Abstract
Lafora disease (LD) is a progressive, lethal, autosomal recessive, neurodegenerative disorder that manifests with myoclonus epilepsy. LD is characterized by the presence of intracellular inclusion bodies called Lafora bodies (LB), in brain, spinal cord and other tissues. More than 50 percent of LD is caused by mutations in EPM2A that encodes laforin. Here we review our recent findings that revealed that laforin regulates autophagy. We consider how autophagy compromise may predispose to LB formation and neurodegeneration in LD, and discuss future investigations suggested by our data.
Key words: autophagy, glycogen metabolism, Lafora disease, laforin, malin, neurodegeneration
Lafora Disease
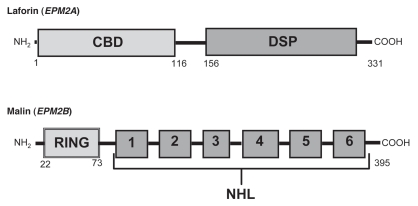
The neurodegenerative disorder Lafora progressive myoclonus epilepsy or Lafora disease (OMIM 254780) was first described in 1911 by the Spanish neurologist Gonzalo Rodríguez Lafora. LD is an autosomal recessive disorder caused in the vast majority of cases by mutations in one of two genes (Fig. 1): EPM2A and EPM2B. EPM2A, mutated in about 58% of LD cases, encodes a protein of 331 amino acids, called laforin, with a dual-specificity phosphatase domain and a carbohydrate-binding domain (CBD). Laforin binds glycogen and can dephosphorylate both phosphoserine/phosphothreonine and phosphotyrosine substrates, as well as complex carbohydrates. EPM2B, mutated in about 40% of Lafora cases, encodes a protein called malin, with six NHL domains, implicated in protein-protein interactions and an N-terminal RING-finger domain characteristic of E3 ubiquitin ligases. Malin and laforin interact and have been proposed to operate through common mechanisms.
Figure 1.
Simplified view of the protein domains of laforin (EPM2A gene) and malin (EPM2B gene). The various protein domains and their approximate localizations are indicated. CBD: carbohydrate binding domain; DSP: dual specificity phosphatase domain; RING: really interesting new gene (RING) finger domain; NHL: a domain present in NCL1, HT2A (Trim32) and LIN-41 proteins, involved in protein-protein interactions.
LD is characterized by the presence of intracellular inclusion bodies called Lafora bodies, in brain, spinal cord and other tissues. LB are mainly composed of insoluble, starch-like and poorly branched glycogen molecules, called polyglucosans, but also contain around 6% protein and are decorated by anti-ubiquitin antibodies, suggesting accumulation of undegraded proteins. Therefore, LD may be a disorder of both carbohydrate metabolism and protein clearance. Although LBs are a hallmark of LD, it is still unclear whether or not they cause the pathology or are simply a consequence.
Possible Functions of Laforin and Malin
Two different, not mutually exclusive, roles have been suggested for laforin. First, laforin could act as a glycogen phosphatase, removing phosphates from glycogen, thus preventing the soluble glycogen molecules from becoming insoluble polyglucosans. However, the role of the E3 ubiquitin ligase malin in this process is less clear, particularly because it interacts with laforin and catalyzes its polyubiquitination and degradation. Thus, defects in malin, which produce a neurological and histological phenotype indistinguishable from defects in laforin, would elevate the levels and presumably the activity, of laforin. However, it has been also suggested that in the absence of malin, laforin becomes sequestered in an inactive form. Second, laforin could act as an ancillary protein for the E3 ubiquitin ligase malin, placing specific substrates related with glycogen metabolism in the proximity of malin to be polyubiquitinated and degraded by the proteasomes.
Possible Roles of Proteasomes and Autophagy in Lafora Disease
Different experimental findings support a possible function of proteolytic processes in LD, and a role of the malin-laforin complex in the degradation of misfolded proteins by the ubiquitin-proteasome pathway has already been proposed. Therefore, we also considered the possibility of altered autophagy in LD. Autophagy sequesters a variety of proteins, lipids, carbohydrates and nucleic acids from the cytoplasm in autophagosomes, which are later degraded in lysosomes. Although glycogen is degraded in the cytosol by glycogen phosphorylase and the glycogen debranching enzyme, glycogen can be also degraded in the lysosomes via lysosomal acid alpha-glucosidase, also called acid maltase. The importance of this lysosomal pathway of glycogen degradation is illustrated by glycogen storage disease type II, also called Pompe disease, in which this single lysosomal enzyme is lacking.
Using laforin-deficient fibroblasts from LD patients and mouse embryonic fibroblasts (MEFs) and liver from epm2a-/- mice that replicate LD, we found decreased levels and impaired formation of LC3-II, indicating compromised autophagosome formation. We also found increased accumulation of polyubiquitinated proteins, which can result from a defect in both the ubiquitin-proteasome system and/or autophagy. This accumulation was even observed in the presence of proteasome inhibitors, suggesting that at least part of this accumulation is proteasome-independent. Finally, there was increased accumulation in liver extracts from laforin null mice of the autophagy substrate p62. However, laforin is not absolutely necessary for autophagy, since the lack of laforin only partially compromises long-lived autophagic protein clearance. Nevertheless, a partial deficit would be sufficient to contribute to many of the protein accumulation pathologies we have observed in these mouse models. In agreement with the laforin null data, overexpression of laforin increased the levels of LC3-II and reduced the amount of protein aggregates in an autophagy-dependent manner. Thus, it appears that laforin positively regulates autophagy.
Unanswered Questions
Since the Akt-mTOR signaling pathway is the main mechanism that negatively regulates macroautophagy, we tested its status. We found increased phosphorylation of the mTOR effector p70 S6 kinase (p70S6K) in laforin-deficient human fibroblasts and in livers from three-month-old epm2a-/- fed mice. Therefore, the observed decrease of macroautophagy in laforin-deficient cells may result from altered signaling by the Akt-mTOR pathway, a possibility supported by the decrease in mTOR activity resulting from laforin overexpression. However, the specific protein(s) responsible for the laforin-mediated modulation of mTOR activity remain to be identified, although our data suggest that they are probably upstream of TSC1/TSC2. It is, however, possible that additional pathways may be affected as well, and future experiments will need to test the role of malin in autophagy.
Finally, what could be the consequences of defective autophagy for formation of LB and for the disease? While autophagy is likely to regulate LB formation, it is possible/likely that other consequences of the mutations causing LD may contribute to LB formation. The observation of an autophagy defect in laforin deficiency suggests that many other autophagy substrates will accumulate, in addition to laforin, as we observed. This would predict the accumulation of various proteins, lipid droplets and also damaged mitochondria, which may all contribute to pathology. These possibilities and their potential contributions to the neurodegeneration in LD will need to be carefully investigated.
Acknowledgements
Work in EK's lab was supported by grants from the Spanish Ministry of Science and Innovation (BFU2008-00186BMC) and the Instituto de Salud Carlos III (Intramural Project, CIBERER). We are also grateful for a Hughes Hall Research Fellowship (S.S. and V.I.K.), a Wellcome Trust Senior Fellowship in Clinical Science (D.C.R.), the Medical Research Council, the EU Framework VI (EUROSCA) and the National Institute for Health Research Biomedical Research Centre at Addenbrooke's Hospital for funding.
Punctum to: Aguado C, Sarkar S, Korolchuk VI, Criado O, Vernia S, Boya P, Sanz P, Rodríguez de Córdoba S, Knecht E, Rubinsztein DC. Laforin, the most common protein mutated in Lafora disease, regulates autophagy. Hum Mol Genet. 2010;19:2867–2876. doi: 10.1093/hmg/ddq190.
Footnotes
Previously published online: www.landesbioscience.com/journals/autophagy/article/13308