Abstract
The human DEAD-box Y (DBY) RNA helicase (aka DDX3Y) gene is thought to be the major azoospermia factor a (AZFa) gene in proximal Yq11. Men with its deletion display no somatic pathologies, but suffer from complete absence of germ cells. Accordingly, DDX3Y protein is expressed only in the germline in spermatogonia, although the transcripts were found in many tissues. Here, we show the complex transcriptional control of a testis-specific DDX3Y transcript class with initiation at different sites upstream of the gene’s open reading frame (5′Untranslated Region; UTR) and with polyadenylation in their proximal 3′UTR. The most distal transcriptional start site (TSS; ∼1 kb upstream) was mapped in MSY2, a Y-specific minisatellite. As this testis-specific 5′UTR was subsequently processed by three alternative splicing events, it has been tentatively designated ‘exon-T’(estis). The MSY2 sequence unit was also found upstream of the mouse Ddx3y gene. However, only after its tandem amplification on the Y chromosome of Platyrrhini (new world monkeys) and Catarrhini (old world monkeys) did MSY2 become part of a novel distal promoter for DDX3Y expression in testis tissue and provides a second transcriptional start site (T-TSS-II) in Catarrhini. We therefore suggest that the development of a novel distal DDX3Y promoter in primates, which is activated only in testis tissue, is probably part of the gene’s germline translation control.
Keywords: AZFa gene expression, human Y chromosome, primates, spermatogenesis, transcriptional control
Introduction
Large rearrangements on the sex chromosomes during mammalian evolution ∼200 million years ago (Mya) have led to distinct structures of the X and Y chromosomes in the human genome including the loss of many genes on the Y chromosome, which originally were present on the ancient sex chromosomes (Charlesworth & Charlesworth, 2000). It can therefore be assumed that the 16 presently still functional human X and homologous Y genes were evolutionarily selected for some male-specific functions (Lahn & Page, 1997; Skaletsky et al., 2003). Examples are the anciently homologous X–Y gene pairs: SOX3/SRY (Marshall Graves, 2006), TSPX/TSPY (Delbridge et al., 2004) and RBMX/RBMY (Elliott, 2004). According to the strata evolution model of Lahn & Page (1999), these X–Y genes belong to strata 1 and started diverging their genomic structures ∼200 Mya. Another functional homologous X–Y gene pair, diverged from regular meiotic recombination ∼80–130 Mya, is the human DDX3X/DDX3Y gene pair. It belongs to strata 3 (Lahn & Page, 1999).
DDX3Y (aka DBY; HGNC: 2699; GenBank accession no.: NM_004660) is one of the two Y genes mapped to the azoospermia factor a (AZFa) deletion interval in the proximal part of the long arm of the Y chromosome (Yq11.1; Vogt et al., 1996; Lahn & Page, 1997; Vogt, 2005); its functional homologue on the X chromosome, DDX3X (aka DBX; HGNC: 2745; GenBank accession no.: NM_001356), was mapped to the proximal part of the short X arm (Xp11.4; Park et al., 1998). Although both genes lie outside the pseudoautosomal regions (PAR1 and PAR2) located at the tips of the sex chromosomes, they have a >94% sequence identity throughout the coding regions and their proteins are reported to be functionally exchangeable in the control of translation initiation in the cytoplasm (Chuang et al., 1997) and of cell cycle progression at the G1-S phase (Fukumura et al., 2003). Despite this functional equivalence and transcriptional activity of DDX3Y and DDX3X in each human tissue analysed (Lahn & Page, 1997; Ditton et al., 2004), DDX3Y proteins were only found in pre-meiotic male germ cells (Ditton et al., 2004). A germline-specific function of the DDX3Y gene was first indicated by absence of any somatic phenotype in men with AZFa deletions that also remove DDX3Y (Vogt et al., 1996). They all only suffer from severe testicular pathologies including the Sertoli-cell-only (SCO) syndrome (Vogt et al., 1996, 2008; Foresta et al., 1998; Kamp et al., 2001; Krausz & Degl’Innocenti, 2006). It has been speculated that DDX3Y might be the major AZFa gene functionally required for male fertility because its deletion was found in men with the SCO syndrome (Foresta et al., 2000). However, these gene deletions were not confirmed by sequence analyses and not yet found in other similar studies.
Deletion of the second AZFa gene, ubiquitin-specific protease 9Y (USP9Y; HGNC: 2633), was found to be compatible with normal sperm function (Luddi et al., 2009), although probably with reduced fertility. Its phenotype looks heterogeneous, as in another case of a familial truncated USP9Y gene impaired spermatogenesis was reported (Krausz et al., 2006). This suggests that the germline USP9Y expression, although not essential, might be able to ‘finetune’ the post-meiotic germ cell differentiation process in human. Indeed, USP9Y proteins were found to be expressed in spermatids (Vogt et al., 2008).
Interestingly, truncation of the USP9Y gene was also found on the Y chromosome of fertile chimpanzee and bonobos (Tyler-Smith, 2008). Male fertility in these apes was obviously not hampered by loss of the USP9Y function. Instead, the DDX3Y gene is conserved on the Y chromosome of primates (Goto et al., 2009) and the DDX3Y protein is expressed in the pre-meiotic germ cells (Ditton et al., 2004). It can thus be assumed that DDX3Y is the major AZFa gene functionally required for human spermatogenesis (Vogt et al., 2007, 2008; Tyler-Smith & Krausz, 2009).
In this article, we present experimental evidence for a large complexity of the human DDX3Y transcripts expressed in testis tissue. They initiate at different sites and the longest 5′UTR starts in a tandem repetitive sequence block known as the Y-specific ‘MSY2 minisatellite’ (DYS440; Bao et al., 2000). Processing for polyadenylation occurred at different sites in the proximal part of the ∼2.5-kb long 3′UTR. We found the basic MSY2 sequence unit also upstream of the mouse Ddx3y gene. Tandem amplification on the Y chromosome was only found in the Platyrrhine (new world monkey) Callithrix jacchus, in the Catarrhine (old world monkey) Macaca mulatta and in the hominids (great apes) Pongo pygmaeus and Pan troglodytes. Testis-specific DDX3Y expression starting from these MSY2 repeats was found in the catarrhine and the hominid primate class. We therefore like to propose that this primate-specific increase in the complexity of DDX3Y transcript variants expressed only in the male germline is part of a germline-specific translational control mechanism in human and most likely also in other primates, but not present in mouse.
Materials and methods
Human blood and tissue samples used for DNA and RNA extractions were only collected by our collaborating clinical partners after medical indication and written consent of the patients. The study was approved by the local ethical commission of the University of Heidelberg. Primate tissue and blood samples were obtained from the German (DPZ) and Dutch (BPRC) primate centres via the EUPRIM network (EU contract: RII3-026155 of the 6th framework programme: http://www.euprim-net.eu). Before DNA and RNA extraction, they were only handled according to the current biosafety guidelines of the university.
RNA isolation and RT-PCR analyses
RNA were isolated from all tissue samples with the Qiagen RNeasy Mini Kit (cat. no. 74106) using the manufacturer’s protocol (Qiagen GmbH, Hilden, Germany). Efficient on-column digestion of DNA during RNA purification was performed with the RNase-free DNAse digestion protocol (Qiagen, cat. no. 79254). For all reverse transcription-polymerase chain reaction (RT-PCR) assays, the first strand cDNA synthesis from the total RNA samples was performed after incubation with Oligo(dT)15 primer (Promega GmbH, Mannheim, Germany; cat. no. C1101) by RT with the Promega M-MLV reverse transcriptase enzyme (cat. no. M3683) following the manufacturer’s recommendations. About 1.3 μg of total RNA (producing ∼26 ng of cDNA from the polyA fraction) was used in each RT reaction. The quality of each cDNA sample was controlled by β-actin RT-PCR with species-specific oligonucleotides; its extension in the 5′UTR of the DDX3Y gene was with gene-specific oligonucleotides as described previously (Ditton et al., 2004).
All RT-PCRs were performed with the Invitrogen recombinant Taq DNA polymerase (cat. no. 10342-020; Invitrogen GmbH, Darmstadt, Germany) and using either 30 or 35 cycles with 1 μL of cDNA (∼1 ng) and optimal melting temperature (usually at 61 °C). The list of the used primer pairs is given in Tables S1 and S3 of the Supporting Information. They were designed from the genomic sequences given in the database to distinguish DDX3Y gene expression starting from different transcriptional start sites (‘TSS-I’ region; ‘T-TSS-I’ and ‘T-TSS-II’ respectively; see Fig. 2a). Primers were synthesized by Thermo Fisher Scientific (Ulm, Germany). For subsequent cloning and sequence analyses, we used the Promega pGEM-T easy vector system (cat. no. A1360) and the BigDye Terminator v1.1 Cycle Sequencing Kit (Applied Biosystems, cat. no. 4337450) with the ABI PRISM 3100 Genetic Analyzer (Applied Biosystems GmbH, Darmstadt, Germany).
Figure 2.
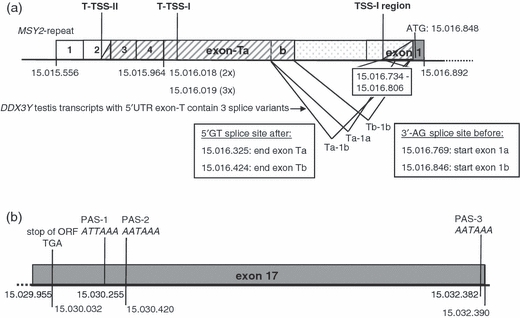
Schematic view on the genomic sequence structure of the DDX3Y gene in the current Y reference sequence (NC_000024; NCBI: build GRCh37) with the complexity of its putative transcriptional start sites (TSS) and polyadenylation sites (PAS) in the common and testis-specific transcripts. (a) 5′RACE and RT-PCR 5′UTR mapping experiments revealed a TSS for the common DDX3Y transcript class in exon 1 between positions 15.016.733 and 15.016.794 of the Y reference sequence (marked here as ‘TSS-I’ region). Two TSS were found for the testis-specific DDX3Y transcript class with long 5′UTR extensions defining ‘exon-T’. The first, tentatively designated ‘T-TSS-I’, was found by inspecting the sequence starts of five capped DDX3Y testis transcripts in the ‘database of TSS’ (http://dbtss.hgc.jp). They mapped downstream of MSY2 in exon-T at the Y reference positions 15.016.018 and 15.016.019 respectively. The second, tentatively designated ‘T-TSS-II’, was mapped to the second repeat of the MSY2 minisatellite (MSY2-2) located with four tandem repeats between Y sequence positions 15.015.556 and 15.015.964 respectively. Below the schematic DDX3Y exon-T-exon 1 structure, the three splicing variants of DDX3Y transcripts starting at ‘T-TSS-I’ or at ‘T-TSS-II’ in exon-T and designated as ‘Ta-1a’, ‘Ta-1b’ and ‘Tb-1b’, are tabled with their end and start positions after their 5′-GT and before their 3′-AG splice sites in exon-T and exon 1 respectively. (b) Schematic view of DDX3Y exon 17 with its three used PAS located between the TGA stop codon of the DDX3Y open reading frame at position 15.030.032 and the end of the 3′UTR at position15.032.390. Our 3′RACE experiments with RNA samples of multiple human tissues indicated that PAS1 and PAS2 were used only for testis transcripts and that PAS3 is the common site of DDX3Y transcripts expressed in all human tissues analysed.
RACE experiments
All 5′- and 3′RACE experiments were performed with the RACE System of Invitrogen for Rapid Amplification of cDNA Ends (version 2.0; cat. no. 18374-058). In short, cDNAs were prepared as described before, but with gene-specific adaptor oligonucleotides containing two restriction sites for subsequent cloning and sequencing experiments. We used primers bridging DDX3Y exons 1–2 as reverse primers in 5′RACE experiments. In 3′RACE experiments, we used gene-specific adaptor oligonucleotides as forward primers upstream of the different 3′UTR polyadenylation sites (PAS) in DDX3Y exon 17 (PAS1, 2, 3; see Fig. 2b). To select subsequently for full-length 5′UTR cDNAs, the single-stranded 5′RACE cDNA products were tailed with dCTP and terminal deoxynucleotidyl transferase at their 3′ end. To select for cDNAs from the 3′UTR polyadenylated mRNA fraction, a oligo(dT) containing adaptor primer was used. In a second nested PCR experiment, an internal set of gene-specific primer pairs was used as reverse (5′RACE) and forward (3′RACE) primers respectively. PCRs were performed with the abridged universal amplification primer that was able to anneal selectively to the dC-tailed 5′ends of the 5′RACE cDNA products respectively, to the dT-tailed 3′ends of the 3′RACE products. Before sequence analyses, all RACE products were purified by cloning in the pCR2.1-TOPO vector (Invitrogen, cat. no. K4510-20). For preparative isolation of the recombinants, we used our standard lab protocols (Ditton et al., 2004) for sequencing the BigDye Terminator v1.1 Cycle Sequencing Kit (Applied Biosystems, cat. no. 4337450) and the ABI PRISM 3100 Genetic Analyzer.
RNA blot analyses
Two Normal multiple Human Adult Northern tissue blots (NmHAN-IIIA and NmHAN-VA; BioCat GmbH, Heidelberg, Germany) loaded with polyadenylated RNA pools (each 3 μg) from different human male and female tissues were hybridized with two different 32P-labelled PCR products generated from the 5′ end of a DDX3Y cDNA produced from transcripts of the exon-Ta-1b splicing variant. Both products were amplified with the same forward primer in 5′UTR exon-T (5′-TCA AGT CTG TCG AGC CTC TG-3′); starting at sequence position 54362 in the Bacterial Artificial Chromosome (BAC) RP11-475I1 sequence (GenBank accession no. AC004474) and one reverse primer bridging exons 2–3 (5′-CCT TTG CTC GCT GTA CTT GC’-3′) starting at sequence position 59572 in the same BAC sequence respectively, one reverse primer (5′-TTGACAAGCTTCAGCAAAGTTG-3′) starting in exon-T at sequence position 54621. RNA samples were run on denaturing 1% formaldehyde agarose gels and transferred to a charge-modified nylon membrane. Pre-hybridization and hybridization experiments were carried out with 5 mL of FastHyb-hybridization solution (cat. no. L1031250; BioCat GmbH, Heidelberg, Germany) according to the manufacturer’s instructions.
‘In silico’ analyses of nucleic acid sequences
Putative transcriptional start sites (TSS’s) of the murine Ddx3y and the human DDX3Y gene transcripts were analysed ‘in silico’ in the database of TSS (DBTSS; release 6.01) at the DBTSS web server (http://dbtss.hgc.jp) and compared with those listed in the GenBank database of the NCBI (http://www.ncbi-nih.gov) and ENSEMBL database (http://www.ensembl.org) respectively. Sequence comparisons and homology searches were performed by MegaBLAST and BLASTN via the BLAST server at NCBI. Multiple sequence alignments were performed using clustalW2, release 2.0.10, through the EMBL-EBI web server (http://www.ebi.ac.uk).
Results
Transcriptional start sites (TSS’s) in DDX3Y exon 1 produce variable 5′UTR lengths
We first explored the complexity of the testis-specific DDX3Y transcript population observed earlier on RNA blots (Ditton et al., 2004) by 5′RACE experiments and sequence analyses of the isolated DDX3Y transcript lengths present in testis tissue and absent in leucocytes. The data are based on the genomic sequence information for the DDX3Y gene in the current Y reference sequence (NCBI database: NC_000024; GRCh37Y reference sequence assembly) with position of its ATG translational start codon at 15.016.848 Mb (Fig. S1).
We identified in the testis and leucocyte RNA samples, the putative commonly used DDX3Y TSS (referred to as ‘TSS-I’ from here onwards) upstream of the ATG codon in exon 1 between positions 15.016.744 and 15.016.794 in the Y reference sequence. We confirmed this result by additional RT-PCR assays with primer sets located around the proposed ‘TSS-I’ region. With cDNA samples from leucocytes and from different human tissues (testes, kidney, brain and spleen), the most distal forward primer with a positive RT-PCR signal in each RNA population was starting at Y sequence position 15.016.733; some further extension to position 15.016.627 was only observed for the testis cDNA sample (Table S1). We, therefore, assume a variable TSS of the common human DDX3Y transcript class defining the ‘TSS-I’ region between positions 15.016.733 and 15.016.794 of the Y reference sequence. The common 5′UTR length of DDX3Y exon 1 can thus be variable between 54 and 115 nucleotides (nt) respectively. In the testis tissue, it might extend further to 221 nt.
Novel testis-specific DDX3Y transcripts with long 5′UTR extension start in MSY2 repeats
With testis RNA samples, some clones of our 5′RACE experiments contained much longer 5′UTR sequences extending significantly beyond the commonly found 5′UTR exon 1 extensions. Sequence analyses found part of the tandemly repetitive MSY2 sequence block in these testis transcripts. MSY2 was described earlier as a human Y-specific minisatellite block with three, or more commonly, four tandem copies of a ∼100-nt sequence block containing a high sequence homology (DYS440; Bao et al., 2000). As MSY2 is located 885 nt upstream of the DDX3Y ATG translation codon (Fig. S1), our data suggest a significantly longer 5′UTR extension for these testis-specific DDX3Y transcripts most likely initiated from a novel distal DDX3Y promoter domain.
As clones containing tandem repetitive sequence blocks are well known to contain inherent instability, we wanted to confirm the transcriptional DDX3Y start region in MSY2 by some additional RT-PCR assays with primer sets amplifying specific regions in the MSY2 repeats (Table S1). By the parallel use of testis genomic DNA and cDNA as templates, amplification products including all MSY2 repeats (‘MARP1–4’ reaction) or the first and second MSY2 repeats (‘MARP2–4’ reaction) were only found in the genomic PCR assays; the primer set amplifying specifically the third and fourth MSY2 repeats (‘MARP3–4’ reaction) found two amplification products with a size difference of ∼100 nt also in the testis cDNA sample (Fig. 1a). Two cDNA amplification products indicated that the longest testis transcript should start upstream of the first site of the MARP3–4 reaction bridging the second and third repeats, thus in the second MSY2 repeat (MSY2–2; Fig. S1). This was confirmed by subsequent sequence analysis; the additional 100 nt shorter RT-PCR product was produced by the second identical MARP3–4 site bridging the border of the third (MSY2–3) and fourth (MSY2–4) repeats. Both testis cDNA amplification products were significantly shorter than those of the corresponding genomic amplification products (Fig. 1a). It suggests that these transcripts were further processed by some splicing events.
Figure 1.
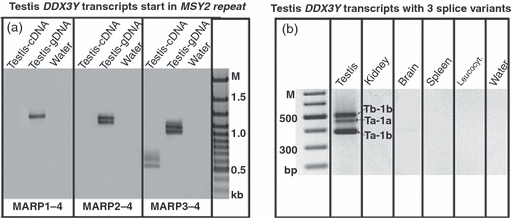
(a) Reverse transcription-polymerase chain reaction (RT-PCR) mapping of putative start site of the testis expressed DDX3Y transcripts with extended 5′UTR sequence in the MSY2 repeat block. For all sequence information, see Table S1. To judge the specificity of the designed MSY2 repeat-specific PCR assays, they were performed in parallel with cDNA samples of the polyadenylated testis mRNA fraction (testis-cDNA lanes) and with genomic DNA samples extracted from the same testis tissue [testis-g(enomic) DNA lanes; positive controls]. The water lanes control absence of any DNA and RNA contamination in the different PCR experiments (negative controls). We found two amplification products in the testis cDNA lanes only with the MARP3-4 reaction, that is, when amplifying specifically the third (MSY2-3) and fourth (MSY2-4) repeats. This result maps the putative start site of DDX3Y testis transcripts into the second MSY2 repeat (MSY2-2). Their different lengths compared with the genomic PCR products suggest some subsequent splicing processes. (b) Three different lengths of DDX3Y transcription products were identified in each RT-PCR assay independent of location of the primer sites along the upstream sequence of DDX3Y exon 1 and always only in the testis tissue, when performed with cDNA samples prepared from the mRNA pools of the kidney, brain and spleen and from leucocytes. Their sequence analyses identified three distinct splice variants as their common molecular origin (see Fig. 1). As no unspliced transcripts were found, this testis-specific 5′UTR was tentatively designated as ‘exon-T(estis)’ and its splicing variants coined ‘Ta-1a’, ‘Ta-1b’ and ‘Tb-1b’ respectively.
To map and sequence the putative splice sites in the long 5′UTR sequence experimentally, we performed RT-PCR assays with a number of primer sets located along the upstream sequence of DDX3Y exon 1 until the MSY2 repeats (Table S1). To study additionally the tissue distribution of these transcripts, we used polyA cDNA samples as templates, not only from leucocytes and from testis tissue but also from the kidney, brain and spleen tissues. Three distinct amplification products were found always, independent of location of the upstream primer site and always only with the cDNA sample of testis tissue (Fig. 1b).
Their sequence analyses revealed the use of two canonical splice donor (5′-GT) and two acceptor (AG-3′) sites in the amplified 5′UTR sequence parts (Fig. S1). We did not identify any unspliced long 5′UTR in these DDX3Y testis transcripts. It was therefore tentatively designated 5′UTR ‘exon-T’(estes) of the DDX3Y gene and its three alternative splicing products: ‘exon-Ta-1a’, -‘Ta-1b’ and -‘Tb-1b’ respectively (Fig. 1b).
Inspecting the ‘DBTSS’ (http://dbtss.hgc.jp) for DDX3Y testis transcripts with long 5′UTR extensions, we found five samples with part of the 5′UTR exon-T sequence. However, their TSS were not in the MSY2 repeats, but always downstream of it, namely at the Y reference positions: 15.016.018 (two samples) and 15.016.019 (three samples) respectively (Fig. S1). They were found by using the RLM-RACE (RNA ligase-mediated rapid amplification of 5′ cDNA ends) protocol (Wakaguri et al., 2008). As we expected that this and our 5′RACE protocol should deliver comparable results, we might have missed this alternative 5′UTR exon-T extension because we did not sequence all our 5′RACE amplification products, but only the longest ones. As independent from their start site, all DDX3Y transcripts with exon-T were expressed only in the testis tissue with the same three alternative splice variants (Fig. 1b), we can assume that this testis-specific DDX3Y transcript class has thus probably two distinct TSS in exon-T located in and downstream of the MSY2 repeats. We tentatively designated them as ‘T-TSS-I’ and ‘T-TSS-II’ respectively (Fig. 2a).
MSY2 starting DDX3Y-exon-T testis transcripts have short 3′UTR lengths
On Northern blots, we and others observed polyadenylated DDX3Y testis transcripts with variable 3′UTR lengths. Besides an RNA population of ∼5 kb expressed in each tissue, the major testis RNA population had a length of ∼3 kb (Lahn & Page, 1997; Ditton et al., 2004). This suggests their processing for polyadenylation in the proximal region of the usually ∼2.5-kb long 3′UTR sequence. We speculated that these testis-specific transcripts might also include those with the afore described 5′UTR exon-T extensions.
To explore this experimentally, two human tissue RNA blot experiments were performed with two DDX3Y cDNA probes both including exon-Ta downstream of ‘T-TSS-I’ (see Materials and methods). Both probes should therefore hybridize to all DDX3Y testis transcripts including exon-T, independent of their start at ‘T-TSS-I’ or at ‘T-TSS-II’ repectively.
In both experiments, we found a strong hybridization signal with the short testis-specific RNA population (Fig. 3a,b). The additional weaker signal with the long transcript population (∼5 kb) in the different male tissues was only observed with the exon-Ta-1b probe including besides exon-Ta, the exon-1b part of exons 1 and 2 completely (Fig. 3a). This part of the probe is also present in all DDX3Y transcripts starting from the commonly used ‘TSS-I’ region of exon 1. The pure exon-Ta probe only cross-hybridized to the short transcript population of the testis tissue (Fig. 3b). Repeating the blot experiment with an exon-Ta probe located distal of ‘T-TSS-I’ and therefore identifying only those testis transcripts starting from ‘T-TSS-II’, we obtained the same result as shown in Fig. 3b. We conclude that all DDX3Y testis transcripts with the 5′UTR exon-T extension should become processed for polyadenylation in the proximal part of their 3′UTR in exon 17.
Figure 3.
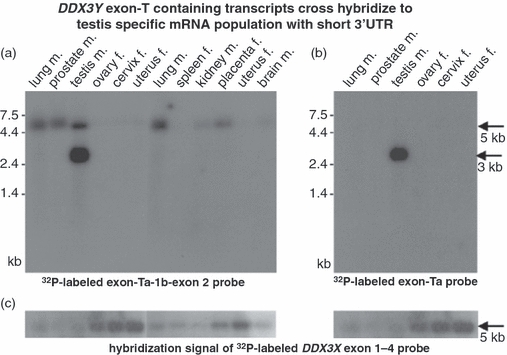
Two different 32P-labelled DDX3Y exon-T probes mainly cross-hybridized to the ∼3-kb long testis-specific mRNA population (marked by an arrow on the right). The Northern blots used in (a) and (b) contain the same quantities of polyadenylated RNA samples in each lane extracted from the tissues indicated (m, male tissue; f, female tissue). As the pure exon-Ta probe in (b) only cross-hybridized to the short testis-specific RNA population, the additional cross-hybridization of the exon-Ta-1b-exon-2 probe in (a) to the 5-kb long RNA class in different male tissues (marked by an arrow on the right) indicates that this additional hybridization signal is because of the DDX3Y exon-1b-exon 2 sequence part of this exon-Ta probe. The absence of cross-hybridization in RNA samples from female tissues confirms that both exon-T probes are only part of male-specific DDX3Y transcripts [the weak female placenta signal in (a) probably indicates some tissue contamination with embryonic male tissue]. (c) Cross-hybridization of a 32P-labelled DDX3X exon 1–4 probe to the ubiquitously expressed 5-kb long DDX3X transcripts (marked by an arrow on the right) is stronger in the female samples than in the male samples. This confirms the expression of DDX3X transcripts from both X chromosomes, and together with (a) and (b), assures the integrity of the RNA samples in each blot lane.
The proximal 3′UTR sequence of DDX3Y exon 17 in the Y reference sequence contains two possible internal PAS according to the rules of Sheets et al. (1990): one (PAS1) with a suboptimal consensus sequence (ATTAAA) and another (PAS2) with an optimal canonical ‘AATAAA’ sequence (Fig. 2b). Processing at both would fit to the calculated lengths of exon-T starting DDX3Y testis transcripts being in a range between 2.8 and 3 kb, depending on the exon-T splice variant included and their start at ‘T-TSS-I’ or at ‘T-TSS-II’ respectively (Fig. 2a,b). As PAS1 and PAS2 were separated by 165 nt in DDX3Y exon 17, some 3′RACE experiments were performed, which distinguished testis DDX3Y transcripts ending after PAS1 or PAS2 respectively (see Materials and methods). Sequence analyses of the cloned oligo(dT)-primed 3′RACE RT-PCR products revealed the use of both PAS, either PAS1 or PAS2, but only in the DDX3Y testis RNA population. In the RNA pools of leucocytes, DDX3Y transcripts ending behind PAS1 or PAS2 were absent and only transcripts containing the complete 3′UTR sequence, that is, using PAS3 (Fig. 2b), were identified.
Our 3′RACE assays thus confirmed that the short testis-specific DDX3Y transcripts are ending in proximal 3′UTR (Fig. 3a,b). Furthermore, we conclude that this transcript class mainly contains transcripts with two distinct long 5′UTR exon-T extensions (depending on their start at ‘T-TSS-I’ or ‘T-TSS-II’ respectively). This conclusion gained support from the complete sequence of an exon-T containing DDX3Y testis transcript published by the Mammalian Gene Collection (MGC) programme team (http://mgc.nci.nih.gov) in the GenBank database (accession no. BC034942). It starts in exon-T at the ‘T-TSS-I’ site and contains the major ‘Ta-1b’ splice variant. Its polyadenylation was found after PAS2 in the proximal 3′UTR.
MSY2 tandem repetition first occurs in primates
It has been reported that two tandem MSY2 repeats (MSY2-1 and MSY2-2) are also present upstream of the DDX3Y gene on the Y chromosome of the chimpanzee and orangutan (Bao et al., 2000). We therefore wanted to know whether the testis-specific start site for DDX3Y transcripts in the MSY2 repeats (T-TSS-II) is also present in these hominids (great apes) or only in human, the only species with three or more commonly four MSY2 repeats (Bao et al., 2000).
In the database, genomic Y sequences upstream of and including the DDX3Y gene were found not only for the hominid, P. troglodytes in BAC clone CH251-128L22 (Genbank accession no. AC146254) but also for the catarrhine primate M. mulatta in BAC clone CH250-541A11 (GenBank accession no. AC213321) and for the platyrrhine primate C. jacchus in BAC clone CH259-161K20 (GenBank accession no. AC225609). Using these genomic BAC sequences, we found two MSY2 tandem repeats located ∼1 kb upstream of DDX3Y, not only in the chimpanzee, as already suggested by Bao et al. (2000) but also in the other primates (Table S2). Most interestingly, comparative sequence alignment of the primates’MSY2 repeats revealed a strong sequence homology to the first and second human MSY2 sequence units in all catarrhine primates (>93%) and somewhat less (>80%) also in the platyrrhine species, C. jacchus (Fig. 4). When we screened the genomic Y sequence upstream of the mouse Ddx3y gene in BAC clone RP24-208N6 (GenBank accession no. AC145393) for MSY2 homology, only one MSY2-like sequence stretch with highest homology to MSY2-1 (54.5%) could be identified. Different deletion and insertion events on the mouse Y chromosome in this sequence region distinguished it from the same region conserved in primates significantly. This suggests that MSY2 tandem repetition started and then became conserved after separation of the rodent and primate lineages, that is, ∼80–90 Mya.
Figure 4.

Sequence homology of the two MSY2 repeats, MSY2-1 (a) and MSY2-2 (b), in different primate species. Alignments were performed with the clustalW2 program through the EMBL-EBI web server (http://www.ebi.ac.uk). Percentage data in brackets at the left indicate grade of sequence homology in the different primate species when compared with the human MSY2-1 and MSY2-2 sequences respectively. Stars underneath the aligned sequences indicate conserved nucleotides in all five primate species, dashes indicate missing nucleotides in comparison with another sequence because of insertion or deletion (indel) events. Sequence blocks boxed “TATATAAT” and grey toned (CA) are described in Discussion as putative conserved sequence elements of the proposed ‘core promoter’ in the MSY2 repeats with ‘T-TSS-II’ location at the ‘CA’ dinucleotide of the MSY2-2 repeat conserved only in the catarrhines.
MSY2 starting testis-specific DDX3Y transcripts with exon-T are only found in Catarrhini
Our comparative genomic Y sequence analyses upstream of the primates’DDX3Y gene identified not only conservation of the duplicated MSY2 sequence unit but also downstream of it along the complete exon-T sequence including conservation of both 5′-GT splice donor sites and their corresponding 3′-AG splice acceptor sites in exon 1. We therefore speculated that the novel distal promoter domain of the DDX3Y gene used for the testis-specific initiation of DDX3Y transcripts with exon-T in human might have evolved already earlier during primate evolution.
To test this assumption experimentally, we extracted RNA samples from the testis, kidney, liver, spleen tissues and leucocytes of the primate species, P. troglodytes, M. mulatta and C. jacchus, the only primates for which currently these tissue samples were available via the EUPRIM network; http://www.euprim-net.eu). We analysed the putative 5′UTR extension of their DDX3Y transcripts along the homologous exon-T sequence by a series of RT-PCR assays that are able to distinguish different exon-T extensions (Table S3). As positive control, we also designed a primer set for analysing the start of the primates’ common DDX3Y transcript class expected to be in the same ‘TSS-I’ region as found for the human DDX3Y gene. A comparative RT-PCR analysis of transcripts from the mouse Ddx3y gene starting at ‘TSS-I’ or upstream of it was used as out-group.
When screening for transcripts with the ‘TSS-I’ homologous sequence region, amplification products of mouse Ddx3y transcripts were found in each cDNA sample and the same result was obtained for these DDX3Y transcripts in all primate species (Table S4). However, amplification of DDX3Y transcripts with 5′UTR extension in exon-T was only found in the testis RNA samples of the primates. In C. jacchus, it suggests a probable beginning of these transcripts in the ‘T-TSS-I’ homologous region downstream of MSY2; in the catarrhine M. mulatta, it suggests a probable transcription start in the ‘T-TSS-II’ homologous region in MSY2; similarly, this was found for the chimpanzee testis transcripts (Table S4).
Moreover, as shown in Fig. 5 for the samples of the macaque, the lengths of the primates’ testis-specific exon-T amplification products suggest that these transcripts are spliced like the human exon-T containing DDX3Y transcripts (Fig. 5). We therefore sequenced these testis-specific RT-PCR products with exon-T extension from all primates; they always displayed the same splice junction sites as found for the major human exon-Ta-1b variant. No indication was found for one of the other human exon-T splice variants (‘Ta-1a’ and ‘Tb-1b’) in the primates’ testis transcripts. The same exon-Ta-1b splice variant was also found for the shorter 5′UTR exon-T sequence of the C. jacchus DDX3Y testis transcripts. Formation of 5′UTR exon-T by this splicing event seems therefore to be conserved in different primate lineages and independent from the exon-T start site, being in a homologous ‘T-TSS-I’ or ‘T-TSS-II’ region respectively.
Figure 5.
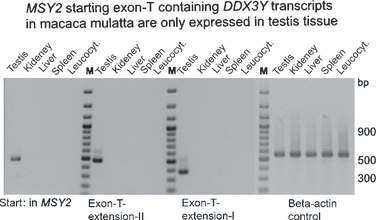
RT-PCR analyses of DDX3Y expression in different Macaca mulatta tissues and leucocytes (indicated above the gel slots) to analyse the putative extension of their homologous 5′UTR exon-T sequence in the corresponding transcripts. Similar quantities of the β-actin control in each RNA sample shown at the right confirm their general integrity. Three distinct primer sets called ‘in MSY2’, ‘exon-T-extension-I and -II’ respectively, were designed to distinguish the putative transcriptional start site (TSS) of DDX3Y transcripts along exon-T (for all sequence information, see Table S3). The first five lanes on the left will display only a positive reaction product if the transcripts would start in MSY2 at a ‘T-TSS-II’ homologous site. The following second five lanes would also display a positive reaction product if the transcripts would start downstream of MSY2 but upstream of ‘T-TSS-I’. The third five lanes would only display a positive reaction product if the transcripts would start downstream of this ‘T-TSS-I’. All cDNAs were obtained from RNA pools extracted from the same fertile male rhesus monkey. The DNA marker given at the right is a 100 bp DNA ladder [GeneRuler 100 bp DNA Ladder Plus; Fermentas; Fermentas GmbH, St. Leon-Rot, Germany]. RT-PCR amplification products were always only identified in the cDNA samples of the testis tissue. Its putative start site in MSY2 (indicated by amplification product in the testis lane on the left) suggests that the longest testis-specific DDX3Y transcripts would start in a similar location of the MSY2 repeats of M. mulatta as found in human.
In summary, we propose that duplication of the MSY2 sequence unit in primates might have triggered the development of a novel distal promoter domain for the DDX3Y gene that is able to initiate a novel testis-specific transcript class with longer 5′UTR extensions and the formation of exon-T by a subsequent splicing process. Most likely, after separation of the Platyrrhini and Catarrhini primate lineages, that is, ∼38 Mya (Goodman et al., 2005), only the catarrhine primate lineage then evolved a second TSS (T-TSS-II) in the duplicated MSY2 sequence block providing the same transcript class with still longer 5′UTR extensions. Besides ‘Ta-1b’, the alternative exon-T splice variants, ‘Ta-1a’ and ‘Tb-1b’, were evolved only recently after divergence of the human and chimpanzee lineages, that is, 6–7 Mya.
Discussion
Novel male germline-specific DDX3Y transcription products in primates
Most genes functionally expressed only during spermatogenesis are known to be under the pressure of sexual selection to maximize reproductive success (Kleene, 2003). This often leads to complex testis-specific patterns of their transcription products which help control their translation in the different testicular cell types. We believe that one example of such a strong gender-related gene in human spermatogenesis is probably the DDX3Y gene. The data presented in this article point to evolution of a novel distal promoter domain for the DDX3Y gene on the Y chromosome of primates, which is not present on that of the mouse upstream of the Ddx3y gene. Most interestingly, this second DDX3Y promoter domain seems to become activated only in the male germline and initiates a complex pattern of transcription products with distinct 5′UTR extensions processed at different splice-junction sites and polyadenylated in the gene’s proximal 3′UTR.
In mouse, Ddx3y is expressed in all tissues analysed; two transcript variants were found distinguished by different lengths of their 3′UTR (Vong et al., 2006). In contrast to human, there is no evidence for requirement of the Ddx3y protein at any stage of mouse spermatogenesis (Mazeyrat et al., 2001). It is believed that this function is fulfilled here by an autosomal retrogene of the homologous Ddx3x gene (PL10/D1Pas1 located on chromosome 1), which is expressed only in the testis tissue (Leroy et al., 1989; Session et al., 2001). In human, the RBMY gene located on the long Y arm in the AZFb deletion interval seems to have followed a similar evolutionary pathway (Elliott et al., 2000). It has a functional X homologue (RBMX) and an autosomal retrogene on chromosome 11 (HNRNPG-T) expressed in the nuclei of meiotic spermatocytes. However, a functional autosomal DDX3Y (or DDX3X) retrogene cannot be found. The two autosomal DDX3X homologous genes on chromosomes 4 and 20 contained truncated protein coding frames defining them as non-functional pseudogenes (Kim et al., 2001).
Instead of this, the genomic Y structure upstream of the primates DDX3Y gene was found to be conserved and developed a novel promoter domain with functional restriction to the male germline. As in human, the non-human primates also express DDX3Y transcripts in the testis tissue, which contains the exon-T 5′UTR (Table S4). Its different extensions in the platyrrhine and catarrhine primate lineages suggest that development of a second start site for these transcripts in the MSY2 repeat (T-TSS-II) was probably a following and independent second evolutionary step in this genomic Y region aimed at further optimizing the DDX3Y expression profile in the catarrhines’ testis tissue. We are not yet aware whether both TSS were indeed used in these catarrhines like in human. Nor are we aware of the cellular distribution of the DDX3Y protein in these primates. However, the evolutionary conservation of the novel distal promoter domain upstream of the DDX3Y gene in different primate lineages, always activated only in testis tissue, led us to speculate that its protein expression in these primates might be restricted to the male germline as in human as well. If this holds true, the distinct 5′UTR extensions of their DDX3Y testis transcripts have developed during primate evolution as an essential part of the gene’s germline-specific translation control.
Is MSY2 tandem duplication involved in development of the germline-specific distal promoter domain for the primates’DDX3Y gene?
Short tandem repetitive sequence units like MSY2– also called minisatellites – are well known in the literature as functional enhancer elements for the transcriptional activation of upstream or downstream protein coding genes (e.g. Paquette et al., 1998 and references therein). However, these regions are also often prone to genomic instability and cause hotspots for meiotic recombination (Jeffreys et al., 1998). Consequently, minisatellite sequence units are usually not conserved beyond closely related species unless their sequence variation is functionally restricted. We can assume that the high sequence conservation, which we found for the MSY2 repeats in primates (Fig. 4), suggests some functional constraints on the complete MSY2 sequence structure. Probably, the already duplicated MSY2 sequence unit on the C. jacchus Y chromosome has evolved some sequence features, which supported the development of a novel distal promoter region for DDX3Y on this primate Y chromosome. These MSY2 repeats might then have served as functional important enhancer domains to control its activation only in the testis tissue.
Short tandem repeats contain an inherent ‘enhancer’ quality to control the transcriptional efficiency of the downstream gene (Lee et al., 2008). Multiple examples are therefore known where variation in sequence or copy number of a minisatellite located upstream of a gene can cause significant human pathologies (e.g. insulin-dependent diabetes mellitus type 2 or IDMM2; Kennedy et al., 1995). We have not yet found any variation in sequence and copy number of the MSY2 repeats in the DNA samples of our European patient collective with idiopathic azoospermia. We also did not find any men with only three MSY2 tandem repeats as described by Bao et al. (2000) in a fertile male population in southern China. Probably, there is a functional need for the complete MSY2 structure in most human populations, which only allows a restricted copy variation without causing some germline pathology. Based on these data, it is therefore most likely that the tandem repetitive structure of MSY2 is indeed a functional part of the novel distal promoter domain of the primates’DDX3Y gene.
There are two distinct testis-specific TSS in human 5′UTR exon-T
Further MSY2 sequence evolution in the Catarrhini lineage has developed a second start site for the testis-specific DDX3Y transcript class in its repetitive sequence structure. In human, the result is six testis specific variants because the two distinct exon-T extensions each are processed with three alternative splice variants, ‘Ta-1b’, ‘Ta-1a’, and ‘Tb-1b’ respectively (Fig. 2a).
Surprisingly, we did not find any 5′-capped DDX3Y human testis transcripts starting with MSY2-2 in the ‘DBTSS’ (http://dbtss.hgc.jp; Wakaguri et al., 2008). Probably, isolating stable minisatellite sequence repeats in the used cloning vectors was not possible. Alternatively, lack of these transcripts in the database would indicate that DDX3Y testis transcripts starting with MSY2-2 might not be capped during their further processing.
Our RT-PCR assays with different primer sets along exon-T (Fig. S1b & Table S1) revealed that independent of these start sites, all exon-T containing DDX3Y transcripts are expressed as testis-specific and all are spliced at the same splice-junction sites. We therefore concluded that they belong to the same testis-specific DDX3Y transcript class, which starts downstream of MSY2 at ‘T-TSS-I’, or at ‘T-TSS-II’ in the second MSY2 repeat respectively. Unfortunately, we did not succeed in mapping the precise location of ‘T-TSS-II’ in the MSY2-2 repeat experimentally because cloning of the corresponding capped 5′UTR sequence by the RLM protocol of Wakaguri et al. (2008) was also not possible.
Extensive analyses for conserved sequence motifs in putative TSS of eukaryotic genes have revealed a ‘YA’ dincucleotide (nucleotide code is according to the IUPAC table given at: http://www.bioinformatics.org/sms/iupac.html; TSS nucleotide marked underlined) as the most important ‘TSS’ sequence element conserved from yeast to man (Smale & Kadonaga, 2003). It is part of the so-called ‘Inr’ motif (consensus: YYANWYY) located – if functional – within a small range downstream of a second conserved sequence motif called the ‘TATA box’ (consensus: TATAWAAR). Both sequence motifs, when spaced in a functional distance to each other (28–34 nts; Ponjavic et al., 2006), have been also coined ‘core promoter’ because this context is sufficient to bind efficiently the general transcription factors, such as the TFIID complex (Juven-Gershon et al., 2008).
When we screened the conserved MSY2 repeats for such sequence motifs, we found a nearly perfect TATA box in the MSY2-1 repeat of all primates linked to a putative initiation start motif (CA) in the MSY2-2 repeat within an optimal distance (30 nt) only in the catarrhines and not in C. jacchus (Fig. 4). Although this ‘in silico’ analysis for a putative ‘core promoter’ structure in MSY2 would nicely confirm the experimental outcome of our RT-PCR assays predicting ‘T-TSS-II’ to be located in MSY2-2, we cannot rule out some variability in the proposed ‘T-TSS-II’ site in these repeats because of the presence of more conserved ‘YA’ dinucleotides within other putative ‘Inr’ motifs in the MSY2 repeats.
Post-transcriptional control of the testis-specific DDX3Y transcript class by alternative exon-T splicing and 3′UTR PAS
In all non-human primates analysed, the testis-specific DDX3Y transcripts with exon-T were further processed by a specific splicing event designated as ‘Ta-1b’ according to the human major splice variant having the same splice-junction sites. RT-PCR assays, which analyse the 3′UTR extension of these primates’ exon-T transcripts, suggested similar PAS in their proximal 3′UTR as we found for the use of human exon-T transcripts (data not shown).
Post-transcriptional alternative splicing events, with or without alternative 3′UTR polyadenylation processes, are found for ubiquitously transcribed genes with functional expression of some of their transcript variants at distinct phase(s) of the male germline (Elliott, 2003; Kleene, 2003; Ehrmann & Elliott, 2005). To ensure their tissue-specific splicing processes, they usually require testis-specific cofactors binding together with the common spliceosome protein complex to the specific target sites. Similarly, to initiate the recognition of testis-specific PAS in the proximal 3′UTR, tissue-specific protein variants are required, which bind to the common 3′UTR cleavage protein complex, thereby directing the binding to specific 3′UTR sequence motifs.
In human and mouse, prominent examples of such testis splicing cofactors are two members of the RNA Binding Motif (RBM) family, RBMY and hnRNPGT (Elliott, 2004). Interestingly, the RBMY protein is encoded by a Y gene on the mouse and human Y chromosome and hnRNPGT by its autosomal retrogene in mouse and human as well. Functional assays for analysis of testis-specific 3′UTR-binding proteins involved in 3′UTR processing with subsequent polyadenylation were performed in the mouse. Most common is probably a variant of the cleavage stimulation factor (CstF)-64, the RNA-binding component of the general CstF binding downstream of the mRNA cleavage site. It is encoded on the X chromosome and has an autosomal paralogue, τCstF-64 (gene name Cstf2t), which is expressed only during meiosis and post-meiotic germ cell development (Dass et al., 2007). The 3′UTR cleavage factor-I (CFIm) also supports the polyadenylation process in the proximal 3′UTR of mouse transcripts expressed during spermatogenesis, most interestingly, with a preference for non-canonical PAS as found here for the testis-specific PAS1 site (Sartini et al., 2008).
It has been also repeatedly argued that there might be some functional interactions between distinct 5′ and 3′ termini of the polyadenylated mRNAs for controlling their translation in distinct cells differentially (Gallie, 1998; Kozak, 2004; Hughes, 2006); and especially in male germ cells (DeJong, 2006; Kolthur-Seetharam et al., 2008). It is therefore most likely that the complex transcriptional control, which we have described here for the DDX3Y gene in human testis tissue, is indeed an essential part of its observed strict translation control mechanism allowing significant expression of the DDX3Y protein only in the pre-meiotic germ cells (Ditton et al., 2004). Further functional analyses of these testis-specific DDX3Y transcript variants will now have to reveal the molecular tools of this germline-specific translation programme.
Acknowledgments
The authors are indebted to the German and Dutch primate centres in Göttingen (http://www.dpz.gwdg.de) and Rijswijk (http://www.bprc.nl), which provided, via the European EUPRIM-net coordination, the precious primate tissue samples for RNA and DNA isolation. They also thank especially Penny Nayudu from the DPZ and Ronald Bontrop, Gaby Doxiadis and Ivanela Kondova from the BPRC for their continuous and generous support during the primate research experiments. Thomas Strowitzki is thanked for his continuous support for this research project with his excellent laboratory infrastructure. This work was financially supported by a grant from the Deutsche Forschungsgemeinschaft to P.H. Vogt (DFG: Vo403/16–1/2).
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Figure S1. Genomic sequence region of the human Y chromosome reference in the NCBI database (accession no. NC_000024; GRCh37) starting from position 15.014.702 upstream of the MSY2 sequence block to position 15.016.892, the end of DDX3Y exon 1.
Table S1. List of oligonucleotides used as primer sets for the three RT-PCR assays described in this article.
Table S2. Sequence positions and extensions of MSY2-1 and MSY2-2 repeats in the genomic BAC clones of the primates.
Table S3. List of the species-specific oligonucleotides designed from genomic BAC sequences containing the homologous 5′UTR exon-T sequence upstream of the human and primates' DDX3Y gene.
Table S4. Outcome of the comparative exon-T expression assays in tissues and leucocytes of some primates and human and of mouse as out-group.
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer-reviewed and may be re-organized for online delivery, but are not copy-edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
References
- Bao W, Zhu S, Pandya A, Zerjal T, Xu J, Shu Q, Du R, Yang H, Tyler-Smith C. MSY2: a slowly evolving minisatellite on the human Y chromosome which provides a useful polymorphic marker in Chinese populations. Gene. 2000;244:29–33. doi: 10.1016/s0378-1119(00)00021-4. [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Charlesworth D. The degeneration of Y chromosomes. Philos Trans R Soc Lond B, Biol Sci. 2000;355:1563–1572. doi: 10.1098/rstb.2000.0717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang RY, Weaver PL, Liu Z, Chang TH. Requirement of the DEAD-Box protein ded1p for messenger RNA translation. Science. 1997;275:1468–1471. doi: 10.1126/science.275.5305.1468. [DOI] [PubMed] [Google Scholar]
- Dass B, Tardif T, Park JY, Tian B, Weitlauf HM, Hess RA, Carnes K, Griswold MD, Small CL, MacDonald CC. Loss of polyadenylation protein τCstF-64 causes spermatogenic defects and male infertility. Proc Natl Acad Sci USA. 2007;104:20374–20379. doi: 10.1073/pnas.0707589104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeJong J. Basic mechanisms for the control of germ cell gene expression. Gene. 2006;366:39–50. doi: 10.1016/j.gene.2005.10.012. [DOI] [PubMed] [Google Scholar]
- Delbridge ML, Longepied G, Depetris D, Mattei MG, Disteche CM, Marshall Graves J, Mitchell M. TSPY, the candidate gonadoblastoma gene on the human Y chromosome, has a widely expressed homologue on the X- implications for Y chromosome evolution. Chromosome Res. 2004;12:345–356. doi: 10.1023/B:CHRO.0000034134.91243.1c. [DOI] [PubMed] [Google Scholar]
- Ditton HJ, Zimmer J, Kamp C, Rajpert-De Meyts E, Vogt PH. The AZFa gene DBY (DDX3Y) is widely transcribed but the protein is limited to the male germ cells by translation control. Hum Mol Genet. 2004;13:2333–2341. doi: 10.1093/hmg/ddh240. [DOI] [PubMed] [Google Scholar]
- Ehrmann I, Elliott DJ. Post-transcriptional control in the male germ line. Reprod Biomed Online. 2005;10:55–63. doi: 10.1016/s1472-6483(10)60804-8. [DOI] [PubMed] [Google Scholar]
- Elliott DJ. Pathways of post-transcriptional gene regulation in mammalian germ cell development. Cytogenet Genome Res. 2003;103:210–216. doi: 10.1159/000076806. [DOI] [PubMed] [Google Scholar]
- Elliott DJ. The role of potential splicing factors including RBMY, RBMX, hnRNPG-T and STAR proteins in spermatogenesis. Int J Androl. 2004;27:328–334. doi: 10.1111/j.1365-2605.2004.00496.x. [DOI] [PubMed] [Google Scholar]
- Elliott DJ, Venables JP, Newton CS, Lawson D, Boyle S, Eperon IC, Cooke HJ. An evolutionarily conserved germ cell-specific hnRNP is encoded by a retrotransposed gene. Hum Mol Genet. 2000;9:2117–2124. doi: 10.1093/hmg/9.14.2117. [DOI] [PubMed] [Google Scholar]
- Foresta C, Ferlin A, Garolla A, Moro E, Pistorello M, Barbaux S, Rossato M. High frequency of well-defined Y-chromosome deletions in idiopathic Sertoli cell-only syndrome. Hum Reprod. 1998;13:302–307. doi: 10.1093/humrep/13.2.302. [DOI] [PubMed] [Google Scholar]
- Foresta C, Ferlin A, Moro E. Deletion and expression analysis of AZFa genes on the human Y chromosome revealed a major role for DBY in male infertility. Hum Mol Genet. 2000;9:1161–1169. doi: 10.1093/hmg/9.8.1161. [DOI] [PubMed] [Google Scholar]
- Fukumura J, Noguchi E, Sekiguchi T, Nishimoto T. A temperature-sensitive mutant of the mammalian RNA helicase, DEAD-BOX X isoform, DBX, defective in the transition from G1 to S phase. J Biochem. 2003;134:71–82. doi: 10.1093/jb/mvg126. [DOI] [PubMed] [Google Scholar]
- Gallie DR. A tale of two termini: a functional interaction between the termini of an mRNA is a prerequisite for efficient translation initiation. Gene. 1998;216:1–11. doi: 10.1016/s0378-1119(98)00318-7. [DOI] [PubMed] [Google Scholar]
- Goodman M, Grossman L, Wildman D. Moving primate genomics beyond the chimpanzee genome. Trends Genet. 2005;21:511–517. doi: 10.1016/j.tig.2005.06.012. [DOI] [PubMed] [Google Scholar]
- Goto H, Peng L, Makova K. Evolution of X-degenerate Y chromosome genes in greater apes: conservation of gene content in human and gorilla, but not chimpanzee. J Mol Evol. 2009;68:134–144. doi: 10.1007/s00239-008-9189-y. [DOI] [PubMed] [Google Scholar]
- Hughes T. Regulation of gene expression by alternative untranslated regions. Trends Genet. 2006;22:119–122. doi: 10.1016/j.tig.2006.01.001. [DOI] [PubMed] [Google Scholar]
- Jeffreys AJ, Neil DL, Neumann R. Repeat instability at human minisatellites arising from meiotic recombination. EMBO J. 1998;17:4147–4157. doi: 10.1093/emboj/17.14.4147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juven-Gershon T, Hsu JY, Theisen J, Kadonaga JT. The RNA polymerase II core promoter – the gateway to transcription. Curr Opin Cell Biol. 2008;20:253–259. doi: 10.1016/j.ceb.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamp C, Huellen K, Fernandes S, Sousa M, Schlegel PN, Mielnik A, et al. High deletion frequency of the complete AZFa sequence occurs only in men with complete germ cell aplasia (Sertoli-cell-only-syndrome) Mol Hum Reprod. 2001;10:987–994. doi: 10.1093/molehr/7.10.987. [DOI] [PubMed] [Google Scholar]
- Kennedy GC, German MS, Rutter WJ. The minisatellite in the diabetes susceptibility locus IDDM2 regulates insulin transcription. Nat Genet. 1995;9:293–298. doi: 10.1038/ng0395-293. [DOI] [PubMed] [Google Scholar]
- Kim Y-S, Lee S-G, Park SH, Song K. Gene structure of the human DDX3 and chromosome mapping of its related sequences. Mol Cells. 2001;12:206–214. [PubMed] [Google Scholar]
- Kleene KC. Patterns, mechanisms, and functions of translation regulation in mammalian spermatogenic cells. Cytogenet Genome Res. 2003;103:217–224. doi: 10.1159/000076807. [DOI] [PubMed] [Google Scholar]
- Kolthur-Seetharam U, Martianov I, Davidson I. Specialization of the general transcriptional machinery in male germ cells. Cell Cycle. 2008;7:3493–3498. doi: 10.4161/cc.7.22.6976. [DOI] [PubMed] [Google Scholar]
- Kozak M. How strong is the case for regulation of the initiation step of translation be elements at the 3′end of eukaryotic mRNAs? Gene. 2004;343:41–54. doi: 10.1016/j.gene.2004.08.011. [DOI] [PubMed] [Google Scholar]
- Krausz C, Degl’Innocenti S. Y chromosome and male infertility: update, 2006. Front Biosci. 2006;11:3049–3061. doi: 10.2741/2032. [DOI] [PubMed] [Google Scholar]
- Krausz C, Degl’Innocenti S, Nuti F, Morelli A, Felici F, Sansone M, Varriale G, Forti G. Natural transmission of USP9Y gene mutations: a new perspective on the role of AZFa genes in male fertility. Hum Mol Genet. 2006;15:2673–2681. doi: 10.1093/hmg/ddl198. [DOI] [PubMed] [Google Scholar]
- Lahn BT, Page DC. Functional coherence of the human Y chromosome. Science. 1997;278:675–680. doi: 10.1126/science.278.5338.675. [DOI] [PubMed] [Google Scholar]
- Lahn BT, Page DC. Four evolutionary strata on the human X chromosome. Science. 1999;286:964–967. doi: 10.1126/science.286.5441.964. [DOI] [PubMed] [Google Scholar]
- Lee LTO, Lam IPY, Chow BKC. A functional variable number of tandem repeats is located at the 5′flanking region of the human secretin gene plays a downregulatory role in expression. J Mol Neurosci. 2008;36:125–131. doi: 10.1007/s12031-008-9083-5. [DOI] [PubMed] [Google Scholar]
- Leroy P, Alzari P, Sassoon D, Wolgemuth D. The protein encoded by a murine male germ cell-specific transcript is a putative ATP-dependent RNA helicase. Cell. 1989;57:549–559. doi: 10.1016/0092-8674(89)90125-6. [DOI] [PubMed] [Google Scholar]
- Luddi A, Margollicci M, Gambera L, Serafini F, Cioni M, De Leo V, Balestri P, Piomboni P. Spermatogenesis in a man with complete deletion of USP9Y. N Eng J Med. 2009;360:881–885. doi: 10.1056/NEJMoa0806218. [DOI] [PubMed] [Google Scholar]
- Marshall Graves JA. Sex chromosome specialization and degeneration in mammals. Cell. 2006;124:901–914. doi: 10.1016/j.cell.2006.02.024. [DOI] [PubMed] [Google Scholar]
- Mazeyrat S, Saut N, Grigoriev V, Mahadevaiah SK, Ojarikre OA, Rattigan A, Bishop C, Eicher EM, Mitchell MJ, Burgoyne PS. A Y-encoded subunit of the translation initiation factor Eif2 is essential for mouse spermatogenesis. Nat Genet. 2001;29:49–53. doi: 10.1038/ng717. [DOI] [PubMed] [Google Scholar]
- Paquette J, Giannoukakis N, Polychronakos C, Vafiadis P, Deal C. The INS 5′ variable number of tandem repeats is associated with IGF2 expression in humans. J Biol Chem. 1998;273:14158–14164. doi: 10.1074/jbc.273.23.14158. [DOI] [PubMed] [Google Scholar]
- Park SH, Lee SG, Kim Y, Song K. Assignment of a human putative RNA helicase gene, DDX3, to human X chromosome bands p11.3 → p11.23. Cytogenet Cell Genet. 1998;81:178–179. doi: 10.1159/000015022. [DOI] [PubMed] [Google Scholar]
- Ponjavic J, Lenhard B, Kai C, Kawai J, Carninci P, Hayashizaki Y, Sandelin A. Transcriptional and structural impact of TATA-initiation site spacing in mammalian core promoters. Genome Biol. 2006;R78:1–8. doi: 10.1186/gb-2006-7-8-r78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartini BL, Wang H, Millette CF, Kilpatrick DL. Pre-messenger RNA cleavage factor I (CFIm): potential role in alternative polyadenylation during spermatogenesis. Biol Reprod. 2008;78:472–482. doi: 10.1095/biolreprod.107.064774. [DOI] [PubMed] [Google Scholar]
- Session DR, Lee GS, Wolgemuth DJ. Characterization of D1Pas1, a mouse autosomal homologue of the human AZFa region DBY, as a nuclear protein in spermatogenic cells. Fertil Steril. 2001;76:804–811. doi: 10.1016/s0015-0282(01)01996-3. [DOI] [PubMed] [Google Scholar]
- Sheets MD, Ogg SC, Wickens MP. Point mutations in AAUAAA and the poly(A) addition site: effects on the accuracy and efficiency of cleavage and polyadenylation in vitro. Nucleic Acids Res. 1990;18:5799–5805. doi: 10.1093/nar/18.19.5799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaletsky H, Kuroda-Kawaguchi T, Minx PJ, Cordum HS, Hillier L, Brown LG, et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature. 2003;423:825–837. doi: 10.1038/nature01722. [DOI] [PubMed] [Google Scholar]
- Smale ST, Kadonaga JT. The RNA polymerase II core promoter. Annu Rev Biochem. 2003;72:449–479. doi: 10.1146/annurev.biochem.72.121801.161520. [DOI] [PubMed] [Google Scholar]
- Tyler-Smith C. An evolutionary perspective on Y-chromosomal variation and male infertility. Int J Androl. 2008;31:376–382. doi: 10.1111/j.1365-2605.2008.00889.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler-Smith C, Krausz C. The Will-o'-the Wisp of Genetics – hunting for the azoospermia factor gene. N Eng J Med. 2009;360:925–927. doi: 10.1056/NEJMe0900301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt PH. Azoospermia factor (AZF) in Yq11: towards a molecular understanding of its function for human male fertility and spermatogenesis. Reprod Biomed Online. 2005;10:81–93. doi: 10.1016/s1472-6483(10)60807-3. [DOI] [PubMed] [Google Scholar]
- Vogt PH, Edelmann A, Kirsch S, Henegariu O, Hirschmann P, Kiesewetter F, et al. Human Y chromosome azoospermia factors (AZF) mapped to different subregions in Yq11. Hum Mol Genet. 1996;5:933–943. doi: 10.1093/hmg/5.7.933. [DOI] [PubMed] [Google Scholar]
- Vogt PH, Ditton HJ, Kamp C, Zimmer J. Structure and function of AZFa locus in human spermatogenesis. In: Lau YF, Chan WY, editors. The Y Chromosome and Male Germ Cell Biology in Health and Diseases. Singapore: World Scientific Publishing Co. Pte. Ltd; 2007. pp. 91–125. [Google Scholar]
- Vogt PH, Falcao CL, Hanstein R, Zimmer J. The AZF proteins. Int J Androl. 2008;31:383–394. doi: 10.1111/j.1365-2605.2008.00890.x. [DOI] [PubMed] [Google Scholar]
- Vong QP, Li Y, Lau Y-FC, Dym M, Rennert OM, Chan W-Y. Structural characterization and expression studies of Dby and ist homologs in mouse. J Androl. 2006;27:653–661. doi: 10.2164/jandrol.106.000471. [DOI] [PubMed] [Google Scholar]
- Wakaguri H, Yamashita R, Suzuki Y, Sugano S, Nakai K. DBTSS: database of transcription start sites, progress report 2008. Nucleic Acids Res. 2008;36:97–101. doi: 10.1093/nar/gkm901. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


