Abstract
Autophagy mediates the degradation of cytoplasmic components in eukaryotic cells and plays a key role in immunity. The mechanism of autophagosome formation is not clear. Here we examined two potential membrane sources for antibacterial autophagy: the ER and mitochondria. DFCP1, a marker of specialized ER domains known as ‘omegasomes,’ associated with Salmonella-containing autophagosomes via its PtdIns(3)P and ER-binding domains, while a mitochondrial marker (cytochrome b5-GFP) did not. Rab1 also localized to autophagosomes, and its activity was required for autophagosome formation, clearance of protein aggregates and peroxisomes, and autophagy of Salmonella. Overexpression of Rab1 enhanced antibacterial autophagy. The role of Rab1 in antibacterial autophagy was independent of its role in ER-to-Golgi transport. Our data suggest that antibacterial autophagy occurs at omegasomes and reveal that the Rab1 GTPase plays a crucial role in mammalian autophagy.
Key words: autophagy, DFCP1, Rab1, Salmonella, ER-to-golgi trafficking
Introduction
Macroautophagy (hereafter referred to as autophagy) targets long-lived proteins and organelles for lysosomal degradation in eukaryotic cells. During this process, portions of the cytoplasm are sequestered into a compartment called an autophagosome, which then fuses with lysosomes. Recently, autophagy has been linked to a variety of disease states, including cancer, myopathies, neurodegeneration, infection and inflammation.1,2
The source of the autophagosomal membrane in both yeast and mammalian cells is not clear. A recent study indicates that in mammalian cells the mitochondria provides membrane for autophagosome formation during starvation in mammalian cells.3 In contrast, other studies have suggested that the endoplasmic reticulum (ER) may be involved.4,5 Studies in yeast have also suggested a requirement for regions near the mitochondria6 and for ER-to-Golgi trafficking in autophagosome formation.7 However, the mechanism of autophagosome formation may be distinct or more complex in mammalian cells than in yeast cells. Specialized phosphatidylinositol 3-phosphate (PtdIns(3)P)-enriched regions of the mammalian ER called ‘omegasomes’ are thought to act as membrane scaffolds for the assembly of autophagy proteins.4 Utilizing 3D electron tomography, two recent studies have demonstrated a physical connection between the ER and the phagophore, the precursor to autophagosomes.8,9 Together, these findings suggest that specialized domains of the ER are involved in mammalian autophagy, although a direct role for ER-to-Golgi trafficking has not been investigated.
Rab1 (Ypt1 in yeast) is a small GTPase that plays a well-established role in mediating ER-to-Golgi protein transport in both yeast and mammalian cells.10–13 Specifically, this GTPase recruits effector proteins to budding COPII vesicles at the ER, forming cis-SNARE complexes that promote targeting to and fusion of these vesicles with the cis-Golgi.14,15 Rab1 is also involved in COPI vesicle formation and other distinct transport pathways, including ER-to-Golgi intermediate compartment (ERGIC)-to-cell periphery trafficking.16,17 We recently demonstrated that Ypt1 is required for specific and nonspecific forms of autophagy in yeast,18 however the role of Rab1 in mammalian autophagy has not been tested.
Autophagy plays an important role in both innate and adaptive immune responses.19,20 Autophagy targets a number of intracellular pathogens during infection.2 For example, Salmonella enterica serovar Typhimurium (S. typhimurium) is a bacterium that typically occupies a membrane-bound compartment within host cells called the Salmonella-containing vacuole (SCV) that permits intracellular bacterial growth.21 However, a population of intracellular S. typhimurium damages the SCV and this population is thought to be targeted by autophagy, which protects the cytosol from bacterial colonization.22 Furthermore, autophagy restricts intracellular growth of these bacteria in Dictyostelium discoideum and Caenorhabditis elegans infection models.23 Autophagy of bacteria, including possibly S. typhimurium, is thought to be impaired in some individuals with inflammatory bowel disease.24–29
The regulation of antimicrobial autophagy has some mechanistic differences from starvation-induced autophagy30 and it is unclear whether the two forms of autophagy utilize the same source(s) of membrane. Here we examined two potential membrane sources for antibacterial autophagy: the ER and mitochondria. We demonstrate that PtdIns(3)P-enriched domains of the ER are sites where antimicrobial autophagy occurs. We also demonstrate a novel role for Rab1 in antimicrobial autophagy, as well as other forms of autophagy in mammalian cells.
Results
DFCP1 associates with bacteria-containing autophagosomes via its PtdIns(3)P and ER-binding domains.
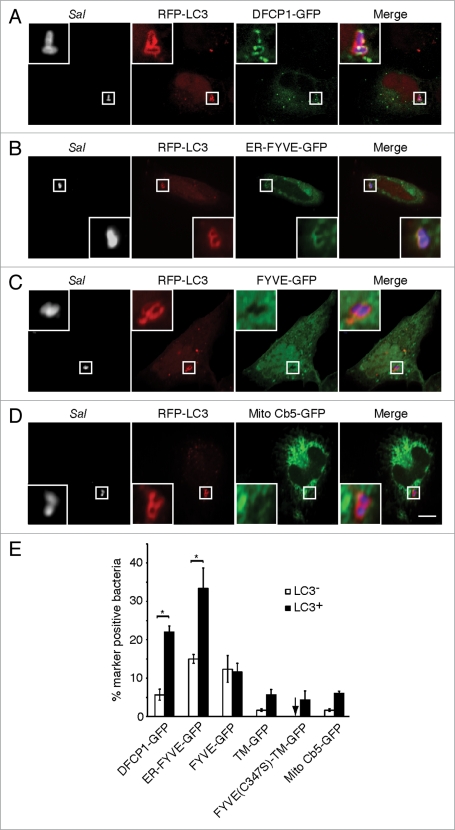
Omegasomes are involved in starvation-induced autophagosome formation in mammalian cells.4 A double FYVE domain-containing protein (DFCP1) is a marker of omegasomes. It targets to the ER via its transmembrane (TM) domain. To determine if omegasomes contribute to autophagy of S. typhimurium, we transfected cells with a GFP-fusion to DFCP1 (DFCP1-GFP) prior to infection with S. typhimurium for 1 h (the time shown to be maximal for autophagy of these bacteria22). Cells were also transfected with a red fluorescent protein-labeled autophagosome marker, microtubule-associated protein 1 light chain 3 (RFP-LC3) to follow autophagosome formation.31 DFCP1-GFP colocalized significantly more with the population of bacteria targeted by autophagy (LC3+) compared to LC3− bacteria (Fig. 1A, E and S1A). Similar colocalization with LC3+ bacteria was observed with an ER-FYVE-GFP construct, which mimics DFCP1 in that it contains both PtdIns(3)P and ER-binding domains4 (Fig. 1B and E). Mutation of a cysteine residue critical for PtdIns(3)P-binding (C347S) in the FYVE domain of DFCP1 prevented colocalization of the mutant DFCP1 construct, FYVE-(C347S)-TM-GFP, with LC3+ bacteria, despite the fact that this construct was targeted to the ER membrane via its TM domain (Fig. 1E and S1B). A PtdIns(3)P-binding FYVE domain alone (FYVE-GFP) (Fig. 1C and E) associated with both LC3+ and LC3− bacteria. However, it should be noted that normal SCV maturation includes PtdIns(3)P production by a RAB5-VPS34 complex.32 An ER-directed transmembrane domain (TM-GFP) did associate with LC3+ bacteria but at relatively low levels (approx. 5%) (Fig. 1E and S1C). These findings demonstrate that DFCP1 associates with bacteria-containing autophagosomes via both its PtdIns(3)P and ER-binding domains.
Figure 1.
DFCP1 associates with bacteria-containing autophagosomes via its PtdIns(3) P and ER-binding domains. HeLa cells were co-transfected with RFP-LC3 and either DFCP1-GFP (A) ER-FYVE-GFP (B) FYVE-GFP (C) or Mito Cb5-GFP (D). Cells were infected with S. typhimurium (Sal) for 1 h and immunostained with a polyclonal antibody to S. typhimurium. Representative confocal z-slices are shown, with the insets representing a higher magnification of the boxed area. Size bar, 10 µm. The percentage of LC3− or LC3+ bacteria co-localizing with each indicated marker was determined by fluorescence microscopy and quantified in (E). Asterisks indicate a significant difference, p < 0.05.
A mitochondrial marker (mitochondrial cytochrome b5-GFP; Mito Cb5-GFP) associates with starvation-induced autophagosomes in mammalian cells.33 Mito Cb5-GFP colocalized at low levels with LC3+ bacteria (approx. 5%), indicating that this organelle has only a minor contribution, if any, to the formation of Salmonella-containing autophagosomes (Fig. 1D and E). Together, these findings suggest that antimicrobial autophagy occurs at PtdIns(3)P-enriched domains of the ER, and are consistent with our previous observation that pharmacological inhibition of PI3-kinases blocks autophagy of S. typhimurium.22
Rab1 is required for autophagy of S. typhimurium.
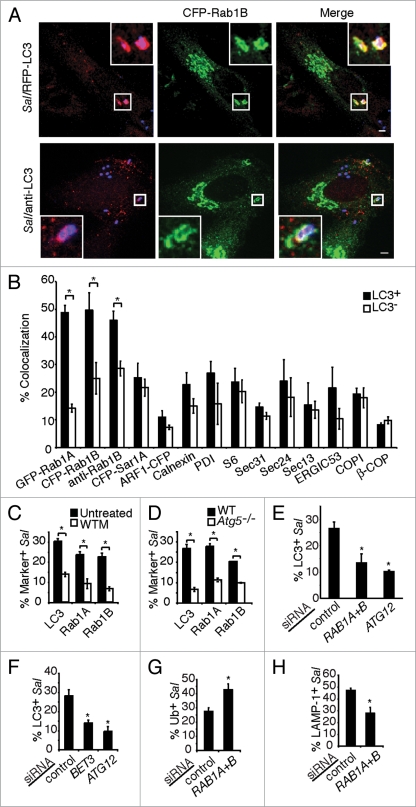
To further characterize the contribution of the ER or ER-derived membrane to antibacterial autophagy, we examined the localization of early secretory pathway markers with autophagy-targeted bacteria. We observed that both fluorescently labeled isoforms of Rab1A and Rab1B colocalized significantly more with LC3+ bacteria than LC3− bacteria (Fig. 2A and B). Similarly, Rab1 also colocalized with endogenous LC3 on bacteria (Fig. 2A). Rab1 colocalization with LC3+ S. typhimurium was confirmed with polyclonal antibodies to endogenous Rab1B (Fig. 2B). None of the other markers examined, including other GTPases involved in ER-to-Golgi trafficking (Sar1A and ARF1), significantly accumulated on LC3+ S. typhimurium (Fig. 2B and S2A).
Figure 2.
Rab1 is involved in autophagy of S. typhimurium. (A) HeLa cells were co-transfected with RFP-LC3 and CFP-Rab1B (upper part) or transfected with CFP-Rab1B alone and immunostained with a rabbit polyclonal antibody against endogenous LC3 (anti-LC3) (lower part), then infected with S. typhimurium for 1 h. Magnified images of boxed areas are shown in the upper right or lower left corners, indicating LC3+ S. typhimurium (Sal) labeling with CFP-Rab1B. Size bars, 5 µm. (B) Quantification of fluorescently-labeled GTPase constructs or immunostaining of indicated protein markers with RFP-LC3+ or LC3− bacteria. (C) Cells were transfected and infected as in (A) and either treated or not treated with wortmannin (WTM) for 30 min prior to infection. The percentage of bacteria positive for the indicated constructs was quantified. (D) Wild-type or Atg5−/− MEFs were transfected and infected as in (A). The percentage of bacteria positive for the indicated constructs was quantified as in (C). Asterisks indicate a significant difference, p < 0.05. (E) HeLa cells were treated with control, RAB1 and ATG12 siRNA or (F) BET3 siRNA, transfected with GFP-LC3 and infected with S. typhimurium expressing RFP for 1 h. The percentage of LC3+ bacteria was quantified. (G) Cells were treated with siRNA and infected as in (E), and then stained for ubiquitinated proteins. The percentage of S. typhimurium colocalizing with ubiquitinated proteins was quantified as in (E). (H) Cells were treated with siRNA and infected as in (E) and stained for the vacuole marker LAMP-1. The percentage of LAMP-1+ bacteria was quantified as in (E). Asterisks indicate a significant difference from control siRNA levels, p < 0.05.
To determine if Rab1 localized to bacteria as a result of autophagy, we examined conditions under which autophagy of S. typhimurium is blocked.22 Rab1 recruitment to bacteria was inhibited by the autophagy inhibitor wortmannin similar to that observed for LC3 recruitment (Fig. 2C). Mouse embryonic fibroblasts (MEFs) lacking the essential autophagy factor Atg534 demonstrated an inhibition of Rab1 and LC3 recruitment to S. typhimurium (Fig. 2D). Live imaging during S. typhimurium infection of HeLa cells showed that GFP-Rab1B and RFP-LC3 are recruited concomitantly to bacteria (Fig. S3 and Suppl. Movie 1). Therefore, Rab1 recruitment to S. typhimurium is specifically associated with autophagy and does not occur as a result of normal SCV maturation. Furthermore, Rab1 localized to DFCP1+ and LC3+ bacteria (Suppl. Movie 2), suggesting that Rab1 acts at PtdIns(3)P-enriched domains of the ER during autophagy of S. typhimurium.
Figure 3.
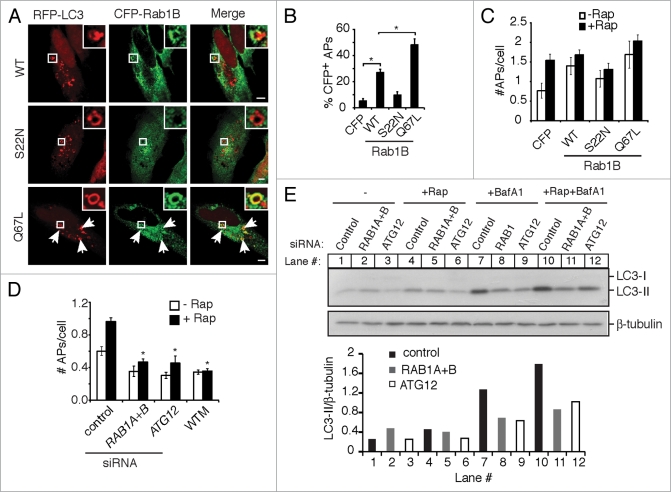
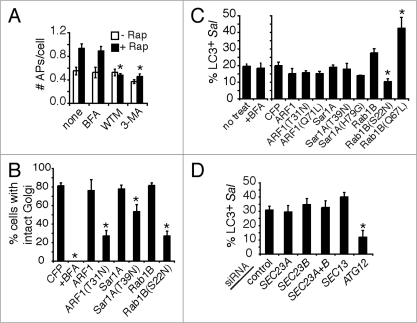
Rab1 is associated with autophagosomes and its activity is required for their formation. (A) HeLa cells were co-transfected with RFP-LC3 and the indicated CFP-Rab1B constructs, then treated with rapamycin for 2 h to induce autophagy. Magnified images of boxed areas are shown in the upper right corners. Arrows indicate further instances of CFP-Rab1B(Q67L) colocalization with autophagosomes. Size bars, 5 µm. (B) Quantification of the colocalization of CFP-Rab1B constructs with autophagosomes (APs) (RFP-LC3+ round vacuoles with clearly visible hollow centers). Asterisks indicate a significant difference, p < 0.05. (C) Cells were transfected as in (A) and either left untreated (−Rap) or treated with rapamycin for 2 h (+Rap). The average number of autophagosomes (APs) per transfected cell was quantified. (D) HeLa cells treated with the indicated siRNAs were transfected with GFP-LC3 and treated with (+) or without (−) rapamycin for 2 h. The number of autophagosomes (APs) per cell was quantified. In a parallel experiment, cells were treated with the autophagy inhibitor WTM during rapamycin treatment. Asterisks indicate a significant difference from control siRNA levels (+Rap), p < 0.05. (E) HeLa cells treated with the indicated siRNAs were incubated in the absence (−) or presence of rapamycin (+Rap), in the presence of bafilomycin A1 (BafA1) alone or rapamycin and bafilomycin A1 together (+Rap+BafA1) for 2 h. Then cells were lysed, harvested and analyzed by western blot. Endogenous LC3-I and LC3-II were detected by a rabbit polyclonal antibody. β-tubulin serves as a loading control. Densitometry was performed using ImageJ software and the ratio of LC3-II to β-tubulin density is plotted below.
To test if Rab1 is required for autophagy of bacteria, HeLa cells were treated with siRNA to target expression of both RAB1 isoforms (Fig. S2B) and then infected with S. typhimurium for 1 h. RAB1 siRNA reduced the percentage of LC3+ bacteria to a level similar to that seen with ATG12 siRNA treatment (Fig. 2E). Transport protein particle (TRAPP) complexes act as guanine nucleotide exchange factors (GEFs) to activate Ypt1 in yeast35–37 and play an important role in autophagy in this organism.18 Treatment with siRNA targeting BET3, which encodes a conserved component of TRAPP complexes,38,39 significantly reduced the percentage of LC3+ bacteria at 1 h (Fig. 2F). These results suggest that Rab1 is necessary for efficient autophagy of S. typhimurium.
Autophagy of S. typhimurium protects the cytoplasm from bacterial colonization. 22 Therefore, in autophagy-deficient cells or in cells treated with autophagy inhibitors, more bacteria escape into the cytoplasm to become associated with ubiquitinated proteins40 and less bacteria are retained in vacuoles.22,41 RAB1 siRNA resulted in significantly more S. typhimurium in the cytosol (marked by bacterial association with ubiquitinated proteins) at 1 h post infection compared to control siRNA (Fig. 2G). Furthermore, RAB1 siRNA resulted in less bacteria retained in lysosome-associated membrane protein-1 (LAMP-1)+ vacuoles compared to control siRNA (Fig. 2H). We further examined the role of Rab1 in bacterial degradation. We found that overall intracellular bacterial replication at 8 h post infection in cells treated with RAB1 siRNA was relatively less when compared to cells treated with control siRNA (Fig. S4A and B). This is not surprising as RAB1 knockdown not only inhibits autophagy, but also impairs ER-to-Golgi trafficking. After invasion, the majority of S. typhimurium are within the LAMP-1+ vacuoles and undergo rapid replication. Intracellular growth of S. typhimurium requires a normal Golgi network, and that Golgi disruption leads to an inhibition of bacterial replication.42 Therefore, RAB1 knockdown causes more cytosolic bacterial growth by impairing autophagy, but leads to an inhibited replication of the bacteria in vacuoles. As a net result, the overall intracellular bacterial growth is less in RAB1 siRNA-treated cells than control siRNA-treated cells. Altogether, Rab1 appears to be required for autophagy of S. typhimurium and protection of the cytosol from bacterial colonization.
Rab1 is associated with rapamycin-induced autophagosomes and its activity is required for their formation.
We next investigated the involvement of Rab1 in other forms of autophagy. HeLa cells were treated with the mTOR inhibitor rapamycin to induce autophagy.43 To distinguish autophagosomes from other nonspecific LC3+ structures,44 we only counted visibly hollow LC3+ rings as autophagosomes. Rab1B(WT) colocalized with rapamycin-induced autophagosomes (Fig. 3A and B). Rab1B(Q67L), which cannot hydrolyze GTP and thus acts as a constitutively active construct, demonstrated an even greater degree of colocalization with autophagosomes. However, dominant negative Rab1B(S22N) showed little association with rapamycin-induced autophagosomes (Fig. 3A and B). Overexpression of Rab1B(WT) and Rab1B(Q67L) increased the number of autophagosomes per cell compared to overexpression of a CFP vector even in the absence of rapamycin (Fig. 3C). Expression of Rab1B(S22N) reduced rapamycin-induced autophagosome formation, although the reduction was not statistically significant (Fig. 3C). Furthermore, RAB1 siRNA significantly decreased the number of autophagosomes formed per cell compared to control siRNA, and this effect was comparable to that seen with siRNA to ATG12 or in the presence of wortmannin (Fig. 3C). In order to determine the effect of Rab1 on autophagic flux, we examined the amount of LC3-II, which is the phosphatidylethanolamine (PE)-conjugated form of LC3,31 in the presence or absence of the lysosomal inhibitor bafilomycin A1. As shown in Figure 3E, in the absence of bafilomycin A1, rapamycin increased the amount of LC3-II in control siRNA-treated cells, but not in RAB1 or ATG12 siRNA-treated cells. In the presence of bafilomycin A1 alone, the amount of LC3-II reflects the basal level of autophagy. In this condition, more LC3-II accumulated in cells treated with control siRNA than RAB1 siRNA, suggesting that RAB1 knockdown decreases basal autophagy, similar to the effect of ATG12 knockdown (Fig. 3E). When bafilomycin A1 and rapamycin were added in the cells together, the amount of LC3-II increased more in control siRNA-treated cells than in RAB1 or ATG12 siRNA-treated cells (Fig. 3E). Altogether, these results indicate that Rab1 plays an important role in rapamycin-induced autophagic flux, specifically in autophagosome formation.
Rab1 is required for autophagy-mediated clearance of ubiquitinated protein aggregates.
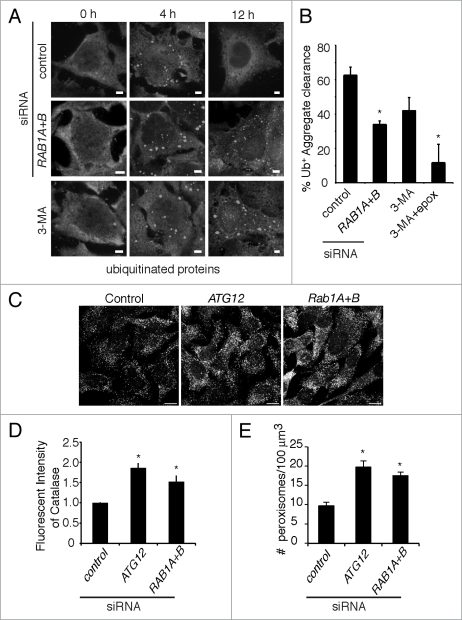
Aggregates of ubiquitinated proteins are specific cargo for macroautophagy in mammalian cells.45 These aggregates can form in response to cellular stress and store misfolded proteins prior to their ubiquitination and degradation.46 These aggregates are cleared synergistically through autophagy-dependent and proteasome-dependent mechanisms.47 Aggregate formation can be induced by treatment with puromycin, a translational inhibitor that induces premature polypeptide chain termination and the subsequent accumulation of misfolded proteins.47 In our aggregate clearance assay, HeLa cells were pulsed with puromycin for 4 h to induce aggregate formation (marked by staining for ubiquitinated proteins). The drug was then removed by extensive washing, followed by an 8 h chase to observe aggregate clearance. As shown in Figure 4A and B, ∼61% of cells treated with control siRNA were able to clear aggregates by 8 h after puromycin removal. Consistent with previous results,47 autophagy inhibition with 3-methyladenine (3MA) reduced aggregate clearance, and inhibition of both autophagy and the proteasome (by epoxomicin treatment) almost completely blocked the clearance of these structures (Fig. 4B). Treatment of cells with siRNA to both isoforms of RAB1 resulted in a reduction in aggregate clearance similar to that observed in the presence of 3MA (Fig. 4A and B). These results are consistent with a role for Rab1 in autophagy-dependent clearance of ubiquitinated protein aggregates in mammalian cells.
Figure 4.
Rab1 is required for autophagy-mediated clearance of ubiquitinated protein aggregates and peroxisomes. (A) HeLa cells were treated with the indicated siRNAs. Puromycin was added to the cells for 4 h to induce ubiquitinated protein aggregate formation, followed by extensive washing. Aggregate clearance was followed for a further 8 h (12 h total). Cells were fixed and stained for ubiquitinated proteins (with the FK2 antibody) to visualize aggregates. In a parallel experiment, cells were treated with the autophagy inhibitor 3-methyladenine (3-MA) immediately after puromycin removal. Size bars, 5 µm. (B) Quantification of the percentage of cells that cleared all aggregates by 8 h after puromycin removal. Where indicated, cells were treated with 3-MA with or without the proteasome inhibitor epoxomicin (epox) immediately after puromycin removal. Asterisks indicate a significant difference from cells treated with control siRNA, p < 0.05. (C) HeLa cells were transfected with indicated siRNAs, then fixed and immunostained for catalase and visualized using anti-rabbit Alexa 488 antibody. Size Bar, 10 µm. (D) The total fluorescent intensity from the fluorescent signal of endogenous catalase was measured as described in the Materials and Methods. The error bars indicate the standard deviations from at least 75 cells. Asterisks indicate a significant increase in catalase intensity compared to control siRNA-treated cells, p < 0.05. (E) In a parallel experiment to D, the number of fluorescently labeled catalase-positive structures or peroxisomes, was quantified as described in the Materials and Methods. The number of perxisomes per 100 µm3 in both isoforms of RAB1 or ATG12 siRNA-treated cells was compared with that in control siRNA-treated cells. Asterisks indicate a significant increase in peroxisome numbers, p < 0.05.
Rab1 is required for autophagy-mediated degradation of peroxisomes.
Mammalian cells have a basal level of constitutive peroxisomal turnover.48 This is autophagy-dependent, though it is not yet clear if the degradation is through selective autophagy or bulk autophagy. To test whether Rab1 is involved in autophagy-mediated degradation of peroxisomes, peroxisome levels were determined under Rab1 knockdown conditions. Previously it has been shown that inhibition of autophagy by ATG12 siRNA resulted in an increased number of peroxisomes.49 This increase in peroxisome number can be determined by quantifying the increase in catalase, an abundant peroxisomal protein. Either fluorescence intensity or the number of catalase-positive structures (representing peroxisomes) were quantified. When RAB1 expression was targeted by siRNA, a similar increase in catalase-positive structures was observed when compared to cells treated with nontargeting siRNA (Fig. 4C–E). Thus, these results are consistent with a role for Rab1 in autophagy-mediated degradation of peroxisomes.
Rab1 is involved in antimicrobial autophagy independent of its role in ER-to-Golgi trafficking.
In mammalian cells, Rab1 is involved in ER-to-Golgi trafficking.11–13 However, treatment of HeLa cells with the ARF1 inhibitor Brefeldin A (BFA) did not inhibit rapamycin-induced autophagy even though the Golgi was disrupted (Fig. 5A, B and S5A), consistent with previous results.50 BFA also had no effect on autophagy of S. typhimurium (Fig. 5C). Therefore, the BFA-sensitive Golgi network appears to be unnecessary for rapamycin-induced autophagosome formation as well as autophagy of S. typhimurium in mammalian cells. To determine the role of ER-to-Golgi trafficking in autophagy of S. typhimurium, we transfected cells with the dominant negative constructs ARF1(T31N), Sar1A(T39N) and Rab1B(S22N). Expression of all of these constructs disrupted the Golgi, but only Rab1B(S22N) significantly affected autophagy of bacteria (Fig. 5B, C and S5A). Expression of wild-type Rab1B or constitutively active Rab1B(Q67L) increased autophagy of bacteria, whereas constitutively active ARF1(Q71L) and Sar1A(H79G) had no effect (Fig. 5C). These results suggest that autophagy is rate limited by Rab1 activity, but not by other ER-to-Golgi transport modulators. Consistent with this observation, overexpression of Ypt1 in yeast increases autophagy.18
Figure 5.
Rab1 is involved in autophagy independent of its role in ER-to-Golgi transport. (A) HeLa cells were transfected with GFP-LC3 and treated with rapamycin for 2 h. Where indicated, cells were also treated with Brefeldin A (BFA), WTM or 3-MA. The number of autophagosomes (APs) per cell was quantified. Asterisks indicate a significant difference from no treatment (none) levels (+Rap), p < 0.05. (B) Cells were transfected with the indicated dominant negative CFP-GTPase constructs or treated with BFA, then fixed and stained with a monoclonal antibody to giantin to visualize the Golgi. The percentage of cells with intact Golgi stacks after the indicated treatment/CFP-GTPase transfection was quantified. Asterisks indicate a significant difference from the CFP-transfected control, p < 0.05. (C) Cells were co-transfected with RFP-LC3 and the indicated CFP-GTPase constructs and infected with S. typhimurium for 1 h. The colocalization of bacteria with LC3 was quantified only in transfected cells. In parallel experiments, cells were treated with BFA during infection. Asterisks indicate a significant difference from the CFP-transfected control, p < 0.05. (D) HeLa cells were treated with the indicated siRNAs, transfected with GFP-LC3 and infected with S. typhimurium expressing RFP for 1 h. The percentage of LC3+ bacteria was quantified. The asterisk indicates a significant difference from control siRNA levels, p < 0.05.
In yeast, certain components of the COPII coat (Sec23/Sec24) are required for autophagy whereas other components (Sec13/Sec31) are not.7 To test if COPII coat components are involved in autophagy of S. typhimurium, we treated HeLa cells with siRNA to SEC23 (both the A and B isoforms) or SEC13. Knockdown of either of the two components resulted in defective ER-to-Golgi trafficking, which was evidenced by Golgi network disruption and dispersing of the Golgi resident protein GM130 (Fig. S5B and C). As shown in Figure 5D, neither SEC23 nor SEC13 siRNA affected autophagy of S. typhimurium. To further test whether the Sec23/Sec24 COPII components are involved in mammalian autophagy, we transfected HeLa cells with the enterohemorrhagic Escherichia coli type III-secreted effector NleA fused to GFP. NleA specifically binds Sec24 and disrupts COPII-dependent protein trafficking.51 Consistent with the siRNA results, the percentage of S. typhimurium colocalizing with LC3 was similar in the presence or absence of NleA-GFP (Fig. S5D and E). These data demonstrate that COPII-dependent ER-to-Golgi trafficking is not required for antimicrobial autophagy in mammalian cells and reveal a novel function for Rab1 in autophagy independent of its well-characterized role in ER-to-Golgi transport.
Discussion
The mechanism of autophagosome formation in mammalian cells is unclear. Here we show that autophagy of S. typhimurium occurs at PtdIns(3)P-enriched domains of the ER. As well, our data suggest that antimicrobial autophagy shares many important similarities with starvation-induced autophagy. Importantly, we do not observe en bloc fusion of the ER with bacteria-containing autophagosomes (as judged by marker analysis e.g., calnexin, PDI; Fig. S2A). Whether ER membrane is used to generate autophagosomes during bacterial infection or starvation is not clear. Connections have been observed linking the ER and the isolation membrane of forming autophagosomes.8,9 However, it is not known if lipids translocate from the ER to the phagophore or whether these specialized ER domains merely serve as scaffolds for autophagosome formation. It is likely that other organelles are involved in autophagosome formation.52 While mitochondria have been linked to starvation-induced autophagy,3 our data suggest that they play a very minor role in antimicrobial autophagy.
Here we reveal a novel role for Rab1 in mammalian autophagy. We show that Rab1 promotes autophagy of S. typhimurium through a pathway that is independent of ER-to-Golgi trafficking. We recently determined a role for the yeast Rab1 homolog Ypt1 in autophagy.18 Therefore, Rab1 plays a conserved role in autophagy and acts in the autophagic degradation of many different cargoes in both yeast and mammalian cells. We have previously identified a novel TRAPP complex (TRAPPIII) in yeast that functions as a Ypt1 guanine nucleotide exchange complex in autophagy.18 These studies are consistent with our finding that Bet3, an essential TRAPP component, is required for antibacterial autophagy in mammalian cells (Fig. 2F). The nature of the TRAPP complexes that mediate Rab1 activation during autophagy in mammalian cells and the upstream signals that induce their targeting, remain important questions for future studies. Together, our results provide new insight into the mechanism of autophagosome formation and establish Rab1 as an important regulator of this process.
Materials and Methods
Mammalian cell culture.
HeLa human epithelial cells, wild-type and autophagy-deficient (Atg5-/-) mouse embryonic fibroblasts (MEFs)34 were maintained in DMEM growth medium (Fisher, SH-3024301) supplemented with 10% FBS (Wisent, 080450) at 37°C in 5% CO2 without antibiotics.
Bacterial strains and infection.
S. typhimurium SL1344,53 and bacteria expressing RFP were previously described.22 Late-log wild-type S. typhimurium cultures were used for infecting cells as previously described.54
Plasmids and transfection.
FuGene 6 (Roche Diagnostics, 11814443001) or GeneJuice (Novagen, CA80511-358) transfection reagents were used according to the manufacturer's instructions. Cells were transfected 16–24 h before infection. GFP-LC3 was from Dr. T. Yoshimori (National Institute of Genetics, Shizuoka-ken, Japan); RFP-LC3 was from Dr. W. Beron (Universidad Nacional de Cuyo, Medoza, Argentina); GFP-Rab1A was from Dr. C. Roy (Yale University School of Medicine, New Haven, CT), GFP-Rab1B was from Dr. C. Alvarez (Cuidad Universitaria, Cordoba, Argentina); Sec23-GFP was from Dr. H. Kai (Kumamoto University, Kumamoto, Japan), NleA-GFP was described previously.51 DFCP1-GFP, ER-FYVE-GFP, FYVE-GFP, TM-GFP and FYVE(C347S)-TM-GFP were described previously.4 Mito Cb5-GFP was described elsewhere.3 The plasmid expressing CFP was from Clontech (U55762).
ARF1-CFP, CFP-Sar1A and CFP-Rab1B constructs were generated as follows: ARF1, SAR1A and RAB1B were isolated from human whole total RNA and human brain total RNA by RT-PCR with gene-specific primers containing the appropriate restriction enzyme sites. The primers used were: ARF1(N)-CGG AAT TCA TGG GGA ACA TCT TCG CC, ARF1(C)-CGG GAT CCT GCT CCT GCG CAC TTC TGG TTCC GGA GC, Sar1A(N)-CGG AAT TCA TGT CTT TCA TCT TTG AGT GGA TCT, Sar1A(C)-GGG ATC CTC AGT CAA TAT ACT GGG AGA GCC, Rab1B(N)-CGG AAT TCA TGA ACC CCG AAT ATG ACT ACC, Rab1B(C)-CGG GAT CCT CAT GAA CGC CTG CTC GA. The PCR products were digested with EcoRI and BamHI and cloned into the pECFP-N1 (for ARF1-CFP) or pECFP-C2 (for CFP-Sar1A and CFP-Rab1B). To generate constitutively active and dominant negative forms of each protein, site-directed mutagenesis was performed with the QuikChange Site-Directed mutagenesis kit (Stratagene, 200519) according to the manufacturer's protocol. Gln was replaced with Leu in the G3 GTP-binding motif for ARF1(Q71L) and Rab1B(Q67L) and His with Leu for Sar1A(H79L) to make the constitutively active forms. Ser or Thr was replaced with Asn in the G1 GTP-binding motif to generate the dominant negative constructs ARF1(T31N), Rab1B(S22N) and Sar1A(T39N). After mutagenesis, each clone was verified by sequencing.
Drug treatments.
The following reagents were added to and maintained in the media as follows unless otherwise indicated: 3-methyladenine (Sigma; M9281; 10 mM), Brefeldin A (Invitrogen; cat.#B7450; 10 µg/mL), epoxomicin (AG Scientific; E-1098; 1 µM), puromycin (Sigma; P8833; 5 µg/mL), rapamycin (BioMol; A275-0005; 25 µg/mL), wortmannin (Sigma; W1628; 100 nM), bafilomycin A1 (Sigma; B 1793; 100 nM).
siRNA treatment.
RAB1A (#D-008283-02), RAB1B (#D-008958-03), ATG12 (GUG GGC AGU AGA GCG AAC A) and BET3 (GGA GAC GGU GUG ACA GAA AUU) siRNA duplexes and SEC13 (#M-012351-01), SEC23A (# M-009582-00) and SEC23B (#M-009592-01) SMARTpool siRNAs were from Dharmacon. The siCONTROL nontargeting siRNA pool (#D-001206-13) was used for control experiments. HeLa cells were transfected with 50 nM of each siRNA 48 h before use. Oligofectamine transfection reagent (Invitrogen, 12252011) was used according to the manufacturer's instructions. Successful Rab1B, Atg12 and Sec13 knockdown was confirmed by western blot. Successful Sec23 knockdown was confirmed by loss of Sec23-GFP expression by western blot.
Antibodies.
The following antibodies were used: rabbit anti-Salmonella (Difco Laboratories), mouse anti-Salmonella (BioDesign, C65336M), rabbit anti-calnexin (Dr. D. Williams, University of Toronto, Toronto, ON), mouse anti-protein disulphide isomerase (SPA-891) and mouse anti-Grp78/BiP (SPA-826) were from Stressgen, rabbit anti-Atg12 (2010) and rabbit anti-ribosomal protein S6 (2212) were from Cell Signaling, rabbit anti-Sec24 (Dr. W. Balch, The Scripps Research Institute, La Jolla, CA), rabbit anti-Sec13 and anti-Sec31 (Dr. F. Gorelick, The University of Tennessee Health Science Center, Memphis, TN), mouse anti-COPI (Dr. J. Rothman, Rockefeller Research Laboratory, New York), rabbit anti-β-COP (Affinity BioReagents, PAI-061), goat anti-Rab1B (Santa Cruz, sc-26541), mouse anti-ERGIC53 (Alexis Biochemicals, ALX-804-602), mouse (A11120) or rabbit (A11122) anti-GFP antibodies were from Molecular Probes, mouse anti-giantin (Dr. S. Grinstein, Hospital for Sick Children, Toronto, ON), mouse anti-GM130 (BD Biosciences Pharmingen, 610822), mouse anti-β-tubulin (Sigma, T4026), mouse anti-ubiquitinated proteins (FK2; BioMol, PW8810-0500),55 rabbit anti-catalase (Calbiochem, 219010), rabbit anti-LC3 for western blot (Novus Biologicals, NB100-2331) and rabbit anti-LC3 for immunofluorescence (Dr. K. Kirkegaard, Stanford University, CA USA).
All fluorescent secondary antibodies were AlexaFluor conjugates from Molecular Probes/Invitrogen (A-11029, A-11031, A-11045, A-11070, A-11036, A-31556, A-21068). Mouse and rabbit anti-horseradish peroxidase secondary antibodies for western blots were from Jackson ImmunoResearch Laboratories Inc. (115-035-146 ) and Sigma (A-6154), respectively.
Western blotting.
Samples were separated on 15% SDS-PAGE gels, then transferred to polyvinylidene fluoride membranes and probed with appropriate antibodies. Densitometry was carried out by using ImageJ software.
Immunofluorescence.
Cells were fixed with 2.5% paraformaldehyde for 10 min at 37°C. Extracellular and total S. typhimurium were differentially stained as previously described.56 For all other staining, permeabilization and blocking were performed with 0.2% saponin and 10% normal goat serum overnight at 4°C and staining performed as previously described.57 Colocalization quantifications were done using a Leica DMIRE2 epifluorescence microscope equipped with a 100x oil objective, 1.4 numerical aperture. Confocal images are shown in figures and were taken using a Zeiss Axiovert confocal microscope equipped with a 63x oil objective and LSM 510 software. Images were imported into Adobe Photoshop and assembled in Adobe Illustrator.
Peroxisome assessment.
HeLa cells were transfected with siRNA for ATG12, both isoforms of RAB1 or nontargeting sequence as above. 72 h after transfection, cells were fixed with 3% paraformaldehyde and a standard immunofluorescence staining protocol was used to detect the endogenous peroxisomal marker catalase. To quantify changes in the number of peroxisomes, cells were probed with rabbit anti-catalase and goat anti-rabbit IgG Alexa 488 antibodies. Fluorescence images were acquired with a laser-scanning confocal microscope (LSM 710; Carl Zeiss MicroImaging, Inc.) using a 40 × 1.3 NA Plan-NeoFluor oil objective. A fully open pinhole was used to obtain the maximal signal from all the planes. The total fluorescence intensity from the catalase staining was measured using Volocity (PerkinElmer). The average of the total intensities was normalized against the mock-transfected control cells. At least 75 cells were analyzed for each condition. To count peroxisomes within the cell, high-resolution z-sections images of the entire cell stained with anti-catalase were acquired on a Zeiss LSM 710. The number of peroxisomes and the volume of the cells were determined using the Volocity 3D analysis software (Perkin-Elmer). In short, the z-sections were used to construct a 3D image, from which the number of peroxisomes was calculated using the intensity and the particle count algorithm. Peroxisome number is presented as the number of peroxisomes in 100 µm3.
Live imaging.
Coverslips seeded with cells were placed in imaging chambers and maintained in RPMI medium with HEPES without bicarbonate. Bacteria were grown as described above and 1 mL harvested by pelleting. To label bacteria with a succinimidyl ester conjugated to AlexaFluor 647 (NHS-647; Invitrogen, A20006), pelleted bacteria were washed twice and resuspended in 250 mL of phosphate buffered saline. 100 mL of the resuspended bacteria was incubated with NHS-647 (0.5 mg/mL) for 5 min, shaking in the dark at 37°C. Labeled bacteria were washed once with phosphate buffered saline, resuspended in RPMI medium and added directly to the cells. Imaging was performed at 37°C on a Quorum spinning disk Leica DMIRE2 confocal microscope equipped with a 63x oil objective using Volocity software (Improvision). When infection was established (25–30 min post infection), gentamicin was added to the medium (100 µg/mL). Images were taken at 1 min intervals with a Hamamatsu Back-Thinned EM-CCD camera and z-stacks acquired as indicated in the figure legends. Fast iterative deconvolution was performed with Volocity software.
Statistics.
Colocalization quantifications were performed by direct visualization with a Leica DMIRE2 epifluorescence microscope and a 100x oil objective. Unless indicated, at least 100 bacteria or cells were counted per condition for each experiment. For all graphs, at least three independent experiments were performed. Unless indicated, the mean ± standard error (SE) is shown in the figures and p-values were calculated using the two-tailed student's t-test.
Acknowledgements
John H. Brumell, Ph.D. holds an Investigators in Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund. Infrastructure for the Brumell laboratory was provided by a New Opportunities Fund from the Canadian Foundation for Innovation and the Ontario Innovation Trust. J.H. holds a postdoctoral fellowship from the Canadian Association of Gastroenterology (CAG)/Canadian Institutes of Health Research/Crohn's and Colitis Foundation of Canada administered by the CAG. C.L.B. held a Canadian Graduate Studentship from the Natural Sciences and Engineering Research Council of Canada. We thank Drs. Cecilia Alvarez, William Balch, Walter Beron, Fred Gorelick, Sergio Grinstein, Hirofumi Kai, Noboru Mizushima, James Rothman, Craig Roy, William Trimble, David Williams and Tamotsu Yoshimori for providing reagents. We also thank Michael Woodside and Paul Paroutis for assistance with confocal microscopy and Drs. Nicola Jones, Mauricio Terebiznik and the Brumell laboratory for critical reading of this manuscript.
Abbreviations
- 3-MA
3-methyladenine
- BFA
brefeldin A
- DFCP1
double FYVE domain-containing protein 1
- ER
endoplasmic reticulum
- ERGIC
ER-to-Golgi intermediate compartment
- GEF
guanine nucleotide exchange factor
- LAMP-1
lysosome-associated membrane protein-1
- LC3
microtubule-associated protein 1 light chain 3
- MEF
mouse embryonic fibroblast
- Mito Cb5, mitochondria cytochrome b5; PE
phosphatidylethanolamine
- PtdIns(3)P
phosphatidylinositol 3-phosphate
- SCV
Salmonella-containing vacuole
- TM
transmembrane
- TRAPP
transport protein particle
- WTM
wortmannin
Footnotes
Previously published online: www.landesbioscience.com/journals/autophagy/article/13840
Supplementary Material
References
- 1.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hailey DW, Rambold AS, Satpute-Krishnan P, Mitra K, Sougrat R, Kim PK, et al. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell. 2010;141:656–667. doi: 10.1016/j.cell.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, et al. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dunn WA., Jr Studies on the mechanisms of autophagy: formation of the autophagic vacuole. J Cell Biol. 1990;110:1923–1933. doi: 10.1083/jcb.110.6.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mari M, Griffith J, Rieter E, Krishnappa L, Klionsky DJ, Reggiori F. An Atg9-containing compartment that functions in the early steps of autophagosome biogenesis. J Cell Biol. 2010;190:1005–1022. doi: 10.1083/jcb.200912089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ishihara N, Hamasaki M, Yokota S, Suzuki K, Kamada Y, Kihara A, et al. Autophagosome requires specific early Sec proteins for its formation and NSF/SNARE for vacuolar fusion. Mol Biol Cell. 2001;12:3690–3702. doi: 10.1091/mbc.12.11.3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hayashi-Nishino M, Fujita N, Noda T, Yamaguchi A, Yoshimori T, Yamamoto A. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat Cell Biol. 2009;11:1433–1437. doi: 10.1038/ncb1991. [DOI] [PubMed] [Google Scholar]
- 9.Ylä-Anttila P, Vihinen H, Jokitalo E, Eskelinen E-L. 3D tomography reveals connections between the phagophore and endoplasmic reticulum. Autophagy. 2009;5:1180–1185. doi: 10.4161/auto.5.8.10274. [DOI] [PubMed] [Google Scholar]
- 10.Bacon RA, Salminen A, Ruohola H, Novick P, Ferro-Novick S. The GTP-binding protein Ypt1 is required for transport in vitro: the Golgi apparatus is defective in ypt1 mutants. J Cell Biol. 1989;109:1015–1022. doi: 10.1083/jcb.109.3.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nuoffer C, Davidson HW, Matteson J, Meinkoth J, Balch WE. A GDP-bound of rab1 inhibits protein export from the endoplasmic reticulum and transport between Golgi compartments. J Cell Biol. 1994;125:225–237. doi: 10.1083/jcb.125.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pind SN, Nuoffer C, McCaffery JM, Plutner H, Davidson HW, Farquhar MG, et al. Rab1 and Ca2+ are required for the fusion of carrier vesicles mediating endoplasmic reticulum to Golgi transport. J Cell Biol. 1994;125:239–252. doi: 10.1083/jcb.125.2.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Plutner H, Cox AD, Pind S, Khosravi-Far R, Bourne JR, Schwaninger R, et al. Rab1b regulates vesicular transport between the endoplasmic reticulum and successive Golgi compartments. J Cell Biol. 1991;115:31–43. doi: 10.1083/jcb.115.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Allan BB, Moyer BD, Balch WE. Rab1 recruitment of p115 into a cis-SNARE complex: programming budding COPII vesicles for fusion. Science. 2000;289:444–448. doi: 10.1126/science.289.5478.444. [DOI] [PubMed] [Google Scholar]
- 15.Moyer BD, Allan BB, Balch WE. Rab1 interaction with a GM130 effector complex regulates COPII vesicle cis-Golgi tethering. Traffic. 2001;2:268–276. doi: 10.1034/j.1600-0854.2001.1o007.x. [DOI] [PubMed] [Google Scholar]
- 16.Monetta P, Slavin I, Romero N, Alvarez C. Rab1b interacts with GBF1 and modulates both ARF1 dynamics and COPI association. Mol Biol Cell. 2007;18:2400–2410. doi: 10.1091/mbc.E06-11-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sannerud R, Marie M, Nizak C, Dale HA, Pernet-Gallay K, Perez F, et al. Rab1 defines a novel pathway connecting the pre-Golgi intermediate compartment with the cell periphery. Mol Biol Cell. 2006;17:1514–1526. doi: 10.1091/mbc.E05-08-0792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lynch-Day MA, Bhandari D, Menon S, Huang J, Cai H, Bartholomew CR, et al. Trs85 directs a Ypt1 GEF, TRAPPIII, to the phagophore to promote autophagy. Proc Natl Acad Sci USA. 2010;107:7811–7816. doi: 10.1073/pnas.1000063107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levine B, Deretic V. Unveiling the roles of autophagy in innate and adaptive immunity. Nat Rev Immunol. 2007;7:767–777. doi: 10.1038/nri2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmid D, Münz C. Innate and adaptive immunity through autophagy. Immunity. 2007;27:11–21. doi: 10.1016/j.immuni.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haraga A, Ohlson MB, Miller SI. Salmonellae interplay with host cells. Nat Rev Microbiol. 2008;6:53–66. doi: 10.1038/nrmicro1788. [DOI] [PubMed] [Google Scholar]
- 22.Birmingham CL, Smith AC, Bakowski MA, Yoshimori T, Brumell JH. Autophagy controls Salmonella infection in response to damage to the Salmonellacontaining vacuole. J Biol Chem. 2006;281:11374–11383. doi: 10.1074/jbc.M509157200. [DOI] [PubMed] [Google Scholar]
- 23.Jia K, Thomas C, Akbar M, Sun Q, Adams-Huet B, Gilpin C, et al. Autophagy genes protect against Salmonella typhimurium infection and mediate insulin signaling-regulated pathogen resistance. Proc Natl Acad Sci USA. 2009;106:14564–14569. doi: 10.1073/pnas.0813319106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuballa P, Huett A, Rioux JD, Daly MJ, Xavier RJ. Impaired autophagy of an intracellular pathogen induced by a Crohn's disease associated ATG16L1 variant. PLoS One. 2008;3:3391. doi: 10.1371/journal.pone.0003391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roberts RL, Hollis-Moffatt JE, Gearry RB, Kennedy MA, Barclay ML, Merriman TR. Confirmation of association of IRGM and NCF4 with ileal Crohn's disease in a population-based cohort. Genes Immun. 2008;9:561–565. doi: 10.1038/gene.2008.49. [DOI] [PubMed] [Google Scholar]
- 26.Rioux JD, Xavier RJ, Taylor KD, Silverberg MS, Goyette P, Huett A, et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat Genet. 2007;39:596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Travassos LH, Carneiro LA, Ramjeet M, Hussey S, Kim YG, Magalhaes JG, et al. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat Immunol. 2010;11:55–62. doi: 10.1038/ni.1823. [DOI] [PubMed] [Google Scholar]
- 28.Cheng JF, Ning YJ, Zhang W, Lu ZH, Lin L. T300A polymorphism of ATG16L1 and susceptibility to inflammatory bowel diseases: a meta-analysis. World J Gastroenterol. 2010;16:1258–1266. doi: 10.3748/wjg.v16.i10.1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hampe J, Franke A, Rosenstiel P, Till A, Teuber M, Huse K, et al. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat Genet. 2007;39:207–211. doi: 10.1038/ng1954. [DOI] [PubMed] [Google Scholar]
- 30.Noda T, Yoshimori T. Molecular basis of canonical and bactericidal autophagy. Int Immunol. 2009;21:1199–1204. doi: 10.1093/intimm/dxp088. [DOI] [PubMed] [Google Scholar]
- 31.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mallo GV, Espina M, Smith AC, Terebiznik MR, Aleman A, Finlay BB, et al. SopB promotes phosphatidylinositol 3-phosphate formation on Salmonella vacuoles by recruiting Rab5 and Vps34. J Cell Biol. 2008;182:741–752. doi: 10.1083/jcb.200804131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hailey DW, Rambold AS, Satpute-Krishnan P, Mitra K, Sougrat R, Kim PK, et al. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell. 2010;141:656–667. doi: 10.1016/j.cell.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 35.Sacher M, Barrowman J, Wang W, Horecka J, Zhang Y, Pypaert M, et al. TRAPP I implicated in the specificity of tethering in ER-to-Golgi transport. Mol Cell. 2001;7:433–442. doi: 10.1016/s1097-2765(01)00190-3. [DOI] [PubMed] [Google Scholar]
- 36.Jones S, Newman C, Liu F, Segev N. The TRAPP complex is a nucleotide exchanger for Ypt1 and Ypt31/32. Mol Biol Cell. 2000;11:4403–4411. doi: 10.1091/mbc.11.12.4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang W, Sacher M, Ferro-Novick S. TRAPP stimulates guanine nucleotide exchange on Ypt1p. J Cell Biol. 2000;151:289–296. doi: 10.1083/jcb.151.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sacher M, Jiang Y, Barrowman J, Scarpa A, Burston J, Zhang L, et al. TRAPP, a highly conserved novel complex on the cis-Golgi that mediates vesicle docking and fusion. EMBO J. 1998;17:2494–2503. doi: 10.1093/emboj/17.9.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamasaki A, Menon S, Yu S, Barrowman J, Meerloo T, Oorschot V, et al. mTrs130 is a component of a mammalian TRAPPII complex, a Rab1 GEF that binds to COPI coated vesicles. Mol Biol Cell. 2009;20:4205–4215. doi: 10.1091/mbc.E09-05-0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perrin AJ, Jiang X, Birmingham CL, So NS, Brumell JH. Recognition of bacteria in the cytosol of mammalian cells by the ubiquitin system. Curr Biol. 2004;14:806–811. doi: 10.1016/j.cub.2004.04.033. [DOI] [PubMed] [Google Scholar]
- 41.Brumell JH, Tang P, Zaharik ML, Finlay BB. Disruption of the Salmonella-containing vacuole leads to increased replication of Salmonella enterica serovar typhimurium in the cytosol of epithelial cells. Infect Immun. 2002;70:3264–3270. doi: 10.1128/IAI.70.6.3264-3270.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Salcedo SP, Holden DW. SseG, a virulence protein that targets Salmonella to the Golgi network. EMBO J. 2003;22:5003–5014. doi: 10.1093/emboj/cdg517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kirkegaard K, Taylor MP, Jackson WT. Cellular autophagy: surrender, avoidance and subversion by microorganisms. Nat Rev Microbiol. 2004;2:301–314. doi: 10.1038/nrmicro865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kirkin V, McEwan DG, Novak I, Dikic I. A role for ubiquitin in selective autophagy. Mol Cell. 2009;34:259–269. doi: 10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- 46.Lelouard H, Ferrand V, Marguet D, Bania J, Camosseto V, David A, et al. Dendritic cell aggresome-like induced structures are dedicated areas for ubiquitination and storage of newly synthesized defective proteins. J Cell Biol. 2004;164:667–675. doi: 10.1083/jcb.200312073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Szeto J, Kaniuk NA, Canadien V, Nisman R, Mizushima N, Yoshimori T, et al. ALIS are stress-induced protein storage compartments for substrates of the proteasome and autophagy. Autophagy. 2006;2:189–199. doi: 10.4161/auto.2731. [DOI] [PubMed] [Google Scholar]
- 48.Huybrechts SJ, Van Veldhoven PP, Brees C, Mannaerts GP, Los GV, Fransen M. Peroxisome dynamics in cultured mammalian cells. Traffic. 2009;10:1722–1733. doi: 10.1111/j.1600-0854.2009.00970.x. [DOI] [PubMed] [Google Scholar]
- 49.Kim PK, Hailey DW, Mullen RT, Lippincott-Schwartz J. Ubiquitin signals autophagic degradation of cytosolic proteins and peroxisomes. Proc Natl Acad Sci USA. 2008;105:20567–20574. doi: 10.1073/pnas.0810611105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Purhonen P, Pursiainen K, Reunanen H. Effects of brefeldin A on autophagy in cultured rat fibroblasts. Eur J Cell Biol. 1997;74:63–67. [PubMed] [Google Scholar]
- 51.Kim J, Thanabalasuriar A, Chaworth-Musters T, Fromme JC, Frey EA, Lario PI, et al. The bacterial virulence factor NleA inhibits cellular protein secretion by disrupting mammalian COPII function. Cell Host Microbe. 2007;2:160–171. doi: 10.1016/j.chom.2007.07.010. [DOI] [PubMed] [Google Scholar]
- 52.Hamasaki M, Yoshimori T. Where do they come from?? Insights into autophagosome formation. FEBS Lett. 2010;584:1296–1301. doi: 10.1016/j.febslet.2010.02.061. [DOI] [PubMed] [Google Scholar]
- 53.Hoiseth SK, Stocker BA. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature. 1981;291:238–239. doi: 10.1038/291238a0. [DOI] [PubMed] [Google Scholar]
- 54.Steele-Mortimer O, Meresse S, Gorvel JP, Toh BH, Finlay BB. Biogenesis of Salmonella typhimurium-containing vacuoles in epithelial cells involves interactions with the early endocytic pathway. Cell Microbiol. 1999;1:33–49. doi: 10.1046/j.1462-5822.1999.00003.x. [DOI] [PubMed] [Google Scholar]
- 55.Fujimuro M, Sawada H, Yokosawa H. Production and characterization of monoclonal antibodies specific to multi-ubiquitin chains of polyubiquitinated proteins. FEBS Lett. 1994;349:173–180. doi: 10.1016/0014-5793(94)00647-4. [DOI] [PubMed] [Google Scholar]
- 56.Birmingham CL, Canadien V, Kaniuk NA, Steinberg BE, Higgins DE, Brumell JH. Listeriolysin O allows Listeria monocytogenes replication in macrophage vacuoles. Nature. 2008;451:350–354. doi: 10.1038/nature06479. [DOI] [PubMed] [Google Scholar]
- 57.Brumell JH, Rosenberger CM, Gotto GT, Marcus SL, Finlay BB. SifA permits survival and replication of Salmonella typhimurium in murine macrophages. Cell Microbiol. 2001;3:75–84. doi: 10.1046/j.1462-5822.2001.00087.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.