Abstract
Programmed nuclear death (PND) in Tetrahymena is a unique process during conjugation, in which only the parental macronucleus is degraded and then eliminated from the progeny cytoplasm, but other co-existing nuclei such as new micro- and macronuclei are unaffected. PND through autophagic elimination is expected to be strictly controlled, considering the significant roles in ciliates such as turnover of disused organelles and production of the next generation. Here we demonstrate that PND in Tetrahymena involves peculiar aspects of autophagy, which differ from mammalian or yeast macroautophagy. Drastic change of the parental macronucleus occurs when differentiation of new macronuclei is initiated. Combined use of monodansylcadaverine and a lysosome indicator LysoTracker Red showed that prior to nuclear condensation, the envelope of the parental macronucleus changed its nature as if it is an autophagic membrane, without the accumulation of a pre-autophagosomal structure from the cytoplasm. Subsequently, lysosomes approached only to the parental macronucleus and localized at the envelope until a final resorption stage. In addition, we found that the parental macronucleus exhibits certain sugars and phosphatidylserine on the envelope, which are possible “attack me” signals, that are not found on other types of nuclei. These findings suggest that PND is a highly elaborated process, different from the typical macroautophagy seen in other systems, and is executed through interaction between specific molecular signals on the parental macronuclear envelope and autophagic/lysosomal machineries.
Key words: Tetrahymena, conjugation, nuclear apoptosis, monodansylcadaverine, macroautophagy, phagocytosis marker, glycoconjugates, phosphatidylserine
Introduction
Ciliates, represented by Tetrahymena and Paramecium, are large unicellular protozoans up to ∼50 µm in length and characterized by maintaining two kinds of differentiated nuclei, macro- and micronuclei, which are spatially separated, within a single cytoplasm. The polyploid macronucleus is extremely large, compared with diploid nuclei of most eukaryotic cells. It is transcriptionally active and is therefore functionally analogous to the somatic nucleus of multicellular organisms. The micronucleus, in contrast, corresponds to a canonical diploid germinal nucleus that is transcriptionally silent, except in meiotic prophase during sexual reproduction (conjugation). Conjugation of ciliates is initiated by the interaction of cells of different mating types and is followed by a complex series of nuclear events, including meiosis, fertilization (formation of a zygotic micronucleus) and postzygotic mitosis. Both types of nuclei are commonly derived from the postzygotic mitotic products, and the macronuclear differentiation involves an RNAi-mediated, heterochromatin formation large-scale DNA rearrangement and amplification process.1
Because parental cell-derived cytoplasm is taken over by a progeny macronucleus after conjugation, the parental macronucleus must be selectively eliminated from the progeny cytoplasm. The degradation of the parental macronucleus behaves in a similar manner to the nucleus in apoptotic cells.2,3 DNA is fragmented into high-molecular-weight DNA (kb-sized DNA), followed by formation of an oligonucleosome-sized ladder and the parental macronucleus is then completely resorbed.4 This process is controlled by specific gene expression,2 referred to as programmed nuclear death (PND) or nuclear apoptosis. In PND, the parental macronucleus must trigger signaling pathways that activate the engulfing machinery of the autodigestive vacuoles to protect other nuclei and cytoplasm from digestive enzymes, when the macronucleus is destined to degrade. In Tetrahymena thermophila, treatment with a phosphoinositide 3-kinase (PI 3-kinase) inhibitor such as wortmannin or 3-methyladenine blocks acidification and final resorption of several types of nuclei such as meiotic products and the parental macronucleus, resulting in the accumulation of extranuclei during conjugation.5 Since these drugs are known to act specifically upon induction and vesicle nucleation of macroautophagy in mammalian cells,6 PND is likely to require the autophagic/lysosomal pathway.
Previous reports suggested that the degenerating macronucleus is sequestered by an autophagosome-like structure from other organelles, in which many dead mitochondria are incorporated.7,8 Mitochondria of Tetrahymena contain apoptosis-inducing factor (AIF) and as yet-unidentified DNases, similar to endonuclease G.7 When the new macronuclei are differentiated, Tetrahymena AIF translocates from the sequestered mitochondria to the parental macronucleus.9 Knockout of Tetrahymena AIF showed delayed progression of PND, that is, delay of nuclear condensation and kb-sized DNA fragmentation, corresponding to the initial stage of the nuclear apoptosis. Furthermore, in vitro assay using AIF-deficient mitochondria revealed that mitochondrial DNase activity is drastically reduced, suggesting that mitochondrial DNase activity depends upon the presence of AIF.9 In a final stage, many lysosomes fuse with the macronucleus in the posterior region of the cell, leading to the eventual resorption of the nucleus via acidification.10 If new macronuclei fail to develop, PND is interrupted before the final resorption stage.2,11 However, no wrapping process of the parental macronucleus with large or small cisterna, represented by a pre-autophagosomal structure (PAS), has been observed throughout conjugation.12 Thus, it is still unclear whether PND involves typical membrane dynamics similar to yeast or mammalian macroautophagy.13,14 At present, a new lysosomal-wrapping model, which differs from the process of mammalian macroautophagy, has been proposed in T. thermophila PND.15 In this model, the parental macronucleus is sequestered from the cytoplasm via mutual fusion of many lysosomes without this sequence of events leading to the formation of a large autophagosome and autolysosome. Based on our previous observations, an autophagic/lysosomal pathway might be present in PND in Tetrahymena,7,8 but it appears to be far different from the mammalian or yeast macroautophagy.6
In animal apoptosis, by analogy, dying cells expose some molecules on the cell surface as “eat-me” signals, thereby the engulfing machinery of macrophages is activated.16–18 These cells are finally degraded in phagocytes with activation of the lysosomal enzymes.19,20 PND also must trigger the signaling pathway for targeting only the parental macronucleus among the different sorts of nuclei, coexisting in the same cytoplasm. Considering the above-mentioned information of animal apoptosis, we can expect similar “eat-me” signals on the parental macronucleus. Relying on the exposed signals, the autophagic/lysosomal pathway might selectively recognize the parental macronucleus destined to die, distinct from other nuclei, such as new micro- and macronuclei. Such signals are unknown to date.
In this study, we show the occurrence of autophagosome-like structures on the envelope of the parental macronucleus during conjugation without accumulation of PAS-like membranes. The fusion of lysosomes also occurs prior to the final stage, and thereby the macronucleus is subjected to acidification, resulting in the final resorption. Furthermore, we demonstrate changes in molecular constitution on the macronuclear envelope during conjugation, and some molecules involved in the membrane change can be possible candidates for the “eat-me” signals, comparable to that in animal apoptosis. These observations lead us to the idea that PND is executed through the interaction between specific molecular marks on the macronuclear envelope and autophagic/lysosomal machineries.
Results
Outline of autophagic-lysosomal process in PND.
There are a number of diagnostic assays for autophagy.6,13 Among them, localization of the LC3 (Atg8) protein has been analyzed in detail using mammalian cells.21 LC3 is an 18 kDa protein and plays an important role for autophagosome formation. It remains on the membrane even after spherical autophagosomes are completely formed.21,22 We initially applied anti-human LC3 polyclonal antibody (MBL, PM036; 1:1,000 dilution) for labeling of autophagic vacuoles in T. thermophila cells. This antibody bound to an approximately 18-kDa protein in western blotting, but it did not show any cross-reaction in a cytological assay (data not shown). Therefore, we used a fluorescent compound, monodansylcadaverine (MDC).
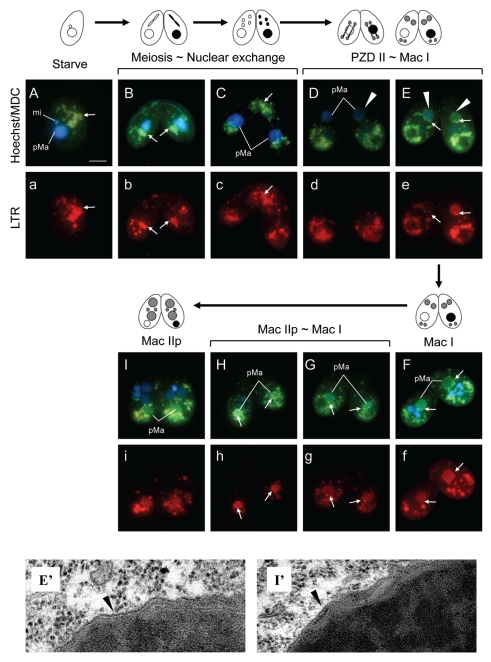
MDC accumulates in autophagic vesicles under in vivo conditions, but does not accumulate in endoplasmic reticulum nor in endosomes.23 MDC has therefore been proposed as a useful tracer for autophagic vesicles.24 To monitor autophagic/lysosomal events during PND, we stained living cells with a combination of MDC and a lysosome indicator LysoTracker Red (LTR), and then observed staining patterns under a fluorescent microscope. As shown in Figure 1A, starved cells showed some MDC-positive vesicles in the cytoplasm and almost all of these were co-localized with LTR-positive dots (Fig. 1a), suggesting that lysosomes are likely to attach on autophagic vesicles at early stages of biogenesis. Since such vesicles were always observed in nonconjugating cells, we could not precisely determine the origin of the content in the vesicles at any given moment. The distribution pattern of these vesicles showed no specificity in nonconjugating cells. When conjugation was induced, however, these vesicles accumulated in the posterior region of the conjugating cells (Fig. 1B and b). At the transition stage from meiosis to nuclear exchange, many MDC/LTR-positive vesicles appeared in the anterior region of the cells, where the conjugating pairs adhere to each other (Fig. 1C and c). These vesicles migrated to the posterior region of cells by the stage of fertilization. When new macro- and micronuclei began to differentiate after two successive postzygotic divisions of a fertilized nucleus (corresponding to PZD II∼Mac I, see Akematsu and Endoh 2010,9), the parental macronucleus somewhat reduced in size, reflecting chromatin condensation of the nucleus (Fig. 1D and E). At the same time, the envelope of the parental macronucleus uniformly became MDC-stainable (Fig. 1D and E arrowhead), but not LTR-stainable (Fig. 1d), indicating formation of an autophagosomal structure on the envelope. If typical membrane dynamics of macroautophagy are involved in PND, the peripheral structure of the degenerating macronucleus at Mac I must consist of four layers of membrane that derived from the pre-autophagosomal structure (PAS) and the nuclear membrane. However, we could not observe such a structure around the degenerating macronucleus, and only nuclear membrane was observed under transmission electron microscopy (Fig. 1E′).
Figure 1.
Autophagic/lysosomal events during PND visualized with a combination of monodansylcadaverine (MDC; upper case) and LysoTracker Red (LTR; lower case). Living cells were stained with MDC and Hoechst 33342 (nuclear stain; upper) and LTR (lower). (A/a) Starved nonconjugating cell. Almost all MDC-sensitive vacuoles co-localize with lysosomes (arrows). (B/b) Meiotic prophase. After initiation of conjugation, these vacuoles accumulate in the posterior region of the cells. (C/c) Nuclear exchange-stage. Many MDC/LTR-staining vacuoles appear in the anterior region of the cells. (D/d and E/e) Transition stage from PZD II to Mac I. Envelope of the parental macronucleus becomes MDC sensitive (arrowheads). (F/f) Mac I-stage cell. Several MDC/LTR-sensitive vacuoles migrate from the posterior region of the cell to the surface of the parental macronucleus (arrows). (G/g and H/h) Transition stage from Mac I to Mac II p. The parental macronucleus translocates to the posterior region of the cell, accompanied by more migration of the vacuoles on their surface (arrows). (I/i) Mac II p-stage. The entire parental macronucleus becomes both MDC- and LTR-sensitive. Transmission electron micrographs of parental macronucleus at two different stages (E′ and I′) are corresponding to (E and I) stages. Black arrowheads in these micrographs indicate the envelope of the parental macronucleus. pMa and mi denote parental macronucleus and micronucleus, respectively. The scale bar in (A) indicates 10 µm.
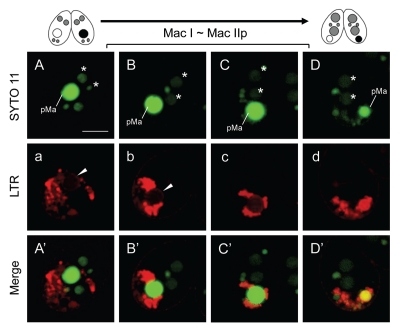
The fluorescence intensity on the envelope gradually increased with time (Fig. 1E), and then several MDC/LTR-positive vesicles migrated from the posterior region of the cell to the surface of the parental macronucleus (Fig. 1E and F and e and f, arrows). The migration of the vesicles caused acidification of the periphery of the nuclear envelope (Fig. 1e and f), whereas the inside of the nucleus seemed to still remain at neutral pH (Fig. 1e and f; see also Kobayashi and Endoh 20057). Confocal live cell imaging, using a combination of SYTO green dye and LTR, more clearly revealed that the inside of the nucleus was LTR-negative at these stages (Fig. 2a, arrowhead). After that, the parental macronucleus condensed more and migrated to the posterior region of the cell, accompanied by further accumulation of the vacuoles on their surface (Fig. 1G and g). These events rapidly occurred within ∼1 h. Just after the migration (corresponding to the early stage of Mac IIp), many MDC/LTR-positive vesicles immediately attached to the parental macronucleus (Fig. 1H and h), but the inside of the nucleus still remained at near neutral pH (Fig. 2b and c). Meanwhile, the whole parental macronucleus became both MDC- and LTR-positive (Fig. 1l and i), indicating that these vesicles entirely fused with the nuclear envelope, into which lysosomal enzymes were released, followed by acidification of the whole nucleus (Fig. 2d). At this stage, further migration of the digestive vesicle complexes on the nucleus does not occur (Fig. 1i). As well as Mac I cells, we could also not confirm the presence of autophagosomal membrane on the outside of the macronuclear envelope in Mac IIp cells (Fig. 1I′).
Figure 2.
Confocal imaging of parental macronuclear acidification during Mac I ∼Mac IIp. Living cells in the transition stage from Mac I to Mac IIp were stained with MDC and SYTO 11 nucleic acid stain (upper case; top part) and LTR (lower case; middle part). The lower parts (upper case′) show a merged image. (A/a/A′ and B/b/B′) The envelope of the parental macronucleus shows LTR-positive staining (arrowhead) but the inside is LTR-negative. (C/c/C′) The inside of the parental macronucleus is gradually acidifying by attachment of lysosomes on the envelope. (D/d/D′) The entire parental macronucleus becomes acidic. Arrows in the middle micrographs indicate the envelope of the parental macronucleus. pMa and asterisk (*) denote the parental macronucleus and the new macronucleus, respectively. The scale bar in (A) indicates 10 µm.
Biogenesis of the autophagosomal property on the macronuclear envelope.
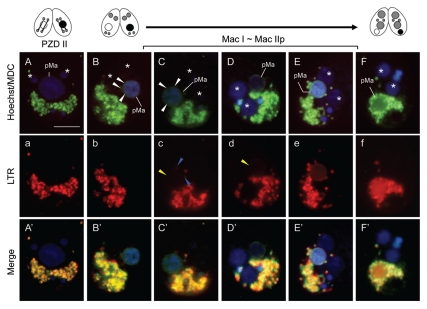
As shown in the previous section, the MDC-stainability of the nuclear envelope initiated at the stage of Mac I without PAS-like double membrane enwrapping. Another possibility, however, would be that the vesicles in the posterior part of the cell fuse together and enwrap the nucleus. To identify the initial change of the macronucleus in detail, the live cell imaging, focusing on the early stage of PND, was carried out with high magnification. Figure 3A indicates PZD II-staged cell and no MDC fluorescence is observed at the periphery of the parental macronucleus. When condensation of the parental macronucleus occurred (corresponding to Mac I), however, a green-colored haze of the MDC fluorescence was hanging over the nucleus without migration and accumulation of the vesicles from the posterior region of the cell (Fig. 3B). As soon as that occurred, the nuclear envelope was clearly bordered with the MDC fluorescence within ∼15 min (Fig. 3C and D). From this observation and taking the results in the previous section into consideration, the nuclear envelope is likely to change directly into an autophagic membrane during this period. The nuclear envelope was LTR-negative at first (Fig. 3a and b) and it became slightly LTR-positive within a few minutes after the beginning of the MDC-stainability (Fig. 3c and d). This change might derive from either the attachment of the several digestive vacuole complexes on the envelope (Fig. 3c and blue arrowheads) or an increase of ion trapping of the membrane as proposed in part for mammalian autophagosomes.25 After that, more migration of the vesicle immediately occurred (Fig. 3E and F) and this allowed the final acidification of the nucleus (Fig. 3e and f).
Figure 3.
Biogenesis of the autophagosomal property on the parental macronuclear envelope. Living cells in the transition stage from PZD II to Mac IIp were stained with MDC and Hoechst 33342 (upper case; top part) and LTR (lower case; middle part). The lower parts show a merged image (upper case′; lower part). (A/a/A′) PZD II-stage cell. (B/b/B′–F/f/F′) Transition stage from Mac I to Mac IIp. Concomitant with parental macronuclear condensation, a green-colored haze of the MDC fluorescence is hanging over the nucleus (white arrowheads in B and C) and then covered the envelope (white arrowhead in D). The envelope was LTR-negative at first (b) and this became slightly LTR-positive after the beginning of the MDC-stainability (yellow arrowheads in c and d) owing to the attachment of several digestive vesicle complexes (blue arrowheads in c). After that, more migration of the digestive vesicle complexes occurred (E and e) and this allowed for acidification of the nucleus (F and f). pMa and asterisk (*) denote the parental macronucleus and the new macronucleus, respectively. The scale bar in (A) indicates 10 µm.
Possible involvement of epigenetic change for commitment of autophagic events and developmental programming.
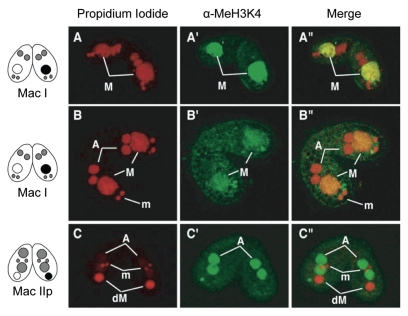
Since ciliates undergo alteration of generation in nuclear level, it has been suggested that an inextricable link exists between new macronuclear differentiation and the degradation of parental macronucleus. In T. thermophila, the level of methylation at Lys4 of histone H3 (H3K4), an epigenetic mark for active transcription, appears to be the same before and after nuclear differentiation.26 Indeed, we found that Lys4 methylation of the new macronucleus occurred just after nuclear differentiation, concomitant with the loss of the methyl mark from the parental macronucleus, which is just beginning nuclear condensation (Fig. 4A′–C′). These observations suggest that the parental macronucleus appears to be committed to death and triggers the autophagic/lysosomal events at the stage of Mac I, once differentiation of the new macronucleus has been initiated successfully.
Figure 4.
Lys 4 methylation of histone H3 in the new and parental macronuclei during conjugation. (A/A′/A″) Early stage of Mac I. (B/B′/B″) Later stage of Mac I. Lys 4 of H3 in the parental macronucleus (M) is methylated, that in the new macronucleus (A/A′/A″) is not at this stage. (C/C′/C″) Mac IIp. Just after nuclear differentiation, the methyl mark is only detected in the developing macronuclei, while it has disappeared in the degenerating parental macronucleus (dM). Lys 4 of H3 in the micronucleus (m) is not methylated throughout the life cycle. Left, middle and right parts show propidium iodide (upper case; DNA), a-MeH3K4 (upper case′) and a merged image (upper case″), respectively.
Drastic change of the macronuclear envelope during PND.
The important observation described above was the change of MDC-stainability of the parental macronucleus. The subsequent approach of the MDC/LTR-positive vesicles occurred in distinct stages, followed by nuclear acidification in the final stage. These observations suggest that it is likely that such vesicles are attracted to the nuclear membrane after the nuclear membrane changed MDC-stainability. Thus, we assume an alteration of the molecular characteristics on the macronucleus has occurred, by which the digestive vesicle complexes could distinguish the nucleus to die from other nuclei to survive. To ask whether the parental macronucleus exhibits any specific molecules on the envelope during PND, we searched for the presence or absence of some glycans associated with the nucleus.
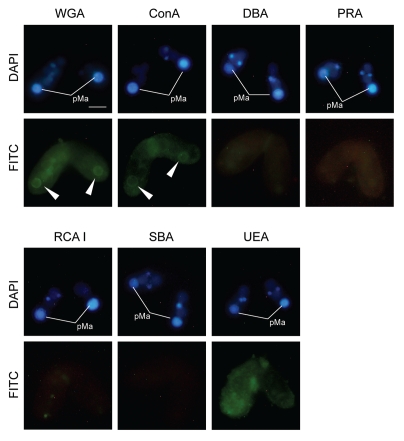
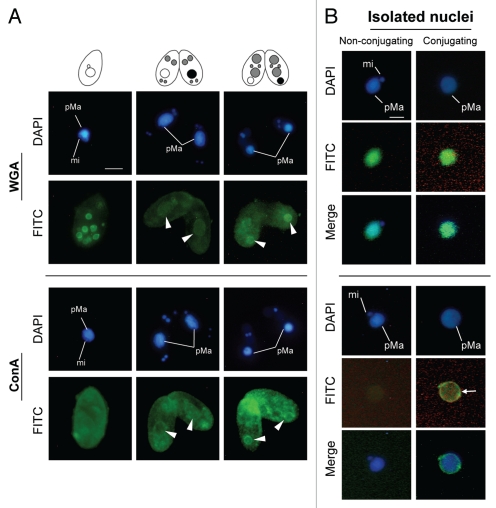
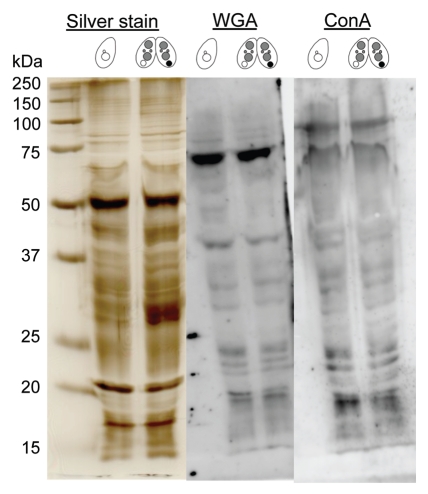
Figure 5 indicates binding specificities of FITC-labeled lectin probes to the nuclei at Mac IIp (16 h after induction of conjugation). Noticeable differences between the parental macronucleus and other nuclei were observed using wheat germ agglutinin (WGA) and concanavalin A (ConA), but never with other lectin probes. Although the labeling with WGA and ConA were slightly observed not only in the parental macronucleus but also in other nuclei, the stainability of the parental macronucleus was specifically increased when the nucleus began to condense during conjugation (Fig. 6A). Note that the envelope of the degenerating macronucleus appeared to be more lectin-positive than the nucleoplasm. These changes occurred with similar timing as the increase of MDC-stainability of the nuclear envelope (Fig. 1D and 3B). Thus, it is reasonable to assume that some glycosylation or sugar modification is induced in the nuclear membrane. To confirm whether the fluorescence specificities originated in the nuclear envelope, we isolated the degenerating macronucleus from Mac IIp cells and observed binding of WGA or ConA to the nucleus. As shown in Figure 6B, WGA crossreacted with the degenerating macronucleus and the nucleoplasm was also stained. In the macronucleus from starved cells, which is used as a control, an almost similar staining pattern was observed. It is likely that an artifact of the nuclear isolation was responsible for the loss of the stage-specificity of WGA staining. ConA, in contrast, crossreacted with only the degenerating macronucleus, especially with the envelope (Fig. 6B and arrow). Different from WGA, ConA-binding specificity did not change during the nuclear isolation procedure. This observation suggests that the nucleus exhibits glycoprotein or glycolipid-carrying alpha-mannosyl oligosaccharides on their surface. To confirm the specific appearance of glycoproteins in the degenerating macronucleus, we isolated nuclear proteins from both nonconjugating and conjugating cells and performed lectin-western blot analysis (Fig. 7). Contrary to our expectation, no significant differences were observed against both ConA and WGA. This suggests that ConA-positive glycoproteins or lipids are usually present, but restricted to the inner leaflet of the macronuclear envelope and are exposed to the outer surface of the nucleus at the beginning of PND.
Figure 5.
Binding of lectin to the parental macronucleus. The Mac IIp-stage cells were fixed and stained with various FITC-labeled lectin probes. WGA: Wheat germ agglutinin. ConA: Concanavalin A. DBA: Dolichos biflorus agglutinin. PRA: Peanut root lectin. RCA I: Ricinus communis agglutinin. SBA: Soybean agglutinin. UEA: Ulex europaeus agglutinin. The FITC fluorescence is concentrated around the parental macronucleus when WGA or ConA are applied (arrow heads). pMa denotes the parental macronucleus. The scale bar in the photograph indicates 10 µm.
Figure 6.
Increase of lectin binding in the parental macronucleus. (A) Cells were fixed and stained with FITC-labeled WGA or ConA probes. FITC fluorescence is concentrated around the parental macronucleus at the nuclear degeneration stage (arrowheads). (B) Isolated nuclei were stained with FITC-labeled WGA or ConA probes. The degenerating macronucleus shows an affinity with ConA in the envelope (arrow). pMa and mi denote the parental macronucleus and micronucleus, respectively. The scale bar in (A) indicates 10 µm.
Figure 7.
Comparative analysis of nuclear glycoprotein present in starved cells and Mac IIp-stage cells. Nuclei were isolated from starved cells or Mac IIp-stage cells and the lysates were analyzed by lectin-western blotting. The pattern of separated proteins that was visualized by silver staining is also shown. Positions of markers are indicated at the left with molecular masses in kilodaltons (kDa).
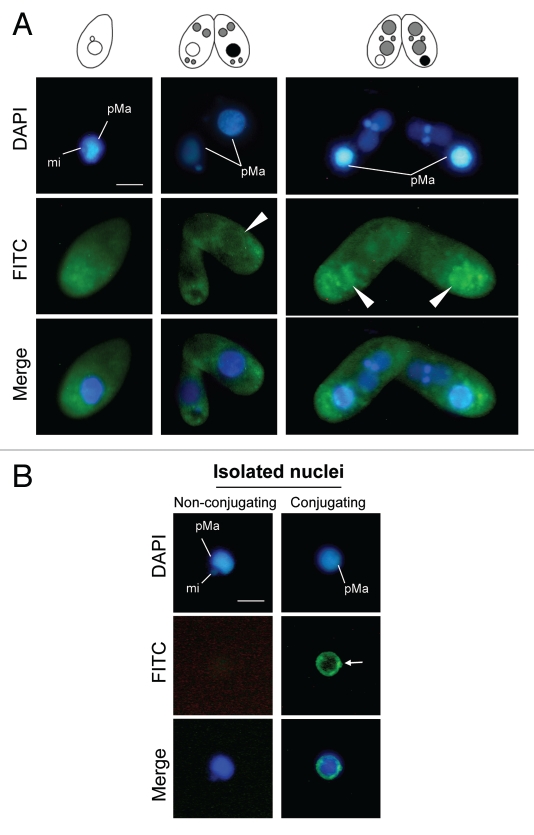
This characteristic is reminiscent of the exposure of phospholipid phosphatidylserine (PS) in animal cells, which is the best characterized phagocytosis marker.16,17 We therefore asked whether PS is exhibited on the degenerating macronucleus. We first examined binding of FITC-labeled annexin V using fixed Tetrahymena cells. As shown in Figure 8A, specific binding of annexin V around the degenerating macronucleus was detected. The specificity appeared when the parental macronucleus began to condense (Fig. 8A and arrowheads). When annexin V was incubated with isolated macronuclei, the specific binding of annexin V was also detected restricted on the envelope of the degenerating macronucleus (Fig. 8B and arrow), but not with the macronucleus from nonconjugating cells. We conclude that PND involves chemical modifications of the macronuclear envelope and these modifications could be the target signals for the autophagic/lysosomal activity in a single cell.
Figure 8.
Exhibition of an apoptotic molecular marker on the parental macronucleus. (A) The cells were fixed and stained with a FITC-labeled annexin V probe. The FITC fluorescence is concentrated around the parental macronucleus reflecting nuclear degeneration (arrowheads). (B) Isolated nuclei were stained with FITC-labeled annexin V probe. The degenerating macronucleus shows an affinity for annexin V in the envelope (arrow). pMa and mi denote the parental macronucleus and micronucleus, respectively. The scale bar in (A) indicates 10 µm.
The frequency of the lectin or annexin V positive staining on fixed cells or isolated nuclei is presented in Table 1.
Table 1.
Cross-reactivities between FITC-labeled indicators and Tetrahymena
| Indicators | Number of observation | Number of positiveness | Percentage of positiveness | |
| WGA | Fixed cells | 66 | 64 | 96.97 |
| Isolated pMa | 42 | 42 | 100 | |
| ConA | Fixed cells | 55 | 55 | 100 |
| Isolated pMa | 38 | 38 | 100 | |
| AnnexinV | Fixed cells | 50 | 11 | 22 |
| Isolated pMa | 72 | 24 | 33.33 | |
Discussion
PND is an essential process to ensure the establishment of the new generation in T. thermophila. Following differentiation of the new macronucleus, the parental macronucleus is committed to death and some remarkable changes occur via a crosstalk between the new and parental macronuclei. For instance, the level of dimethylation of histone H3K4 rapidly increases in the new macronucleus, while loss of the methyl mark occurs from the parental macronucleus when condensation of the parental macronucleus just begins (Fig. 4). AIF and mitochondrial DNases participate in this early stage of PND.9 To understand PND in more detail, we addressed two important questions. The first question was to elucidate autophagic/lysosomal events, which occur around the parental macronucleus. The second question was to know how the digestive vesicle complexes recognize the parental macronucleus. Below, we discuss these two issues.
A unique autophagic-lysosomal process for eliminating the parental macronucleus.
In general, unnecessary organelles or toxic substances in eukaryotic cells are removed from the cytoplasm by at least two intracellular degradation systems. One such system is the ubiquitin-proteasome system, which is responsible for the selective degradation of most proteins.27,28 Another is the autophagic/lysosomal system, in which substances from both inside and outside of cells are engulfed by a double-membrane autophagosome and delivered to resorptive compartments.6 There are multiple pathways to the lysosome for degradation, i.e., endocytosis/phagocytosis, macroautophagy, microautophagy and chaperone-mediated autophagy.13,29 In macroautophagy, regulated particularly by nutrient conditions, a number of autophagy-related (ATG) genes have been identified using yeast genetics.30–32 Most of the gene products function at the steps of autophagosome formation and elongation of the phagophore.
In mammalian or yeast macroautophagy, the formation of a membranous structure enclosing the targeted components, such as aged mitochondria, is crucial for sequestering them from the remaining cytoplasm, called autophagosome formation.32 Although the origin of the membrane is controversial, autophagosomes fuse with lysosomes to form an autolysosome and the sequestered contents are degraded by lysosomal enzymes and then recycled.13,32
In this study, we showed that T. thermophila PND involves a peculiar autophagic/lysosomal process. Unlike the cases of macroautophagy in other systems, the envelope itself of the parental macronucleus appears to directly change to an autophagic membrane (Fig. 1D and 3B–D), concomitant with nuclear condensation, followed by the subsequent approach of the digestive vesicle complexes and the nuclear acidification in distinct stages. Our findings are consistent with a previous cytochemical analysis utilizing acid phosphatase, but do not support the lysosomal-wrapping model proposed by Lu and Wolfe (2001),15 in which model small lysosomal bodies gather around the parental macronucleus and fuse to each other, leading to the formation of a large autophagosome. In fact, our findings rather indicate a possibility that the MDC/LTR-positive vesicles directly contact with the nuclear envelope without mutual fusion of them (Fig. 1E–H and 3E and F). AIF and/or mitochondrial DNase, both of which are major executors in the early stages of PND, translocate from the sequestered mitochondria to the inside of the macronucleus by this stage,9 whereas the inside of the macronucleus is not acidified until the final resorption stage (Fig. 1h and 2b–d). The AIF-mediated DNase activity has an optimal pH at around 6.5,7 which is responsible for the kb-sized DNA fragmentation and nuclear condensation.
These observations suggest that some MDC-positive vesicles contain permeabilized mitochondria and that only the vesicles fuse with the macronuclear membrane in the early stage of PND, but the attached lysosomes remain at the periphery of the nucleus. Two sorts of digestive vesicles might fuse with the nuclear envelope in a stepwise fashion and could differentially release their contents into the nucleus at distinct stages. Previously, fine-structure observations on the degenerating macronucleus in T. thermophila revealed that an initial event of nuclear condensation involves transformation of the nuclear envelope, e.g., the losses of nuclear pore complexes, attached ribosomes associated with the outer membrane and continuities with the endoplasmic reticulum.12 These observations on the nuclear membrane and our present findings strongly support the idea that PND involves drastic changes in the nuclear envelope (discussed below) and sequestration of the parental macronucleus from the cytoplasm and other organelles is achieved by direct change of the parental macronuclear membrane itself.
Possible “attack-me” signals on the nuclear membrane.
The second issue is how the digestive vesicle complexes recognize the parental macronucleus. Some specific signals should be indispensable for the selective elimination of the targeted nucleus. The presence of such specific signals on the nuclear membrane ensures the reliable death of the parental macronucleus despite the coexistence of the different kinds of nuclei in the same cytoplasm.
Early assumptions were that autophagy was a random process. During the last decade, however, increasing evidence has been changing this view to one in which macroautophagy is a selective process rather than a random process.33,34 As mentioned previously, the mechanism of mitochondrial turnover predominantly consists of macroautophagic sequestration and delivery to lysosomes for hydrolytic degradation, a process also called mitophagy.35 In this process, dysfunctional or aged mitochondria are specifically targeted by PAS, depending on the loss of membrane potential.33,35 Such timely and selective elimination of disused organelles is essential to protect cells from the harm of disordered metabolism and release of hazardous proteins. The yeast cytoplasm-to-vacuole targeting pathway (Cvt), is one such well-understood example.36,37
In Tetrahymena, the digestive vesicle complexes seem to approach the parental macronucleus by targeting MDC-positive signals of the nuclear envelope (Fig. 1D–H and d–h), comparable to a pinpoint attack by guided missiles. We propose to call such a signal an “attack-me” signal. Our findings also indicate that the envelope of the parental macronucleus becomes lectin- (ConA or WGA) and annexin V-positive as well as being MDC-stainable (Fig. 6 and 8), when the nuclear condensation, the fine-structural transformation of the nuclear envelope,12 and the loss of methyl mark from the parental macronucleus are initiated (Fig. 4). This type of change in lipid/protein composition and distribution in the nuclear membrane is quite similar to the mechanism underlying phagocytosis in apoptosis or enucleation in animals,38–40 suggesting that it is a prerequisite for guiding to the nucleus that is to be eliminated. Along with the intracellular degradation systems, phagocytosis in animal apoptosis is essential not only for clearance of unnecessary cells from the body, but also for the maintenance of tissue homeostasis.41,42 Apoptosis involves a drastic change of the plasma membrane, and it causes migration and change of distribution of PS and variation of glycoconjugates from the inner to the outer leaflet.38,43 These exposed molecules are recognized by phagocytes and are referred to as the “eat-me” signal.17 Enucleation, on the other hand, is an entirely different apoptotic process, in which only a nucleus is eliminated without degradation of cells. Enucleation has been observed in differentiation of erythrocytes41 and lens periphery cells,42 and in platelet biogenesis.46,47 For instance, as lens periphery cells differentiate to lens fiber cells, they lose nuclei and most organelles to increase transparency, an essential attribute of the lens.48 The nucleus to be expelled rapidly exposes PS on the surface, providing an “eat-me” signal as apoptotic cells do.39,40 Some molecules on the macronuclear envelope identified in this study would be candidates of the “attack-me” signals in Tetrahymena by the digestive vesicle complexes.
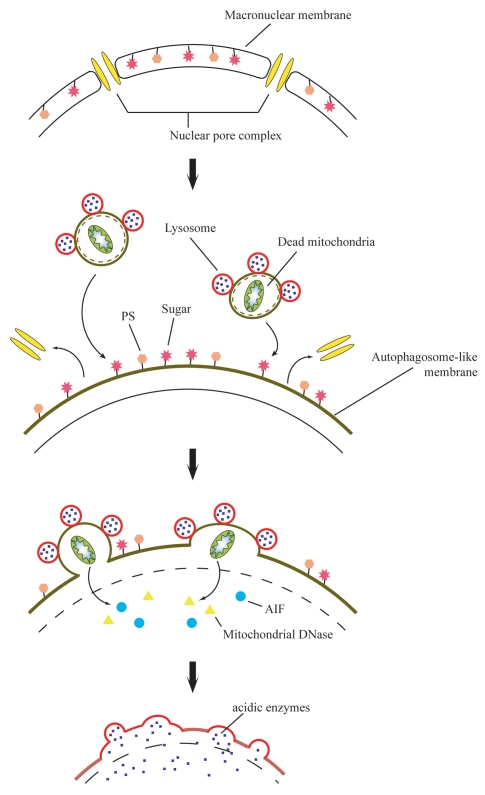
The overall autophagic/lysosomal process on the parental macronucleus during PND is summarized in Figure 9. Concomitant with nuclear condensation and loss of the methyl mark, alteration of the macronuclear envelope into an autophagosomal structure occurs by itself, corresponding to autophagosome formation in mammalian or yeast macroautophagy. Since a specificity of MDC-staining is derived from interaction with lipid molecules found in high concentration in autophagosomes,25 membrane lipid composition may be modified at the early stage of PND. The nuclear alteration involves the exposing of sugars and PS previously restricted in the inner leaflet on their surface. These exposed molecules act as an “attack-me” signal, which is responsible for attraction of digestive vacuole complexes. The digestive vesicle complexes fuse with the nuclear envelope in a stepwise fashion and release their contents into the nucleus at distinct stages, the initial stage of DNA degradation and the final resorption stage.
Figure 9.
Cartoon illustrating a possible sequence of events leading to the unique atophagic/lysosomal nuclear degradation in Tetrahymena. Concomitant with nuclear condensation, alteration of the macronuclear envelope occurs, which is similar to autophagosome formation. Sugars and phosphatidylserine (PS) restricted in the inner leaflet are exposed on their surface at the nuclear alteration stage. These act as an “attack-me” signal, which is responsible for the attraction of the digestive vacuole complexes. The two sorts of digestive vesicles fuse with the nuclear envelope stepwise and release their contents into the nucleus at distinct stages, the initial stage of DNA degradation and the final resorption stage.
While ciliates may have developed a unique pathway for executing PND, many original parts of the autophagic and apoptotic machineries seem to have been diverted to the unique autophagy of Tetrahymena. To obtain a better understanding of the regulatory pathway that controls PND, the specific “attack-me” signals, which target the parental macronucleus for autophagic/lysosomal degradation, need to be specified more precisely. Obtaining such information will provide new insights into intracellular signaling and recognition system in ciliates.
Materials and Methods
Culture methods and the induction of conjugation.
T. thermophila strains CU428 and B2086 (wild-type), were distributed from the National Tetrahymena Stock Center (Cornell University; http://tetrahymena.vet.cornell.edu/). Cells were cultured at 26°C in 2% proteose peptone (Difco, 211684), 1% yeast extract (Difco, 212750) and 0.5% glucose. To induce mating, the cells were incubated in 0.25% proteose peptone, 0.25% yeast extract and 4% glucose at room temperature. At mid-log phase, the cells were washed with 10 mM Tris-HCl (pH 7.2) and incubated overnight. To induce conjugation, equal numbers of both strains were mixed and kept at 26°C.
Labeling of autophagic vacuoles and acidic compartments.
Autophagic vacuoles and acidic compartments were labeled with 0.1 mM monodansylcadaverine (MDC; BioChemika, 30432) and 1 µM LysoTracker Red (LTR; Molecular Probes Inc., L-7528), respectively. Hoechst 33342 (Dojindo, H342; 5 µg/ml) was also used to stain nuclei. Living cells in 10 mM Tris-HCl were incubated with these fluorescence compounds at room temperature for 10 minutes. After incubation, the cells were washed with 10 mM Tris-HCl (pH 7.2) and immediately analyzed by fluorescence microcopy. For photography, cells were anesthetized with NiCl2 (Nacalai Tesque, 24224-1; 100 µM).
Confocal live cell imaging.
To monitor the acidic condition of the parental macronucleus, living cells in 10 mM Tris-HCl (pH 7.2) were incubated with 1 µM LTR and 0.1 µM SYTO 11 nucleic acid stain (Molecular Probes Inc., S-7572) at room temperature for 30 minutes. After incubation, the cells were washed with 10 mM Tris-HCl and immediately analyzed by confocal microscopy (OLYMPUS, BX51). For photography, cells were anesthetized with 100 µM NiCl2.
Preparation of cells for electron microscopy.
Conjugating cells were fixed with 2% glutaraldehyde and kept for 30 min. Glutaraldehyde was washed away by centrifugation and the samples were resuspended in 0.1 M cacodylate buffer containing 0.5% OsO4, 50 mM KCl and 2.5 mM MgCl2. The samples were then washed three times in the 0.1 M cacodylate buffer, postfixed with 2% OsO4 in distilled water for 1 h, dehydrated with ethanol and embedded in Epon (Oken 02-1001). The ultrathin sections were stained with 5% uranyl acetate and lead citrate. Observations were performed on a Hitachi H-7100 transmission electron microscope.
Cytological analysis.
Lectin binding. To examine binding of lectin to the parental macronucleus, cells were fixed and membrane-permeabilized by successive treatment with 4% paraformaldehyde and 0.1% Tween 20. The samples were then washed with PBS (0.2 g KCl, 0.24 g KH2PO4, 8 g NaCl, 1.44 g Na2HPO4, adjusted to pH 7.4 and brought to 1 L with distilled water) and incubated with 10 µg/ml of FITC-labeled lectin probes (wheat germ agglutinin (WGA)/concanavalin A (ConA)/dolichos biflorus agglutinin (DBA)/peanut root lectin (PRA)/ricinus communis agglutinin (RCA I)/soybean agglutinin (SBA)/ulex europaeus agglutinin (UEA); Vector Laboratories, FLK-2100) on ice for 1 h. DAPI (Dojindo 340-7971; 1 µg/µl) was also used to stain nuclei. After incubation, the samples were washed with PBS and analyzed by fluorescence microcopy.
Annexin V binding. An annexin V apoptosis assay kit (BioVision, K101-100) was used to detect phosphatidylserine on the parental macronucleus. To examine binding of annexin V to the macronucleus, cells were fixed and membrane-permeabilized as described above. The samples were then washed with attached reaction buffer and incubated with FITC-labeled annexin V (1:100 dilution) at room temperature for 10 min. DAPI (1 µg/µl) was also used to stain nuclei. After incubation, the samples were washed with reaction buffer and analyzed by fluorescence microcopy.
Nuclear isolation.
To isolate nuclei from Mac IIp cells, conjugating cells were harvested by centrifugation and washed with 10 mM Tris-HCl (pH 7.2). The washed cell pellets were resuspended in cold nuclear isolation solution (NIS) containing 3% sucrose, 5 mM MgCl2 and 10 mM Tris-HCl (pH 7.4). To lyse cells, 0.63% octanol was added with 100 µM PMSF and homogenized on ice. The nuclei were purified by centrifugation through a 2.2 M sucrose solution containing 5 mM MgCl2, 5 mM CaCl2, 25 mM KCl and 10 mM Tris-HCl (pH 7.2) in 13 PET centrifuge tubes. The sample solution was layered onto the sucrose and centrifuged at 22,500 rpm for 1 h at 4°C using an RPS40T rotor in an SCP70H ultracentrifuge (Hitachi). After centrifugation, the supernatants were discarded and the sedimented nuclei were re-suspended in NIS. For use in cytological analysis, the nuclei were treated according to the procedure above without fixation and membrane-permeabilization.
Lectin-western blot analysis.
Isolated nuclei, purified as described above, were used for lectin-western blot analysis. NIS was washed by centrifugation and the samples were lysed with a buffer consisting of 150 mM NaCl, 100 mM EDTA, 1% TritonX-100, 2.5% 2-mercaptoethanol, 10 mM HEPES (pH 8.0) and incubated at 4°C for 2 h. The resulting lysates were centrifuged at 10,000× g for 10 min and the supernatants were collected. Proteins in this fraction (15 µg) were boiled at 98°C for 5 min in 2X SDS buffer consisting of 4% SDS, 5% 2-mercaptoethanol, 20% glycerol and 0.1 M Tris-HCl (pH 6.8), separated by electrophoresis through 12% polyacrylamide-SDS (SDS-PAGE) and electrophoretically transferred onto a polyvinylidene fluoride membrane (Bio-Rad 162-0255). The membrane was washed with a blocking buffer containing 150 mM NaCl, 0.05% Tween 20 and 10 mM Tris-HCl (pH 7.4), incubated with biotin-conjugated WGA (Vector Laboratories, T0924) or ConA (Vector Laboratories, B1005) for 1 h and washed. It was then reacted with alkaline phosphatase-conjugated avidin D (Vector Laboratories, V0414) and the signals were visualized using the CDP-star Detection Reagent (Bio-Rad, N7001S).
Indirect immunofluorescence.
To image Lys 4 methylation of histone H3 in the new and parental macronuclei during conjugation, cells were fixed in 4% paraformaldehyde and membrane-permeabilized with cold acetone for 20 min. After washing with PBS, the cells were blocked with 1% bovine serum albumin and incubated with the rabbit α-Me[Lys4]-H3 (MILLIPORE, DAM1570816; 1:500 dilution) for 3 h. After washing twice with PBS, the cells were incubated with secondary antibody, (Molecular Probes Inc., A-11034; 1:500 dilution), for 1 h. Following antibody labeling, the cells were stained with propidium iodide (1 µg/µl) for 1 min, washed and observed.
Acknowledgements
We thank Toshinobu Suzaki and Yasuhiro Fukuda for technical support with electron microscopy and Karen Rethoret for technical support with confocal microscopy. Thanks also to Atsushi Mukai for providing the fluorescence micrograph in Figure 4. We are very grateful to Eriko Osada and Jyoti Garg for helpful discussion and encouragement. This study was supported by Grant-in-Aid for JSPS Fellows (21-2589) to T.A. and a grant from the Canadian Institutes for Health Research (MOP 97799) to R.E.P.
Footnotes
Previously published online: www.landesbioscience.com/journals/autophagy/article/13287
References
- 1.Mochizuki K, Gorovsky MA. Small RNAs in genome rearrangement in Tetrahymena. Curr Opin Gent Dev. 2004;14:181–187. doi: 10.1016/j.gde.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 2.Davis MC, Ward JG, Herrick G, Allis CD. Programmed nuclear death: apoptotic-like degradation of specific nuclei in conjugating Tetrahymena. Dev Biol. 1992;154:419–432. doi: 10.1016/0012-1606(92)90080-z. [DOI] [PubMed] [Google Scholar]
- 3.Mpoke S, Wolfe J. DNA digestion and chromatin condensation during nuclear death in Tetrahymena. Exp Cell Res. 1996;225:357–365. doi: 10.1006/excr.1996.0186. [DOI] [PubMed] [Google Scholar]
- 4.Kobayashi T, Endoh H. Caspase-like activity in programmed nuclear death during conjugation of Tetrahymena thermophila. Cell Death Differ. 2003;10:634–640. doi: 10.1038/sj.cdd.4401216. [DOI] [PubMed] [Google Scholar]
- 5.Yakisich JS, Kapler GM. The effect of phosphoinositide 3-kinase inhibitors on programmed nuclear degradation in Tetrahymena and fate of surviving nuclei. Cell Deth Differ. 2004;11:1146–1149. doi: 10.1038/sj.cdd.4401473. [DOI] [PubMed] [Google Scholar]
- 6.Klionsky DJ, Cuervo AM, Seglen PO. Methods for monitoring autophagy from yeast to human. Autophagy. 2007;3:181–206. doi: 10.4161/auto.3678. [DOI] [PubMed] [Google Scholar]
- 7.Kobayashi T, Endoh H. A possible role of mitochondria in the apoptotic-like programmed nuclear death of Tetrahymena thermophila. FEBS J. 2005;272:5378–5387. doi: 10.1111/j.1742-4658.2005.04936.x. [DOI] [PubMed] [Google Scholar]
- 8.Endoh H, Kobayashi T. Death harmony played by nucleus and mitochondria: nuclear apoptosis during conjugation of Tetrahymena. Autophagy. 2006;2:129–131. doi: 10.4161/auto.2.2.2368. [DOI] [PubMed] [Google Scholar]
- 9.Akematsu T, Endoh H. Role of apoptosis-inducing factor (AIF) in programmed nuclear death during conjugation in Tetrahymena thermophila. BMC Cell Biol. 2010;11:13. doi: 10.1186/1471-2121-11-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mpoke S, Wolfe J. Differential staining of apoptotic nuclei in living cells: application to macronuclear elimination in Tetrahymena. J Histochem Cytochem. 1997;45:675–683. doi: 10.1177/002215549704500505. [DOI] [PubMed] [Google Scholar]
- 11.Ward JG, Davis MC, Allis CD, Herrick G. Effects of nullisomic chromosome deficiencies on conjugation events in Tetrahymena thermophila: insufficiency of the parental macronucleus to direct postzygotic development. Genetics. 1995;140:989–1005. doi: 10.1093/genetics/140.3.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weiske-Benner A, Eckert WA. Differentiation of nuclear structure during the sexual cycle in Tetrahymena thermphila; II. Degeneration and autolysis of macro-and micronuclei. Differentiation. 1987;34:1–12. [Google Scholar]
- 13.Mizushima N. Methods for monitoring autophagy. Int J Biochem Cell Biol. 2004;36:2491–2502. doi: 10.1016/j.biocel.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 14.Suzuki K, Kubota Y, Sekito T, Ohsumi Y. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells. 2007;12:209–218. doi: 10.1111/j.1365-2443.2007.01050.x. [DOI] [PubMed] [Google Scholar]
- 15.Lu E, Wolfe J. Lysosomal enzymes in the macronucleus of Tetrahymena during its apoptosis-like degradation. Cell Death Differ. 2001;8:289–297. doi: 10.1038/sj.cdd.4400807. [DOI] [PubMed] [Google Scholar]
- 16.Depraetere V. “Eat me” signals of apoptotic bodies. Nat Cell Biol. 2000;2:104. doi: 10.1038/35014098. [DOI] [PubMed] [Google Scholar]
- 17.Savill J, Gregory C. Apoptotic PS to phagocyte TIM-4: eat me. Immunity. 2007;27:830–832. doi: 10.1016/j.immuni.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 18.Eda S, Yamanaka M, Beppu M. Carbohydrate-mediated phagocytic recognition of early apoptotic cells undergoing transient capping of CD43 glycoprotein. J Biol Chem. 2004;279:5967–5974. doi: 10.1074/jbc.M310805200. [DOI] [PubMed] [Google Scholar]
- 19.Baker KP, Baron WF, Henzel WJ, Spencer SA. Molecular cloning and characterization of human and murine DNase II. Gene. 1998;215:281–289. doi: 10.1016/s0378-1119(98)00280-7. [DOI] [PubMed] [Google Scholar]
- 20.Yasuda T, Takeshita H, Iida R, Nakajima T, Hosomi O, Nakashima Y, et al. Molecular cloning of the cDNA encoding human deoxyribonuclease II. J Biol Chem. 1998;273:2610–2616. doi: 10.1074/jbc.273.5.2610. [DOI] [PubMed] [Google Scholar]
- 21.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mizushima N, Yamamoto A, Hatano M, Kobayashi Y, Kabeya Y, Suzuki K. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J Cell Biol. 2001;152:657–667. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Biederbick A, Kern HF, Elsasser HP. Monodansylcadaverine (MDC) is a specific in vivo marker for autophagic vacuoles. Eur J Cell Biol. 1995;66:3–14. [PubMed] [Google Scholar]
- 24.Munafó DB, Colombo MI. A novel assay to study autophagy: regulation of autophagosome vacuole size by amino acid deprivation. J Cell Sci. 2001;114:3619–3629. doi: 10.1242/jcs.114.20.3619. [DOI] [PubMed] [Google Scholar]
- 25.Niemann A, Takatsuki A, Elsässer HP. The lysosomotropic agent monodansylcadaverine also acts as a solvent polarity probe. J Histochem Cytochem. 2000;48:251–258. doi: 10.1177/002215540004800210. [DOI] [PubMed] [Google Scholar]
- 26.Taverna SD, Coyne RS, Allis CD. Methylation of histone h3 at lysine 9 targets programmed DNA elimination in Tetrahymena. Cell. 2002;110:701–711. doi: 10.1016/s0092-8674(02)00941-8. [DOI] [PubMed] [Google Scholar]
- 27.Hochstrasser M. Protein degradation or regulation: Ub the judge. Cell. 1996;84:813–815. doi: 10.1016/s0092-8674(00)81058-2. [DOI] [PubMed] [Google Scholar]
- 28.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 29.Dice JF. Peptide sequences that target cytosolic proteins for lysosomal proteolysis. Trends Biochem Sci. 1990;15:305–309. doi: 10.1016/0968-0004(90)90019-8. [DOI] [PubMed] [Google Scholar]
- 30.Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993;333:169–174. doi: 10.1016/0014-5793(93)80398-e. [DOI] [PubMed] [Google Scholar]
- 31.Kamada Y, Funakoshi T, Shintani T, Nagano K, Ohsumi M, Ohsumi Y. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol. 2000;150:1507–1513. doi: 10.1083/jcb.150.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suzuki K, Kirisako T, Kamada Y, Mizushima N, Noda T, Ohsumi Y. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 2001;20:5971–5981. doi: 10.1093/emboj/20.21.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim I, Rodriguez-Enriquez S, Lemasters JJ. Selective degradation of mitochondria by mitophagy. Arch Biochem Biophys. 2007;462:245–253. doi: 10.1016/j.abb.2007.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kanki T, Klionsky DJ. Mitophagy in yeast occurs through a selective mechanism. J Biol Chem. 2008;283:32386–32393. doi: 10.1074/jbc.M802403200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Elmore SP, Qian T, Grissom SF, Lemasters JJ. The mitochondrial permeability transition initiates autophagy in rat hepatocytes. FASEB J. 2001;15:2286–2287. doi: 10.1096/fj.01-0206fje. [DOI] [PubMed] [Google Scholar]
- 36.Klionsky DJ, Ohsumi Y. Vacuolar import of proteins and organelles from the cytoplasm. Annu Rev Cell Dev Biol. 1999;15:1–32. doi: 10.1146/annurev.cellbio.15.1.1. [DOI] [PubMed] [Google Scholar]
- 37.Yorimitsu T, Klionsky DJ. Eating the endoplasmic reticulum: quality control by autophagy. Trends Cell Biol. 2007;17:279–285. doi: 10.1016/j.tcb.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 38.Savill J, Fadok V, Henson P, Haslett C. Phagocyte recognition of cells undergoing apoptosis. Immunol Today. 1993;14:131–136. doi: 10.1016/0167-5699(93)90215-7. [DOI] [PubMed] [Google Scholar]
- 39.Yoshida H, Kawane K, Koike M, Mori Y, Uchiyama Y, Nagata S. Phosphatidylserine-dependent engulfment by macrophages of nuclei from erythroid precursor cells. Nature. 2005;437:754–758. doi: 10.1038/nature03964. [DOI] [PubMed] [Google Scholar]
- 40.Miyanishi M, Tada K, Koike M, Uchiyama Y, Kitamura T, Nagata S. Identification of Tim4 as a phosphatidylserine receptor. Nature. 2007;450:435–439. doi: 10.1038/nature06307. [DOI] [PubMed] [Google Scholar]
- 41.Kerr JF, Wyllie AH, Currie AR. Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br J Cancer. 1972;26:239–257. doi: 10.1038/bjc.1972.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jacobson MD, Weil M, Raff MC. Programmed cell death in animal development. Cell. 1997;88:347–354. doi: 10.1016/s0092-8674(00)81873-5. [DOI] [PubMed] [Google Scholar]
- 43.Hart SP, Haslett C, Dransfield I. Recognition of apoptotic cells by phagocytes. Cell Mol Life Sci. 1996;52:950–956. doi: 10.1007/BF01920103. [DOI] [PubMed] [Google Scholar]
- 44.Daugas E, Candé C, Kroemer G. Erythrocytes: death of a mummy. Cell Death Differ. 2001;8:1131–1133. doi: 10.1038/sj.cdd.4400953. [DOI] [PubMed] [Google Scholar]
- 45.Wride MA. Minireview: apoptosis as seen through a lens. Apoptosis. 2000;5:203–209. doi: 10.1023/a:1009653326511. [DOI] [PubMed] [Google Scholar]
- 46.Kaluzhny Y, Ravid K. Role of apoptotic processes in platelet biogenesis. Acta Haematol. 2004;111:67–77. doi: 10.1159/000074487. [DOI] [PubMed] [Google Scholar]
- 47.Gordge MP. Megakaryocyte apoptosis: sorting out the signals. Br J Pharmacol. 2005;145:271–273. doi: 10.1038/sj.bjp.0706202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ishizaki Y, Jacobson MD, Raff MC. A role for caspases in lens fiber differentiation. J Cell Biol. 1998;140:153–158. doi: 10.1083/jcb.140.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]