Abstract
Atg4 cysteine proteases (autophagins) play crucial roles in autophagy by proteolytic activation of Atg8 paralogs for targeting to autophagic vesicles by lipid conjugation, as well as in subsequent deconjugation reactions. However, the means to measure the activity of autophagins is limited. Herein, we describe two novel substrates for autophagins suitable for a diversity of in vitro assays, including (i) fluorogenic tetrapeptide acetyl-Gly-L-Thr-L-Phe-Gly-AFC (Ac-GTFG-AFC) and (ii) a fusion protein comprised of the natural substrate LC3B appended to the N-terminus of phospholipase A2 (LC3B-PLA2), which upon cleavage releases active PLA2 for fluorogenic assay. To generate the synthetic tetrapeptide substrate, the preferred tetrapeptide sequence recognized by autophagin-1/Atg4B was determined using a positional scanning combinatorial fluorogenic tetrapeptide library. With the LC3B-PLA2 substrate, we show that mutation of the glycine proximal to the scissile bond in LC3B abolishes activity. Both substrates showed high specificity for recombinant purified autophagin-1/Atg4B compared to closely related proteases and the LC3B-PLA2 substrate afforded substantially higher catalytic rates (kcat/Km 5.26 × 105 M−1/sec−1) than Ac-GTFG-AFC peptide (0.92 M−1/sec−1), consistent with substrate-induced activation. Studies of autophagin-1 mutants were also performed, including the protease lacking a predicted autoinhibitory domain at residues 1 to 24 and lacking a regulatory loop at residues 259 to 262. The peptide and fusion protein substrates were also employed for measuring autophagin activity in cell lysates, showing a decrease in cells treated with autophagin-1/Atg4B siRNA or transfected with a plasmid encoding Atg4B (Cys74Ala) dominant-negative. Therefore, the synthetic substrates for autophagins reported here provide new research tools for studying autophagy.
Key words: autophagin, fluorogenic assay, tetrapeptide, phospholipase A2, LC3
Introduction
Autophagy is an evolutionarily conserved lysosome-dependent process whereby cells catabolize proteins and various macromolecules as well as various organelles, serving a general “housekeeping” function to avoid toxic accumulation of unfolded proteins and senescent organelles. Autophagy also provides a means of generating substrates for sustaining ATP production during times of nutrient deprivation (reviewed in ref. 1 and 2). The process of macroautophagy involves formation of double-membrane vesicles engulfing cytosol and organelles, delivering their contents to lysosomes for digestion. Microautophagy and chaperone-mediated autophagy are alternative routes for achieving lysosome-dependent degradation of proteins and other cellular components. The genes responsible for autophagy (ATG) have been identified, largely through genetic analysis of the yeast, Saccharomyces cerevisiae,3 and are conserved in mammals, plants and essentially all eukaryotes.4,5 Dysregulation of autophagy may contribute to various diseases, including cancer and neurodegeneration, as well as playing important roles in responses to hypoxia and microbial infection.6
Two ubiquitin-like conjugation systems are critical to autophagy, which have been well characterized in yeast, involving conjugation of Atg8 to phosphatidylethanolamine (PE) and conjugation of Atg12 to Atg5.7,8 Atg7 is a common E1-like activating enzyme required for both ubiquitin-like proteins Atg8 and Atg12, whereas Atg3 and Atg10 are the E2-like enzymes specific for Atg8 and Atg12, respectively. Conjugation of Atg12 to Atg5 involves formation of a covalent isopeptide bond via its C-terminal Gly residue,4 analogous to the ubiquitin system. In contrast, Atg8 must undergo proteolytic processing to produce a C-terminal Gly appropriate for conjugation. In the case of Atg8, however, conjugation is to phospholipids rather than a protein acceptor, resulting in covalent attachment of Atg8 to autophagic vesicles.9
Atg8 is proteolytically activated through removal of a C-terminal extension by the cytoplasmic cysteine protease Atg4, converting pro-Atg8 to its active form. Atg4 is also required for deconjugating PE from the C-terminal Gly of Atg8-PE. Atg8-PE attached to the cytoplasmic side of autophagosomes and precursor membranes is required for clustering and hemifusion of autophagosomes with lysosomes,9,10 upon which Atg8-PE residing on the surface of autophagosomes is liberated by Atg4-mediated hydrolysis, allowing recycling of cleaved Atg8 molecules.11
The human genome contains four independent genes encoding Atg4 orthologs (termed autophagins), including Atg4A/autophagin-2, Atg4B/autophagin-1, Atg4C/autophagin-3 and Atg4D/autophagin-4.12 Human Atg4B and Atg4C rescue Atg4 deletion mutant yeast,12 suggesting functional equivalence. Humans also have at least four proteins (excluding isoform variants) that operate analogous to Atg8, including (i) microtubule-associated light chain 3 (LC3) (which has A, B and C isoforms), (ii) Golgi-associated ATPase enhancer of 16 kDa (GATE-16, also known as GABA(A) receptor-associated protein-like 2), (iii) GABA(A) receptor-associated protein (GABARAP) and (iv) GABA(A) receptor-associated protein-like 1 (GABARAP-L1).13–15 Atg4A appears to have activity specifically for proGATE-16 in vitro and (like many cysteine proteases) is negatively regulated by reactive oxygen species (ROS).16 Atg4B cleaves LC3, GATE-16 and GABARAP, while also delipidating LC3-II and GABARAP-II.14,17 In contrast, overexpressed Atg4C has little or no activity against lipidated form of LC3 and GABARAP-L1 in HeLa cells,13 and Atg4C knockout in cells results in very minor inhibitory effects on autophagy under starvation conditions.18 Mammalian expressed Atg4C, however, shows hydrolytic activity against the synthetic fluorescence-quenched peptide, 7-(methoxycoumarin-4-yl)-acetyl-ThrPhe-Gly-Met-N-3-(2,4-dinitrophenyl)-l-α,β-diamino propionyl-NH2.12 Recently, it was reported that Atg4D is cleaved at Asp63 by caspase-3, triggering in turn its cleavage of GABARAP-L1 and GATE-16.13 Caspase-cleaved Atg4D is cytotoxic via exposure of a C-terminal Bcl-2 homology 3 (BH3) domain that neutralizes anti-apoptotic members of the Bcl-2 family. Of the four human autophagins, only Atg4B has been shown to efficiently cleave LC3.13,19
Little is known about the molecular events that regulate autophagin (Atg4) activity in cells, including whether upstream activators, downstream inhibitors or post-translational modifications control the activities of these proteases. Assays for monitoring autophagin/Atg4 activity thus are needed to assist with advancing research on this unique family of intracellular cysteine proteases. In this study, we devised two types of reporter substrates for measuring autophagin-1 (Atg4B) activity, including a fluorogenic synthetic tetrapeptide and a LC3B-PLA2 fusion protein, which couples released PLA2 to a convenient fluorogenic assay.20,21 Altogether, the fluorogenic assays reported here are the first developed for Atg4B, providing powerful tools for high throughput screening (HTS) or for addressing fundamental questions about the regulation of these proteases.
Results
Recombinant autophagin-1 (Atg4B) cleaves Atg8 paralogs in a time- and concentration-dependent manner.
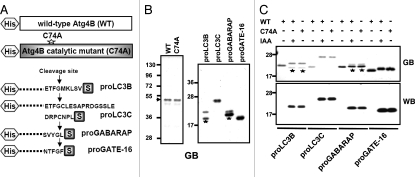
The cleavage of recombinant substrate proteins by Atg4B was tested in vitro. For these experiments, wild-type Atg4B, catalytic mutant C74A and precursor forms of ubiquitin-like substrates, including Atg8 para-logs proLC3B, proLC3C, proGABARAP and proGATE-16 were expressed in bacteria and purified. Atg4B and Atg4B C74A were produced with N-terminal His6-tags, while Atg8 substrates were produced with both N-terminal His6-tags and C-terminal S-tags (Fig. 1A). The recombinant proteins were purified by Ni-NTA-agarose and analyzed by SDS-PAGE to verify protein purity using GelCode Blue staining (Fig. 1B). Prior to treatment with Atg4B, single bands were observed for substrates LC3C and GATE-16, whereas additional partially degraded bands were found in preparations of recombinant LC3B and GABARAP.
Figure 1.
Recombinant Atg4B protease cleaves Atg8 substrates in vitro. (A) Schematic of recombinant wild-type Atg4B protease, catalytic mutant Atg4B C74A and its substrate Atg8 paralogs are depicted. Atg4B and a mutant C74A were expressed in bacteria fused with His6-tag at N-terminus. Atg8 paralogs were expressed in bacteria with fused His6-tag and S-tag at N- and C-terminus, respectively. (B) His6-tagged Atg4B, catalytic mutant Atg4B C74A and His- and S-tagged Atg8 paralogs including LC3B, LC3C, GABARAP and GATE-16 were expressed in E. coli BL21 DE3 cells. Proteins were purified by Ni-NTA resin. The purity of proteins was assessed by SDS-PAGE analysis and staining with GelCode Blue. Stars indicate partially degraded bands in preparations of recombinant LC3B and GABARAP (C), 100 nM recombinant Atg4B wild-type (WT) or C74A mutant was incubated with 400 nM of the various substrates in reaction buffer (50 mM Tris, pH 8.0, 150 mM NaCl and 1 mM DTT) with or without 10 mM iodoacetic acid (IAA) at 37°C for 1 h. Cleavage of protein substrates was visualized by SDS-PAGE analysis with GelCode Blue staining (GB, top part) and by immunoblotting using anti-S-tag antibody (WB, bottom part). Molecular weight markers are indicated in kilodaltons (kDa). Stars indicate partially degraded bands in preparations of recombinant LC3B and GABARAP.
Wild-type Atg4B cleaved the Atg8 paralogs LC3B and LC3C, as demonstrated by shifts in band size to lower apparent molecular mass in SDS-PAGE experiments where proteins were visualized by staining (Fig. 1C). Mobility shifts were also seen with GABARAP and GATE-16 following treatment with Atg4B, though the shift was very slight for GABARAP, as predicted (Fig. 1A). In contrast, substrate proteins remained intact when incubated either with Atg4B C74A catalytic mutant or with wild-type Atg4B treated with iodoacetamide (IAA), which blocks activity through modification of the active site cysteine (Fig. 1C, top). Cleavage was also confirmed by immunoblotting, using the C-terminal S-tags as markers. (Fig. 1C and bottom), showing essentially complete removal of the S-tag from all substrates following exposure to active Atg4B.
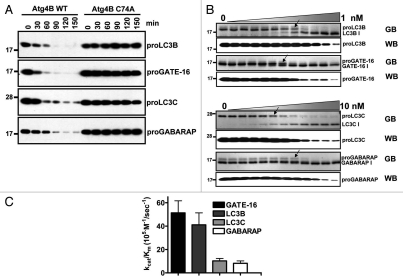
The cleavage in vitro of Atg4B substrate proteins was time- and concentration-dependent (Fig. 2A and B). Of the four natural protein substrates tested, the catalytic efficiency of Atg4B appeared to be most efficient for proGATE-16, which reached a kcat/Km value of 6.16 × 106 M−1sec−1 (Fig. 2C). The catalytic efficiency for substrates proLC3B, proLC3C and proGABARAP was estimated at 3.08 × 106, 1.23 × 106 and 6.16 × 105 M−1sec−1, respectively (Fig. 2C).
Figure 2.
Estimating activity of recombinant Atg4B using full-length Atg8 paralog substrates. (A) Atg4B WT or C74A mutant (1 nM) was incubated with 400 nM Atg8 paralogs at 37°C for various times as indicated. Cleavage of substrates was determined by immunoblotting using anti-S-tag antibody. (B) Atg8 paralogs (400 nM) were incubated with two-fold serial dilutions of wild-type Atg4B in reaction buffer (50 mM Tris, pH 8.0, 150 mM NaCl and 1 µM DTT) at 37°C for 1 h (where maximum final concentration of Atg4B is either 1 or 10 nM). Proteins were subjected to SDS-PAGE and stained with GelCode Blue (GB, top part for each pair). Arrows indicate dilution at which cleavage is detectable. The cleavage of Atg8 paralogs was further analyzed by immunoblotting using anti-S-tag antibody (WB, bottom part for each pair). EC50 for different substrates was indicated with arrowhead. (C), EC50 values for Atg4B cleavage of LC3B were used for calculating catalytic efficiency kcat/Km. Molecular weight markers are indicated in kilodaltons (kDa). Data represent the mean ± S.D. of three experiments.
Generation of optimized fluorogenic tetrapeptide substrates for Atg4B.
Previous reports suggest that low molecular weight synthetic substrates can be cleaved by Atg4C. To test if this is true for Atg4B, we synthesized fluorogenic tetrapeptides, Ac-NTFG-ACC (7-amino-4-carbamoylmethylcoumarin) and Ac-ETFG-ACC, which correspond to the last four amino acids proximal to the cleavage site of the natural substrates, GATE-16 and LC3B, respectively (Suppl. Data, Fig. S1). These substrates were hydrolyzed by Atg4B, although not very efficiently.
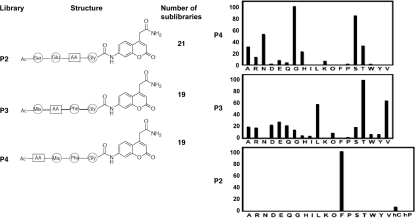
To identify alternative tetrapeptides for improving assay sensitivity, a PS-SCL was prepared with Gly fixed in the P1 position and P2, P3 and P4 varying among 19 L-amino acids (cysteine excluded), conjugated to the fluorogenic ACC leaving group (Fig. 3). Only glycine was tested at P1 due to structural information showing that this is the only amino acid tolerated (Suppl. Data, Fig. S2). Atg4B showed very high selectivity for Phe at the P2 position, whereas the P3 and P4 positions showed more promiscuity in terms of the amino acids tolerated. In the P3 position, the most preferred amino acid is threonine (consistent with natural substrates), followed by valine and leucine. In the P4 position, small amino acids are preferred, with glycine the most active, followed closely by serine. Less accepted residues at P4 included basic amino acids and those with hydrophobic side-chains (Fig. 3B).
Figure 3.
Synthetic substrate specificity of Atg4B. (A) The positional scanning strategy is depicted for using combinatorial peptide library to define optimal substrate sequence for Atg4B. (B) Summary of cleavage of fluorogenic tetrapeptides is presented. The enzyme concentration was 6–8 µM. ACC production was monitored with assay times varying from 15–60 min. Standard deviation for each measurement shown is <20%. The x-axis indicates the amino-acid tested at the P2, P3 or P4 positions (standard single letter code for natural L-amino acids; O, nor-leucine, hC, cyclohexylalanine, hP, homo-phenylalanine). The y-axis represents the average relative activity expressed as a percent of the best amino acid. ACC fluorescence was monitored using an fmax multiwell fluorescence plate reader at excitation wavelength of 355 nm and an emission wavelength of 460 nm for 15–60 mins.
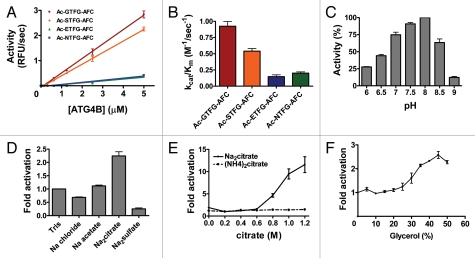
To further verify the preferred peptide sequence obtained from library screening, individual AFC (7-amino-4-trifluoromethylcoumarin) tetrapeptides were synthesized bearing the optimal sequences identified from PS-SCL (Ac-GTFGACC, Ac-STFG-ACC) as well as sequences from known natural substrates (Ac-NTFG-ACC, Ac-ETFG-ACC) and compared with respect to hydrolysis by Atg4B (Fig. 4A and Suppl. Data, Fig. S3). The catalytic efficiency of Atg4B on Ac-GTFG-AFC was almost twice that of Ac-STFG-AFC and 6 and 4.5 fold better than Ac-ETFG-AFC or Ac-NTFG-AFC, respectively (Fig. 4B). These results thus independently confirm the PS-SCL determined substrate preferences for Atg4B/autophagin-1.
Figure 4.
Characterization of Atg4B activity using synthetic peptide substrates. (A) Various concentrations of recombinant Atg4B were incubated with 100 µM synthetic tetrapeptides conjugated with AFC in 50 mM Tris-HCl, pH 8.0, 5 mM DTT at 37°C for 30 min. Data are expressed at RFU per sec (mean ± S.D.; n = 3). (B) 1 µM Atg4B was incubated with two-fold serial dilutions of 100 µM synthetic tetrapeptide substrates at 37°C for 30 min. kcat/Km was calculated using AFC standard curve. (C) Activity of Atg4B was assayed in buffers (50 mM) titrated to various pH values as indicated in the presence of 150 mM NaCl and 5 mM DTT. The concentrations of Atg4B and Ac-GTFG-AFC were 1 µM and 50 µM, respectively. (D) Activity of Atg4B in 50 mM Tris-HCl buffer, pH 8.0, 5 mM DTT with different Hofmeister salts was assayed at 37°C for 30 min. The non-Hofmeister salt NaCl was used as a control to rule out effects from alteration of ionic strength. (E) 1 µM Atg4B was assayed with 50 µM Ac-GTFG-AFC in 50 µM Tris-HCl buffer, pH 8.0, 5 mM DTT in the presence of Na2Citrate (solid line) or (NH4)2Citrate (dashed line). (F) 1 µM Atg4B was assayed with 50 µM Ac-GTFG-AFC in 50 mM Tris-HCl buffer, pH 8.0, 5 mM DTT in the presence of various amounts of glycerol. Effect of different Hofmeister salts or glycerol on activity was expressed as fold increase. AFC production was measured with an excitation wavelength of 400 nm and an emission wavelength of 505 nm. Data represent the mean ± S.D. of four experiments.
To further optimize protease assay conditions using the preferred fluorogenic peptide substrate, Ac-GTFG-AFC, we determined the optimal buffer conditions using a range of pH values, Hofmeister salts and glycerol for recombinant purified Atg4B/autophagin-1. Protease activity was maximal at pH 8.0 (Fig. 4C), consistent with the notion that ionization of C74-H278-D280 triad in active site promotes catalytic activity, similar to other cysteine proteases.22 Among the Hofmeister salts, only sodium citrate increased Atg4B activity (Fig. 4D and E). Atg4B activity was also elevated in the presence of chemical chaperone glycerol, suggesting both sodium citrate and glycerol may stabilize an active conformation of Atg4B (Fig. 4F).
Experiments with cleavable reporter LC3B-PLA2.
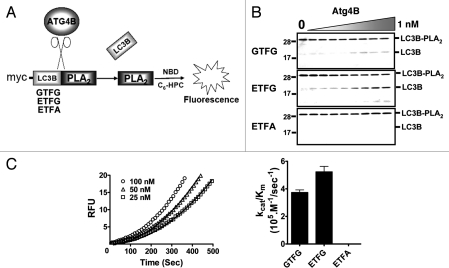
A free N-terminus is required for activity of PLA2, which provides the basis for creating substrates for endopeptidases.20,21 We devised a novel reporter fusion protein LC3B-PLA2 to measure Atg4B/autophagin-1 activity (Fig. 5A). PLA2 fused with natural substrate LC3B at its N-terminus is inactive. Cleavage of LC3B by Atg4B removes LC3B from the N-terminus of PLA2, restoring PLA2 activity. In addition to creating a LC3B-PLA2 fusion protein with wild-type LC3B as the fusion partner, we also created variants of LC3B in which the natural ETFG cleavage sequence was replaced by the optimal sequence deduced from peptide library screening (GTFG) or with the non-cleavable sequence ETFA, where the Gly proximal to the scissile bond was replaced with Ala (Fig. 5B). As determined by SDS-PAGE analysis, Atg4B cleaved the wild-type and GTFG-variant of the LC3B-PLA2 fusion protein substrate in protease concentration-dependent manner. In contrast, the Gly/Ala mutant of LC3B-PLA2 was not cleaved by Atg4B (Fig. 5B).
Figure 5.
Generation and characterization of a novel LC3B-PLA2 reporter substrate for Atg4B. (A) Scheme depicting basis for the LC3B-PLA2 reporter assay for monitoring Atg4B activity. Atg4B cleaves Gly117 site of proLC3B to release catalytically active PLA2, which hydrolyzes NBD-C6-HPC, generating fluorescence. LC3B-PLA2 fusion proteins were generated with various cleavage sites, containing the natural ETFG, altered GTFG or noncleavable ETFA sequences. (B) two-fold serial dilutions of Atg4B (maximum final concentration 1 nM) were incubated with 400 nM wild-type (ETFG) or mutant (GTFG, ETFA) LC3B-PLA2 fusion proteins at 37°C for 1 h. Cleavage of LC3B-PLA2 proteins was determined by SDS-PAGE with GelCode Blue staining. (C) Atg4B (0.05 nM) was mixed with two-fold dilutions of substrates to give a concentration range from ∼1.5–100 nM LC3B-PLA2 wild-type (ETFG) fusion protein in PLA2 reaction buffer at 37°C. PLA2 activity was monitored by hydrolysis of NBD-C6-HPC, measuring fluorescence with excitation and emission filters of 485 nm and 530 nm, respectively and reporting results as RFU (left part). Data points obtained with 25, 50 or 100 nM substrate are represented by the symbols of square, triangle and circle, respectively. Computer-generated curves were fit to the data for kcat/Km calculation of Atg4B. The deduced catalytic efficiency kcat/Km of Atg4B as measured with the various LC3B-PLA2 reporter substrates is shown in the right part. Data represent the mean ± S.D. of three experiments.
Treatment of the wild-type and GTFG variant LC3B-PLA2 fusion proteins with Atg4B also generated substrate concentration-dependent PLA2 activity (Fig. 5C). In contrast, essentially no PLA2 activity was detected in reactions containing the Gly/Ala mutant of LC3B/PLA2 (Suppl. Data, Fig. S4). Estimates of the catalytic efficiency of Atg4B using the wild-type (ETFG) and mutant (GTFG) LC3B-PLA2 substrates showed that the GTFG mutant is approximately 1.4-fold less efficient as a substrate than the wild-type ETFG (Fig. 5C), with estimated catalytic efficiencies of kcat/Km 3.8 × 105 and 5.3 × 105 M−1/sec−1 for mutant and wild type, respectively (Fig. 5C). Thus, in the context of the LC3B-PLA2 fusion protein, unlike the tetrapeptides, the wild-type ETFG sequence is preferred over GTFG by Atg4B/autophagin-1.
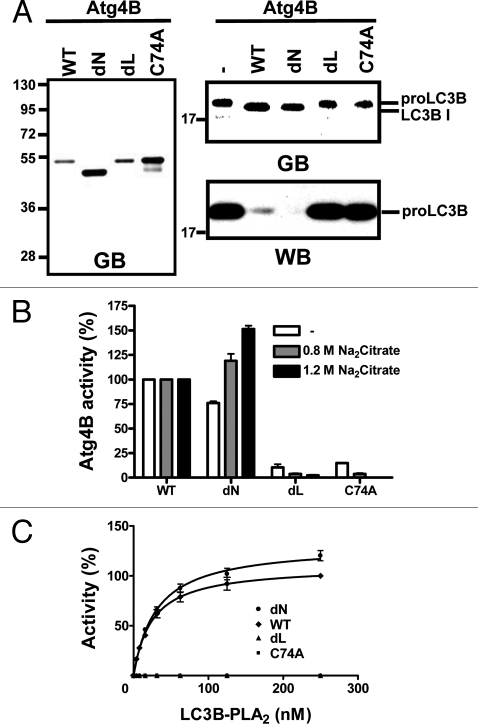
Structure-function analysis of Atg4B/autophagin-1.
The reported 3D-structure of the Atg4B-LC3 complex reveals a loop (amino acids 259–262) that covers the active site of Atg4B and also suggests that the N terminus (amino acids 1–24) impairs full access of substrates into or cleaved products out of the active site of the protease.23 Upon binding with substrate proLC3, the structural data suggest that the two regulatory regions of Atg4B significantly undergo conformational changes to expose the active site. Using the synthetic substrates, tetrapeptide Ac-GTFG-AFC and fusion protein LC3B-PLA2, we undertook a structure-function analysis of Atg4B, comparing the activity of the wild-type Atg4B protein with deletion mutants lacking the aforementioned N-terminal (dN) or loop regions (dL). Accordingly, the Atg4B wild-type and mutant proteins were expressed in bacteria, purified and their activities were compared by cleavage in vitro of full-length proLC3B (Fig. 6A). Atg4B (dN) retained activity, whereas the Atg4B (dL) mutant almost entirely lost activity. Similar results were obtained using the Ac-GTFG-AFC peptide substrate to monitor enzyme activity (Fig. 6B) with dN retaining activity and dL largely lacking activity. In fact, the N-terminal deletion mutant was slightly more active than the wild-type Atg4B when assayed in the presence of Na2-citrate (Fig. 6B), which could be a reflection that the region deleted is predicted to obstruct exit of substrates from the active site.23 Similar results were also obtained using the LC3B-PLA2 reporter substrate (Fig. 6C). When combined with the available structural data,25 these functional studies suggest that the regulatory loop of Atg4B/autophagin-1 is required for activity or for conformational stability of the active protease, while the N-terminal region appears to play a minor role in regulating protease activity. With respect to analysis of the Atg4B mutants, it is reassuring that similar conclusions were reached using all substrates tested, including the natural substrate proLC3B, the fluorogenic tetrapeptide substrate Ac-GTFG-AFC and the LC3B-PLA2 fusion protein. Also, though the dL mutant lacked activity, it was still capable of binding LC3B in vitro, suggesting that the protein was nevertheless folded (Suppl. Data, Fig. S5).
Figure 6.
Structure-function analysis of Atg4B. (A) His6-tagged recombinant wild-type Atg4B (WT), N-terminal deletion (dN), regulatory loop deletion (dL) or catalytic mutant (C74A) proteins were expressed in bacteria and purified by Ni-NTA resin. Purified Atg4B proteins (1 nM) were incubated with 400 nM LC3B at 37°C for 1 h. The purity of recombinant proteins (left) and cleavage of LC3B (right) were accessed by SDS-PAGE analysis and staining with GelCode Blue (GB). In addition, LC3B cleavage was assessed by immunoblotting using anti-S-tag antibody as described for Figure 1 (WB). (B) Activity of wild-type and mutant Atg4B proteins (1 µM) was assayed using 100 µM Ac-GTFG-AFC in 50 mM Tris-HCl, pH 8.0, 5 mM DTT, with 0, 0.8 M or 1.2 M sodium citrate, at 37°C. The activity of Atg4B mutants was normalized relative to activity of wild-type Atg4B for each buffer condition and expressed as % control. (C), WT (diamonds), dN (circles), dL (triangles) and C74A mutant (squares) Atg4B proteins (0.1 nM) were assayed using various concentrations of LC3B-PLA2 in PLA2 reaction buffer at room temperature for 30 min. Data are expressed as % relative to WT Atg4B and represent mean ± S.D. of four experiments.
Applications of Ac-GTFG-AFC and LC3B-PLA2 substrates for measuring activity of endogenous autophagins.
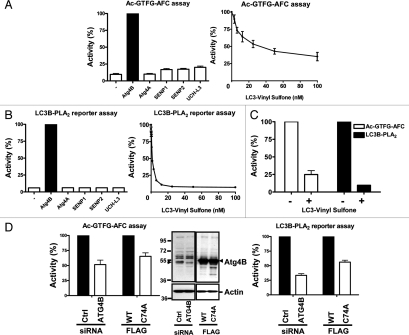
Prior to employing the autophagin synthetic substrates for cellular activity assays, we explored the specificity of the synthetic peptide Ac-GTFG-AFC and LC3B-PLA2 fusion protein substrates in terms of hydrolysis by other classes of cysteine proteases that cleave ubiquitin or ubiquitin-like proteins. To this end, the peptide and fusion protein substrates were mixed with autophagins (Atg4A or Atg4B), deSumolating enzymes (SENP-1 or -2) or deubiquitinating enzyme (UCHL3) in reaction buffer (Fig. 7A and B, left part). Note that prior to analysis the preparation of recombinant proteases were tested for ability to cleave in vitro natural substrates (Suppl. Data, Fig. S6), which confirmed the activity of Atg4A, Atg4B, SENP-1, SENP-2 and UCH-L3. The synthetic tetrapeptide Ac-GTFG-AFC was susceptible to hydrolysis by Atg4B, but not the other enzymes tested. The specificity of the assay was further determined with LC3-vinyl sulfone, which is expected to selectively inhibit autophagin family proteases, reacting with the active site cysteine to irreversibly inhibit these enzymes.14 LC3-vinyl sulfone inhibited activity of recombinant purified Atg4B in a dose-dependent manner at nanomolar concentrations (Fig. 7A and B right part). The potency and efficiency of the LC3 vinyl sulfone inhibitor was greater when measuring Atg4B activity using the LC3B-PLA2 fusion protein as the substrate compared to the tetrapeptide Ac-GTFG-AFC, probably because of direct competition with the LC3 component of the fusion protein and because the catalytic efficiency of the LC3-PLA2 substrates is much higher, allowing ∼50-times less enzyme to be used for assays. The inhibitory effect of LC3-vinyl sulfone on autophagin activity was confirmed in cell lysates as containing the endogenous proteases (Fig. 7C). Thus, LC3-vinyl sulfone is a suitable research tool for monitoring Atg4B activity using our LC3B-PLA2 fusion protein substrate, though less reliable when using Ac-GTFG-AFC peptide substrate due to residual protease activity.
Figure 7.
Specificity of tetrapeptide and LC3B-PLA2 substrates. (A) 1 µM of recombinant Atg4 enzymes (Atg4A or Atg4B), desumolating enzymes (SENP-1 or -2) or deubiquitinating enzyme (UCH-L3) was mixed with 50 µM Ac-GTFG-AFC at 37°C for 45 min to determine the specificity of the synthetic tetrapeptide (left part). Data represent percentage activity relative to Atg4B (mean ± std dev, n = 4). In addition, 1 µM Atg4B was combined with various concentrations of 100 nM Atg4B inhibitor LC3-vinyl sulfone (right part), measuring Atg4B activity using Ac-GTFG-AFC peptide as above. Data represent % activity relative to reactions without LC3-vinyl sulfone (mean ± std dev; n = 4). (B) Various enzymes (0.1 nM) were assayed using 20 nM LC3B-PLA2 as the substrate (left part). Atg4B was further assayed with Atg4B inhibitor LC3-vinyl sulfone (right part). (C) HeLa S3 cells were lysed by sonication to extract soluble proteins (1 [LC3B-PLA2] or 50 [Ac-GTFG-AFC] µg per reaction) for assaying Ac-GTFG-AFC (1 µM) or LC3B-PLA2 (100 nM) hydrolytic activity in the presence (+) or absence (−) of 1 µM (Ac-GTFG-AFC) or 100 nM (LC3B-PLA2) LC3-Vinyl Sulfone. (D) siRNA against Atg4B or Flag tagged dominant-negative Atg4B C74A was transfected into HeLa S3 cells. Efficiency of Atg4B siRNA knockdown or Atg4B C74A overexpression was determined by immunoblotting (middle part) using 20 µg total protein per lane and probing blots with antibodies to Atg4B (top) and b-actin (bottom). Cellular proteins extracted as above were incubated with 50 µM Ac-GTFG-AFC in 50 mM Tris-HCl, pH 8.0, 0.8 M sodium citrate, 5 mM DTT buffer at 37°C to measure Atg4B activity (left part). Extracted cellular proteins were also assayed with 200 nM LC3B-PLA2 fusion protein in PLA2 assay buffer in the presence of 20 µM NBD C6-HPC at room temperature for 30 min (right part). Data represent mean ± S.D. of three experiments.
We then explored the utility of our Atg4B substrates for measuring Atg4B activity in cell lysates. For these experiments, HeLa S3 cells were transfected either with plasmids encoding Atg4B or a catalytic mutant Atg4B C74A or with siRNA reagents corresponding to Atg4B targeting or scrambled control double-strand synthetic RNAs (Fig. 7C and D). Knockdown of endogenous Atg4B by the specific siRNA and overexpression of Flag-tagged Atg4B was evaluated by immunoblotting (Fig. 7D). In contrast to control cells, both siAtg4B knockdown cells and Atg4B C74A catalytic mutant-expressing cells displayed diminished autophagin activity in lysates prepared from these genetically engineered cells, as measured using either the Ac-GTFG-AFC (Fig. 7C) or LC3B-PLA2 fusion protein (Fig. 7D) substrates. Interestingly, treatment of cells with various inducers of autophagy prior to generation of lysates did not result in an increase in Atg4B activity (Suppl. Data, Fig. S7), suggesting that activity of this protease is unlikely to represent the rate-limiting step in autophagy. Taken together, these experiments illustrate applications of the synthetic substrates for monitoring activity of endogenous autophagins in cell lysates, thus aiding cell biological investigations of mechanisms regulating Atg4-family proteases.
Discussion
Autophagins play a crucial role in conjugation and deconjugation of Atg8 paralogs to autophagic vesicles and are indispensable for the process of autophagy.12,14,17 For the most part, analysis of the in vitro activity of autophagins has been limited to assessing cleavage of natural substrates such as LC3 by SDS-PAGE and immunoblotting,11,13,24 which are laborious and nonquantitative methods for assessing protease activity. The synthetic peptide 7-(methoxycou-marin-4-yl)-acetyl-Thr-Phe-Gly-Met-N-3-(2,4-dinitrophenyl)-l-α,β-diamino propionyl-NH2 is reportedly hydrolyzed by mammalian autophagin-3 (Atg4C), but its activity for other members of the autophagin family has not been assessed.12 In this study, we have devised two fluorogenic assays for measuring activity of autophagin-1 (Atg4B). These synthetic substrates permit the design of simple, homogeneous solution assays and are appropriate for high throughput screening (HTS) applications (manuscript in preparation).
Fluorogenic tetrapeptides have previously been generated for several cysteine protease families, including caspases25 and deubiquitinating proteases,26 prompting us to explore their utility for autophagins. We employed the method of PS-SCL to identify tetrapeptide substrates optimized for Atg4B, holding fixed the Gly at the P1 position that is found in all known endogenous substrates of autophagins. Similar to the natural substrates LC3 and GATE-16, the preferred amino acids at P2 and P3 were Phe and Thr, respectively, suggesting that the positional scanning method of library deconvolution successfully recapitulated the endogenous substrate preferences of Atg4B. Interestingly, Gly was the most highly preferred residue at P4, an amino acid that is not represented in any of the known autophagin substrates at this position. Recombinant purified Atg4B showed catalytic efficiency for the GTFG tetrapeptide substrate that is approximately six-fold greater than the “natural” ETFG tetrapeptide sequence derived from the LC3B and LC3C substrates. Besides Gly, the other amino acids (Ala, Ser, Thr and Asn) with relatively small side chains were preferred at P4 position. This observation is consistent with a recent report showing that mutation of the corresponding Glu(117) to Ala in recombinant LC3 did not result in substantial changes in proteolytic processing compared with the wild-type substrate.23 Analysis of the crystal structure bound substrate with Atg4B shows that the Glu residue in P4 position makes some contacts with enzyme surface, but the side chain of this amino acid is oriented away from surface of Atg4B (Suppl. data, Fig. S8). In the case of the SENPs and DUBs, this feature results in broad amino acid tolerance for substrates at the P4 posi tion.23,26,27 Alternatively for UCH-L3 protease (DUBs family), while equal preference for Gly and Val was observed when employing short peptides as substrates, much stronger preference for Gly was observed in the context of the full-length substrate due to the conformational changes of UCH-L3 upon binding to its full-length substrate. We thus hypothesize that similar effects occur for Atg4B, especially when considering that binding of LC3 to Atg4B results in substantial conformational changes around the active center of the enzyme.23,26
When tested against natural substrates in vitro, Atg4B showed a rank-order catalytic efficiency of GATE-16 > LC3B > LC3C > GABARAP. In this regard, GATE-16 contains the tetrapeptide sequence at the cleavage site of NTFG rather than the ETFG found in LC3B and LC3C. The preference of Asn over Glu revealed by PS-SCL agrees with the preference displayed by Atg4B for cleavage in vitro of full-length natural protein substrates. Overall, however, the catalytic efficiency of Atg4B for the optimal tetrapeptide substrate GTFG is only 0.92 ± 0.07 M−1/sec−1, which is much lower than tetrapeptide substrates reported for most DUBs and SENPs.26,27
In addition to tetrapeptide substrates, we developed a cleavable LC3B-PLA2 fusion protein as a synthetic substrate for Atg4B. The catalytic efficiency kcat/Km of Atg4B for this substrate reaches 5.3 × 105 M−1/sec−1, which provides an extremely sensitive assay. The structure of LC3B in complex with Atg4B reveals that LC3B substrate binding changes the conformation of the enzyme and may help to expose the active site.23 Thus, the LC3B-PLA2 substrate presumably is far more efficient than peptide substrates of Atg4B due to: (a) additional interactions that natural substrates such as LC3B make with autophagins and (b) a very large degree of substrate-induced activation by the LC3 domain, consistent with observations in crystal structures. We did not detect significant cleavage of the LC3B-PLA2 substrate by Atg4A, Atg4C or Atg4D, in contrast to Atg4B. Of the autophagins, only Atg4B has been shown definitively to cleave LC3 in vitro. However, the strategy of fusing autophagin substrates with PLA2 as a means of generating assay reagents appears to be extensible to other members of the protease family as shown by preliminary results with a GATE-16-PLA2 substrate, which is activated by Atg4A (Suppl. Data, Fig. S9).
In terms of practicalities of employing the LC3B-PLA2 substrate for autophagy research, HTS and other applications, we routinely prepare LC3B-PLA2 substrate by expression in bacteria with yields of 6 mg per liter, indicating that access to sufficient amounts of material is not an issue. In fact, we have formatted a robust HTS assay using this highly sensitive substrate, which requires only 6 ng of LC3B-PLA2 protein per well of 1,536-well plates in 10 µL reaction volumes (unpublished observations).
We used both the fluorogenic tetrapeptide and LC3B-PLA2 substrates to perform a limited structure-function analysis of Atg4B. Deletion of the N-terminal domain previously speculated to play an autoinhibitory role had relatively little effect on Atg4B activity as measured against natural substrate proLC3, tetrapeptide substrate or PLA2-fusion protein substrate. In contrast, eradication of the regulatory loop essentially completely abolished Atg4B activity as measured on all three classes of substrates. Thus, the N-terminal first 24 amino-acids of Atg4B are largely dispensable, whereas the regulatory loop at residues 259 to 262 is essential for activity.
We employed the optimized tetrapeptide sequence derived from PS-SCL to generate a novel LC3B-PLA2 substrate in which the naturally occurring ETFG sequence at which cleavage occurs was converted to GTFG. However, unlike the tetrapeptide context, the LC3B-PLA2 E117G (GTFG) mutant fusion protein was a less efficient substrate compared to the wild-type version. This result raises the possibility that contact of portions of LC3 distal to the cleavage site may induce conformational changes in Atg4B that influence the conformation of the active site. This observation is similar to results obtained previously for deubiquitinating (DUB) enzyme, UCH-L3, which in PS-SCL screening experiments showed almost equal preference for the wild-type Gly residue (ubiquitin) and non-natural Val at the P2 position.26 Subsequent analysis of wild-type and mutant (Gly/Val) full-length ubiquitin revealed that the natural substrate is several times more efficiently cleaved compared to the Gly/Val P2 mutant. Altogether, these findings emphasize the importance of using natural substrates in assays for identifying chemical inhibitors of autophagins and DUBs, as the geometry of the active site is likely to be altered and thus the structures of chemicals that score as inhibitors significantly impacted by choice of substrate. Further attesting to this notion is our finding that the activity of Atg4B measured against the GTFG tetrapeptide substrate was only partially suppressed by LC3-vinyl sulfone, whereas this reagent essentially completely suppressed Atg4B activity when measured using the LC3B-PLA2 fusion protein. Thus, using this prototypical inhibitor (LC3-vinyl sulfone), different degrees of inhibition were obtained when measured using a tetrapeptide as opposed to a LC3B fusion protein substrate, likely reflecting the different conformations of the active site in the LC3-bound and unbound forms characteristic of differences between the two substrate classes.
To explore the potential utility of the substrates reported here for measuring cellular autophagin activity, we experimentally manipulated levels of activity of Atg4B in cells. Both siRNA and Cys/Ala catalytic-negative mutant of Atg4B reduced cellular levels of autophagin activity as measured with either the optimized tetrapeptide substrate or the LC3B-PLA2 fusion protein substrate. The extent of reduction as defined by percentage decrease in Atg4B activity was greater when measured using the LC3B-based substrate than the tetrapeptide. In terms of residual protease activity observed in these lysates from siRNA-treated or Atg4B C74A-expressing cells, it should be noted that the knockdown of Atg4B by siRNA was not complete and that catalytic mutants, acting in a dominant-negative manner, typically only partially compete with endogenous wild-type proteins. Also, other members of the autophagin family may cleave these substrates in extracts where the endogenous proteases may have post-translational modifications or cofactors not present when testing recombinant purified Atg4 family proteases. In this regard, because Atg4B (authophagin-1) showed the highest in vitro protease activity against natural substrates among the four human autophagins expressed in bacteria as recombinant proteins, our studies were performed with this particular protease. Thus, the suitability of our substrates for other members of the autophagin family remains to be clarified. Interestingly, treatment of cells with various autophagy inducers did not stimulate an increase in LC3-PLA2 hydrolyase activity (Suppl. Data). This finding suggests that total cellular levels of Atg4B activity do not represent a rate-limiting step in autophagy induction and implies perhaps that intracellular targeting of this autophagin to specific cellular compartments rather than its specific activity may be more relevant. Alternatively, it is possible that endogenous inhibitors of Atg4B control the net activity of this protease in cells and that association of these hypothetical autophagin inhibitors is lost during cell lysis.
While autophagy is critical for cell survival in the context of nutrient deprivation, circumstances where autophagy and cell death occur in tandem have been identified, especially in tumor biology. However, it is unclear whether autophagy is truly responsible for cell death versus merely a concomitant of cell death, representing a failed attempt at survival.1 Moreover, apoptotic cell death and autophagy appear to be mutually antagonistic. Agents that suppress apoptosis, such as chemical inhibitors of caspase-family apoptotic proteases, induce autophagy in some cell types.28 Conversely, the antiapoptotic protein Bcl-2 binds to and suppresses the autophagy protein Beclin 1.29 Thus, autophagy and apoptosis appear to be incompatible. Our assays provide useful tools to address questions about the regulation of autophagy in health and disease.
Materials and methods
Plasmids construction.
Autophagin-1 (Atg4B) and N-terminal deletion mutant (Atg4B dN, comprised of amino acids 25–393) were amplified from pcDNA3-Atg4B, kindly provided by Dr. Carlos Lopez-Otin,12 using PCR. The substrate proteins LC3B, GABARAP, GATE-16 genes were amplified from human fetal brain cDNA library (Open Biosystems, LHS1207), whereas LC3C was amplified from a human fetal heart cDNA library (Open Biosystems, LHS1601). PCR products were cloned into bacterial expression vector pETDuet-1 (Novogen, 71146-3), in-frame with N-terminal His tag. The mature human PLA2 group X G10 (amino acid 43–165) was amplified from a human fetal brain cDNA library and fused with LC3B in pETDuet-1. To generate the Atg4B loop deletion mutant (Atg4B dL, 259–262 amino acid deletion), catalytic mutant Atg4B C74A, LC3B-PLA2/GTFG gain-of-function mutant and LC3B-PLA2/ETFA loss-of-function mutant, site-directed mutagenesis was carried out by the QuickChange kit according to the manufacturer's instructions (Stratagene, 200518). Atg4B and Atg4B C74A were subcloned into FLAG-tagged mammalian expression vector pcDNA3 (Invitrogen, A150228). All the clones were verified by DNA sequencing. Primer sequences are available upon request.
Protein expression and purification in Escherichia coli.
Expression plasmids were transformed into E. coli BL21 (DE3, Invitrogen, C6010-03). The proteins were induced by 0.1 mM Isopropyl β-D-1-thiogalactopyranoside (IPTG) at 15°C for 6 h. His-tagged proteins were purified using Ni-NTA-agarose (Qiagen, 30250) and eluted with a 20∼250 mM gradient imidazole in 10 mM HEPES, pH 7.4, 150 mM NaCl, 10 mM β-mercaptoethanol. His-tagged LC3B-PLA2/ETFG, GTFG and ETFA proteins were induced by 1 mM IPTG at 37°C for 4 h to obtain inclusion bodies. The inclusion bodies were solubilized with 20 mM Tris buffer, pH 8.0 containing 0.5 M NaCl, 5 mM imidazole, 6 M guanidine hydrochloride, 1 mM β-mercaptoethanol and loaded onto a Ni-NTA column. The denatured proteins were refolded with refolding buffer (20 mM Tris, pH 8.0, 0.5 M NaCl, 20 mM imidazole, 1 mM β-mercaptoethanol) in the presence of linear 6-0 M gradient urea. The refolded proteins were eluted with a 20–500 mM gradient imidazole in 20 mM Tris, pH 8.0, 150 mM NaCl, 10 mM β-mercaptoethanol. DeSUMOylating and deubiquitinating enzymes were produced and purified as previously described.26,27
Atg4B cleavage of natural and synthetic protein substrates.
Atg4B (wild-type [WT]) or catalytic mutant Atg4B C74A was incubated with 400 nM protein substrates in 50 µl reaction buffer containing 50 mM Tris, pH 8.0, 150 mM NaCl and 1 mM DTT at 37°C for 1 or 2 h. Reactions were stopped by addition of 50 µl of 2X Laemmli SDS-sample buffer and subjected to 13% SDS-PAGE gel followed by GelCode Blue (Thermo Fisher Scientific, 24592) staining or transfer to nitrocellulose membrane (Signa-Aldrich, Z613673) for immunoblot analysis using anti-S-tag antibody (Novogen, 71549) or anti-myc antibody (Roche, 11667203001). Proteins were detected using peroxidase-conjugated anti-mouse IgG secondary antibody (GE Healthcare, NA931) by Super Signal (West Pico) and chemiluminescence substrate (Pierce, 34080) with exposure to x-ray film. The EC50 was defined as the enzyme concentration required for cleaving 50% precursor of substrates and this value was used to calculate kcat/Km value using the equation: 0.693/(t × EC50) where E is the enzyme concentration.30
Peptide library synthesis.
Synthesis of the positional scanning-synthetic combinatorial peptide library (PS-SCL) was carried out using semiautomatic FlexChem Peptide Synthesis System (model 202) and is based on related procedures.26,27 After completing synthesis steps, each sub-library (P4, P3 and P2) was dissolved in biochemical grade dried DMSO [Sima-Aldrich, D2650] at a concentration of 10 mM and stored at −20°C until use.
Assay of the PS-SCL.
Atg4B was assayed at 37°C in 25 mM Tris buffer (pH 8.0). Enzyme was preincubated for 30 min at 37°C before being added to the wells containing substrate. Standard enzyme assay conditions in 100 µl reactions included 250 µM total final substrate mixture concentration (assuming approximately 13 µM per single substrate) and enzyme concentration of 6.0 µM. Release of fluorophore was monitored continuously with excitation at 355 nm and emission at 460 nm with an assay time of 60 min using an fMax fluorimeter (Molecular Devices). Only the linear portion of progress reactions was used for further analysis, which was based on total RFU (relative fluorescence unit) for every sub-library. Final data were obtained by setting the highest value to 100% and adjusting the other results accordingly. Data were normalized to the highest rate obtained for each position.
Synthesis of individual fluorogenic substrates.
Both solution and solid-phase syntheses were carried out according to established methods. Boc-Gly-AFC (tert-butoxycarbonylglycyl-AFC) was synthesized using the method of Alves et al.27 and subsequently the Boc group was deprotected using 4 M HCl in dioxan. N-terminal acetylated and side chain protected peptides (P2-P4 position) were synthesized using chlorotrityl resin (Novabiochem, 855017) and subsequently coupled with Gly-AFC using 2-(1H-Benzotriazole-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate (HBTU, Novabiochem, 851006), DIEA (diisopropylethylamine) in DMF. Subsequent purification and TFA (trifluoroacetic acid) deprotection yielded raw fluorogenic substrates, which were purified using preparative HPLC (Beckman-Coulter System Gold 126P) and re-analyzed by analytical HPLC (Beckman-Coulter System Gold 125). Purity and molecular weight of the peptides were confirmed by LC-MS.
Atg4B activity assays using synthetic tetrapeptide substrates.
For recombinant Atg4B, the enzyme assay used either mixed tetrapeptide-ACC or single tetrapeptide-AFC substrates in 100 µl reaction volumes containing 50 mM Tris, pH 8.0, 150 mM NaCl and 5 mM DTT at 37°C. Release of fluorophore was monitored kinetically with excitation/emission wavelength 355/460 (ACC) 400/505 (AFC) using microplate spectrofluorometer (Molecular Devices).
Effect of pH, salts and glycerol on Atg4B activity.
The pH optimization of Atg4B was determined by incubating 50 µM Ac-GTFG-AFC substrate in 50 mM of reaction buffer, containing MES buffer (pH 6.0–6.5), PIPES buffer (pH 6.5–7.0), HEPES buffer (pH 7.0–7.5), Tris-HCl (pH 7.5–8.0) or Bistris propane buffer (pH 8.5–9.0), in the presence of 200 mM NaCl and 5 mM DTT. To determine the effect of salts and glycerol, recombinant Atg4B was incubated with Ac-GTFG-AFC in 50 mM Tris-HCl, pH 8.0, 5 mM DTT in the presence of various concentrations of salts and glycerol at 37°C for 30 min.
Atg4B activity measured using LC3B-PLA2 substrate.
The reporter assay was modified from a previous report.21 Briefly, recombinant Atg4B was mixed with 100 nM LC3B-PLA2 fusion protein in 20 µl PLA2 reaction buffer containing 20 mM Tris-HCl, pH 8.0, 2 mM CaCl and 1 mM DTT and 20 µM 2-(6-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino) hexanoyl-1-hexadecanoyl-sn-glycero-3-phosphocholine (NBD-C6-HPC) (Invitrogen, N-3786). Fluorescence intensity was measured within 30–60 min using an Analyst™ HT (Molecular Devices Corp.,) at room temperature with excitation and emission wavelength of 485 and 530, respectively. The kcat/Km of Atg4B on LC3B-PLA2 was determined using nonlinear regression fitting of a series of progress curves captured at different LC3B-PLA2 concentrations, using the sequential activation model described in detail previously.31
Cell culture and transfections.
HeLa S3 cells were cultured in DMEM medium with 10% FBS, 100 µg/ml streptomycin, 100 IU penicillin and 1% L-glutamine at 37°C in 5% CO2: 95% air. HeLa S3 cells (2 × 106) were seeded into 6 cm plates and cultured overnight, then transfected with 2 µg pcDNA-FLAG Atg4B WT or C74A expression plasmids using 2.5 µl Lipofectamine 2000 (Invitrogen, 11668-027) for 30 h. Alternatively, cells were transfected with 10 nM scramble siRNA (siCtrl, Ambion) or siRNA targeting human Atg4B (siAtg4B, Ambion) for 60 h using Lipofectamine RNAimax (Invitrogen, 13778-150). Transfected cells were harvested and washed with PBS once, then resuspended in lysis buffer containing 25 mM Tris-HCl, pH 8.0, 100 mM NaCl, 1 mM CaCl2, 5 mM MgCl, 5% glycerol, 0.1% NP-40, 1 mM PMSF and 1 mM DTT on ice followed by sonication 2 × 2 sec to fracture cells. Lysates were centrifuged at 16,000x g for 15 min and 1–50 µg of supernatant proteins were used for either Ac-GTFG-AFC tetrapeptide assay or LC3B-PLA2 reporter assay.
Acknowledgements
This work was supported by the National Institutes of Health (R01-AI-082629), a fellowship grant to Dr. Shu from Sass Foundation for Medical Research, Inc., Roslyn, New York and grants to Dr. Drag from the Foundation for Polish Science. We also thank Tessa Siegfried and Melanie Hanaii for manuscript preparation and Dr. Carlos Lopez-Otin for kindly providing human Atg4B cDNA.
Abbreviations
- Atg
autophagy-related
- LC3
microtubule-associated proteins 1A/1B/1C light chain 3
- GATE-16
Golgi-associated ATPase enhancer of 16 kDa
- GABARAP
GABA(A) receptor-associated protein
- Ac
acetyl
- PS-SCL
positional scanning substrate combinatorial library
- ACC
7-amino-4-carbamoylmethylcoumarin
- AFC
7-amino-4-trifluoromethylcoumarin
- PLA2
phospholipase A2
- NBD-C6-HPC
2-(6-(7-nitrobenz-2-oxa-1,3-diazol-4-yl) amino)hexanoyl-1-hexadecanoyl-sn-glycero-3-phosphocholine
- SENP
sentrin/SUMO-specific protease
Footnotes
Previously published online: www.landesbioscience.com/journals/autophagy/article/13075
Supplementary Material
References
- 1.Marx J. Autophagy: is it cancer's friend or foe? Science. 2006;312:1160–1161. doi: 10.1126/science.312.5777.1160. [DOI] [PubMed] [Google Scholar]
- 2.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993;333:169–174. doi: 10.1016/0014-5793(93)80398-e. [DOI] [PubMed] [Google Scholar]
- 4.Mizushima N, Noda T, Yoshimori T, Tanaka Y, Ishii T, George MD, et al. A protein conjugation system essential for autophagy. Nature. 1998;395:395–398. doi: 10.1038/26506. [DOI] [PubMed] [Google Scholar]
- 5.Mizushima N, Yamamoto A, Hatano M, Kobayashi Y, Kabeya Y, Suzuki K, et al. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J Cell Biol. 2001;152:657–668. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol. 2009;10:458–467. doi: 10.1038/nrm2708. [DOI] [PubMed] [Google Scholar]
- 8.Thumm M, Egner R, Koch B, Schlumpberger M, Straub M, Veenhuis M, et al. Isolation of autophagocytosis mutants of Saccharomyces cerevisiae. FEBS Lett. 1994;349:275–280. doi: 10.1016/0014-5793(94)00672-5. [DOI] [PubMed] [Google Scholar]
- 9.Nakatogawa H, Ichimura Y, Ohsumi Y. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell. 2007;130:165–178. doi: 10.1016/j.cell.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 10.Kirisako T, Baba M, Ishihara N, Miyazawa K, Ohsumi M, Yoshimori T, et al. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J Cell Biol. 1999;147:435–446. doi: 10.1083/jcb.147.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kirisako T, Ichimura Y, Okada H, Kabeya Y, Mizushima N, Yoshimori T, et al. The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J Cell Biol. 2000;151:263–276. doi: 10.1083/jcb.151.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marino G, Uria JA, Puente XS, Quesada V, Bordallo J, Lopez-Otin C. Human autophagins, a family of cysteine proteinases potentially implicated in cell degradation by autophagy. J Biol Chem. 2003;278:3671–3678. doi: 10.1074/jbc.M208247200. [DOI] [PubMed] [Google Scholar]
- 13.Betin VM, Lane JD. Caspase cleavage of Atg4D stimulates GABARAP-L1 processing and triggers mitochondrial targeting and apoptosis. J Cell Sci. 2009;122:2554–2566. doi: 10.1242/jcs.046250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hemelaar J, Lelyveld VS, Kessler BM, Ploegh HL. A single protease, Apg4B, is specific for the autophagy-related ubiquitin-like proteins GATE-16, MAP1-LC3, GABARAP and Apg8L. J Biol Chem. 2003;278:51841–51850. doi: 10.1074/jbc.M308762200. [DOI] [PubMed] [Google Scholar]
- 15.Tanida I, Tanida-Miyake E, Ueno T, Kominami E. The human homolog of Saccharomyces cerevisiae Apg7p is a Protein-activating enzyme for multiple substrates including human Apg12p, GATE-16, GABARAP and MAP-LC3. J Biol Chem. 2001;276:1701–1706. doi: 10.1074/jbc.C000752200. [DOI] [PubMed] [Google Scholar]
- 16.Scherz-Shouval R, Shvets E, Fass E, Shorer H, Gil L, Elazar Z. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J. 2007;26:1749–1760. doi: 10.1038/sj.emboj.7601623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tanida I, Sou YS, Ezaki J, Minematsu-Ikeguchi N, Ueno T, Kominami E. HsAtg4B/HsApg4B/autophagin-1 cleaves the carboxyl termini of three human Atg8 homologues and delipidates microtubule-associated protein light chain 3- and GABAA receptor-associated protein-phospholipid conjugates. J Biol Chem. 2004;279:36268–36276. doi: 10.1074/jbc.M401461200. [DOI] [PubMed] [Google Scholar]
- 18.Marino G, Salvador-Montoliu N, Fueyo A, Knecht E, Mizushima N, Lopez-Otin C. Tissue-specific autophagy alterations and increased tumorigenesis in mice deficient in Atg4C/autophagin-3. J Biol Chem. 2007;282:18573–18583. doi: 10.1074/jbc.M701194200. [DOI] [PubMed] [Google Scholar]
- 19.Kabeya Y, Mizushima N, Yamamoto A, Oshitani-Okamoto S, Ohsumi Y, Yoshimori T. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J Cell Sci. 2004;117:2805–2812. doi: 10.1242/jcs.01131. [DOI] [PubMed] [Google Scholar]
- 20.Dijkstra BW, Drenth J, Kalk KH. Active site and catalytic mechanism of phospholipase A2. Nature. 1981;289:604–606. doi: 10.1038/289604a0. [DOI] [PubMed] [Google Scholar]
- 21.Nicholson B, Leach CA, Goldenberg SJ, Francis DM, Kodrasov MP, Tian X, et al. Characterization of ubiquitin and ubiquitin-like-protein isopeptidase activities. Protein Sci. 2008;17:1035–1043. doi: 10.1110/ps.083450408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mikolajczyk J, Drag M, Bekes M, Cao JT, Ronai Z, Salvesen GS. Small ubiquitin-related modifier (SUMO)-specific proteases: profiling the specificities and activities of human SENPs. J Biol Chem. 2007;282:26217–26224. doi: 10.1074/jbc.M702444200. [DOI] [PubMed] [Google Scholar]
- 23.Satoo K, Noda NN, Kumeta H, Fujioka Y, Mizushima N, Ohsumi Y, et al. The structure of Atg4B-LC3 complex reveals the mechanism of LC3 processing and delipidation during autophagy. EMBO J. 2009;28:1341–1350. doi: 10.1038/emboj.2009.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tanida I, Ueno T, Kominami E. Human light chain 3/MAP1LC3B is cleaved at its carboxyl-terminal Met121 to expose Gly120 for lipidation and targeting to autophagosomal membranes. J Biol Chem. 2004;279:47704–47710. doi: 10.1074/jbc.M407016200. [DOI] [PubMed] [Google Scholar]
- 25.Thornberry NA, Rano TA, Peterson EP, Rasper DM, Timkey T, Garcia-Calvo M, et al. A combinatorial approach defines specificities of members of the caspase family and granzyme B. Functional relationships established for key mediators of apoptosis. J Biol Chem. 1997;272:17907–17911. doi: 10.1074/jbc.272.29.17907. [DOI] [PubMed] [Google Scholar]
- 26.Drag M, Mikolajczyk J, Bekes M, Reyes-Turcu FE, Ellman JA, Wilkinson KD, et al. Positional-scanning fluorigenic substrate libraries reveal unexpected specificity determinants of DUBs (deubiquitinating enzymes) Biochem J. 2008;415:367–375. doi: 10.1042/BJ20080779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Drag M, Mikolajczyk J, Krishnakumar IM, Huang Z, Salvesen GS. Activity profiling of human deSUMOylating enzymes (SENPs) with synthetic substrates suggests an unexpected specificity of two newly characterized members of the family. Biochem J. 2008;409:461–469. doi: 10.1042/BJ20070940. [DOI] [PubMed] [Google Scholar]
- 28.Yu L, Alva A, Su H, Dutt P, Freundt E, Welsh S, et al. Regulation of an ATG7-beclin 1 program of autophagic cell death by caspase-8. Science. 2004;304:1500–1502. doi: 10.1126/science.1096645. [DOI] [PubMed] [Google Scholar]
- 29.Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 30.Stennicke HR, Salvesen GS. Caspase assays. Methods Enzymol. 2000;322:91–100. doi: 10.1016/s0076-6879(00)22010-7. [DOI] [PubMed] [Google Scholar]
- 31.Zhou Q, Salvesen GS. Activation of pro-caspase-7 by serine proteases includes a non-canonical specificity. Biochem J. 1997;324:361–364. doi: 10.1042/bj3240361. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.