Abstract
Background
Prediction of protein subcellular localization generally involves many complex factors, and using only one or two aspects of data information may not tell the true story. For this reason, some recent predictive models are deliberately designed to integrate multiple heterogeneous data sources for exploiting multi-aspect protein feature information. Gene ontology, hereinafter referred to as GO, uses a controlled vocabulary to depict biological molecules or gene products in terms of biological process, molecular function and cellular component. With the rapid expansion of annotated protein sequences, gene ontology has become a general protein feature that can be used to construct predictive models in computational biology. Existing models generally either concatenated the GO terms into a flat binary vector or applied majority-vote based ensemble learning for protein subcellular localization, both of which can not estimate the individual discriminative abilities of the three aspects of gene ontology.
Results
In this paper, we propose a Gene Ontology Based Transfer Learning Model (GO-TLM) for large-scale protein subcellular localization. The model transfers the signature-based homologous GO terms to the target proteins, and further constructs a reliable learning system to reduce the adverse affect of the potential false GO terms that are resulted from evolutionary divergence. We derive three GO kernels from the three aspects of gene ontology to measure the GO similarity of two proteins, and derive two other spectrum kernels to measure the similarity of two protein sequences. We use simple non-parametric cross validation to explicitly weigh the discriminative abilities of the five kernels, such that the time & space computational complexities are greatly reduced when compared to the complicated semi-definite programming and semi-indefinite linear programming. The five kernels are then linearly merged into one single kernel for protein subcellular localization. We evaluate GO-TLM performance against three baseline models: MultiLoc, MultiLoc-GO and Euk-mPLoc on the benchmark datasets the baseline models adopted. 5-fold cross validation experiments show that GO-TLM achieves substantial accuracy improvement against the baseline models: 80.38% against model Euk-mPLoc 67.40% with 12.98% substantial increase; 96.65% and 96.27% against model MultiLoc-GO 89.60% and 89.60%, with 7.05% and 6.67% accuracy increase on dataset MultiLoc plant and dataset MultiLoc animal, respectively; 97.14%, 95.90% and 96.85% against model MultiLoc-GO 83.70%, 90.10% and 85.70%, with accuracy increase 13.44%, 5.8% and 11.15% on dataset BaCelLoc plant, dataset BaCelLoc fungi and dataset BaCelLoc animal respectively. For BaCelLoc independent sets, GO-TLM achieves 81.25%, 80.45% and 79.46% on dataset BaCelLoc plant holdout, dataset BaCelLoc plant holdout and dataset BaCelLoc animal holdout, respectively, as compared against baseline model MultiLoc-GO 76%, 60.00% and 73.00%, with accuracy increase 5.25%, 20.45% and 6.46%, respectively.
Conclusions
Since direct homology-based GO term transfer may be prone to introducing noise and outliers to the target protein, we design an explicitly weighted kernel learning system (called Gene Ontology Based Transfer Learning Model, GO-TLM) to transfer to the target protein the known knowledge about related homologous proteins, which can reduce the risk of outliers and share knowledge between homologous proteins, and thus achieve better predictive performance for protein subcellular localization. Cross validation and independent test experimental results show that the homology-based GO term transfer and explicitly weighing the GO kernels substantially improve the prediction performance.
Background
As an important research field in molecular cell biology and proteomics, protein subcellular localization is closely related to protein function, metabolic pathway, signal transduction and biological process, and plays an important role in drug discovery, drug design, basic biological research and biomedicine research. Experimental determination of subcellular localization is time-consuming and laborious, and in some cases, it is hard to determine some subcellular compartments by fluorescent microscopy imaging techniques. Computational methods may help biologist select target proteins and design experiments.
Recent years have witnessed much progress in protein subcellular localization prediction [1-35]. Machine learning methods for predicting protein subcellular localization involve two major aspects: one is to derive protein features and the other is to design predictive model. State-of-art feature extraction methods are data- and model- dependent. We should guarantee that the features not only capture rich biological information but also should be discriminative enough to construct an effective classifier for prediction. On one hand, high throughout sequencing technique makes protein sequences cheaply available, and many computational models are based on protein primary sequences only in computational proteomics. On the other hand, data integration has become a popular method to integrate diverse biological data, including non-sequence information, such as GO annotation, protein-protein interaction network, protein structural information, cell image features etc.
There are many effective protein features extracted specifically for protein subcellular localization prediction. Amino acid composition (AA) has close relation with protein subcellular localization [16] and is the most frequently-used features. PseAA [4,10,12,13,17-32] encodes the pair-wise correlation of two amino acids at λ intervals using amino acid physiochemical properties. Sliding-window based k-mer feature representation is often used to capture the contextual information of amino acid and the conserved motif information, such as gapAA, di-AA, and motif kernel [35,36], etc. Since the dimensionality of k-mer feature space (20n for 20 amino acids) expands exponentially with the window size n, some researches [37,38] compress 20 amino acids into 7 groups according to amino acid physiochemical properties. Sorting signal and anchoring signal are important information for protein subcellular localization [39,40], but with the disadvantages that the cleavage sites vary substantially with proteins and the signal peptides may be missing.
Sequence profile is also important information for protein subcellular localization. Marcotte E et al. (2000) [41] revealed the relation between phylogenetic profile distribution and protein subcellular localization pattern. Sequence profile reveals the approximately true amino acid distribution for each amino acid residue along protein sequence, and thus can be naturally used as evolutionary distance between amino acids for measuring the similarity between two protein sequences. Through deliberate design, the similarity between two protein profile distributions can lead to a valid Mercer kernel [14,15,42-46]. Mak M et al. (2008) [42] derived the alignment score between two protein profile distributions using dynamic programming, based on which to derive a valid profile alignment kernel. Profile kernels [43,44] used PSSM & PSFM to derive the similarity score between any two k-mers, based on which to measure the similarity between two protein sequences. Kuang R et al. (2005) [44] designed a profile kernel, a variant mismatch kernel [45], which allowed a k fragment to match its corresponding k-mer if the fragment fell within the positional mutation neighbourhood defined by k-mer self-entropy. Kuang R et al. (2009) [46] extended the profile kernel by simple kernel fusion for prediction of malaria degradomes. Spectrum kernel [47] is based on exact k-mer match while (k, l) mismatch kernel [45] allows l mismatches within each k-mer, both of which are based on protein sequence only without profile incorporation. Actually, we can derive multiple kernels from multi-aspect knowledge about protein and then combine the kernels for more accurate definition of protein similarity. Alexander Z et al. [36] used semi-infinite linear programming to derive the optimal kernel weights for motif kernels combination. Mei S et al. (2010) [48] derived multiple motif kernels from diverse physiochemical constraints on amino acid substitution and combined the kernels for protein subnuclear localization. Kernel method is a good approach for heterogeneous data integration in computational biology.
Although protein sequence contains all the information for proteins to be transported to due compartments, to form correct folding, to form proper 3-D structural conformation and to function properly, etc., quality feature extraction from protein sequence is still a challenging problem because there is no general law or complete knowledge for effective feature extraction from protein sequence. However, large amount of biological experiments and computational inference have accumulated reliable multi-aspect local knowledge about genes and gene products, which has been well organized in the biological knowledgebase: gene ontology (GO). Gene ontology is a controlled vocabulary that describes biomolecules and gene products in terms of biological process, function and components. With the rapid progress of experimental and electronic annotation, gene ontology has become a general feature of proteomics that can be used to boost the predictive performance of protein subcellular localization [49-60]. In what follows, we briefly review the GO-based predictive models for protein subcellular localization from three viewpoints: (1) from the viewpoint of GO term extraction, the previous models can be classified into three categories. The first type of method directly uses protein accession number to query GO terms against GOA database [61], fast but not applicable to novel proteins [4-12,49-53]. The second type of method uses PSI-Blast to transfer the GO terms of homologous proteins to the target protein [54,55]. The third method uses InterProScan [56] to transfer the GO terms of signature proteins to the target proteins [57,58], which may be more reliable than the PSI-Blast transfer. Tung T et al. (2009) enlarged the GO term coverage by transferring to the target protein the GO terms of physically interacting partners in yeast interacting network [59]. (2) From the viewpoint of GO feature construction, the previous models also can be classified into three categories. The first way of GO feature construction is to simply turn all GOA GO terms into a flat binary feature vector to represent proteins [49-53,57-60]. This method has large GO term coverage but introduces many GO terms irrelevant to the problem concerned. The second type of method uses genetic algorithm to select the most informative GO component terms to minimize the irrelevant GO terms [54,55], but low GO term coverage may be highly likely to turn the test proteins to be null feature vector, so that the effect of PSI-Blast GO term transfer would be counteracted. The third type of method does not use explicit GO feature representation but designs an implicit kernel function to measure the semantic similarity between two GO terms [62]. Actually, the three aspects of gene ontology have different discriminative abilities, but the aforesaid three types of method assume equal feature weight. (3) From the viewpoint of data integration, the previous models can be classified into two categories. The first type of method uses ensemble learning to combine protein sequence with gene ontology, such as k-NN ensemble [52], fuzzy k-NN [59], and SVM ensemble [62]. The second type of method concatenates all the heterogeneous feature space (e.g. AA, di-AA, gap-AA, chem-AA, GO, PPI, etc) into a highly sparse high-dimension feature space [60].
In this paper, we design an explicitly weighted kernel learning system to transfer the known knowledge in terms of GO terms from related homology to the target problem, called Gene Ontology Based Transfer Learning Model (GO-TLM), for the purpose of sharing knowledge between closely-evolved protein families and achieving better model performance for protein subcellular localization. We use InterProScan to conduct multiple homologous signatures based queries against the InterPro database, and then transfer the homologous GO terms to the target protein. The transferred GO terms are potentially prone to errors, partly because of possibly noisy annotations from fluorescent microscopy experiments, electronic annotations using text mining, computational inference, etc. [49], or partly because of the outliers from homology transfer, that is, the homologous proteins actually have distinct function, process and subcellular localization patterns due to evolutionary divergence. Therefore, we should further construct a learning system that is trained on the transferred GO terms for reliable prediction. Such a scenario of borrowing knowledge in terms of GO terms from homologous proteins for further learning can be viewed as a case of Transfer Learning [63-66], where knowledge is transferred between well-correlated domains for better learning in the target domain. Dai W et al. (2007) [63] proposed an instance-based knowledge transfer learning method, where auxiliary data were drawn in to augment the target training set using AdaBoost weighing system to reduce the unfavourable impact of auxiliary data that are subjected to different distribution. Dai W et al. (2008) [64] proposed a feature-based translated transfer learning method, where a translator was constructed between text feature space and image feature space for knowledge transfer from text data to image data. Yang Q et al. (2009) [65] proposed a parameter-based knowledge transfer learning method, where the knowledge contained in annotated image of heterogeneous social web was transferred for target image clustering. Pan S et al. (2010) [66] reviewed the recent progress in transfer learning modelling. Because of the unbalanced knowledge about protein, the three aspects of gene ontology may have distinct discriminative abilities. For this reason, we derive GO process features, GO function features and GO component features individually, and then derive three individual GO kernels from the three types of GO feature representation. Besides the three GO kernels, we further derive another two sequence kernels from amino acid composition (AA) and di-pepetide (di-AA), which are actually spectrum kernel. These heterogeneous feature representations then are then merged into one kernel using linear kernel combination, a classical scenario of multiple kernel learning [36,67]. To reduce the computational cost of parameter optimization for multiple kernel learning, we use simple non-parametric cross validation to estimate the kernel weights instead. The model GO-TLM is evaluated against three baseline models on three eukaryotic benchmark datasets using cross validation and independent test.
Methods
GO feature construction
The InterPro database [68] integrates into a single source the most frequently-accessed signature databases including PROSITE [69], PRINTS [70], PFAM [71], ProDom [72], SMART [73] and TIGRFAMs [74]. PROSITE uses regular expression to represent significant amino acid patterns or uses profile (weight matrices) to detect structural and functional domains; PRINTS collects protein family fingerprints (motif); PFAM is a database of protein domain families that contains curated multiple sequence alignments for each family and corresponding profile hidden Markov models (HMMs); ProDom provides automatic domain query that is based on recursive use of PSI-BLAST homology search; SMART collects domains that are extensively annotated with respect to phyletic distributions, functional class, tertiary structures and functionally important residues; TIGRFAMs are a collection of protein families that are characteristic of curated multiple sequence alignments, Hidden Markov Models (HMMs) and associated information supporting functional identification of proteins by sequence homology. InterProScan [61] combines different protein signature recognition methods into one resource and provides a uniform web service interface to query signatures against the integrative InterPro database. InterProScan provides an option "--goterms" that enables GO term query using protein sequence only, which can collect more reliable GO terms than Blast transfer [54,55]. Parallel access and fast B-tree indexing make InterProScan practicable to large problem. For the reason, we use the perl script InterProScan.pl as a GO term extraction tool. The GO term set consists of three subsets: process, function and component. The three GO term subsets are organized as three individual binary feature vectors: (xP,1, xP,2,...,xP,l); (xF,1, xF,2,...,xF,m); (xC,1, xC,2,...,xC,n). It should be noted that InterProScan can overcome the problem of data unavailability to a certain degree. If we set high threshold to query more reliable GO terms with higher confidence, or the homology also is unannotated, InterProScan could neither transfer GO terms to the target proteins.
Kernel weight
K-mer occurrence patterns can reveal some conserved sub-sequences (e.g. motif) and k-spectrum kernel can be used to define the similarity between protein sequences. Since the feature space expands exponentially with window size |Σ|k, we only use 1-mer (AA) and 2-mer (di-AA) as protein sequence feature representation, thus we can derive kernels KAA, KdiAA. Based on the GO feature representation, we define GO process kernel KP, GO function kernel KF and GO component kernel KC. The 5 kernels are fused into single kernel for more accurate protein similarity definition. Kernel fusion is equivalent to the kernel that is computed in the concatenated feature space, but kernel fusion has the advantage of explicitly weighing the importance of feature subsets. The information content transferred from GO kernels to sequence kernels is measured by GO kernel weights. The weights of feature subsets vary with problems and should be derived from data. The final kernel is defined as the following linear combination of sub-kernels:
| (1) |
Lanckriet G et al. (2004) [75] used semi-definite programming to solve the problem, and Alexander Zien et al. (2007) [36] used semi-indefinite linear programming to derive the optimal weights. Both methods have rather large time & space complexity. Here, we use simple non-parametric cross validation to derive the kernel weights we,e ∈ {P, F, C, AA, diAA}. Given a training data X, derive kernels KAA, KdiAA, KP, KF, KC and split X into K folds, then conduct K-fold cross validation, we can estimate the recall rate or sensitivity (SE) for each kernel. Sensitivity reflects the discriminative ability of kernel or feature subset, but sensitivity is highly biased towards predominant class in the case of unbalanced data, so we should include Matthew's correlation coefficient (MCC) into performance measure to objectively estimate the kernel weights:
| (2) |
For denotation simplicity, the subscript e is omitted. Assume confusion matrix M for some kernel (KAA, KdiAA, KP, KF, KC), Mi,j records the counts that class i is classified to class j. Given the following variables that can be derived from the confusion matrix M:
| (3) |
We can derive the kernel's SE and MCC measure as follows:
| (4) |
Where, superscript L denotes subcellular locations.
As regards with Ke, e ∈ {AA, diAA, P, F, C}, Gaussian kernel is used here:
| (5) |
γ should be fine tuned by experiments.
Results
Dataset description
We choose three highly unbalanced eukaryotic benchmark datasets to evaluate GO-TLM performance. The first dataset MultiLoc collects 5859 proteins that are unevenly distributed to 10 subcellular locations for Plant data and 9 subcellular locations for Fungi data and Animal data [58]; the second dataset BaCelLoc, originally from the work [76], collects 491 proteins for Plant, 1198 proteins for Fungi and 2597 proteins for Animal that are unevenly located in 5 subcellular locations for Plant and 4 subcellular location for Fungi and Animal [58,77]; the third dataset Euk-mPLoc collects 5618 proteins that are unevenly located in 22 subcellular locations, the largest dataset as far in terms of number of subcellular locations [50]. To overcome overestimation of model performance, a cut-off threshold of 25% sequence similarity is generally accepted in current researches [5-7,13,15,33,34]. In this paper, to allow more training data and as conducted as the baseline models do, 30% threshold of sequence similarity is adopted on all the benchmark datasets, except 40% threshold of sequence similarity for MultiLoc plant dataset and 25% threshold of sequence similarity for Euk-mPloc dataset.
Model evaluation and model selection
Among the independent dataset test, sub-sampling (e.g. 5 or 10-fold cross-validation) test and jackknife test (leave-one-out cross validation), the jackknife test is deemed the most objective model evaluation method, as elucidated in [13,15]. Therefore, the jackknife test has been increasingly adopted and widely recognized by investigators to test the power of various prediction methods [1-34]. 5-fold cross validation is a commonly-accepted model evaluation approach in computational biology for large dataset or complex learning models, whereas leave-one-out cross validation (LOOCV) (i.e. jackknife test) is a better choice for small data or simple computational model. We use 5-fold cross validation to evaluate GO-TLM on data MultiLoc, BaCelLoc and Euk-mPLoc, and evaluate GO-TLM on BaCelLoc independent test as MultiLoc-GO did. For 5-fold cross validation, the protein dataset is randomly split into five disjoint parts with equal size. The last part may have 1-4 more examples than the former 4 parts in order for each example to be evaluated on the model. One part of the dataset is used as test set and the remained parts are jointly used as training set. The procedure iterates for five times, and each time a different part is chosen as test set. The independent test is actually hold-out test that randomly partition the dataset into training set and test set. As performance measure, hold-out set is not so objective as cross validation because it does not ensure that each data point is chosen to be tested. For the sake of comparison, we also conduct performance evaluation on BaCelLoc independent sets.
As regards to the cross validation for kernel weight evaluation, we select the cvK from {3, 5, 10} that achieves best overall accuracy. We use four commonly-adopted measures: Sensitivity (SE), Specificity (SP), Matthew's correlation coefficient (MCC) and Overall Accuracy. MCC is often used to evaluate the performance balance of model prediction. As compared to MCC, Overall Accuracy is a better candidate performance measure for model selection, because it has taken MCC into account. The overall MCC is not given, now that we pay more attention to the bias comparison between sub-categories. LIBSVM (http://www.csie.ntu.edu.tw/~cjlin/libsvm/) is used together with the model GO-TLM. The regularization parameter C is selected within {21, 22, 23, 24, 25, 26, 27, 28, 29, 210, 211} and the kernel parameter γ is selected within {2-1, 2-2, 2-3, 2-4}. We adopt the cvK, γ, C combination that achieves the best overall accuracy.
Comparison with baseline model
We choose MultiLoc-GO and Euk-mPLoc as baseline models for performance comparison. Both the baseline models incorporated gene ontology information to boost the model's predictive performance. MultiLoc-GO used InterProScan to draw in GO terms while Euk-mPLoc used protein accession to directly query GO terms against GOA database. We use Specificity (SP), Sensitivity (SE), MCC and Overall Accuracy as performance measures.
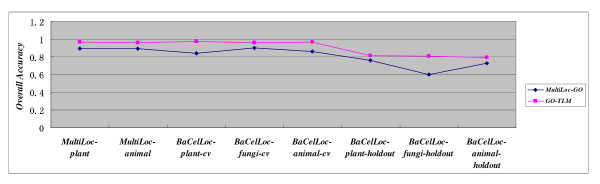
The baseline model MultiLoc-GO gave overall accuracy only for cross validation estimation on MultiLoc dataset and BaCelLoc dataset without detailed SP, SE and MCC. For intuitive illustration of eight comparison experiments between GO-TLM and MultiLoc-GO, we give performance comparison in a separate chart Figure 1. As can be seen from Figure 1GO-TLM significantly outperforms MultiLoc-GO on all benchmark datasets. GO-TLM achieves quite satisfactory performance for cross validation but significant decrease on BaCelLoc independent sets. The accuracy decrease may be caused by the subjective partition of training set and test set. From Figure 1 we can see that GO-TLM demonstrates more stable performance than MultiLoc-GO. GO-TLM's detailed performance measures see Table 1, 2, 3.
Figure 1.
Performance comparison between MultiLoc-GO and GO-TLM.
Table 1.
Performance comparison on 5859 MultiLoc protein dataset
| Subcellular location | Size | MultiLoc | MultiLoc-GO | GO-TLM | |||||
|---|---|---|---|---|---|---|---|---|---|
| SP | SE | MCC | - | SP | SE | MCC | |||
|
Fungi (5407) |
cytoplasm | 1411 | 0.68 | 0.85 | 0.70 | - | 0.9181 | 0.9773 | 0.9306 |
| endoplasmic | 198 | 0.71 | 0.59 | 0.63 | - | 0.9611 | 0.8737 | 0.9136 | |
| extracellular | 843 | 0.73 | 0.81 | 0.73 | - | 0.9915 | 0.9656 | 0.9749 | |
| golgi | 150 | 0.71 | 0.53 | 0.60 | - | 0.9530 | 0.9467 | 0.9485 | |
| mitochondria | 510 | 0.88 | 0.82 | 0.83 | - | 0.9723 | 0.9647 | 0.9655 | |
| nucleus | 837 | 0.81 | 0.74 | 0.73 | - | 0.9754 | 0.9486 | 0.9558 | |
| peroxisome | 157 | 0.68 | 0.30 | 0.43 | - | 0.9735 | 0.9363 | 0.9535 | |
| plasmamembrane | 1238 | 0.76 | 0.89 | 0.78 | - | 0.9869 | 0.9774 | 0.9774 | |
| vacuole (Astrik) | 63 | 0.76 | 0.24 | 0.42 | - | 0.9355 | 0.9206 | 0.9273 | |
|
Plant (5856) |
chloroplast (Astrik) | 449 | 0.88 | 0.85 | 0.85 | - | 1.0000 | 0.9911 | 0.9952 |
| Overall Accuracy | 74.60% | 89.60% | 96.65% | ||||||
|
Animal (5547) |
cytoplasm | 1411 | 0.67 | 0.85 | 0.68 | - | 0.9221 | 0.9809 | 0.9340 |
| endoplasmic | 198 | 0.68 | 0.56 | 0.60 | - | 0.9667 | 0.8788 | 0.9189 | |
| extracellular | 843 | 0.79 | 0.83 | 0.77 | - | 0.9891 | 0.9692 | 0.9753 | |
| golgi | 150 | 0.71 | 0.43 | 0.53 | - | 0.9527 | 0.9400 | 0.9448 | |
| mitochondria | 510 | 0.88 | 0.82 | 0.83 | - | 0.9723 | 0.9627 | 0.9642 | |
| nucleus | 837 | 0.82 | 0.73 | 0.73 | - | 0.9826 | 0.9438 | 0.9566 | |
| peroxisome | 157 | 0.71 | 0.31 | 0.44 | - | 0.9799 | 0.9299 | 0.9533 | |
| plasmamembrane | 1238 | 0.73 | 0.90 | 0.76 | - | 0.9765 | 0.9750 | 0.9687 | |
| lysosome (Astrik) | 103 | 0.69 | 0.36 | 0.48 | - | 0.9592 | 0.9126 | 0.9344 | |
| Overall Accuracy | 74.60% | 89.60% | 96.27% | ||||||
Table 2.
Performance comparison on BaCelLoc protein dataset
| Subcellular Location | Size | Cross validation | Independent test | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MultiLoc-GO | GO-TLM |
Size (train/test) |
MultiLoc-GO | GO-TLM | |||||||||
| - | SP | SE | MCC | SP | SE | MCC | SP | SE | MCC | ||||
|
Plant (491) |
Nucleus | 121 | - | 0.9516 | 0.9752 | 0.9514 | 60/61 | 0.91 | 0.90 | 0.77 | 1.0000 | 0.6500 | 0.7252 |
| Cytoplasm | 58 | - | 0.9434 | 0.8772 | 0.8986 | 52/6 | 0.41 | 0.94 | 0.38 | 0.7500 | 1.0000 | 0.8590 | |
| Extracellular | 41 | - | 0.9762 | 1.0000 | 0.9869 | 35/6 | 0.83 | 0.95 | 0.58 | 1.0000 | 0.6667 | 0.8100 | |
| Mitochondria | 67 | - | 0.9552 | 0.9552 | 0.9482 | 57/10 | 0.67 | 0.96 | 0.51 | 0.6429 | 0.9000 | 0.7386 | |
| Chloroplast | 204 | - | 0.9951 | 0.9951 | 0.9916 | 158/46 | 0.77 | 0.94 | 0.72 | 0.7302 | 1.0000 | 0.7777 | |
| Overall Accuracy | 83.70% | 97.14% | 76.00% | 81.25% | |||||||||
|
Fungi (1198) |
Nucleus | 711 | - | 0.9641 | 0.9817 | 0.9354 | 589/122 | 0.63 | 0.79 | 0.36 | 1.0000 | 0.7213 | 0.7246 |
| Cytoplasm | 211 | - | 0.9880 | 0.9318 | 0.9564 | 181/30 | 0.54 | 0.78 | 0.27 | 0.4839 | 1.0000 | 0.6311 | |
| Extracellular | 88 | - | 0.9130 | 0.8957 | 0.8849 | 72/16 | 0.78 | 0.98 | 0.60 | 1.0000 | 0.9375 | 0.9653 | |
| Mitochondria | 188 | - | 0.9780 | 0.9570 | 0.9616 | 177/11 | 0.68 | 0.94 | 0.62 | 0.7857 | 1.0000 | 0.8786 | |
| Overall Accuracy | 90.10% | 95.90% | 60.00% | 80.45% | |||||||||
|
Animal (2597) |
Nucleus | 1166 | - | 0.9646 | 0.9854 | 0.9551 | 803/363 | 0.62 | 0.93 | 0.57 | 0.7965 | 0.9945 | 0.7876 |
| Cytoplasm | 439 | - | 0.9402 | 0.8952 | 0.9017 | 302/137 | 0.72 | 0.82 | 0.43 | 0.7095 | 0.9270 | 0.7648 | |
| Extracellular | 804 | - | 0.9900 | 0.9900 | 0.9856 | 632/172 | 0.97 | 0.97 | 0.89 | 1.0000 | 0.2326 | 0.4406 | |
| Mitochondria | 188 | - | 0.9783 | 0.9574 | 0.9653 | 153/35 | 0.89 | 0.97 | 0.81 | 0.9714 | 0.9714 | 0.9699 | |
| Overall Accuracy | 85.70% | 96.85% | 73.00% | 79.46% | |||||||||
Table 3.
Performance comparison on 5618 Euk-mPLoc protein dataset
| Subnuclear location | size | GO-TLM | ||
|---|---|---|---|---|
| SP | SE | MCC | ||
| Acrosome | 17 | 0.9286 | 0.7647 | 0.8422 |
| Cell | 53 | 0.8085 | 0.7170 | 0.7593 |
| Centriole | 64 | 0.8958 | 0.6719 | 0.7737 |
| Chloroplast | 501 | 0.9681 | 0.9681 | 0.9650 |
| Cyanelle | 85 | 1.0000 | 0.9882 | 0.9940 |
| Cytoplasm | 1060 | 0.6356 | 0.7983 | 0.6475 |
| Cytoskeleton | 74 | 0.2500 | 0.0877 | 0.1431 |
| Endoplasmic | 364 | 0.7453 | 0.7790 | 0.7461 |
| Endosome | 89 | 0.6591 | 0.3867 | 0.4999 |
| Extracell | 640 | 0.7895 | 0.6402 | 0.7034 |
| Golgi | 254 | 1.0000 | 0.9231 | 0.9607 |
| Hydrogenosome | 13 | 0.7872 | 0.5968 | 0.6825 |
| Lysosome | 80 | 1.0000 | 0.4615 | 0.6789 |
| Melanosome | 13 | 0.6000 | 0.3103 | 0.4295 |
| Microsome | 31 | 0.9349 | 0.8865 | 0.9020 |
| Mitochondrion | 535 | 0.8071 | 0.8145 | 0.7689 |
| Nucleus | 1333 | 0.9412 | 0.8696 | 0.9032 |
| Peroxisome | 97 | 0.8059 | 0.7781 | 0.7658 |
| Plasma | 725 | 0.9260 | 0.8410 | 0.8694 |
| Spindle pole | 36 | 0.8750 | 0.4118 | 0.5995 |
| Synapse | 15 | 1.0000 | 0.5385 | 0.7334 |
| Vacuole | 102 | 0.9176 | 0.8571 | 0.8851 |
| Overall Accuracy | 80.38% | |||
| Baseline: Euk-mPLoc model | 67.40% | |||
| Baseline: Fuzzy K-NN model | 62.25% | |||
On MultiLoc plant dataset with 10 subcellular compartments, the best parameter combination is cvK = 5, γ = 2-2, C = 28 and the best Overall Accuracy is 96.55%, 7.05% increase from MultiLoc-GO 89.60% [58], 22.05% sharp increase from MultiLoc 74.60% [76]. As can be seen from Table 1GO-TLM demonstrates quite satisfactory performances on all the subcellular locations, with SP, SE and MCC all greater than 90%, far better than sequence-based MultiLoc. MultiLoc-GO gave no detailed cross validation performance measures on each subcellular location. The performance measures SP, SE and MCC demonstrate that GO-TLM shows no bias towards large subcellular locations, e.g. the smallest vacuole SP: 0.9355, SE: 0.9206, MCC: 0.9273 on MultiLoc plant. Similar conclusions can be drawn on MultiLoc animal. The best parameter combination is cvK = 5, γ = 2-2, C = 28 for MultiLoc animal. MultiLoc fungi dataset shares most proteins with MultiLoc plant, without chloroplast compartment, so we don't give results on MultiLoc fungi dataset.
We conduct two sets of experiments on the second dataset BaCelLoc. As can be seen from Table 2 the cross validation experiments show that GO-TLM achieves best overall accuracy 97.14%, 95.90% and 96.85% on BaCelLoc plant, BaCelLoc fungi and BaCelLoc animal, respectively against MultiLoc-GO 83.70%, 90.10% and 85.70%, with accuracy increase 13.44%, 5.8% and 11.15%, respectively. The performance measures SP, SE and MCC demonstrate that GO-TLM shows no bias towards large subcellular locations, e.g. the smallest extracellular SP: 0.9762, SE: 1.0000, MCC: 0.9869 on BaCelLoc plant; the smallest extracellular SP: 0.9130, SE: 0.8957, MCC: 0.8849 on BaCelLoc fungi; and the smallest Mitochondria SP: 0.9783, SE: 0.9574, MCC: 0.9653 on BaCelLoc animal. The best parameter combination is cvK = 5, γ = 2-2, C = 27 for BaCelLoc plant; cvK = 5, γ = 2-2, C = 27for BaCelLoc fungi; and cvK = 5, γ = 2-2, C = 26 for BaCelLoc animal. MultiLoc-GO gave no detailed SP, SE and MCC performance.
As can be seen in Table 2 the independent test on BaCelLoc datasets show that GO-TLM achieves 81.25%, 80.45% and 79.46% on plant, fungi and animal, respectively, as compared against MultiLoc-GO 76%, 60.00% and 73.00%, with accuracy increase 5.25%, 20.45% and 6.46%, respectively. As can be seen from MCC performance, GO-TLM generally shows less bias towards large subcellular locations than MultiLoc-GO, e.g. Cytoplasm (0.8590 vs. 0.38), Extracellular (0.8100 vs. 0.58) on plant; Nucleus (0.7246 vs. 0.36), Cytoplasm (0.6311 vs. 0.27) on fungi; and Nucleus (0.7876 vs. 0.57), Cytoplasm (0.7648 vs. 0.43) on animal. The improvement on MCC measure may indicate the significance of incorporating MCC measure into GO-TLM kernel weight estimation as illustrated in formula (1). At the same time, GO-TLM also shows a little performance decrease on several measure values (in bold italic).
On Euk-mPLoc data with 22 subcellular compartments, the best parameter combination is cvK = 5, γ = 2-3, C = 27 and the best Overall Accuracy is 80.38%, 12.98% substantial increase from Euk-mPLoc 67.40% [50] and 18.13% sharp increase from Fuzzy K-NN 62.25% [59]. Fuzzy K-NN was evaluated on the old version of Euk-mPLoc with 22 subcellular locations and 4708 proteins. The multi-location proteins are excluded and only its single-location Measure I is taken as the comparative baseline here. Euk-mPLoc and Fuzzy K-NN gave no detailed performance. As can be seen from Table 3GO-TLM shows quite satisfactory MCC performance on most subcellular locations, including most small compartments such as Acrosome 0.8764, Microsome 0.8923, Hydrogenosome 0.7747, etc. There are two small compartments that achieve poor MCC performance: Cytoskeleton (MCC: 0.1431) &Melanosome (MCC: 0.5523). As compared to the previous models, GO-TLM can help reduce the bias towards the subcellular locations with larger number of training proteins.
Kernel weight distribution
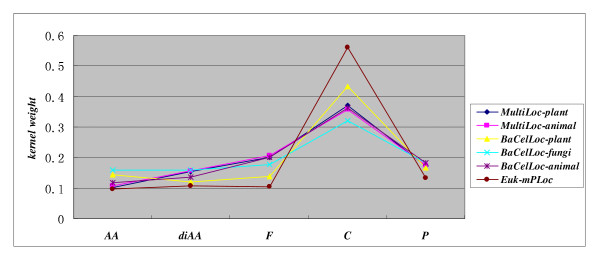
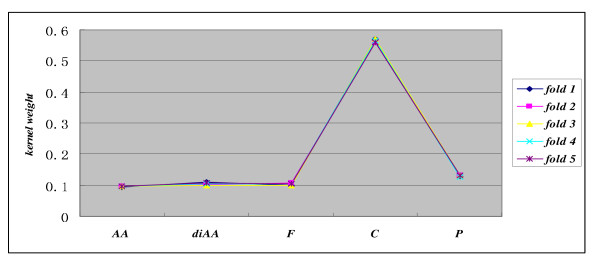
The weights for kernel KAA, KdiAA, KP, KF, KC on the benchmark datasets are illustrated in Figure 2. For each fold of cross validation, the training set is further subjected to cvK-fold cross validation to estimate the five kernels' performance measures (SP, SE and MCC), based on which to further estimate the kernels' weights using formula (1). Experiments shows that the kernel weights for 5-fold cross validation vary slightly (take Euk-mPLoc dataset for instance, see Figure 3). As can be seen from Figure 2GO-TLM demonstrates similar kernel weight distribution on all the benchmark datasets. GO features show much stronger discriminative ability than sequence features and the GO component terms from signature proteins contribute most to the predictive performance, GO process terms the second and GO function terms the third. The results may imply that GO component terms are more directly indicative of subcellular location than GO function terms and GO process terms, or the training proteins have less component term missing rate than function and process term missing rate. Take Euk-mPLoc dataset for example, there are 658 proteins without GO process terms, accounting for 11.71% missing rate; there are 755 proteins without GO function terms, accounting for 13.44% missing rate; and there are 31 proteins without GO component terms, accounting for 0.56%, far less than the missing rate of function terms and process terms. On the other hand, the weights for KAA, KdiAA vary little with datasets, while the weights for KP, KF, KC vary widely with datasets, the higher for KC weight, the lower for KP, KF weights. GO-TLM achieves the highest KC weight on Euk-mPLoc and the lowest KC weight on BaCelLoc-fungi. The result may also be explained by the missing rate of GO terms, e.g. 0 missing rate for BaCelLoc-fungi component terms, while 0.56% missing rate for Euk-mPLoc component terms. BaCelLoc-fungi has less missing rate of process term and function term, and has process weight and function weight slightly increased. We can see that the unbalanced GO term distribution contributes much to the variation of GO kernel weights.
Figure 2.
Kernel weight estimation using 5-fold cross validation.
Figure 3.
Kernel weights estimation on Euk-mPLoc dataset using 5-fold cross validation.
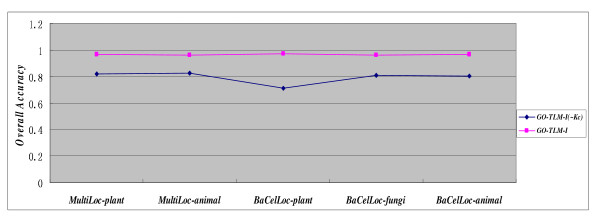
Now that KC weight is much higher than the other kernel weights, we had better further study the predictive performance of the model that is trained on all the kernels except KC , referred to as GO-TLM(~KC). The performance comparison between GO-TLM and GO-TLM(~KC) is illustrated in Figure 4 which shows that the removal of kernel KC leads to substantial 14.67%~26.12% performance decrease. The result demonstrates that the GO component terms play a critical role in protein subcellular localization. However, the model GO-TLM-I(~KC) achieves over 80% overall accuracy on datasets MultiLoc-plant, MultiLoc-animal, BaCelLoc-fungi and BaCelLoc-animal, which demonstrates that the other 4 kernels also benefit the protein subcellular localization prediction. Lu Z et al. (2005) has elucidated that GO function terms are good indicator of protein subcellular localization [78].
Figure 4.
Performance comparison between GO-TLM and GO-TLM(~Kc).
Discussion
Traditionally, the knowledge in terms of GO terms about homology can be directly transferred to the target proteins based on signature or homology search. Such a way of knowledge transfer generally benefits the research on unknown domain, species or family in biology. However, this process may be prone to introducing noise and outlier, partly because sequence similarity unnecessarily implies similar subcellular localization pattern, molecular function or biological process; or partly because the annotations in themselves may be noisy. For the reason, we design a transfer learning system to share knowledge between homology for reliable protein subcellular localization, called Gene Ontology Based Transfer Learning Model (GO-TLM). GO-TLM collects GO terms based on signature or homology search against the integrative database InterPro, and then transfer the GO terms to the target proteins for further learning. All the transferred GO terms are used to train a kernel-based SVM classifier, which can effectively reduce the risk of outliers by allowing larger training error to achieve maximum margin between two-class separating hyperplanes. Thus, the quite different GO terms (e.g. extracell GO term is transferred to nuclear proteins) would be viewed as outlier after SVM training. Such a way of constructing a learning system based on the transferred knowledge between related domains or data may benefit computational biology in many aspects. As compared to concatenation of heterogeneous feature subspace, multiple kernel learning has the advantage of explicitly weighing the feature subset/kernel contribution to the classification task. GO-TLM uses simple non-parametric cross validation to estimate the kernel weights, serially one kernel in memory at a time, such that it requires much less time and space than the complicated semi-definite/semi-indefinite linear programming. Simple non-parametric cross validation is used to estimate the kernel weights. Meanwhile, the kernel weight estimation allows for both sensitivity and unbalanced measure MCC, such that GO-TLM would work better in the scenario of unbalanced training dataset. Experiments reveal that GO component feature play more important role than GO process feature and GO function feature. With less missing rate, GO function terms and GO process terms would further increase the predictive performance.
GO-TLM only uses those GO terms that belong to the problem concerned, thus no irrelevant GO term is into the GO feature vector. However, this method of GO feature construction may cause low GO term coverage, that's, a test GO term (GO term that belongs to a test protein) may find no match in the training GO term set. In such a scenario, we should include the test GO term into the training GO term set to re-train the well-trained learning system. Re-training is generally time-consuming for large data and complex model selection. We had better pull in more statistically correlated GO terms for those proteins with very few evidences. To avoid re-training, we had better use statistically correlated GO term to replace the GO term that may not hit the training GO terms. Lastly, there is still a large chance for InterProScan to miss capturing GO terms from homology because of the unevenly distribution of GO terms. In such a scenario, we can lower the threshold for InterProScan to draw in the GO terms from remote homology. Since user-friendly and publicly accessible web-servers represent the future direction for developing practically more useful predictors [79], we shall make efforts in our future work to provide a web-server for the method presented in this paper.
Conclusions
In this paper, we design an explicitly weighted kernel learning system to transfer the known knowledge in terms of GO terms from related homology to the target problem, called Gene Ontology Based Transfer Learning Model (GO-TLM), to reduce the risk of outlier and achieve better model performance. On one hand, homology or signature based GO term transfer enables reliable knowledge share between homology, protein subfamily or protein family. On the other hand, GO-TLM uses simple and effective non-parametric cross validation to explicitly weigh the contribution of the three aspects of gene ontology. The explicitly weighted kernel combination can better cope with the different missing rates and different discriminative abilities between the three aspects of gene ontology. The kernel weight estimation takes into account MCC measure, such that GO-TLM could perform better in the scenario of unbalanced data distribution among subcellular locations. Experiments on three benchmark datasets show that GO-TLM significantly outperforms the previous models.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
MSY conducted the survey and the computational modelling. WF and SGZ reviewed the study. All authors read and approved the final manuscript.
Contributor Information
Suyu Mei, Email: 061021053@fudan.edu.cn.
Wang Fei, Email: wangfei@fudan.edu.cn.
Shuigeng Zhou, Email: sgzhou@fudan.edu.cn.
Acknowledgements
Thanks for the anonymous reviewers' helpful comments and the editorial office's help.
References
- Chou KC, Elrod DW. Protein subcellular location prediction. Protein Eng. 1999;12:107–118. doi: 10.1093/protein/12.2.107. [DOI] [PubMed] [Google Scholar]
- Chou KC, Cai YD. Using functional domain composition and support vector machines for prediction of protein subcellular location. J Biol Chem. 2002;277:45765–45769. doi: 10.1074/jbc.M204161200. [DOI] [PubMed] [Google Scholar]
- Xiao X, Shao SH, Ding YS, Huang ZD, Chou KC. Using cellular automata images and pseudo amino acid composition to predict protein subcellular location. Amino Acids. 2006;30:49–54. doi: 10.1007/s00726-005-0225-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen HB, Chou KC. A top-down approach to enhance the power of predicting human protein subcellular localization: Hum-mPLoc 2.0. Anal Biochem. 2009;394:269–274. doi: 10.1016/j.ab.2009.07.046. [DOI] [PubMed] [Google Scholar]
- Chou KC, Shen HB. Hum-PLoc: A novel ensemble classifier for predicting human protein subcellular localization. Biochem Biophys Res Commun. 2006;347:150–157. doi: 10.1016/j.bbrc.2006.06.059. [DOI] [PubMed] [Google Scholar]
- Shen HB, Chou KC. Virus-PLoc: A fusion classifier for predicting the subcellular localization of viral proteins within host and virus-infected cells. Biopolymers. 2007;85(3):233–240. doi: 10.1002/bip.20640. [DOI] [PubMed] [Google Scholar]
- Chou KC, Shen HB. Large-scale plant protein subcellular location prediction. J Cell Biochem. 2007;100:665–678. doi: 10.1002/jcb.21096. [DOI] [PubMed] [Google Scholar]
- Chou KC, Shen HB. Predicting eukaryotic protein subcellular location by fusing optimized evidence-theoretic K-nearest neighbor classifiers. Journal of Proteome Research. 2006;5:1888–1897. doi: 10.1021/pr060167c. [DOI] [PubMed] [Google Scholar]
- Shen HB, Chou KC. Gneg-mPLoc: A top-down strategy to enhance the quality of predicting subcellular localization of Gram-negative bacterial proteins. Journal of Theoretical Biology. 2010. in press . [DOI] [PubMed]
- Chou KC, Shen HB. Large-scale predictions of gram-negative bacterial protein subcellular locations. Journal of Proteome Research. 2006;5:3420–3428. doi: 10.1021/pr060404b. [DOI] [PubMed] [Google Scholar]
- Shen HB, Chou KC. Gpos-mPLoc: A top-down approach to improve the quality of predicting subcellular localization of Gram-positive bacterial proteins. Protein & Peptide Letters. 2009;16:1478–1484. doi: 10.2174/092986609789839322. [DOI] [PubMed] [Google Scholar]
- Chou KC, Shen HB. Review: Recent progresses in protein subcellular location prediction. Anal Biochem. 2007;370:1–16. doi: 10.1016/j.ab.2007.07.006. [DOI] [PubMed] [Google Scholar]
- Chou KC. Pseudo amino acid composition and its applications in bioinformatics, proteomics and system biology. Current Proteomics. 2009;6(4):262–274. doi: 10.2174/157016409789973707. [DOI] [Google Scholar]
- Liu H, Yang J, Liu DQ, Shen HB, Chou KC. Using a new alignment kernel function to identify secretory proteins. Protein & Peptide Letters. 2007;14(2):203–208. doi: 10.2174/092986607779816087. [DOI] [PubMed] [Google Scholar]
- Wang M, Yang J, Chou KC. Using string kernel to predict signal peptide cleavage site based on subsite coupling model. Amino Acids. 2005;28:395–402. doi: 10.1007/s00726-005-0189-6. [DOI] [PubMed] [Google Scholar]
- Cedano J, Aloy P, P'erez-Pons J, Querol E. Relation between amino acid composition and cellular location of proteins. Journal of Molecular Biology. 1997;266:594–600. doi: 10.1006/jmbi.1996.0804. [DOI] [PubMed] [Google Scholar]
- Chou K. Prediction of protein subcellular locations by incorporating quasi-sequence-order effect. Biochemical and Biophysical Research Communications. 2000;278:477–483. doi: 10.1006/bbrc.2000.3815. [DOI] [PubMed] [Google Scholar]
- Nanni L, Lumini A. Genetic programming for creating Chou's pseudo amino acid based features for submitochondria localization. Amino Acids. 2008;34:653–660. doi: 10.1007/s00726-007-0018-1. [DOI] [PubMed] [Google Scholar]
- Qiu JD, Huang JH, Liang RP, Lu XQ. Prediction of G-protein-coupled receptor classes based on the concept of Chou's pseudo amino acid composition: an approach from discrete wavelet transform. Analytical Biochemistry. 2009;390(1):68–73. doi: 10.1016/j.ab.2009.04.009. [DOI] [PubMed] [Google Scholar]
- Lin H. The modified Mahalanobis discriminant for predicting outer membrane proteins by using Chou's pseudo amino acid composition. J Theor Biol. 2008;252:350–356. doi: 10.1016/j.jtbi.2008.02.004. [DOI] [PubMed] [Google Scholar]
- Zeng YH, Guo YZ, Xiao RQ, Yang L, Yu LZ, Li ML. Using the augmented Chou's pseudo amino acid composition for predicting protein submitochondrialocations based on auto covariance approach. J Theor Biol. 2009;59:366–372. doi: 10.1016/j.jtbi.2009.03.028. [DOI] [PubMed] [Google Scholar]
- Ding YS, Zhang TL, Gu Q, Zhao PY, Chou KC. Using maximum entropy model to predict protein secondary structure with single sequence. Protein & Peptide Letters. 2009;16:552–560. doi: 10.2174/092986609788167833. [DOI] [PubMed] [Google Scholar]
- Zhou XB, Chen C, Li ZC, Zou XY. Using Chou's amphiphilic pseudo-amino acid composition and support vector machine for prediction of enzyme subfamily classes. J Theor Biol. 2007;248:546–551. doi: 10.1016/j.jtbi.2007.06.001. [DOI] [PubMed] [Google Scholar]
- Ding YS, Zhang TL. Using Chou's pseudo amino acid composition to predict subcellular localization of apoptosis proteins: an approach with immune genetic algorithm-based ensemble classifier. Pattern Recognition Letters. 2008;29:1887–1892. doi: 10.1016/j.patrec.2008.06.007. [DOI] [Google Scholar]
- Chen C, Chen L, Zou X, Cai P. Prediction of protein secondary structure content by using the concept of Chou's pseudo amino acid composition and support vector machine. Protein & Peptide Letters. 2009;16(1):27–31. doi: 10.2174/092986609787049420. [DOI] [PubMed] [Google Scholar]
- Ding H, Luo L, Lin H. Prediction of cell wall lytic enzymes using Chou's amphiphilic pseudo amino acid composition. Protein & Peptide Letters. 2009;16:351–355. doi: 10.2174/092986609787848045. [DOI] [PubMed] [Google Scholar]
- Jiang X, Wei R, Zhang TL, Gu Q. Using the concept of Chou's pseudo amino acid composition to predict apoptosis proteins subcellular location: an approach by approximate entropy. Protein & Peptide Letters. 2008;15:392–396. doi: 10.2174/092986608784246443. [DOI] [PubMed] [Google Scholar]
- Li FM, Li QZ. Predicting protein subcellular location using Chou's pseudo amino acid composition and improved hybrid approach. Protein & Peptide Letters. 2008;15(6):612–616. doi: 10.2174/092986608784966930. [DOI] [PubMed] [Google Scholar]
- Lin H, Ding H, Feng-Biao Guo FB, Zhang AY, Huang J. Predicting subcellular localization of mycobacterial proteins by using Chou's pseudo amino acid composition. Protein & Peptide Letters. 2008;15(No.7):739–744. doi: 10.2174/092986608785133681. [DOI] [PubMed] [Google Scholar]
- Esmaeili M, Mohabatkar H, Mohsenzadeh S. Using the concept of Chou's pseudo amino acid composition for risk type prediction of human papillomaviruses. J Theor Biol. 2010;263(2):203–209. doi: 10.1016/j.jtbi.2009.11.016. [DOI] [PubMed] [Google Scholar]
- Qiu JD, Huang JH, Shi SP, Liang RP. Using the concept of Chou's pseudo amino acid composition to predict enzyme family classes: an approach with support vector machine based on discrete wavelet transform. Protein & Peptide Letters. 2010;17:715–712. doi: 10.2174/092986610791190372. [DOI] [PubMed] [Google Scholar]
- Gu Q, Ding YS, Zhang TL. Prediction of g-protein-coupled receptor classes in low homology using Chou's pseudo amino acid composition with approximate entropy and hydrophobicity patterns. Protein Pept Lett. 2010;17(5):559–567. doi: 10.2174/092986610791112693. [DOI] [PubMed] [Google Scholar]
- Chou KC, Shen HB. A new method for predicting the subcellular localization of eukaryotic proteins with both single and multiple sites: Euk-mPLoc 2.0. PLoS ONE. 2010;5(4):e9931. doi: 10.1371/journal.pone.0009931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou KC, Shen HB. Plant-mPLoc: a top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS ONE. 2010;5(6):e11335. doi: 10.1371/journal.pone.0011335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhasin M, Raghava G. ELSpred: SVM-based method for subcellular localization of eukaryotic proteins using dipeptide composition and PSI-BLAST. Nucleic Acid Res. 2004. pp. W414–W419. [DOI] [PMC free article] [PubMed]
- Alexander Z, Cheng S. An automated combination of kernels for predicting protein subcellular localization. NIPS 2007, workshop on Machine Learning in Computational Biology.
- Dijk A, Bosch D, Braak C, Krol A, Ham R. Predicting sub-Golgi localization of type II membrane proteins. Bioinformatics. 2008;24(16):1779–1786. doi: 10.1093/bioinformatics/btn309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Zhang J, Luo X, Zhu W, Yu K, Chen K, Li Y, Jiang H. Predicting protein-protein interactions based only on sequences information. PNAS. 2007;104(11):4337–4341. doi: 10.1073/pnas.0607879104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider G, Fechner U. Review advances in the prediction of protein targeting signals. Proteomics. 2004;4:1571–1580. doi: 10.1002/pmic.200300786. [DOI] [PubMed] [Google Scholar]
- Hoglund A, Donnes P, Blum T, Adolph H, Kohlbacher O. MultiLoc: prediction of protein subcellular localization using N-terminal targeting sequences, sequence motifs and amino acid composition. Bioinformatics. 2006;22(10):1158–1165. doi: 10.1093/bioinformatics/btl002. [DOI] [PubMed] [Google Scholar]
- Marcotte E, Xenarios I, van Der Bliek A, Eisenberg D. Localizing proteins in the cell from their phylogenetic profiles. Proc Natl Acad Sci. 1997. pp. 12115–12120. [DOI] [PMC free article] [PubMed]
- Mak M, Guo J, Kung S. PairProSVM: protein subcellular localization based on local pairwise profile alignment and SVM. IEEE/ACM Transactions on Computational Biology and Bioinformatics. 2008;5(3):416–422. doi: 10.1109/TCBB.2007.70256. [DOI] [PubMed] [Google Scholar]
- Rangwala H, Karypis G. Profile-based direct kernels for remote homology detection and fold recognition. Bioinformatics. 2005;21(23):4239–4247. doi: 10.1093/bioinformatics/bti687. [DOI] [PubMed] [Google Scholar]
- Kuang R, Ie E, Wang K, Siddiqi M, Freund Y, Leslie C. Profile-based string kernels for remote homology detection and motif extraction. J Bioinform Comput Biol. 2005;3:527–550. doi: 10.1142/S021972000500120X. [DOI] [PubMed] [Google Scholar]
- Leslie C, Eskin E, Cohen A, Weston J, Noble W. Mismatch string kernels for discriminative protein classification. Bioinformatics. 2004;20(4):467–476. doi: 10.1093/bioinformatics/btg431. [DOI] [PubMed] [Google Scholar]
- Kuang R, Jianying Gu, Cai Hong, Wang Yufeng. Improved prediction of malaria degradomes by supervised learning with SVM and profile kernel. Genetica. 2009;136:189–209. doi: 10.1007/s10709-008-9336-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie C, Eskin E, Noble W. The spectrum kernel: a string kernel for SVM protein classification. Proc Pac Biocomput Symp. 2002;7:566–575. [PubMed] [Google Scholar]
- Mei S, Wang Fei. Amino acid classification based spectrum kernel fusion for protein subnuclear localization. BMC Bioinformatics. 2010;11(Suppl 1):S17. doi: 10.1186/1471-2105-11-S1-S17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Yanq J, Chou KC. Euk-PLoc: an ensemble classifier for large-scale eukaryotic protein subcellular location prediction. Amino Acids. 2007;33:57–67. doi: 10.1007/s00726-006-0478-8. [DOI] [PubMed] [Google Scholar]
- Chou KC, Shen HB. Euk-mPLoc: a fusion classifier for large-scale eukaryotic protein subcellular location prediction by incorporating multiple sites. J Proteome Res. 2007;6:1728–1734. doi: 10.1021/pr060635i. [DOI] [PubMed] [Google Scholar]
- Shen HB, Chou KC. Hum-mPLoc: an ensemble classifier for largescale human protein subcellular location prediction by incorporating samples with multiple sites. Biochem Biophys Res Commun. 2007;355:1006–1011. doi: 10.1016/j.bbrc.2007.02.071. [DOI] [PubMed] [Google Scholar]
- Chou KC, Shen HB. Cell-PLoc: A package of web-servers for predicting subcellular localization of proteins in various organisms. Nature Protocols. 2008;3:153–162. doi: 10.1038/nprot.2007.494. [DOI] [PubMed] [Google Scholar]
- Chou K, Cai Y. A new hybrid approach to predict subcellular localization of proteins by incorporating Gene Ontology. Biochem Biophys Res Commun. 2003;311:743–747. doi: 10.1016/j.bbrc.2003.10.062. [DOI] [PubMed] [Google Scholar]
- Huang W, Tunq C, Ho S, Hwang S, Ho S. ProLoc-GO: utilizing informative gene ontology terms for sequence-based prediction of protein subcellular localization. BMC Bioinformatics. 2008;9:80. doi: 10.1186/1471-2105-9-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Tung C, Huang H, Ho S. Predicting protein subnuclear localization using GO-amino-acid composition features. BioSystems. 2009. [DOI] [PubMed]
- Zdobnov EM, Apweiler R. InterProScan - an integration platform for the signature-recognition methods in InterPro. Bioinformatics. 2001;17:847–848. doi: 10.1093/bioinformatics/17.9.847. [DOI] [PubMed] [Google Scholar]
- Chou K, Cai Y. Prediction of protein subcellular locations by GO-FunD-PseAA predictor. Biochem Biophys Res Commun. 2004;320:1236–1239. doi: 10.1016/j.bbrc.2004.06.073. [DOI] [PubMed] [Google Scholar]
- Blum T, Briesemeister S, Kohlbacher O. MultiLoc2: integrating phylogeny and Gene Ontology terms improves subcellular protein localization prediction. BMC Bioinformatics. 2009;10:274. doi: 10.1186/1471-2105-10-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung T, Lee D. A method to improve protein subcellular localization prediction by integrating various biological data sources. BMC Bioinformatics. 2009;10(Suppl 1):S43. doi: 10.1186/1471-2105-10-S1-S43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Chuang H, Beyer A, Sung M, Huh W, Lee B, Ideker T. Protein networks markedly improve prediction of subcellular localization in multiple eukaryotic species. Nucleic Acids Research. 2008;36(20):e136. doi: 10.1093/nar/gkn619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM. et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Z, Dai Y. Assessing protein similarity with Gene Ontology and its use in subnuclear localization prediction. BMC Bioinformatics. 2006;7:491. doi: 10.1186/1471-2105-7-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai W, Yang Q, Xue G, Yu Y. Boosting for Transfer Learning. Proceedings of the 24 th International Conference on Machine Learning. 2007.
- Dai W, Chen Y, Xue G, Yang Q, Yu Y. Translated Learning: Transfer Learning across Different Feature Spaces. NIPS. 2008.
- Yang Q, Chen Y, Xue G, Dai W, Yu Y. Heterogeneous Transfer Learning for Image Clustering via the Social Web. Proceedings of the 47th Annual Meeting of the ACL and the 4th IJCNLP of the AFNLP. 2009. pp. 1–9.
- Pan S, Yang Q. A Survey on Transfer Learning. IEEE Transactions on Knowledge and Data Engineering. 2010;22(10):1345–1359. doi: 10.1109/TKDE.2009.191. [DOI] [Google Scholar]
- Alexander Z, Cheng S. Multiclass Multiple Kernel Learning. Proceedings of the 24th International Conference on Machine Learning.
- Apweiler R, Attwood T, Bairoch A, Bateman A, Birney E, Biswas M. et al. The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic Acids Research. 2001;29(1):37–40. doi: 10.1093/nar/29.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann K, Bucher P, Falquet L, Bairoch A. The Prosite Database, Its Status in 1999. Nucleic Acids Res. 1999;27(1):215–219. doi: 10.1093/nar/27.1.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attwood TK, Croning MD, Flower DR, Lewis AP, Mabey JE, Scordis P. et al. The Database Formerly Known as Prints. Nucleic Acids Res. 2000;28(1):225–227. doi: 10.1093/nar/28.1.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A, Birney E, Durbin R, Eddy SR, Howe KL, Sonnhammer EL. The Pfam Protein Families Database. Nucleic Acids Res. 2000;28(1):263–266. doi: 10.1093/nar/28.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpet F, Gouzy J, Kahn D. Recent Improvements of the Prodom Database of Protein Domain Families. Nucleic Acids Res. 1999;27(1):263–267. doi: 10.1093/nar/27.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz J, Copley RR, Doerks T, Ponting CP, Bork P. A Web-Based Tool for the Study of enetically Mobile Domains. Nucleic Acids Res. 2000;28(1):231–234. doi: 10.1093/nar/28.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haft DH, Loftus BJ, Richardson DL, Yang F, Eisen JA, Paulsen IT, White O. TIGRFAMs: a protein family resource for the functional identification of proteins. Nucleic Acids Res. 2001;29(1):41–3. doi: 10.1093/nar/29.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanckriet G, DeBie T, Cristianini N, Jordan M, Noble W. A statistical framework for genomic data fusion. Bioinformatics. 2004;20(16):2626–2635. doi: 10.1093/bioinformatics/bth294. [DOI] [PubMed] [Google Scholar]
- Hoglund A, Donnes P, Blum T, Adolph H, Kohlbacher O. MultiLoc: prediction of protein subcellular localization using N-terminal targeting sequences, sequence motifs and amino acid composition. Bioinformatics. 2006;22(10):1158–1165. doi: 10.1093/bioinformatics/btl002. [DOI] [PubMed] [Google Scholar]
- Pierleoni A, Luigi P, Fariselli P, Casadio R. BaCelLo: a balanced subcellular localization predictor. Bioinformatics. 2006;22(14):e408–e416. doi: 10.1093/bioinformatics/btl222. [DOI] [PubMed] [Google Scholar]
- Lu Z, Hunter L. GO molecular function terms are predictive of subcellular localization. Pac Symp Biocomput. 2005. pp. 151–61. full_text. [DOI] [PMC free article] [PubMed]
- Chou KC, Shen HB. Review: recent advances in developing web-servers for predicting protein attributes. Natural Science. 2009;2:63–92. doi: 10.4236/ns.2009.12011. http://www.scirp.org/journal/NS/ (openly accessible at) [DOI] [Google Scholar]