y. et al. Reduction of allergenic proteins by the effect of the ripening inhibitor (rin) mutant gene in an F1 hybrid of the rin mutant tomato. Biosci Biotechnol Biochem. 2006;70(5):1227–1233. doi: 10.1271/bbb.70.1227. [DOI] [PubMed] [Google Scholar]
- Hecht A, Grunstein M. Mapping DNA interaction sites of chromosomal proteins using immunoprecipitation and polymerase chain reaction. Methods Enzymol. 1999;304:399–414. doi: 10.1016/s0076-6879(99)04024-0. full_text. [DOI] [PubMed] [Google Scholar]
- Gomez-Mena C, de Folter S, Costa MM, Angenent GC, Sablowski R. Transcriptional program controlled by the floral homeotic gene AGAMOUS during early organogenesis. Development. 2005;132(3):429–438. doi: 10.1242/dev.01600. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Ren N, Wang H, Stromberg AJ, Perry SE. Global identification of targets of the Arabidopsis MADS domain protein AGAMOUS-Like15. Plant Cell. 2009;21(9):2563–2577. doi: 10.1105/tpc.109.068890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann K, Muino JM, Jauregui R, Airoldi CA, Smaczniak C, Krajewski P, Angenent GC. Target genes of the MADS transcription factor SEPALLATA3: integration of developmental and hormonal pathways in the Arabidopsis flower. PLoS Biol. 2009;7(4):e1000090. doi: 10.1371/journal.pbio.1000090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgault R, Bewley JD. Variation in its C-terminal amino acids determines whether endo-β-mannanase is active or inactive in ripening tomato fruits of different cultivars. Plant Physiol. 2002;130(3):1254–1262. doi: 10.1104/pp.011890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa M, Ito H, Shiina T, Nakamura N, Inakuma T, Kasumi T, Ishiguro Y, Yabe K, Ito Y. Characterization of tomato fruit ripening and analysis of gene expression in F-1 hybrids of the ripening inhibitor (rin) mutant. Physiol Plantarum. 2005;123(3):331–338. doi: 10.1111/j.1399-3054.2005.00460.x. [DOI] [Google Scholar]
- Treisman R. Identification of a protein-binding site that mediates transcriptional response of the c-fos gene to serum factors. Cell. 1986;46(4):567–574. doi: 10.1016/0092-8674(86)90882-2. [DOI] [PubMed] [Google Scholar]
- Pollock R, Treisman R. Human SRF-related proteins: DNA-binding properties and potential regulatory targets. Genes Dev. 1991;5(12A):2327–2341. doi: 10.1101/gad.5.12a.2327. [DOI] [PubMed] [Google Scholar]
- Vrebalov J, Pan IL, Arroyo AJ, McQuinn R, Chung M, Poole M, Rose J, Seymour G, Grandillo S, Giovannoni J. et al. Fleshy fruit expansion and ripening are regulated by the Tomato SHATTERPROOF gene TAGL1. Plant Cell. 2009;21(10):3041–3062. doi: 10.1105/tpc.109.066936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blume B, Barry CS, Hamilton AJ, Bouzayen M, Grierson D. Identification of transposon-like elements in non-coding regions of tomato ACC oxidase genes. Mol Gen Genet. 1997;254(3):297–303. doi: 10.1007/s004380050419. [DOI] [PubMed] [Google Scholar]
- Theissen G, Saedler H. Plant biology. Floral quartets. Nature. 2001;409(6819):469–471. doi: 10.1038/35054172. [DOI] [PubMed] [Google Scholar]
- de Folter S, Shchennikova AV, Franken J, Busscher M, Baskar R, Grossniklaus U, Angenent GC, Immink RG. A Bsister MADS-box gene involved in ovule and seed development in petunia and Arabidopsis. Plant J. 2006;47(6):934–946. doi: 10.1111/j.1365-313X.2006.02846.x. [DOI] [PubMed] [Google Scholar]
- Egea-Cortines M, Saedler H, Sommer H. Ternary complex formation between the MADS-box proteins SQUAMOSA, DEFICIENS and GLOBOSA is involved in the control of floral architecture in Antirrhinum majus. EMBO J. 1999;18(19):5370–5379. doi: 10.1093/emboj/18.19.5370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leseberg CH, Eissler CL, Wang X, Johns MA, Duvall MR, Mao L. Interaction study of MADS-domain proteins in tomato. J Exp Bot. 2008;59(8):2253–2265. doi: 10.1093/jxb/ern094. [DOI] [PubMed] [Google Scholar]
- Hileman LC, Sundstrom JF, Litt A, Chen M, Shumba T, Irish VF. Molecular and phylogenetic analyses of the MADS-box gene family in tomato. Mol Biol Evol. 2006;23(11):2245–2258. doi: 10.1093/molbev/msl095. [DOI] [PubMed] [Google Scholar]
- Honma T, Goto K. Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature. 2001;409(6819):525–529. doi: 10.1038/35054083. [DOI] [PubMed] [Google Scholar]
- Goto K, Meyerowitz EM. Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA. Genes Dev. 1994;8(13):1548–1560. doi: 10.1101/gad.8.13.1548. [DOI] [PubMed] [Google Scholar]
- Hill TA, Day CD, Zondlo SC, Thackeray AG, Irish VF. Discrete spatial and temporal cis-acting elements regulate transcription of the Arabidopsis floral homeotic gene APETALA3. Development. 1998;125(9):1711–1721. doi: 10.1242/dev.125.9.1711. [DOI] [PubMed] [Google Scholar]
- Schwarz-Sommer Z, Hue I, Huijser P, Flor PJ, Hansen R, Tetens F, Lonnig WE, Saedler H, Sommer H. Characterization of the Antirrhinum floral homeotic MADS-box gene deficiens: evidence for DNA binding and autoregulation of its persistent expression throughout flower development. EMBO J. 1992;11(1):251–263. doi: 10.1002/j.1460-2075.1992.tb05048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itkin M, Seybold H, Breitel D, Rogachev I, Meir S, Aharoni A. TOMATO AGAMOUS-LIKE 1 is a component of the fruit ripening regulatory network. Plant J. 2009;60(6):1081–1095. doi: 10.1111/j.1365-313X.2009.04064.x. [DOI] [PubMed] [Google Scholar]
- Kende H. Ethylene Biosynthesis. Annu Rev Plant Phys. 1993;44:283–307. doi: 10.1146/annurev.pp.44.060193.001435. [DOI] [Google Scholar]
- Lin Z, Hong Y, Yin M, Li C, Zhang K, Grierson D. A tomato HD-Zip homeobox protein, LeHB-1, plays an important role in floral organogenesis and ripening. Plant J. 2008;55(2):301–310. doi: 10.1111/j.1365-313X.2008.03505.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery J, Pollard V, Deikman J, Fischer RL. Positive and negative regulatory regions control the spatial distribution of polygalacturonase transcription in tomato fruit pericarp. Plant Cell. 1993;5(9):1049–1062. doi: 10.1105/tpc.5.9.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholass FJ, Smith CJ, Schuch W, Bird CR, Grierson D. High levels of ripening-specific reporter gene expression directed by tomato fruit polygalacturonase gene-flanking regions. Plant Mol Biol. 1995;28(3):423–435. doi: 10.1007/BF00020391. [DOI] [PubMed] [Google Scholar]
- Sitrit Y, Bennett AB. Regulation of tomato fruit polygalacturonase mRNA accumulation by ethylene: A Re-examination. Plant Physiol. 1998;116(3):1145–1150. doi: 10.1104/pp.116.3.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeller PW, Lu MW, Taylor LP, Pike DA, Theologis A. Reversible inhibition of tomato fruit senescence by antisense RNA. Science. 1991;254(5030):437–439. doi: 10.1126/science.1925603. [DOI] [PubMed] [Google Scholar]
- Giovannoni JJ, DellaPenna D, Bennett AB, Fischer RL. Expression of a chimeric polygalacturonase gene in transgenic rin (ripening inhibitor) tomato fruit results in polyuronide degradation but not fruit softening. Plant Cell. 1989;1(1):53–63. doi: 10.1105/tpc.1.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell AL, Kalamaki MS, Kurien PA, Gurrieri S, Bennett AB. Simultaneous transgenic suppression of LePG and LeExp1 influences fruit texture and juice viscosity in a fresh market tomato variety. J Agric Food Chem. 2003;51(25):7450–7455. doi: 10.1021/jf034165d. [DOI] [PubMed] [Google Scholar]
- Collin V, Lamkemeyer P, Miginiac-Maslow M, Hirasawa M, Knaff DB, Dietz KJ, Issakidis-Bourguet E. Characterization of plastidial thioredoxins from Arabidopsis belonging to the new y-type. Plant Physiol. 2004;136(4):4088–4095. doi: 10.1104/pp.104.052233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz KJ, Jacob S, Oelze ML, Laxa M, Tognetti V, de Miranda SM, Baier M, Finkemeier I. The function of peroxiredoxins in plant organelle redox metabolism. J Exp Bot. 2006;57(8):1697–1709. doi: 10.1093/jxb/erj160. [DOI] [PubMed] [Google Scholar]
- Rey P, Cuine S, Eymery F, Garin J, Court M, Jacquot JP, Rouhier N, Broin M. Analysis of the proteins targeted by CDSP32, a plastidic thioredoxin participating in oxidative stress responses. Plant J. 2005;41(1):31–42. doi: 10.1111/j.1365-313X.2004.02271.x. [DOI] [PubMed] [Google Scholar]
- Lamkemeyer P, Laxa M, Collin V, Li W, Finkemeier I, Schottler MA, Holtkamp V, Tognetti VB, Issakidis-Bourguet E, Kandlbinder A. et al. Peroxiredoxin Q of Arabidopsis thaliana is attached to the thylakoids and functions in context of photosynthesis. Plant J. 2006;45(6):968–981. doi: 10.1111/j.1365-313X.2006.02665.x. [DOI] [PubMed] [Google Scholar]
- Barski A, Zhao K. Genomic location analysis by ChIP-Seq. J Cell Biochem. 2009;107(1):11–18. doi: 10.1002/jcb.22077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16(6):276–277. doi: 10.1016/S0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL. et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55(4):611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- Florea L, Hartzell G, Zhang Z, Rubin GM, Miller W. A computer program for aligning a cDNA sequence with a genomic DNA sequence. Genome Res. 1998;8(9):967–974. doi: 10.1101/gr.8.9.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salamov AA, Solovyev VV. Ab initio gene finding in Drosophila genomic DNA. Genome Res. 2000;10(4):516–522. doi: 10.1101/gr.10.4.516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exposito-Rodriguez M, Borges AA, Borges-Perez A, Perez JA. Selection of internal control genes for quantitative real-time RT-PCR studies during tomato development process. BMC Plant Biol. 2008;8:131. doi: 10.1186/1471-2229-8-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primer pairs specific to the CArG-box sites used for enrichment test. Primer pairs used in this study. The melting temperature (Tm) of each pair for qChIP-PCR is indicated in the right column.
Primer pairs specific to the tomato ESTs found in this study used for expression analysis and results of similarity searches for the ESTs. Primer pairs used in this study. Known proteins with the most significant similarity to proteins encoded by the ESTs are shown.
A checklist outlining the DNA to qChIP-PCR quality/methodology as described in [59].
A checklist outlining the RNA to qRT-PCR quality/methodology as described in [59].