Abstract
Loss of p53 tumor suppressor function is a key event in the genesis of most human tumors. This observation has prompted efforts to restore p53 activity as an anticancer therapeutic approach. Recent developments that have extended our understanding of how p53 activity is regulated and how mutations disrupt that regulation have provided the insight needed to develop therapeutic strategies that take advantage of this knowledge. In this article, we review the strategies for restoring p53 function and some of the new compounds that show promise as antitumor agents in preclinical models.
Keywords: 37AA, apoptosis, CP-31398, mdm2, Nutlin, p63, p73, PRIMA-1
p53 functions & mechanisms of activation
The p53 protein is a tumor suppressor that sits at the crux of a web of signal transduction pathways that respond to stress and it is activated in response to genotoxic and non-genotoxic insults to the cell [1]. Stress signals induced by DNA damage, oncogene activation and hypoxia lead p53 to stimulate the expression of a set of downstream target genes that can induce apoptosis [2], facilitate DNA repair [3] or halt progression through the cell cycle [4]. Collectively, the activation of these pathways act to suppress genetic mutations and to guard against the emergence of hyperproliferative cells with a damaged genome. Not surprisingly, mutational inactivation of p53 is a frequent event in human tumors [5]. In fact, it has been estimated that 50% of all human tumors have sustained a mutation in the p53 gene, and it has been suggested that the p53 network is inactivated in tumors where the p53 gene remains unmutated. These observations have generated considerable interest in understanding how activation of p53 leads to suppression of tumorigenesis and has prompted efforts to find ways to restore p53 function in tumor cells that lack it.
The p53 protein is a transcription factor, and activation of its transactivation properties is essential for its tumor suppressor function. The crucial event in the activation of p53 is the release of p53 protein from its association with mdm2. mdm2 functions as an E3 ligase that promotes ubiquitinylation and proteosome-mediated degradation of p53, which keeps p53 protein levels low in unstressed cells [6]. The interaction between p53 and mdm2 is abolished when DNA damage, incurred by radiation or anticancer agents, results in activation of ATM and/or ATR. ATM and ATR are members of the PI3K-like kinase family that coordinate the signaling networks that are activated in response to DNA damage [7]. When activated, ATM phosphorylates p53 at serine 15 in the N-terminus, which interferes with mdm2 binding [8]. As a consequence, p53 stability is increased and the protein accumulates in the nucleus, leading to induction of p53-mediated gene expression. The power of mdm2 to suppress p53 activity is exemplified by the observation that, in some tumors, p53 is nonfunctional, even though the p53 gene remains wild-type, since the mdm2 protein is present at abnormally high levels [9]. Hence, mdm2 negatively regulates p53, and genotoxic stress alleviates this by suppressing interaction between the two proteins.
In human tumors, elimination of p53 function typically occurs as a consequence of genetic mutations that result in a nonfunctional protein. Cataloging of the p53 mutations in human tumors revealed that the mutations that occur most frequently are missense mutations that fall within the DNA binding domain of p53. These mutations typically change the identity of the amino acids that are directly involved in contacting DNA or result in a protein with an abnormal 3D structure [10]. Wild-type p53 exerts its tumor suppressor effect through its actions as a transcription factor. Hence, mutant proteins that are incapable of binding DNA lose the capacity to suppress tumorigenesis [11,12]. Similarly, proteins that have an abnormal structure are incapable of binding DNA and also lack tumor suppressor activity. These observations demonstrate that p53 exerts its effects primarily through its ability to interact with DNA and that the wild-type protein has a specific 3D structure that is critical for its function.
Which of the several activities of p53 are most responsible for tumor suppression? This argument continues to be debated; however, the evidence favors the ability of p53 to induce apoptosis as being crucial for its tumor suppressor activity. Consistent with this, knock-in mice expressing transcriptionally inactive but DNA binding-proficient p53 are defective for DNA damage-induced apoptosis and show a propensity for tumor development that is similar to p53-null mice [13]. Furthermore, overexpression of bcl-2, a potent antiapoptotic protein, leads to increased lymphomagenesis in mice, even though these tumors retain a wild-type p53 gene [14]. These and other studies indicate that loss of p53-mediated apoptosis enhances survival of cells that would otherwise be eliminated in response to cellular stresses. Importantly, loss of p53 function also has consequences for patient responses to anticancer therapy. The cytotoxic drugs typically used to treat human solid tumors activate p53-induced apoptosis as a result of the DNA damage they cause [15]. As a consequence, loss of p53 function can lead to chemoresistance since tumor cells are insensitive to the DNA damage caused by anticancer agents [16]. Hence, p53 is important both for the suppression of tumorigenesis and the response to chemotherapeutic treatment.
Although p53 was initially thought to be the only protein of its type, the discovery of two new family members, p63 and p73, has extended our understanding of p53-induced apoptosis. All three proteins share a similar structure that consists of an N-terminal transactivation domain, a central DNA binding domain and a C-terminal oligomerization domain, and the three family members share 60% of their identity in their DNA binding domains [17]. Both new family members function as transcription factors and can stimulate the expression of some of the same proapoptotic genes as p53, and both p63 and p73 can be activated by DNA-damaging agents, prompting speculation that these new family members may also be tumor suppressors [18]. However, in spite of their similarity to p53 in both structure and function, neither p63 nor p73 have been found to be mutated to any significant extent in human tumors. Nevertheless, the discovery of p63 and p73 provides an additional approach to restoring p53-related antitumor activity.
Strategies for restoring p53: taking advantage of what we know
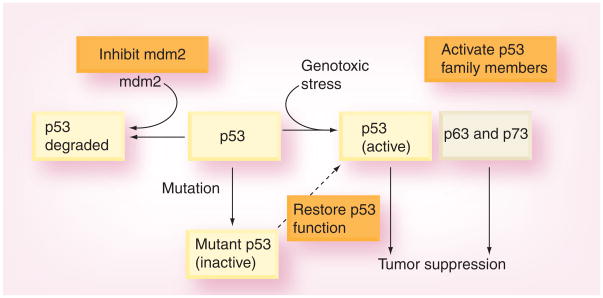
Since loss of p53 function is a key step in the development of tumors, it was reasoned that restoration of p53 activity might suppress or even reverse tumorigenesis. Indeed, reintroduction of wild-type p53 using a replication-defective adenoviral vector showed efficacy in reversing the growth of several human tumor types, demonstrating that restoration of p53 activity is a viable anticancer therapeutic approach [19]. However, as regulation of p53 activity becomes better understood, approaches that exploit our deeper understanding of the biochemistry of p53 activation have led to the identification of small molecules that can manipulate the endogenous nonfunctional protein that is so often expressed in tumor cells. As a result, strategies have focused on restoring wild-type activity to the mutant p53 protein, restoring functionality of the p53 pathway or activating one of the p53 family members (Figure 1).
Figure 1.
Strategies for restoring wild-type p53 function in human tumor cells.
Restoring wild-type activity
The overwhelming majority of genetic mutations that nullify p53 function are missense mutations that substitute one amino acid for another in the protein’s primary amino acid sequence. Most often these mutations result in a protein that has an abnormal structure and is incapable of sequence-specific DNA binding [20]. In addition, tumor cells frequently continue to express these misshapen p53 molecules, and these are often present at elevated levels. Hence, one promising strategy has focused on identifying small molecules that restore the wild-type conformation to mutant proteins by aiding in its refolding or in preventing abnormal folding in the first place. One such molecule, CP-31398 – a qinazoline-based molecule that was found to produce an active p53 in cancer cells – is postulated to act by increasing the thermodynamic stability of the mutant protein [21]. Treating cancer-derived cells in vitro with CP-31398 resulted in increased p53-mediated gene expression. Importantly, the drug also slowed xenograft tumor growth in mice, demonstrating that it could also suppress tumor cell growth in vivo. However, characterization of the ability of CP-31398 to rescue p53 function suggests that it may not act by binding the mutant p53 protein. Instead, it appears that CP-31398 stabilizes the wild-type conformation in a newly synthesized p53 protein [22].
By contrast, p53 reactivation and induction of massive apoptosis (PRIMA)-1 is a small molecule that binds covalently with thiol groups in mutant p53 and restores DNA-binding activity to some mutant p53 proteins [23]. PRIMA-1 preferentially suppresses the growth of tumor cell lines containing mutant p53, indicating that it functions by acting on mutant p53 [24]. PRIMA-1 (ARP-246) is currently in a Phase I clinical trial. Two other small molecules, ellipticine [25] and p53R3 [26], have also been shown to produce a functional p53 in tumor cells that express a mutant p53 protein. These results suggest that rescuing wild-type function from mutant proteins is a promising therapeutic approach.
A second strategy for rescuing wild-type p53 activity relies on the observation that the C-terminus of the protein can regulate sequence-specific DNA binding [27]. Two models have been proposed to explain this. In the steric hindrance model, the C-terminus of p53 is proposed to interact with DNA nonspecifically, thus blocking sequence-specific DNA binding by the core domain. In the second model, the C-terminus of p53 is thought to interact with the core domain, which locks the p53 tetramer into a configuration that is incapable of binding DNA [28]. In either case, DNA binding activity can be unblocked by deletion of 30 amino acids from the C-terminus or through interaction with the monoclonal antibody PAb421 or other cellular proteins such as 53BP2 that bind with the C-terminal domain of p53. Capitalizing on these observations, Friedler and coworkers synthesized a peptide derived from the region of 53BP2 that binds with the C-terminus of p53 (residues 490–498) and showed that it could stabilize a p53 mutant and increase its capacity to bind DNA [29]. Similarly, a peptide derived from residues 361–382 of the C-terminus of p53 itself was shown to activate sequence-specific DNA binding of wild-type p53 and restored DNA-binding in some p53 mutants [30]. These studies demonstrate that modulation of the COOH domain of p53 can activate DNA binding activity in some mutant p53 proteins.
Inhibiting the inhibitor: anti-mdm2 agents
As mentioned earlier, not all tumors have sustained a mutation in the p53 gene and instead have lost p53 function owing to nonmutational changes in the cell. For example, in some tumors, principally soft tissue sarcomas and some breast cancers, p53 remains wild-type, but the protein is nonfunctional because of the actions of mdm2 [31]. Hence, overexpression of mdm2 leads to reduced p53 levels and elimination of its tumor suppressor activities.
The key to regulation of p53 by mdm2 is the physical interaction between the two proteins. Structural analysis showed that a region in the N-terminal transactivation domain of p53 bound with mdm2 via a well-defined hydrophobic cleft in mdm2 [32]. Because the cleft in mdm2 is contacted by only three amino acids in p53 (Phe19, Leu26 and Trp23), it was reasoned that small molecules that mimicked these amino acids might be able to disrupt the p53–mdm2 complex in tumor cells where overexpression of mdm2 inactivates p53. Screening for compounds that disrupt the p53–mdm2 complex led to the identification of a group of compounds named Nutlins [33]. Biochemical analysis confirmed that Nutlins displaced p53 from the p53–mdm2 complex, and crystal structure analysis confirmed that Nutlins bind to the p53 binding site on mdm2. Treating tumor cells that express a wild-type p53 resulted in decreased p53 degradation and increased expression of downstream p53 target genes such as p21 and mdm2. Importantly, Nutlins had no effect on cells that lacked p53, demonstrating that their activity is manifested through wild-type p53. Nutlin-3 is currently in Phase I clinical trials.
Two other small molecules in this category are worth mentioning. TDP665759 is a benzodiazepienedione that interferes with the interaction between mdm2 and p53 and acts synergistically with doxorubicin in the suppression of xenograft tumor growth [34]. MI-219 is a spiro-oxindole that was designed to mimic the p53 residues that bind to mdm2 [35]. MI-219 binds to the p53 binding pocket in mdm2 and disrupts the mdm2–p53 complex, which leads to activation of p53, induction of growth arrest and apoptosis and suppression of xenograft tumor growth.
Alternative targets
Previous approaches for restoring p53 tumor suppressor function have all relied on the presence of a p53 protein. However, the discovery that the p53 family members p63 and p73 have similar structures and have similar biological activities has provided an additional antitumor strategy. Both p63 and p73 can induce apoptosis and do so by inducing some of the same proapoptotic targets that p53 activates [17]. Importantly, mutational inactivation of p63 and p73 is rare in human tumors and they are widely expressed, making these proteins attractive chemotherapeutic targets. Indeed, results from two recent studies demonstrate that targeting these proteins may be a useful anticancer approach. In one of these studies, a screen for small molecules by El-Deiry and coworkers that could activate apoptosis in p53-null cells isolated several agents, some of which were shown to function by activating p73 [36]. Several of the agents caused increased expression of p53 target genes such as DR5 and p21, and in some cases, this was associated with increased p73 protein levels. Moreover, knockdown of p73 neutralized induction of p53-responsive gene expression. Hence, at least some of the compounds stimulate the p53 pathway by stimulating p73-mediated transactivation of p53 target genes.
Similarly a p53-derived peptide termed 37AA was isolated, which could drive cell death through activation of p73. The amino acids in 37AA comprise the evolutionarily conserved domains II and III of p53 fused together in a single peptide, and 37AA evolved with the ability of p53 to induce transactivation-independent apoptosis. Testing demonstrated that 37AA was itself transcriptionally inert, but that it had the effect of stimulating the transactivation capability of p73. Screens for proteins that interact with p53 have identified a family of proteins, termed ASPP, which can augment the ability of p53 to stimulate the expression of proapoptotic genes [37]. One of these family members, iASPP, suppresses the activity of p53, p63 and p73 by interacting with their DNA binding domains [38]. Characterization of the activity of 37AA demonstrated that it functioned by interfering with iASPP binding with p73 and promoted its ability to stimulate the expression of proapoptotic genes such as PUMA and NOXA.
Conclusion
The reactivation of p53 in tumors holds great promise as an anticancer approach. Initial studies that demonstrated tumor cells could be killed by introducing a wild-type p53 validated this concept. However, as our understanding of the regulation of p53 and the p53 pathway has deepened, strategies based on this new knowledge have been developed that take advantage of the proteins that are already resident within the tumor cell.
Future perspective
Although restoration of p53 activity is a promising therapeutic approach, there are several issues that need to be resolved in order to maximize the efficacy of these therapies for future use in the clinic. One relates to the biological outcome (cell cycle arrest, apoptosis, senescence or differentiation) resulting from an activated p53, which appears to depend on the cell type and environment. A recent advance in this area occurred with the discovery of the ASPP family of proteins that bind with the DNA binding domain of p53 and augment its transactivation of proapoptotic genes [39]. Therefore, the presence or absence of these cofactors may be crucial in determining the success of p53-reactivating drugs in suppressing or reversing tumor growth. Nevertheless, it appears unlikely that p53reactivating drugs will be used as a monotherapy. Rather, it appears more likely that they will be used in conjunction with conventional DNA damage-inducing chemotherapeutic agents to increase sensitivity to these highly tested drugs. Indeed, Nutlins have been shown to act synergistically with other anticancer agents [40,41]. Consequently, future studies that identify the most effective combinations will be needed to bring the full promise of p53-restoring drugs to realization.
While most of the attention devoted to p53 has focused on the beneficial effects that restoring its activity would have, there is some surprising evidence indicating that chronic activation of p53 may be as deleterious as its inactivation. Hyperactivation of p53 has been associated with cell death in degenerative diseases such as arthritis, multiple sclerosis [42] and neuropathies [43], as well as in the ischemic damage that occurs from strokes or cardiac arrest [44]. In each of these conditions, it was found that apoptotic death leading to the destruction of key cell types was due to upregulation of the p53 pathway and that suppression of p53 function was protective. At present, it is uncertain whether the several new agents that attempt to promote p53 activity have these deleterious effects in whole animals. However, as p53-restoring drugs make their way into the clinic, it seems prudent to test for potential unintended effects in nontarget tissues that could be caused by p53-restoring drugs.
Footnotes
For reprint orders, please contact: reprints@futuremedicine.com
Financial & competing interests disclosure
Jesse D Martinex received the NIH grant CA090776 for the production of this work. The author has no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Bibliography
Papers of special note have been highlighted as:
▪ of interest
▪▪ of considerable interest
- 1.Vazquez A, Bond EE, Levine AJ, Bond GL. The genetics of the p53 pathway, apoptosis and cancer therapy. Nat Rev Drug Discov. 2008;7(12):979–987. doi: 10.1038/nrd2656. [DOI] [PubMed] [Google Scholar]
- 2.Fridman JS, Lowe SW. Control of apoptosis by p53. Oncogene. 2003;22(56):9030–9040. doi: 10.1038/sj.onc.1207116. [DOI] [PubMed] [Google Scholar]
- 3.Sengupta S, Harris CC. p53: traffic cop at the crossroads of DNA repair and recombination. Nat Rev Mol Cell Biol. 2005;6(1):44–55. doi: 10.1038/nrm1546. [DOI] [PubMed] [Google Scholar]
- 4.Giono LE, Manfredi JJ. The p53 tumor suppressor participates in multiple cell cycle checkpoints. J Cell Physiol. 2006;209(1):13–20. doi: 10.1002/jcp.20689. [DOI] [PubMed] [Google Scholar]
- 5.Hollstein M, Rice K, Greenblatt MS, et al. Database of p53 gene somatic mutations in human tumors and cell lines. Nucleic Acids Res. 1994;22(17):3551–3555. [PMC free article] [PubMed] [Google Scholar]
- 6.Marine JC, Lozano G. mdm2-mediated ubiquitylation: p53 and beyond. Cell Death Differ. 17(1):93–102. doi: 10.1038/cdd.2009.68. [DOI] [PubMed] [Google Scholar]
- 7.Lavin MF. ATM and the Mre11 complex combine to recognize and signal DNA double-strand breaks. Oncogene. 2007;26(56):7749–7758. doi: 10.1038/sj.onc.1210880. [DOI] [PubMed] [Google Scholar]
- 8.Canman CE, Lim DS, Cimprich KA, et al. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science. 1998;281(5383):1677–1679. doi: 10.1126/science.281.5383.1677. [DOI] [PubMed] [Google Scholar]
- 9▪▪.Oliner JD, Kinzler KW, Meltzer PS, George DL, Vogelstein B. Amplification of a gene encoding a p53-associated protein in human sarcomas. Nature. 1992;358(6381):80–83. doi: 10.1038/358080a0. First observation in human tumors that p53 activity can be suppressed by overexpression of mdm2. [DOI] [PubMed] [Google Scholar]
- 10.Cho Y, Gorina S, Jeffrey PD, Pavletich NP. Crystal structure of a p53 tumor suppressor–DNA complex: understanding tumorigenic mutations. Science. 1994;265(5170):346–355. doi: 10.1126/science.8023157. [DOI] [PubMed] [Google Scholar]
- 11.Raycroft L, Wu HY, Lozano G. Transcriptional activation by wild-type but not transforming mutants of the p53 anti-oncogene. Science. 1990;249(4972):1049–1051. doi: 10.1126/science.2144364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fields S, Jang SK. Presence of a potent transcription activating sequence in the p53 protein. Science. 1990;249(4972):1046–1049. doi: 10.1126/science.2144363. [DOI] [PubMed] [Google Scholar]
- 13.Jimenez GS, Nister M, Stommel JM, et al. A transactivation-deficient mouse model provides insights into Trp53 regulation and function. Nat Genet. 2000;26(1):37–43. doi: 10.1038/79152. [DOI] [PubMed] [Google Scholar]
- 14.Schmitt CA, Fridman JS, Yang M, Baranov E, Hoffman RM, Lowe SW. Dissecting p53 tumor suppressor functions in vivo. Cancer Cell. 2002;1(3):289–298. doi: 10.1016/s1535-6108(02)00047-8. [DOI] [PubMed] [Google Scholar]
- 15▪.Gasco M, Crook T. p53 family members and chemoresistance in cancer: what we know and what we need to know. Drug Resist Updat. 2003;6(6):323–328. doi: 10.1016/j.drup.2003.11.001. Summary of the role that the activity of p53 and p53 family members play in determining resistance to chemotherapeutic agents. [DOI] [PubMed] [Google Scholar]
- 16.Lowe SW, Bodis S, McClatchey A, et al. p53 status and the efficacy of cancer therapy in vivo. Science. 1994;266:807–810. doi: 10.1126/science.7973635. [DOI] [PubMed] [Google Scholar]
- 17.Pietsch EC, Sykes SM, Mcmahon SB, Murphy ME. The p53 family and programmed cell death. Oncogene. 2008;27(50):6507–6521. doi: 10.1038/onc.2008.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flores ER, Tsai KY, Crowley D, et al. p63 and p73 are required for p53-dependent apoptosis in response to DNA damage. Nature. 2002;416(6880):560–564. doi: 10.1038/416560a. [DOI] [PubMed] [Google Scholar]
- 19.Gabrilovich DI. Ingn 201 (advexin): adenoviral p53 gene therapy for cancer. Expert Opin Biol Ther. 2006;6(8):823–832. doi: 10.1517/14712598.6.8.823. [DOI] [PubMed] [Google Scholar]
- 20▪.Joerger AC, Fersht AR. Structure–function–rescue: the diverse nature of common p53 cancer mutants. Oncogene. 2007;26(15):2226–2242. doi: 10.1038/sj.onc.1210291. Review of how p53 gene mutations impact the 3D structure of the p53 protein and how this provides the rational basis for the design of drugs aimed at restoring normal function. [DOI] [PubMed] [Google Scholar]
- 21▪▪.Foster BA, Coffey HA, Morin MJ, Rastinejad F. Pharmacological rescue of mutant p53 conformation and function. Science. 1999;286(5449):2507–2510. doi: 10.1126/science.286.5449.2507. Describes the isolation of CP-31398 and the characterization of how it interacts with mutant p53 proteins. [DOI] [PubMed] [Google Scholar]
- 22▪▪.Rippin TM, Bykov VJ, Freund SM, Selivanova G, Wiman KG, Fersht AR. Characterization of the p53-rescue drug CP-31398 in vitro and in living cells. Oncogene. 2002;21(14):2119–2129. doi: 10.1038/sj.onc.1205362. Demonstrates that CP-31398 acts by stabilizing the wild-type conformation in newly synthesized p53. [DOI] [PubMed] [Google Scholar]
- 23▪▪.Lambert JM, Gorzov P, Veprintsev DB, et al. PRIMA-1 reactivates mutant p53 by covalent binding to the core domain. Cancer Cell. 2009;15(5):376–388. doi: 10.1016/j.ccr.2009.03.003. Demonstrates that p53 reactivation and induction of massive apoptosis (PRIMA)-1 deactivates mutant p53 by modification of thiol groups on the protein. [DOI] [PubMed] [Google Scholar]
- 24.Bykov VJ, Issaeva N, Shilov A, et al. Restoration of the tumor suppressor function to mutant p53 by a low-molecular-weight compound. Nat Med. 2002;8(3):282–288. doi: 10.1038/nm0302-282. [DOI] [PubMed] [Google Scholar]
- 25.Garbett NC, Graves DE. Extending nature’s leads: the anticancer agent ellipticine. Curr Med Chem Anticancer Agents. 2004;4(2):149–172. doi: 10.2174/1568011043482070. [DOI] [PubMed] [Google Scholar]
- 26.Sugikawa E, Hosoi T, Yazaki N, Gamanuma M, Nakanishi N, Ohashi M. Mutant p53 mediated induction of cell cycle arrest and apoptosis at G1 phase by 9-hydroxyellipticine. Anticancer Res. 1999;19(4B):3099–3108. [PubMed] [Google Scholar]
- 27.Selivanova G, Kawasaki T, Ryabchenko L, Wiman KG. Reactivation of mutant p53: a new strategy for cancer therapy. Semin Cancer Biol. 1998;8(5):369–378. doi: 10.1006/scbi.1998.0099. [DOI] [PubMed] [Google Scholar]
- 28.Hupp TR, Meek DW, Midgley CA, Lane DP. Regulation of the specific DNA binding function of p53. Cell. 1992;71(5):875–886. doi: 10.1016/0092-8674(92)90562-q. [DOI] [PubMed] [Google Scholar]
- 29.Friedler A, Hansson LO, Veprintsev DB, et al. A peptide that binds and stabilizes p53 core domain: chaperone strategy for rescue of oncogenic mutants. Proc Natl Acad Sci USA. 2002;99(2):937–942. doi: 10.1073/pnas.241629998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30▪▪.Selivanova G, Iotsova V, Okan I, et al. Restoration of the growth suppression function of mutant p53 by a synthetic peptide derived from the p53 C-terminal domain. Nat Med. 1997;3(6):632–638. doi: 10.1038/nm0697-632. Describes the biological effects of a peptide derived from the C-terminus of p53 on the functional activation of mutant p53 proteins. [DOI] [PubMed] [Google Scholar]
- 31.Momand J, Jung D, Wilczynski S, Niland J. The mdm2 gene amplification database. Nucleic Acids Res. 1998;26(15):3453–3459. doi: 10.1093/nar/26.15.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kussie PH, Gorina S, Marechal V, et al. Structure of the mdm2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science. 1996;274(5289):948–953. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- 33▪▪.Vassilev LT. Small-molecule antagonists of p53–mdm2 binding: research tools and potential therapeutics. Cell Cycle. 2004;3(4):419–421. Describes the isolation of Nutlins and how their binding to p53 interferes with its interaction with mdm2. [PubMed] [Google Scholar]
- 34.Koblish HK, Zhao S, Franks CF, et al. Benzodiazepinedione inhibitors of the Hdm2:p53 complex suppress human tumor cell proliferation in vitroand sensitize tumors to doxorubicin in vivo. Mol Cancer Ther. 2006;5(1):160–169. doi: 10.1158/1535-7163.MCT-05-0199. [DOI] [PubMed] [Google Scholar]
- 35.Shangary S, Qin D, McEachern D, et al. Temporal activation of p53 by a specific mdm2 inhibitor is selectively toxic to tumors and leads to complete tumor growth inhibition. Proc Natl Acad Sci USA. 2008;105(10):3933–3938. doi: 10.1073/pnas.0708917105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang W, Kim SH, El-Deiry WS. Small-molecule modulators of p53 family signaling and antitumor effects in p53-deficient human colon tumor xenografts. Proc Natl Acad Sci USA. 2006;103(29):11003–11008. doi: 10.1073/pnas.0604507103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bergamaschi D, Samuels Y, Jin B, Duraisingham S, Crook T, Lu X. ASPP1 and ASPP2: common activators of p53 family members. Mol Cell Biol. 2004;24(3):1341–1350. doi: 10.1128/MCB.24.3.1341-1350.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robinson RA, Lu X, Jones EY, Siebold C. Biochemical and structural studies of ASPP proteins reveal differential binding to p53, p63, and p73. Structure. 2008;16(2):259–268. doi: 10.1016/j.str.2007.11.012. [DOI] [PubMed] [Google Scholar]
- 39.Slee EA, Lu X. The ASPP family: deciding between life and death after DNA damage. Toxicol Lett. 2003;139(2–3):81–87. doi: 10.1016/s0378-4274(02)00421-6. [DOI] [PubMed] [Google Scholar]
- 40.Secchiero P, Zerbinati C, Di Iasio MG, et al. Synergistic cytotoxic activity of recombinant trail plus the non-genotoxic activator of the p53 pathway Nutlin-3 in acute myeloid leukemia cells. Curr Drug Metab. 2007;8(4):395–403. doi: 10.2174/138920007780655432. [DOI] [PubMed] [Google Scholar]
- 41.Ribas J, Boix J, Meijer L. (R)-roscovitine (CYC202, Seliciclib) sensitizes SH-SY5Y neuroblastoma cells to Nutlin-3-induced apoptosis. Exp Cell Res. 2006;312(12):2394–2400. doi: 10.1016/j.yexcr.2006.04.021. [DOI] [PubMed] [Google Scholar]
- 42.Wosik K, Antel J, Kuhlmann T, Bruck W, Massie B, Nalbantoglu J. Oligodendrocyte injury in multiple sclerosis: a role for p53. J Neurochem. 2003;85(3):635–644. doi: 10.1046/j.1471-4159.2003.01674.x. [DOI] [PubMed] [Google Scholar]
- 43.Mattson MP, Duan W, Pedersen WA, Culmsee C. Neurodegenerative disorders and ischemic brain diseases. Apoptosis. 2001;6(1–2):69–81. doi: 10.1023/a:1009676112184. [DOI] [PubMed] [Google Scholar]
- 44▪.Komarova EA, Gudkov AV. Chemoprotection from p53-dependent apoptosis: potential clinical applications of the p53 inhibitors. Biochem Pharmacol. 2001;62(6):657–667. doi: 10.1016/s0006-2952(01)00733-x. Describes a novel clinical use for p53 inhibitors. [DOI] [PubMed] [Google Scholar]