Abstract
Autophagy is the degradative process by which eukaryotic cells digest their own components using acid hydrolases within the lysosome. Originally thought to function almost exclusively in providing starving cells with nutrients taken from their own cellular constituents, autophagy is in fact involved in numerous cellular events including differentiation, turnover of macromolecules and organelles and defense against parasitic invaders. During the past 10–20 years, molecular components of the autophagic machinery have been discovered, revealing a complex interactome of proteins and lipids, which, in a concerted way, induce membrane formation to engulf cellular material and target it for lysosomal degradation. Here, our emphasis is autophagy in protists. We discuss experimental and genomic data indicating that the canonical autophagy machinery characterized in animals and fungi appeared prior to the radiation of major eukaryotic lineages. Moreover, we describe how comparative bioinformatics revealed that this canonical machinery has been subject to moderation, outright loss or elaboration on multiple occasions in protist lineages, most probably as a consequence of diverse lifestyle adaptations. We also review experimental studies illustrating how several pathogenic protists either utilize autophagy mechanisms or manipulate host-cell autophagy in order to establish or maintain infection within a host. The essentiality of autophagy for the pathogenicity of many parasites, and the unique features of some of the autophagy-related proteins involved, suggest possible new targets for drug discovery. Further studies of the molecular details of autophagy in protists will undoubtedly enhance our understanding of the diversity and complexity of this cellular phenomenon and the opportunities it offers as a drug target.
Key words: autophagy; ubiquitination; pexophagy; evolution; free-living protist; parasitic protist; life-cycle differentiation, Trypanosomatidae; Apicomplexa; drug discovery
Introduction
In the 1950s, Christian de Duve discovered lysosomes in mammalian cells as acidic organelles containing a large number of hydrolases responsible for intracellular degradation of proteins and other macromolecules, either taken up from outside the cell by endocytosis or hydrolysis of the cell's own constituents (reviewed in ref. 1). In 1963, de Duve coined the word autophagy for the process by which the cell's own proteins are degraded.2,3 Subsequent research has identified lysosomes in many other eukaryotic organisms or similar organelles called vacuoles in yeasts, and has revealed various pathways by which cytosolic components as well as organelles are delivered to the lysosomes. Although autophagy was initially only detected by electron microscopy, major advances in the last decade have enabled a detailed understanding of the process at the molecular level.4 Many of the molecular components involved in autophagy have been identified in the budding yeast Saccharomyces cerevisiae, owing to its tractable genetics. Currently ATG (standing for AuTophaGy-related) genes encoding more than 30 proteins involved in these processes have been identified and characterized.5 For many of them, orthologues have been identified in mammals, plants, other yeasts and fungi.6 Moreover, detailed cellular and molecular biological studies have provided information about the steps of various autophagy pathways at which the different Atg proteins act, thereby revealing a defined molecular mechanism.7
Autophagy plays different roles in a cell (reviewed in ref. 8). First, it serves as a survival mechanism under conditions of “extracellular stress” such as nutrient starvation, hypoxia and high temperatures or as a housekeeping device under conditions of “intracellular stress” by removing damaged, redundant or otherwise unwanted cytoplasmic components which may include organelles. In yeasts, autophagy allows survival during nutrient deprivation by recycling the cytoplasmic constituents, as well as playing a major role in cellular remodeling. In particular, autophagy is important in adapting the cell's morphology and metabolic repertoire to changes of nutritional conditions. In addition, in mammals, autophagy plays a crucial role in health and disease.9 It is not only involved in the removal of damaged or malfunctioning cell components, but it is important, too, for cell differentiation, tissue remodeling and defense against pathogenic organisms. As a result, links exist between, for example, autophagy and growth or the development of innate immunity. Autophagy has also been implicated in both promoting and preventing diseases such as cancer, neurodegenerative disorders, lysosomal storage diseases, viral, bacterial or parasitic infections and aging. Moreover, autophagy is one of three main cell death mechanisms: necrosis, apoptosis and cell death with autophagy.10
Different autophagic processes have been described (reviewed in ref. 6 and 11). The most common form is macroautophagy, a form of nonselective autophagy in which a bowl-shaped membrane is formed randomly around portions of the cytoplasm, bulk cytosol and other cytoplasmic components such as organelles, creating a structure called the phagophore or isolation membrane that develops into a double-membrane vesicle, the autophagosome. In yeast, a structure is observed called the phagophore-assembly site (PAS, also known as the preautophagosomal structure), which is important for different stages of the autophagy process including autophagosome formation. The PAS concentrates a large number of autophagy components, some Atg proteins and developing vesicles, probably to organize vesicle formation in a concerted manner. In yeast only a single, perivacuolar PAS is found, whereas mammalian cells contain multiple loci of Atg complexes. Once the autophagosome has been formed, its outer membrane fuses with the lysosomal/vacuolar membrane, forming the autophagic body or autolysosome. Due to the action of lysosomal hydrolases, the inner membrane of the autolysosome is ruptured before its content is degraded. The degradation products are recycled to the cytosol through the action of permeases within the lysosomal membrane. In addition, random uptake of cytoplasmic components may also occur by microautophagy, in which the uptake of a portion of the cytoplasm occurs by direct engulfment by the lysosomal membrane. The different mechanisms of macro- and microautophagy have distinct effects on the size of the lysosomal membrane. In the former process, its size increases as a result of fusion with the incoming autophagosomal membrane. In contrast, in microautophagy part of the lysosomal membrane is used to engulf cytoplasmic material resulting in a vesicle bounded by a single membrane of lysosomal origin. This vesicle, including both its membrane and its contents, is degraded. As a result, the size of the lysosomal membrane is decreased. Since newly digested material will be exported from the lysosome, until and unless new lysosomal membrane is synthesized, microautophagy may lead to a reduction in lysosomal volume and may play a homeostatic role in balancing membrane that is delivered to the lysosome via autophagy and other pathways that terminate at this organelle. A third form, a chaperone-mediated autophagy has been described.12 Defined proteins, usually containing the KFERQ or a related pentapetide motif, bind to a cytosolic chaperone that targets the complex to bind to a lysosomal membrane receptor, where—with the help of additional chaperones—the respective protein is taken up and degraded.
Instead of directly fusing with the lysosome, the autophagosome may also be indirectly routed to the lysosome by first fusing with the endosomes resulting from endocytosis. The organelle formed by this process, designated the amphisome, will subsequently fuse with the lysosome.
Processes related to macroautophagy are also used for the selective routing of cytoplasmic macromolecules or organelles to the lysosome/vacuole. The selectivity of the system is in all cases endowed by the specific recognition of material to be degraded. In a few cases, proteins and cofactors responsible for this recognition have been identified, and some of these molecules undergo post-translational modification in order to provide the signal for degradation. However, the molecular basis of the recognition events remains rather elusive. The best-studied selective process is the cytoplasm-to-vacuole targeting (Cvt) pathway, which has only been identified in the yeasts S. cerevisiae and Pichia pastoris. This process does not serve to route proteins for degradation, but to deliver the resident vacuolar lumenal hydrolytic enzymes aminopeptidase I and mannosidase to their destination. These enzymes, after their synthesis in the cytosol, become incorporated into a double-membrane Cvt vesicle that fuses with the vacuole. The inactive precursor dodecameric aminopeptidase (prApe1) assembles into a large complex in the cytosol and is recruited by a receptor protein, Atg19 (originally called Cvt19). Atg19 also binds oligomeric-mannosidase. The Atg19-prApe1-α-mannosidase complex is selectively targeted to the Cvt vesicle through the action of Atg11 and the subsequent interaction between Atg19 and Atg8. After delivery of the vesicle to the vacuole and the disruption of the inner membrane of the internalized Cvt vesicle, the complex is released into the vacuolar lumen, prApe1 processed into an active enzyme and Atg19 degraded. Other selective autophagy-like processes involve the degradation of redundant or damaged organelles. Among these processes are (i) pexophagy, the selective degradation of peroxisomes, (ii) mitophagy, the selective degradation of (part) of mitochondria, (iii) ribophagy, the selective degradation of ribosomes and (iv) ER-phagy or reticulophagy, the degradation of part of the endoplasmic reticulum (ER). Moreover, part of the nucleus may be pinched off and degraded by microautophagy in a process called piecemeal microautophagy of the nucleus (PMN) or micronucleophagy. Selective pexophagy and mitophagy may also occur by either micro- or macroautophagic mechanisms.
As mentioned above, over 30 genes have been identified encoding proteins specifically involved in autophagy. About half of them participate in the core machinery, essential for formation of autophagosomes or Cvt vesicles, respectively, whereas the others complement this machinery according to the requirements of the specific process executed. Although autophagy occurs constitutively at a basal level, it is dramatically enhanced in response to stimuli from within the cell or from the environment. Such stimuli include starvation or other forms of stress, physiological signals such as hormones and growth factors and viral, bacterial or parasitic infection. Induction of autophagy involves common signaling cascades also involved in other cellular processes. For example, the serine/threonine protein kinase TOR (target of rapamycin) is crucial in the regulation of autophagy induction. The TOR protein occurs in two complexes, TORC1 and TORC2, each in control of a distinct signaling pathway. The rapamycin-sensitive TORC1 is involved in regulation of autophagy by responding to various sensors. On the one hand, it responds to the availability of nutrients, notably amino acids, by stimulating temporal cell growth. This is achieved, via a variety of downstream effectors, by orchestrating ribosome biosynthesis, amino acid uptake and translation initiation. On the other hand, nutrient or energy depletion causes TORC1 to maintain cellular viability via other effectors. This latter response includes use of the Atg proteins as effectors to activate autophagy and so increase the recycling of cellular components. Additionally some other enzymes are shared between autophagy pathways and other signaling cascades, such as the phosphatidylinositol 3-kinase (PtdIns3K) domain-containing Vps34, Vps15 and Vps30 (also termed Atg6 or Beclin 1 in mammals). These kinases are located at the PAS and are involved in autophagosome formation. A late step in the autophagy process involves fusion of autophagosomes or Cvt vesicles with the lysosome/vacuole. Other proteins involved in this fusion process are Vac8, SNAP and SNARE, the latter two of which are generally involved in membrane fusion.13
Autophagy processes are not restricted to mammalian cells and yeasts, but have been found in representatives of all eukaryotic phyla: vertebrates (mammals, birds, amphibians, fish), invertebrates such as the nematode Caenorhabditis elegans and the cnidarian polyp Hydra, insects (Drosophila, Lepidoptera and ticks), plants, fungi (different yeasts and filamentous fungi) and several protists.6,14,15 This identification has been done experimentally, often by morphological studies of cells subjected to starvation or other forms of stress or by homology searches in sequence databases with yeast or human Atg sequences. Previously, we have presented genomic and experimental evidence for autophagy and pexophagy in parasitic protists belonging to the family Trypanosomatidae and genomic evidence that autophagy also occurs in a diverse variety of other protists, including some free-living taxa.14,16–23 For some of the species examined so far, genomic signatures are complemented by experimental demonstrations of autophagy and/or analyses of candidate protein function. Significantly, however, bioinformatics surveys of protist genomes reveal some taxa in which autophagy, at least as defined by current paradigms, does not occur and others in which the canonical autophagy pathway has been subject to lineage-specific elaboration or moderation. Thus, even though only a limited spectrum of the many unicellular eukaryotes belonging to the 30–40 disparate phyla grouped in the Kingdom Protista can currently be readily subjected to genome or direct experimental analysis, one can anticipate considerable variation from the pathways described in yeast and animals during the past 10–20 years.
Human and animal pathogenic protists have received much more attention in biological research than either free-living protists or the parasites of plants. This is particularly true for parasites causing devastating human and animal diseases in tropical and subtropical parts of the world and for which no adequate treatment is available, such as the protists belonging to the family Trypanosomatidae that cause African sleeping sickness, Latin-American Chagas' disease and various manifestations of leishmaniasis and the order Apicomplexa responsible for malaria and theileriosis, but also for diseases occurring in parts of the world with a moderate climate such as toxoplasmosis. Some of these parasites (Trypanosoma brucei, responsible for sleeping sickness) live extracellularly in the blood of their human host. Others (like the Chagas' disease parasite Trypanosoma cruzi, Leishmania species and the apicomplexans), spend a major part of their pathogenic life inside host mammalian cells. The host's autophagy may play a major role in the infection process of some of these latter parasites. The process may either be used to eliminate live or dead pathogens or it may be exploited by the parasite to favor invasion. On the other hand, intracellular parasites may block host cell autophagy as a survival strategy, for example by reducing parasite-derived peptide presentation by host cell MHC to CD4+ and CD8+ T cells.
In this review, we will summarize the current knowledge about autophagy in protists, and present some new data. We will provide a rationalization for the differences found between protists and other phylogenetic groups and between different protist lineages with regard to the occurrence or variations in the process, the different pathways and the repertoire of ATGs. This will be done on the basis of evolutionary developments and habitats in which the different organisms live. We will also describe the details and relevance of the host autophagy process for invasion, survival or removal of pathogenic protists.
Molecular Analysis of Autophagy in Protists—Some Practical Considerations
Yeast as the model organism for autophagy studies.
As mentioned above, autophagy is a process that has been found in representatives of all major eukaryotic phyla, including protists. Indications of autophagy in some unicellular eukaryotes were initially obtained by electron microscopy. However, in the last decade most information about the distribution of autophagy in the highly diverse kingdom of protists has come from bioinformatic analyses based on homology searches in sequence databases using mammalian and particularly yeast Atg sequences as queries. Subsequently, the occurrence of the process in some of the better-known, often pathogenic protists has been experimentally confirmed. Since autophagy has been most intensively studied in S. cerevisiae, and this organism thus serves as a model to which other organisms are compared, we will first summarize the respective steps in yeast (reviewed in ref. 1, 24 and 25).
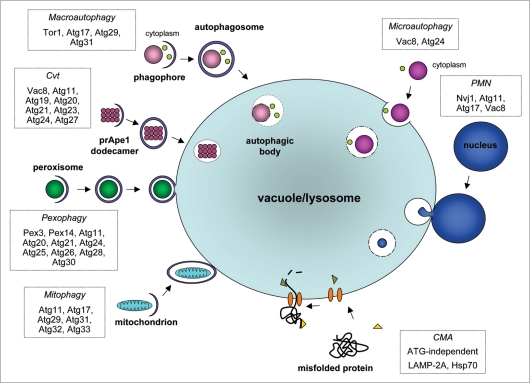
Macroautophagy, and the very similar Cvt pathway can conceptually be divided into a number of consecutive steps mediated by specific proteins (Fig. 1): (1) nutrient sensing and induction of autophagy; (2) cargo selection and packaging; (3) vesicle nucleation (formation of the phagophore); (4) vesicle expansion and completion (autophagosome or Cvt vesicle formation); (5) retrieval; (6) docking and fusion; (7) vesicle breakdown in the vacuole.
Figure 1.
Schematic overview of autophagy-related processes. The main steps of the various pathways, as summarized in the main text of the paper, are indicated. Proteins shown to be specifically required for the different pathways are boxed.
(1) Nutrient sensing and induction of autophagy. Under nutrient-rich conditions, when macroautophagy occurs at a basal level, the protein kinase TOR of the TORC1 complex maintains a hyperphosphorylated state of Atg13 that, together with Atg1, forms the core of the so-called Atg1 complex. When nutrients become limited, Atg13 is partially dephosphorylated causing an increased affinity for Atg1. The less phosphorylated Atg1-Atg13 complex induces, through the Ser/Thr kinase activity of Atg1, an increase of macroautophagy. This kinase activity appears to be also essential for the constitutive Cvt pathway. However, despite the similarity between the macroautophagy and Cvt pathways, and the fact that most components are shared between them, there are differences, too. Some Atgs have been shown to be essential only for the former pathway, whereas others only for the latter one. Atg1 interacts directly or indirectly via Atg13 or Atg11 with Atg20, Atg24 and Vac8 for the Cvt pathway, and with Atg17, Atg29 and Atg31 for macroautophagy. Atg11 is also essential for pexophagy, mitophagy and PMN, whereas Atg24 is additionally required for pexophagy in methylotrophic yeasts. These differences indicate a role of the Atg1 complex as a switch between nonselective autophagy and selective processes such as Cvt and pexophagy. A detailed discussion of the current knowledge of the elaborate regulation of autophagy by TOR and signaling pathways in S. cerevisiae, the nature of all above-mentioned Atgs involved in the induction step, and the temporal and spatial aspects of their interactions, can be found in reviews by Diaz et al.26 and Jung et al.27
(2) Cargo selection and packaging. In contrast to nonselective macroautophagy, where bulk cytoplasm components are randomly sequestered, a defined cargo selection occurs in the cases of Cvt and selective autophagy processes. As mentioned in the introduction, the protein Atg19 or Cvt19 plays a crucial role in cargo selection for the Cvt pathway. The Cvt complex (consisting of prApe1 and α-mannosidase bound to Atg19) binds to Atg11 that brings the complex to the PAS where the Cvt vesicle will nucleate. All components of the core autophagy machinery (see below) are involved in the formation of the Cvt vesicle, notably phosphatidylethanolamine (PE)-conjugated Atg8 that binds to Atg19 to ensure the correct sequestering of the complex within the vesicle.
For the selective process of pexophagy, several proteins are important in S. cerevisiae and/or the methylotrophic yeasts P. pastoris and Hansenula polymorpha (reviewed in ref. 28). Atg30 has been identified in P. pastoris as the peroxisome receptor for pexophagy. It is expressed and associates with peroxisomes already during proliferation of the organelles, but it becomes phosphorylated by an as yet unknown kinase only after induction of pexophagy. The interaction with peroxisomes occurs through two peroxins, Pex3 and Pex14, proteins with a major function in peroxisome biogenesis. In methylotrophic yeasts, Pex14 has been detected in both an unmodified and a phosphorylated form, and only the latter one appears to interact with Atg30. Atg30, after its association with peroxisomes, also interacts with the autophagy machinery at the PAS, and so targets the organelles for degradation to the vacuole.
A few other Atg proteins have been shown to be required for pexophagy: i.e., Atg25 in H. polymorpha, Atg26 in P. pastoris and the plant pathogenic fungus Colletotrichum orbiculare and Atg28 in P. pastoris. These proteins act at the later stages of pexophagy, but their precise role remains to be established.
Pexophagy in methylotrophic yeasts occurs not only by macropexophagy, but also by micropexophagy, depending on the carbon source and the ATP level.29 Macropexophagy usually involves the sequestering of only a single peroxisome by the phagophore, whereas in micropexophagy often a cluster of organelles is engulfed by the vacuolar membrane. Micropexophagy is also a selective process, because the vacuolar membrane contains proteins that recognize the peroxisomes; the signaling is independent of Atg30 phosphorylation. In P. pastoris, the glycolytic enzyme phosphofructokinase plays a role in triggering micropexophagy. The morphological changes observed during micropexophagy are different from those in common microautophagy; in the latter, tubules invaginate into the vacuole lumen, whereas in the former, arm-like extensions of the vacuolar membrane are formed. To complete the sequestration by the arms, they fuse, in an Atg24-dependent manner, with a peculiar cup-shaped double-membrane structure called the micropexophagic membrane apparatus (MIPA). The origin of the MIPA is still unknown.
Autophagy of mitochondria in yeast occurs at a basic rate for regular turnover, but is induced upon change from a respiratory carbon source to a fermentative one. It may also be important upon organelle damage caused by reactive oxygen species (ROS) originating from a malfunctioning respiratory activity. Degradation of the organelle may occur by selective macro- or micromitophagy. Thus far, only two proteins specifically required for mitophagy have been identified in S. cerevisiae, Atg32, which appears to function as an outer membrane tag to mark the organelle,30,31 and Atg33.32 The selective nature of the organelle's degradation has been confirmed by the involvement of Atg11 (that binds Atg32, similar to its interaction with Atg19 in the Cvt pathway), Atg17 and Atg29.
Only limited information about induction, mechanism and the requirement of some Atg proteins involved in selective reticulophagy, ribophagy and piecemeal microautophagy of the nucleus is also available for yeast. However, it will not be discussed here because it is outside the scope of this review as no information about these processes in protists is as yet available.
(3) Vesicle nucleation. The nucleation of vesicles for cargo encapsulation involves the recruitment or formation of a membrane to generate a beginning for a vesicle (Cvt vesicle or autophagosome). The origin of the membrane for these vesicles has been debated for years. Several possibilities have been proposed, such as maturation from the ER or mitochondrion or de novo membrane formation by localized synthesis or transport. No unambiguous data are available for yeast, but recent studies on mammalian cells strongly suggest that both the ER and outer mitochondrial membrane can be the source of these membranes.33–35 Cup-shaped ER protrusions enriched in phosphatidylinositol 3-phosphate [PtdIns(3)P] recruit autophagic effectors and expand for the phagophore to sequester cargo. Formation of PtdIns(3)P is dependent on the activity of a membrane-associated class III PtdIns3K complex comprising the Ser/Thr protein kinase Vps15, the PtdIns3K Vps34, as well as Atg6 and Atg14, which functions at the PAS. In yeast, Atg18, Atg20, Atg21 and Atg24, which all bind PtdIns(3)P, are recruited to the PAS. In H. polymorpha, but not in S. cerevisiae, Atg21 is essential for pexophagy.
(4) Vesicle expansion and completion. The expansion of the phagophore into an autophagosome or Cvt vesicle involves a process reminiscent of protein ubiquitination. Two sets of ubiquitination-like conjugation systems are implicated (reviewed in ref. 36). The first one, in which Atg8 as the ubiquitin-like modifier is conjugated to PE as the substrate, employs Atg7, Atg3 and the complex Atg12-Atg5 as E1, E2 and E3-like proteins, respectively; in the second system Atg12 plays the ubiquitin-like role, while Atg5 is the substrate and Atg7 and Atg10 exert the E1 and E2-like activities, respectively; here the E3 function is unknown. Atg16 can oligomerize and binds to the Atg12-Atg5 conjugate, usually resulting in the formation of a dimeric complex. Together, these activities lead to the following processes at the PAS: First, Atg8 is proteolytically processed at its C terminus by Atg4, exposing a glycine residue. This Atg8 as well as Atg12, also having a C-terminal Gly residue, are activated independently by the E1-like enzyme Atg7 by forming a thioester bond with it and subsequently transferred via E2-like Atg3 or Atg10, respectively, to membrane-associated PE or Atg5. As a result, Atg8 becomes covalently linked to PE and so anchored to both sides of the expanding, curved phagophore. The dimeric Atg12-Atg5-Atg16 complex covers mostly the outer surface of the curved organelle. After closure of the expanded phagophore, resulting in a double-membrane bounded vesicle, its outer surface loses its coating of Atg12-Atg5-Atg16 complex, and Atg4 acts once more on Atg8, releasing it from the outer surface by cleavage of the Atg8-PE linkage. The result is a cargo-filled vesicle with Atg8-PE at its inner membrane surface, now competent for fusion with the vacuole.
(5) Retrieval. Only Atg8-PE and Atg19 remain associated with the autophagosome or Cvt vesicle and are directed to the vacuole. Most other components involved in the autophagy and Cvt pathways are soluble or peripheral membrane proteins and are easily retrieved from the phagophore/autophagosome/Cvt vesicle to the PAS to be reused in further cycles. Only retrieval of the integral membrane protein Atg9 requires PtdIns3K, the PtdIns3K-binding protein Atg18 and several other Atg proteins: 1, 2, 11, 13, 23 and 27. Atg23 is essential only for the Cvt pathway, but loss of this protein does result in a reduced number of autophagosomes.
(6) Docking and fusion. Vesicle targeting to the vacuole/lysosome, its docking to the degradative organelle and membrane fusion involve proteins that are generally used within the cell for other membrane docking/fusion processes and are not specific to autophagy and related processes. Therefore, they will not be further discussed in this review.
(7) Vesicle breakdown in the vacuole. Following fusion of the autophagosome with the vacuole and formation of the autophagic body, the membrane of this body (and that of organelles in it and the Cvt body) is ruptured to release and degrade its content. Vesicle lysis requires the vacuole's acidic pH and peptidase B (Prb1). Probably, this peptidase activates various vacuolar zymogens responsible for breakdown processes. In S. cerevisiae Atg15, an integral membrane protein with presumed lipase activity, is specifically involved in the degradation of this membrane. Another autophagy-related protein operating in the vacuole is Atg22, an integral protein of the organelle's boundary membrane which bears sequence similarity to permeases of the major facilitator superfamily. It is probably involved in export of regenerated amino acids to the cytosol.
Ubiquitination and selective autophagy.
For many years, the ubiquitin-proteasome system has been considered as the main pathway responsible for the removal of misfolded proteins in all eukaryotic cells. Recently, however, it has been observed in mammalian cells that ubiquitination of cytosolic protein aggregates may also function as a specific signal to induce their selective degradation by autophagy (reviewed in ref. 37). By analogy with the proteasome system, where unfolded ubiquitinated proteins are recognized by ubiquitin-binding receptors that deliver them to the proteasome, ubiquitin-dependent selective autophagy involves a ubiquitin-binding receptor and its associated proteins, p62 and NBR1, respectively. Their simultaneous interaction with LC3 or GABARAP (mammalian proteins which belong to two ATG8 subfamilies), mediate the sequestering of ubiquitinated substrates (misfolded single proteins, inclusion bodies and aggresomes) in the autophagosome for their subsequent lysosomal degradation. The ability of p62 and NBR1 to bind ubiquitinated substrate depends on the presence of a C-terminal UBA domain, whereas the interaction with the ubiquitin-like family proteins LC3/GARABAP depends on a short linear motif called ‘LC3-interacting region’ (LIR) that is characterized by an acidic cluster and a conserved aromatic residue. A similar motif is present in Atg19 and is responsible for the interaction with Atg8 in the yeast's Cvt pathway. Nevertheless, functional yeast orthologues of p62/NBR1 have not been identified yet.
Regarding the regulatory mechanism that determines which pathway will be followed for the degradation of a specific ubiquitinated substrate, evidence has been reported that the specific architecture of the ubiquitin moieties (branched K48-linked polyubiquitin chains for proteasomal or mono-ubiquitin and linear K63-linked polyubiquitin chains for autophagy), together with the relative levels of co-chaperones BAG1 and BAG3 in the case of protein aggregates, are elements that select between the two pathways. However, p62 competes for ubiquitinated cargo with proteasomal receptors and is able to directly interact with the proteasome, which suggests the existence of additional regulatory mechanisms.
Ubiquitination is also involved in the selective autophagy of organelles like mitochondria, peroxisomes, ribosomes and intracellular bacteria. In the case of pexophagy, mono-ubiquitination of the integral membrane protein PMP34 is sufficient to induce autophagic degradation of mammalian peroxisomes in a p62-dependent manner.38
In general, most experimental evidence suggests that ubiquitination, too, might function as a signal for selective autophagic degradation of protein aggregates, organelles and pathogenic cells through a common mechanism involving autophagosome formation.
Terminology and methodology appropriate for the bioinformatics analysis of autophagy in protists.
Comparative genomics provides a rapid way to exploit experimental data garnered for one species in order to elucidate the structure and operation of cellular pathways in other species. By compiling ‘parts lists’ for species for which experimental data may be sparse or hard to obtain, similarities and differences compared to model species can be visualized and their implications for physiology considered. Furthermore, mapping presences and absences of pathway components onto phylogenetic trees allows for a deeper understanding of the mechanisms by which the pathway evolved. Nevertheless, genome-mining exercises suffer from some fundamental and practical limitations and the risk of overinterpretation of data is ever present. A brief consideration of key terminology and common bioinformatic methodology illuminates some of the limitations of the area and explains some apparent inconsistencies in the literature.
Comparative genomics exercises cannot pinpoint genes or proteins with particular functions directly. Instead they seek statistically similar protein sequences to a query of known function: Where the similarity is strong enough to indicate homology (common evolutionary ancestry) then there is the possibility that the hit sequence has the same or similar function to the query. The improved statistics of the database search program BLAST39,40 provide a reliable guide to significance, although certain kinds of sequences, such as those exhibiting strong compositional bias, e.g., coiled-coil regions, can still cause problems for the more sensitive, iterated search protocols of PSI-BLAST,39 where search results can become ‘contaminated’ with unrelated sequences. The reliability of all searches is obviously dependent on the completeness of the protein sequence database and nucleotide databases should also be searched to allow for potential inaccuracies in gene calling. It is also prudent to employ query sequences from different species, where possible, since homologous queries may differ in their ability to retrieve divergent relatives. Even with all these efforts, however, it must be recognized that some divergent homologues may resist detection with presently available queries and sequence databases.
Assuming homologues have been reliably found, the harder question emerges as to whether any of them have the same or similar function to the query. Here it is helpful to define two classes of homologue, orthologue and paralogue. Orthologues are defined as homologues that have arisen from a speciation event, i.e., their lineage traces back to a single gene in the last common ancestor of their respective species. Immediately after speciation these independent copies have the same function and, if selection pressure applies thereafter, will tend to retain that function. This consideration leads to the alternative definition of orthologues, as homologues in different species carrying out the same function. Very frequently, the two definitions are equivalent although there are exceptions where one of a pair of orthologues can alter or even lose its function.41 Also, members of large families may have converged on the same function through more complicated evolutionary histories: These would be orthologues by the alternative definition but not by the original. Paralogues are those homologues deriving from gene duplication events within a single species. These are recognized as an important source of functional novelty during evolution and can be expected to differ, at least subtly, in their function(s).
The relationship between sequence identity and functional similarity of protein pairs is complicated and family-dependent.42 However, there are several ways in which detected homologues can be justifiably reclassified as probable candidate orthologues. An accepted computational shortcut to prediction of orthology is the reciprocal BLAST search:43 If two proteins, in different genomes, are each the top of the hit list of the other when queried by cross-genome BLAST, then they are likely to be orthologous and hence probably share the same function. Clearly, any experimental data regarding sequence characteristics key for activity should also be considered. These predictions are significantly enhanced by the availability of structural data but, as yet, rather few autophagy proteins have been fully structurally characterized.18 Many autophagy proteins contain multiple domains18 and these can cause problems, especially in the case of short domains and large inter-domain separations. Since BLAST will separate out individual domain matches in such cases, two proteins may pass the orthology prediction test yet differ in their domain composition. Nevertheless, domains may also be a useful filter in the search for likely orthologues. Some domains, such as those with catalytic activity, will be integral to a protein's molecular function, and less capable of substitution by alternative domains: Any putative orthologue should possess those core domains. Where candidate sequences require manual scrutiny, it is important to employ more sensitive domain annotation tools such as HHSEARCH,44 since domain databases such as Pfam45 and CDD46 may fail to annotate divergent domain sequences, particularly short ones.
Therefore, different criteria may be used to consider a homologous sequence as a strong putative orthologue. For example, it has been initially suggested that the second Atg12-based conjugation system was absent in trypanosomatids.18 However, later reports14,15,23 identify Atg5 and Atg10 orthologues, recently experimentally characterized.23 Interestingly, despite the identification in this last work of a Leishmania protein functionally resembling Atg12, and indeed now named ATG12, this is not likely to be an orthologue, strictly speaking, of the yeast protein. Instead, sequence comparisons suggest a closer, albeit still distant, resemblance to yeast Atg8.14
Finally, another challenge of genome mining is the possibility of gene displacement. Thus, the absence of a putative orthologue to a given protein in a certain organism need not necessarily mean that that function does not exist: an evolutionarily unrelated protein may have taken over the role. Autophagy provides a good example of this: The roles of the vacuolar hydrolases Pep4 (an aspartic peptidase) and Prb1 (a serine peptidase) in S. cerevisiae have been taken over by cysteine peptidases in trypanosomatids.17 However, this resembles more the situation in humans, although the aspartic peptidase cathepsin D also plays an important role there.47
Genomic Predictions Regarding Autophagy and its Evolution in Protists
Independent secondary losses of, and variations on, ‘canonical’ autophagy.
In current views of eukaryotic evolution most organisms are placed within one of six supergroups.48 This classification is based upon results from large-scale phylogenetic analyses and careful comparisons of cell morphology, although some of the relationships between super-groups are uncertain and the identity of the deepest branch(es) that lie(s) closest to the root of the eukaryotic tree remains a hotly-debated topic.49–52 The experimental characterization of autophagy in taxa widely accepted to have last shared a common ancestor at a very early stage in eukaryotic evolution—animals, yeast, trypanosomes and plants—strongly suggests that the basis for macroautophagy was already established prior to the divergence of the last common ancestor of known eukaryotes (or LECA). Using bioinformatics to map the distribution of known autophagy-related proteins in extant eukaryotes, however, indicates that among the protists canonical autophagy has been subjected to elaboration, moderation and even loss on multiple occasions (Fig. 2).14 As exemplified by the experimental characterization of autophagy in malarial parasites (discussed under the heading ‘Autophagy in Apicomplexa’), or the molecular characterization of ATG8 paralogues in Leishmania major23 the cellular consequences of likely moderation to canonical autophagy, or the paralogue expansion of core ATGs are already under investigation in some species. The availability of reverse genetic tools for many of the other protists in which core ATGs have either been lost or subject to paralogue expansion, means that other variations in autophagy that are apparent at the genomic level should be amenable to experimental investigation, too.
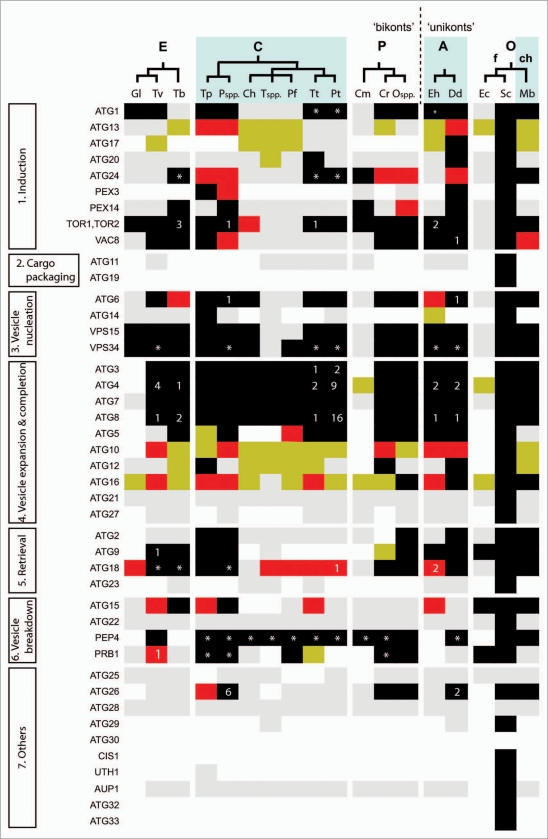
Figure 2.
Predicted distribution of ATG orthologues in protists. Modified from Rigden et al.14 BLAST and PSI-BLAST were used to identify sequences in protists that were either homologous or putatively orthologous to known autophagy components. Proteins are grouped according to the stage of autophagy at which they principally function (see Fig. 1). RPS-BLAST and HHSEARCH were used to check the domain composition of identified hits. Putative orthologues were those corresponding to the query by being reciprocal top hits in BLASTs between complete sets of predicted genome-encoded proteins and by bearing similar domain composition. By this definition there can clearly be only a single orthologue per genome. Additional putative paralogues were defined as further protist hits in the same BLAST searches, which also retrieved the query in top place and had appropriate domain composition. Black cells indicate the presence of a putative orthologue identified using a yeast Atg query; red, a putative orthologue identified using a non-yeast ATG query; light grey, homologous sequences detected using a yeast Atg query (but without indications of orthology from reciprocal BLAST results and domain composition; reviewed in ref. 14 for details); yellow, homologous sequences detected using a non-yeast ATG query; blank, no homologues detected. Numbers denote additional predicted paralogues in an individual species. The figure, for consistency, comes from a uniform and purely computational analysis, and does not necessarily reflect experimental information. However, experimental data suggest that some of the putative paralogues in this figure have apparently very similar functions to the confirmed orthologues' functions. For example, T. brucei, by this methodological definition, contains one predicted orthologue and three paralogues of S. cerevisiae Tor1/Tor2. Nevertheless, experimental data show that two T. brucei proteins have TOR activity116 and therefore, by some definitions based on functional analysis (see text), may both be considered as orthologues. Asterisks indicate the presence of homologous sequences that could not be reliably predicted as paralogous or not. Accepted relationships within eukaryotic ‘supergroups’ for the protists surveyed here are shown. There are competing hypotheses for how the divergence of these supergroups relates to the root of eukaryotic evolution. The last common ancestor of all eukaryotes has been suggested to have existed at the point of unikont-bikont divergence.199 Alternatively, although molecular and morphological evidence for the monophyly of Excavata exist (e.g., reviewed in ref. 60), these data are equivocal and several excavate groups continue to be proposed as extant descendants from the earliest diverging eukaryotic lineage.52,59 Abbreviations for eukaryotic ‘supergroups’: A, Amoebozoa; C, Chromalveolata; E, Excavata; O, Opisthokonta; P, Plantae. Abbreviations for other groupings: ch, choanoflagellates; f, fungi. The choanoflagellates represent the closest known unicellular relatives of animals. Species abbreviations: Ch, Cryptosporidium hominis; Cm, Cyanidioschyzon merolae; Cr, Chlamydomonas reinhardtii; Dd, Dictyostelium discoideum; Ec, Encephalitozoon cuniculi; Eh, Entamoeba histolytica; Gl, Giardia lamblia; Mb, Monosiga brevicolis; Ospp., Ostreococcus species; Pf, Plasmodium falciparum; Pspp., Phytophthora species; Pt, Paramecium tetraurelia; Sc, Saccharomyces cerevisiae; Tb, Trypanosoma brucei; Tp, Thalassiosira pseudonana; Tspp., Theileria species; Tt Tetrahymena thermophila; Tv, Trichomonas vaginalis.
Striking observations to emerge from a comparative genomic census of ATG genes are the loss of autophagy on at least three occasions during eukaryotic evolution and the moderation or divergence of autophagy in the Apicomplexa, such that, other than the ubiquitin-like conjugation systems, many readily recognizable ATGs are either absent or have diverged beyond obvious recognition. The absence of ATG8- or ATG12-related conjugation systems from the complete genome sequences of the microsporidian parasite Encephalitozoon cuniculi,53 the red alga Cyanidioschyzon merolae,54 and the human pathogen Giardia intestinalis,55 however, provides very strong evidence that the process of macroautophagy has been secondarily lost in a recent ancestor of these species. Significantly, the absence of macro-autophagy is not the only illustration of where organellar cell biology has been subject to streamlining in these eukaryotes. For instance, neither microsporidia nor Giardia contain peroxisomes or related microbodies; molecular and cytological analyses indicate that the endomembrane networks are perhaps less complex than in other organisms. Mitochondrial function has also degenerated in both parasites and the relic organelles that remain (known as mitosomes) have no capacity for ATP production.56 In the case of Giardia, the absence of autophagy also correlates with the absence of lysosomes. Instead, one contiguous tubular vesicular network appears to combine ER, endosome and lysosome functions.57 Yet, that is not to suggest that differentiation and intracellular remodelling do not occur. For instance, Giardia and microsporidians form cysts and spores, respectively. In Giardia the appearance of the secretory network is induced to facilitate the deposition of cyst wall components.58 It is not clear whether any turnover of macromolecular structures during the final stages of cyst formation or excystation is dependent upon a process resembling microautophagy.
In contrast to Giardia and the microsporidia, the extremophile red alga C. merolae contains a peroxisome, an aerobic mitochondrion and a photosynthetic chloroplast. Thus, secondary loss of macroautophagy does not always correlate with the extreme loss of metabolic functions that often accompanies the adaptation to parasitism, although niche adaptation to acidic, volcanic hot springs nonetheless likely facilitated the loss of ATGs in C. merolae. A wider comparison with the green algae reveals an extensive genomic footprint for canonical autophagy in the freshwater alga Chlamydomonas reinhardtii and in various oceanic phytoplankton (e.g., the diatom Thalassiosira pseudonana and the picoeukaryote Ostreococcus) (Fig. 2). This clearly indicates that in contrast to C. merolae, fluctuations in nutrient supply, competition for nutrients, other ecological pressures or developmental programs (e.g., sexual reproduction) necessitate the retention of autophagy pathways.
It is worthwhile to note that some might view the absence of macroautophagy from Giardia not as the product of secondary loss, but as a reflection of Giardia's last common ancestor with other eukaryotes possibly diverging prior to the appearance of the ATGs in eukaryotic evolution. Similar arguments have been put forward to explain the minimalism that pervades other aspects of Giardia's molecular cell biology.55 For many years there was a longstanding belief that Giardia was an amitochondriate descendent of a primitive eukaryote that diverged before the α-proteobacterial endosymbiosis that gave rise to the protomitochondrion occurred. This now seems not to be the case: relic mitochondria are present in Giardia,56 and although it is as yet difficult to refute entirely the suggestion that Giardia belongs to the earliest diverging lineage,59 much of the divergent cell biology that characterizes this important human parasite is more often considered to be a product of reductive evolution. In molecular phylogenetic analyses Giardia is often recovered in a clade containing another human parasite Trichomonas (e.g., reviewed in ref. 60) which, in contrast to Giardia, contains obvious ATG homologues. Published thin section electron micrographs of Trichomonas are also suggestive of autophagosome formation following serum deprivation, hydroxyurea treatment or berberine sulphate treatment.61,62 Like Giardia, Trichomonas has a streamlined metabolism:63 In Trichomonas, cellular energy generation is dependent upon carbohydrate metabolism and, to a lesser extent, the catabolism of some amino acids, but peroxisomes and many biosynthetic pathways are absent and its anaerobic mitochondria (known as hydrogenosomes) have lost the capacity for cytochrome-dependent respiration and oxidative phosphorylation. Thus, the example of Trichomonas vaginalis indicates that a streamlined metabolism is not necessarily a prelude to moderation or loss of macroautophagy.
In other protists the genomic survey points towards either moderation or divergence from the autophagy pathway that is broadly conserved in animals, plants, Dictyostelium (see below), and fungi. Experimental studies are required to distinguish between these possibilities. In the malarial parasite Plasmodium falciparum, and other apicomplexans, the differences with other eukaryotes are likely to be more extreme. Here, there are likely to be lineage-specific adaptations to the mechanisms by which autophagy is induced (e.g., a typical TOR-ATG1-ATG13 complex is not predicted) and the expansion of the phagophore membrane occurs (e.g., homologues of neither Atg9, which is an otherwise well-conserved integral membrane protein, nor Atg2, required for retrieval and recycling of Atg9 during phagophore expansion, are readily predicted by bioinformatics searches, although likely Atg18 orthologues were readily identified in P. falciparum and Theileria). Intriguingly, Atg9 orthologues are also not detected in ciliates, which are a sister group to the Apicomplexa.
Of course, an absence of readily detectable Atg orthologues in evolutionarily diverse protists is as likely, if not more likely, to reflect gene divergence, as it is to be due to gene displacement or gene loss. Yet, comparative genomics also throws up curious examples of ATG gene conservation. An unexpected example of conservation is the unambiguous example of an Atg17 orthologue in the amoeba Dictyostelium discoideum. Although proteins with low amino-acid identity to either yeast or the putative Dictyostelium ATG17 can be detected in a range of other protists, including Entamoeba histolytica and the choanoflagellates Monosiga brevicolis and Protereospongia, none is confidently predicted by the reciprocal BLAST to be a likely orthologue. The possible evolutionary significance of these observations was discussed previously.14 ATG17 remains one of the rare examples of an autophagy gene that potentially appeared early in evolution (before the divergence of the Amoebozoa and fungi), but which was subsequently lost or diverged beyond easy recognition during the evolution of lineages, including metazoans, where autophagy is an essential feature of development and normal cell physiology.
Paralogue expansions of some ATGs are evident in evolutionarily related and unrelated protists. For instance, there is a species-specific expansion of ATG8 paralogues into four distinct families totaling 25 genes in Leishmania, but not Trypanosoma species as was noted previously.23 Preliminary localization of some of these paralogues using GFP-tagging approaches suggests that some ATG8-family members have cellular functions in addition to or other than autophagy (reviewed in ref. 23; see also below, ‘Autophagy in Trypanosomatidae as inferred from bioinformatics studies of genome sequences–). From the wider comparative genomic survey of autophagy summarized in Figure 2, the most notable examples of paralogue expansion are ATG4 and ATG8, duplicated several times in Paramecium tetraurelia, but to a much lesser extent in another ciliate, Tetrahymena thermophila. Phylogenetic analyses suggest that the expansions in these two species occurred largely independently of each other (data not shown). The Paramecium ATG4 and ATG8 homologues are not identical. Paralogue expansion of other Paramecium gene families has been reported previously, and it appears that genome architecture and gene content in Paramecium have been shaped by multiple rounds of whole genome duplicaton.64 In instances where examples of unexpected paralogue expansion in Paramecium have been subject to experimental examination (e.g., actin-related proteins65 and vacuolar ATPase66), the different gene products have often been observed to have distinct cellular functions. This suggests that future study of Paramecium ATG4 and ATG8 paralogues will be of merit.
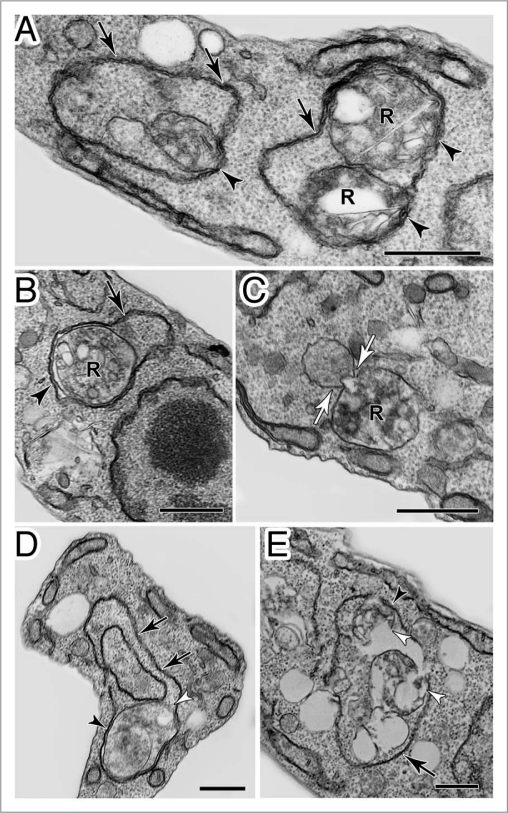
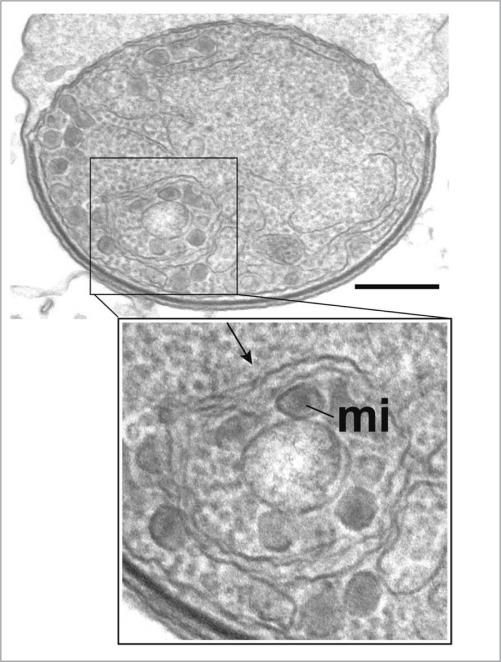
In protists, selective autophagy of organelles—such as glycosomes, the peroxisome equivalents of trypanosomes (see section below ‘Autophagy in Trypanosomatidae’; Trypanosoma brucei)—has been experimentally observed. Some proteins involved in these pathways in yeasts also participate in other autophagy-related processes or entirely different processes. These include Atg24, the peroxin Pex14 and the glycolytic enzyme phosphofructokinase, all involved in pexophagy in methylotrophic yeasts and Atg11, Atg17 and Atg29 involved in mitophagy in S. cerevisiae. Putative orthologues of these have been identified in protists but their role in organelle degradation in protists remains to be proven. Importantly, however, no genes have yet been detected in protists encoding proteins homologous to those found in yeasts that are specifically devoted to recognition/initiation of pexophagy (such as Atg25 and Atg30) or mitophagy (Atg32, Atg33). Thus, Atg25 and Atg30 are probably fungi-specific additions to the core autophagy pathways and their functions may be exerted by different proteins in other organisms.
Ubiquitination and autophagy in protists.
Since the connection between these two processes has only recently been clarified, the proteins involved have not been included in genomic surveys. We initially used the same methodology as previously14 to look for putative orthologues of p62 and NBR1 in a set of complete protist genome sequences. Using human sequences as queries, no putative orthologues were determined by the pipeline. However, p62 and NBR1 both contain relatively short domains separated by regions of intrinsic disorder, compositional bias and some coiled-coil regions (NBR1 alone). As mentioned above, these characteristics complicate automated genome mining. We therefore took a different approach and searched for proteins in the same protist genomes that contained the same PB1-ZZ-UBA domain combination shared by p62 and NBR1. The hits were also analyzed for the presence of coiled-coil and possible LIR motifs for interaction with small ubiquitin-like modifiers represented by [WY]-x-x-[LI] (as in Atg32).67,68
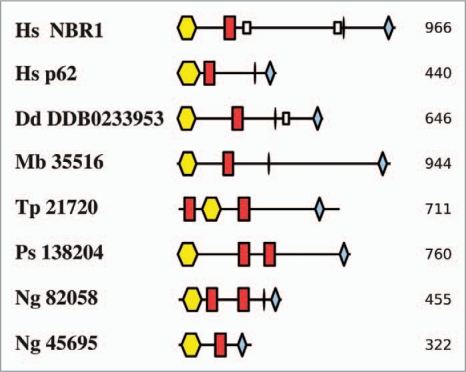
By this approach we identified proteins in five protists that each contain PB1, ZZ and UBA domains including two in Naegleria gruberi (Fig. 3). Clearly, the presence of these three domains in the protist sequences is no guarantee that they share a p62/NBR1-like function, and not all contain a consensus LIR between ZZ and UBA domains. However, recent data for the D. discoideum protein are at least consistent with its sharing a similar function to its human counterpart.69 Thus, Vmp1− D. discoideum cells accumulated large ubiquitinated protein aggregates, including the p62-like and ATG8 proteins, in a way that resembled the results of mutating Atg5 or Atg7 genes in mouse models of neurodegenerative diseases.70,71 Furthermore, this domain combination seems relatively rare: No other human proteins appear to contain all three characteristic domains, for example. It is also interesting to note that the domain order is retained (albeit with some additional domains) and, with the exception of the Thalassiosira pseudonana protein, the PB1 domain maintains its N-terminal location, while the UBA domain is invariably positioned at the very C terminus, as in p62 and NBR1. Interestingly, we found two p62/NBR1-like proteins in the amoeboflagellate N. gruberi, which belongs to the supergroup Excavata. Although Naegleria was not included in our larger scale census of ATGs, we were mindful of the identification of both Atg17 and p62/NBR1-like homologues in Dictyostelium and wished to ask whether homologues might have been retained in evolutionarily distant amoebae (in the case of Naegleria, not likely to have shared an ancestor with Dictyostelium since LECA). No clear candidate for an Atg17 orthologue in Naegleria emerged from this analysis, although the p62/NBR1-like proteins described here all represent interesting candidates for further study.
Figure 3.
Domain architectures of human NBR1 and p62 compared to protist proteins with similar domain compositions. See text for a discussion of these proteins. Each protein is labeled with its species of origin (Hs, Homo sapiens; Dd, Dictyostelium discoideum; Mb, Monosiga brevis; Tp, Thalassiosira pseudonana; Ps, Phytophthora spp.; Ng, Naegleria gruberi) and a genome identifier. Domains are PB1 (yellow hexagons), ZZ (red rectangles) and UBA (blue diamonds), small boxes represent coiled-coil regions and ellipses mark LIR motifs. Numbers to the right are sequence lengths, and the diagram is approximately to scale.
Functional Analyses of Autophagy in Free-Living Protists
Amoebae.
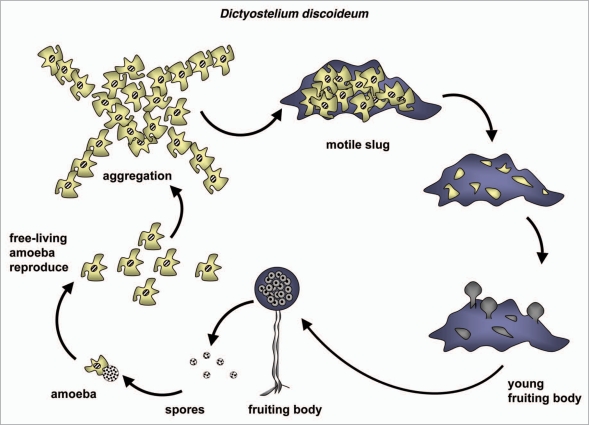
Amoebae are a polyphyletic group of protists that have radiated into many terrestrial and aquatic habitats, and also include some medical pathogens72—see below `Functional analyses of autophagy in parasitic protists'. Some species are known for their ability to undergo dramatic morphological changes in response to appropriate environmental cues. Perhaps the most extreme of these developmental programs are found among the social amoebas which aggregate together to form multicellular fruiting bodies when starved.73 (For a scheme of the Dictyostelium life cycle, see Fig. 4). This developmental process is best described in the soil-dwelling D. discoideum, which normally hunts bacteria and yeast within aerobic leaf litter. Significantly, D. discoideum also serves as an experimentally tractable model phagocyte for the study of bacterial pathogenesis and host-cell dependent killing mechanisms.74 Autophagy is important in both multicellular development and in the phagocytosis and digestion of bacterial prey. Intriguingly, the molecular analysis of autophagy in D. discoideum suggests that in this protist the core features of macro-autophagy retain some interactions that are otherwise thought to occur only in yeast (Atg17-dependency,14 see the previous section), to occur only in yeast and plants (Atg13-dependency) or to occur in mammals but not yeast (VMP1-dependency75,76). In addition, some Dictyostelium ATG proteins exhibit structural architectures that are more reminiscent of animal homologues than they are of yeast homologues (ATG1, ATG16).77,78 Finally, membrane whorls similar to the autophagosomes seen in mammalian cells are evident following cytological examination of ATG5− and ATG7− Dictyostelium mutants, but not wild-type amoebae, thereby providing further evidence that the cell biology of autophagy in social amoebae is more similar to mammals than it is to yeast.79
Figure 4.
Diagram of the life cycle of Dictyostelium discoideum. For a detailed discussion of the life cycles of this and each of the organisms in Figures 6–8 and 10, see the text.
Thus, genomic analyses have identified most of the core ATG components that have been experimentally characterized in yeast or animals, including candidate orthologues of ATG13 and ATG17 that were missed in the earliest studies of autophagy in Dictyostelium.14,78,79 In response to starvation, cAMP-dependent signaling leads to the aggregation of many thousands of adhering cells that transform into a motile slug, which undergoes morphogenesis to produce a 1 to 2 mm high fruiting body containing a spore mass at the tip of a thin stalk. Within the stalk, cells are highly vacuolated; under normal circumstances the induction of starvation-induced autophagy is followed by extensive vacuolization and DIF-1-dependent autophagic cell death.80 (DIF-1 is a chlorinated small molecule morphogen synthesized by Dictyostelium). Autophagic cell death is also dependent upon inositol 1,4,5-triphosphate signaling and Ca2+ release from the ER, and for vacuolization and the subsequent autophagy-dependent cell death to proceed normally a UDP-glucose derivative is required.81,82 Phenotypic analysis of the various ATG gene knock-outs that have been made in D. discoideum reveals that the absence of various ATG components either compromises or results in failure of fruiting body formation, but that failure of the developmental program occurs at different points depending upon the ATG gene that is inactivated.78,79,83,84 Following ATG gene inactivation, the morphological defect that occurs during development can vary depending upon the experimental protocol used to induce multicellular development. Thus, amoebae can be grown axenically and then starved on nitrocellulose filters, induced for development within plaques on bacterial lawns or grown on bacterial lawns and then transferred to nitrocellulose for starvation-induced development. Autophagy is also induced, and readily scored using microscopy, when amoebae are maintained in amino acid-deficient media. Vegetative growth of Dictyostelium appears to be largely unaffected by ATG gene inactivation, at least for the genes examined thus far;78,79,83,84 a result which mirrors the phenotypes seen in parasitic protists where autophagy is also important during cellular differentiation—see next main section.
Following induction of autophagy, transcription of the Dictyostelium ATG1 homologue (DdATG1) is upregulated (as are the ATG8 and ATG9 homologues discussed below, reviewed in ref. 79); the ATG1 protein subsequently colocalizes with the ‘classical’ autophagosome marker ATG8,78 and presumably interacts with a hypophosphorylated form of the Dictyostelium ATG13 homologue (DdATG13). ATG1 null mutants die rapidly in amino acid-free media, are unable to form mature autophagosomes, and fail to aggregate properly when induced for multicellular development on nitrocellulose filters.78 Use of a temperature-sensitive ATG1 mutant reveals that the requirement for ATG1 in autophagy is retained throughout development.78 Moreover, additional mutagenesis studies revealed that the kinase activity of DdATG1 and the C-terminal region of DdATG1 that contains a 122 amino-acid domain conserved in animal, but not yeast, homologues are both required for autophagy to occur in Dictyostelium.77 However, no homologue of the C. elegans protein UNC-14,85 which interacts with the C. elegans ATG1 orthologue UNC-51 through a C-terminal domain conserved in animal and amoebal ATG1, is evident in the D. discoideum genome.
DdATG5− and DdATG7-deficient amoebae are unable to undergo ATG12-ATG5 dependent conjugation and, when developed for multicellularity on nitrocellulose filters, aggregate to form slugs that form multiple, small, abnormal fruiting bodies with thick stalks and empty sori (Fig. 5).79 However, in contrast to the phenotypes seen in DdATG5− and DdATG7− amoebae, the requirements for ATG8 conjugation to PE and for PtdIns3K signaling during autophagosome expansion are less critical. Thus, while autophagosomes are formed and development proceeds to fruiting body formation in Dictyostelium mutants that lack either the DdATG8 or the DdATG6 isoforms characterized by Kessin and co-workers, these mutants form fruiting bodies with atypical morphologies (depending upon whether mutants were developed on nitrocellulose filters or bacterial plaques) and produce fewer spores than wild-type amoebae.83 Interestingly, there are paralogues of DdATG6 and DdATG8 encoded in the nuclear genome,14 perhaps pointing to the possibility that redundancy provides an explanation for the less severe developmental phenotypes seen in ATG6- and ATG8-deletion mutants compared to the other ATG gene deletions examined in Dictyostelium thus far. However, the lack of cytoplasmic turnover or recognizable autophagosomes in amino acid-starved DdATG6− or DdATG8− amoebae83 indicates that these paralogues cannot fully compensate for the absence of their experimentally characterized counterparts.
Figure 5.
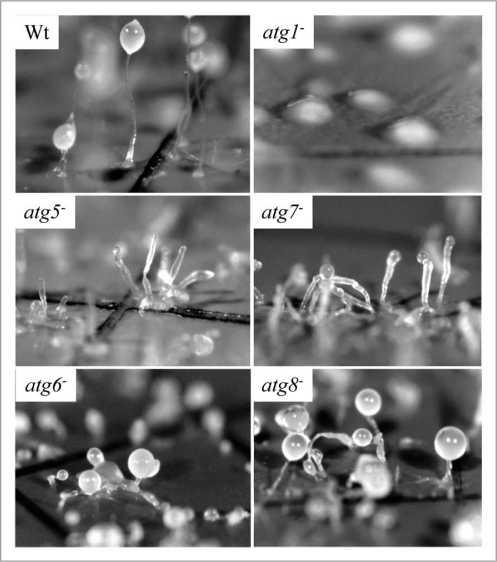
Dictyostelium autophagy mutants vary in the severity of their developmental phenotype. Amoebae were photographed following multicellular development on nitrocellulose filters. Wild-type (Wt) Dictyostelium elaborate fruiting bodies. Amoebae lacking ATG1 form loose mounds that rarely develop further. Amoebae lacking either ATG5 or ATG7 form multi-tipped aggregates, which form abnormal, small fruiting bodies at the end of thickened stalks; viable, detergentresistant spores are not produced. Mutants lacking either ATG6 or ATG8 form multi-tipped aggregates with small, but otherwise normal fruiting bodies; some viable, detergent-resistant spores are formed by these mutants. This research was originally published in the Journal of Biological Chemistry: parts Wt, atg5− and atg7− as Figure 3A, C and E, respectively, in Otto et al. Macroautophagy is required for multicellular development of the social amoeba Dictyostelium discoideum. J Biol Chem 2003; 278:17636–45 and parts atg1−, atg6− and atg8− as Figure 3B, G and H, respectively, in Otto, et al. Dictyostelium macroautophagy mutants vary in the severity of their developmental defects. J Biol Chem 2004; 279:15621–9. ©The American Society for Biochemistry and Molecular Biology.
The most recent ATG gene to be experimentally characterized in Dictyostelium is DdATG9.84 The protein encoded by this gene contains the conserved ‘ATG9’ domain, six putative transmembrane helices, and when fused to green fluorescent protein (GFP) localizes in vesicles that plausibly track along microtubules to the site of autophagosome formation, as in mammalian cells. As with other eukaryotes, ATG9 is a candidate molecule involved in the delivery of lipids to the expanding autophagosome and, akin to other Dictyostelium ATG gene mutants, amoebae lacking DdATG9 are attenuated in development. The ability of DdATG9− mutants to form slugs and occasionally form abnormal-looking fruiting bodies when developed on nitrocellulose filters suggests a phenotype more severe than DdATG6 or DdATG8 deficiency, but less pronounced than loss of DdATG1, DdATG5 or DdATG7.84 Retrieval of Atg9 from the phagophore membrane in yeast requires Atg1 activity.86 Assuming this dependency is evolutionarily conserved, the striking differences in phenotype severity between DdATG1- and DdATG9-deficient amoebae suggest DdATG1 is likely to most critically function at an early, as opposed to late (ATG9-interacting) stage in the autophagy process. Another intriguing phenotype arising from DdATG9 deletion is the impairment of phagocytosis, providing unexpected evidence of a link between the function of an ATG gene product and microbial engulfment.84 The role of autophagy in microbial digestion by Dictyostelium has been studied using bacteria that are either generally readily cleared by the amoebae (Salmonella typhimurium87) or which are able to replicate intracellularly (Legionella pneumophila84). The data from these experiments echo the results from studies of autophagy in mammalian cells and underscore the importance to some intracellular microbes of manipulating the autophagy process in order to replicate.
In line with work on mammalian cells, recent work has also revealed that the orthologue of vacuole membrane protein 1 or VMP1, is required for autophagy in Dictyostelium.69,75,76 As in mammalian cells, this protein, which is absent from yeast, is multifunctional and found at different intracellular locations making it difficult to assess whether the effect of VMP1 deletion on autophagy is direct or a pleiotropic consequence of losing a protein involved in numerous cellular processes. There is, however, some evidence that VMP1 in Dictyostelium is present in autophagosomes.69 Following the resuspension of amoebae in minimal salts solution, the detection of p62-related protein in VMP1−, DdATG1−, DdATG5− and DdATG7− mutants within ubiquitin-containing protein aggregates that colocalize with GFP-ATG8 indicates conservation of ubiquitination-dependent delivery of cytosolic protein aggregates to autophagosomes, as observed in mammalian cells.69 The limited data regarding the function and mechanisms of autophagy in other amoebae are cited in the section on ‘Experimental analyses of autophagy in parasites.’
Other heterotrophs and phototrophs.
Although reverse genetic approaches necessary to facilitate molecular studies of autophagy are available for a diverse assortment of free-living heterotrophic and phototrophic protists, it is only Dictyostelium that has been subject to extensive molecular analysis. However, in addition to genomic signatures that suggest intriguing variations on canonical macroautophagy (e.g., the puzzling expansion of ubiquitin-like protein conjugation components in Paramecium tetraurelia—see above ‘Genomic predictions regarding autophagy and its evolution in protists’), there are various ultrastructural descriptions consistent with roles for autophagy or similar processes in a variety of species. Here, cell cytology not only points towards the importance of autophagy in development (e.g., in ciliates where autophagy is implicated in degradation of the macronucleus during conjugation88), differentiation (e.g., during encystment of Tetrahymena;89 long-term ‘gametic’ survival of plate-grown Chlamydomonas reinhardtii, a ubiquitous green alga90,91), and programmed cell death (PCD) (reviewed in ref. 92 and 93), but also towards the upregulation of basal levels of autophagy in response to the spatially and temporally heterogeneous environments in which many protists are found (reviewed in ref. 94). Given the paucity of relevant molecular data we consider experimental studies relating to autophagy in free-living protists other than Dictyostelium and related amoebae in one combined section.
An involvement of autophagic-like processes in possible turnover or remodeling of metabolic organelles (mitochondria, chloroplasts) was revealed by studies with C. reinhardtii mutants deficient for chlorophyll production (due to the absence of phytoene synthase),95 and Tetrahymena thermophila mutants in which mitochondrial function and fission were compromised by either overexpression or deletion of septin.96 In the Tetrahymena mutants, far higher numbers of mitochondria were seen in association with autophagosome-like vesicles than in wild-type or starved ciliates. Following treatment with trans-resveratrol, inclusion of mitochondria within autophagosome-like structures was also seen in another ciliate species, Philasterides dicentrarchi.97
In the Chlamydomonas phytoene synthase mutant,95 an increased frequency of cytosolic autophagosome-like vesicles correlated with aberrant plastid structure where thylakoid stacks and starch-containing pyrenoid bodies were missing. This increase in the number of autophagic vesicles could have been due to either the pleiotrophic consequence of a defect in chloroplast function and the general poor health of a mutant that could only grow heterotrophically in the dark,95 or because of autophagy during attempted remodeling of the abnormally functioning chloroplast present in these cells. The dramatic remodeling of organellar architecture that follows N-limitation and the entry of Chlamydomonas cultures into stationary phase (reviewed in refs. 90 and 98) provides a more likely example of where autophagy is important in C. reinhardtii. However, it is also relevant to remember that we speculated previously on the possible importance of intra-organellar protein turnover rather than autophagy for remodeling of the single mitochondrion and/or plastids found in many protists (the mitochondrion and chloroplast are single copy organelles in C. reinhardtii).14 The conservation of many ATG homologues in algal and ciliate genomes14,99 suggests that organelle and protein turnover is more likely to occur by ‘classicala’ macroautophagy, than it is by the substantially moderated process implicated in the turnover of organelles in malarial parasites and perhaps other apicomplexan parasites, too (see below, ‘Autophagy in Apicomplexa’).
In the green alga Micrasterias denticulata, stress-induced autophagy can be accompanied by a PCD-like process in which the presence of autophagosomes is readily observed by transmission electron microscopy.93 On the one hand, for some free-living unicellular opportunists the benefits conferred by altruistic cell death of an individual are not necessarily obvious. Yet, in other instances, such as in the seasonal cycling of phytoplankton blooms, the spread of viral infection through an algal population or following excessive ROS-induced damage,100 the benefits of (autophagy-dependent) PCD on a large-scale appear plausible (and even provide a rational explanation for ill-understood examples of phytoplankton cultures that crash in the laboratory).101 The possible ecological significance of PCD, and its link with autophagy, therefore provides interesting parallels with parasitic protists where occurrence of PCD in vector-borne parasites has been proposed as an adaptation that aids life-cycle progression (reviewed in ref. 102). A need for altruistic PCD, coupled with an accommodation of seasonal variation in nutrient availability, provides obvious selective pressures for the retention of ‘classical’ ATG genes in the genomes of ubiquitous pico-phytoplankton, such as the Ostreococcus species. In contrast, autophagy-dependent remodeling of cellular metabolism has either been lost or severely moderated in the extremophile Cyanidioschyzon merolae (Fig. 2), which is found only within acidic warm springs. We speculated previously that limited variation in the supply of or competition for, core nutrients may have, at least in part, facilitated loss of ATG homologues and a capacity for macroautophagy.14 More obvious, clear-cut examples of autophagic cell death in algae are perhaps likely to be found during the development of species such as Volvox carteri, a multicellular green alga and a close evolutionary relative of C. reinhardtii.103
Molecular characterization of the Chlamydomonas ATG8 homologue has recently been reported.104 Here, stress- and rapamycin-induced processing and relocalization of CrATG8 were described, consistent with morphological and genomic studies suggesting the occurrence of a typical autophagy pathway. Interestingly, the rapamycin-sensitive TOR-LST8 complex implicated in the regulation of autophagy initiation in Chlamydomonas104,105 localizes to the peri-basal body region of the alga.106 The overlapping localization of TOR and LST8 homologues in C. reinhardtii within the vicinity of the flagellar basal bodies not only strengthens existing data linking flagellum function and intracellular cell signalling in this alga (reviewed in ref. 107), but also draws to mind the increase in mTOR activity in renal epithelial cells from human patients with polycystic kidney disease or mouse models of polycystic kidney disease where intracellular signalling is perturbed due to defects or loss in function of immotile primary (or sensory) cilia.108,109
Functional Analyses of Autophagy in Parasitic Protists
Parasitic protists such as the organisms responsible for African sleeping sickness (Trypanosoma brucei), Chagas' disease (Trypanosoma cruzi), leishmaniasis (Leishmania), malaria (Plasmodium), toxoplasmosis (Toxoplasma), East Coast Fever and Tropical Theileriosis (Theileria) and various kinds of diarrheal diseases (e.g., Entamoeba), have received considerable interest from the scientific community, notably biochemists and molecular and cell biologists, because of the serious diseases they cause, often to people and domestic animals in developing countries. Since safe and efficient drugs are still not available or are undermined by drug resistance, there is considerable pressure to identify new drug targets and develop lead compounds for therapeutic intervention. Autophagy could represent a valuable drug target, as it may lead to autophagy-induced cell death. Beside this, protists may serve as model organisms, because in many respects they represent early exponents of evolution and may be easier to analyze than higher eukaryotes. Roles for autophagy during exponential growth and encystment in Entamoeba histolytica and the encystment of another amoebal pathogen, the opportunist Acanthamoeba castellanii, have been reported previously.110,111 However, most of the experimental analysis regarding autophagy and parasitic protists has been performed with trypanosomatid and apicomplexan parasites. This work is discussed below.
Autophagy in Trypanosomatidae.
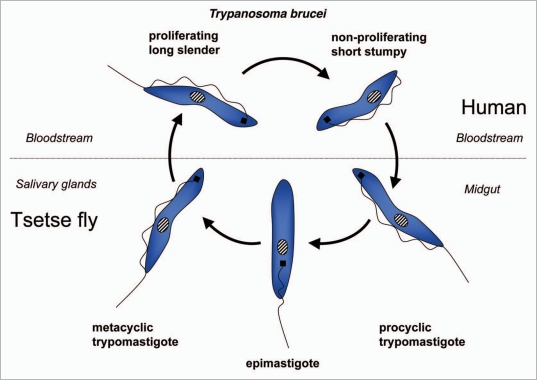
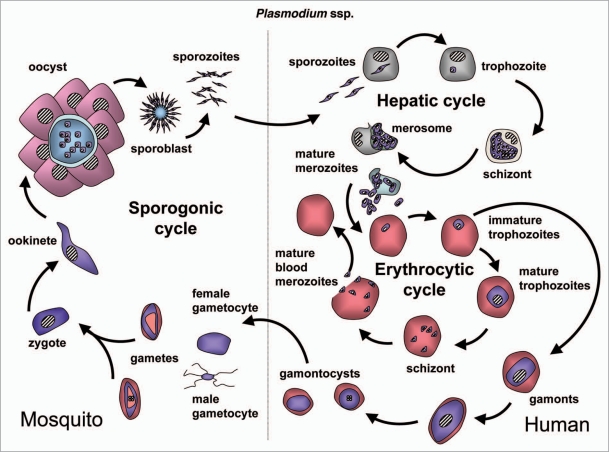
Trypanosomatids: life cycle, morphology and metamorphosis. Trypanosomatidae belong to the order Kinetoplastida that comprises flagellated protists characterized by the presence of a DNA-containing compartment, the kinetoplast, at the base of the flagellum. This compartment is actually part of the single mitochondrion present in these organisms, and the kinetoplast DNA is a complex of standard mitochondrial DNA with other DNA, having a unique structure and function (reviewed in ref. 112). All known species of the Trypanosomatidae are pathogenic for human, animals or crops. Best studied are the human pathogens T. brucei, responsible for sleeping sickness in sub-Saharan Africa, T. cruzi, the causative agent of Chagas' disease in Latin America, and various Leishmania species causing a variety of disorders in tropical and subtropical areas worldwide. These are all so-called neglected diseases (because of the little attention they receive from pharmaceutical companies due to the limited market potential in the countries of affected people) that often are fatal without adequate treatment, and for which efficacious, nontoxic drugs are not generally available. Each of these parasites has a complicated life cycle (Figs. 6–8). They are transmitted between human (or other mammals) by insects and undergo in each of their hosts sequential developmental changes resulting in alterations in their morphology which may be considerable, i.e., major changes in cell size and form; repression or derepression of the mitochondrion; expression of a flagellum providing high motility or immotile forms without flagellum; position of organelles, etc.
Figure 6.
Diagram of the life cycle of Trypanosoma brucei.
Figure 8.
Diagram of the life cycle of Leishmania spp.
T. brucei is an extracellular parasite that is introduced as a metacyclic form in the mammalian bloodstream, from the tsetse fly's saliva when the insect takes a blood meal (Fig. 6). Once in the blood, parasites proliferate as a form that has a slender morphology. During the course of infection, the parasites differentiate into nonproliferating stumpy trypanosomes, which are pre-adapted to life in the tsetse fly's midgut once taken up during a blood meal. Stumpy trypanosomes differentiate into procyclic forms before they migrate to the salivary glands, while undergoing several additional developmental changes to form new metacyclic forms that are infective to mammals.
T. cruzi infections are caused by reduviid bugs when they defecate during feeding. Infective metacyclic trypomastigotes from the feces enter the host through skin lesions and mucous tissues (Fig. 7). The trypomastigote stages do not divide but spread through the blood infecting any tissue, but especially muscle tissue of the heart and alimentary tract, where they develop into proliferating amastigotes that are responsible for the pathogenic symptoms and, after several rounds of multiplication, develop into new trypomastigotes that are released in the blood and will invade new cells. The trypomastigotes can also be taken up again by an insect and develop in its gut into epimastigote forms. In the rectum, where the insect's urine is discharged, the epimastigotes differentiate into human infective metacyclic trypomastigotes, induced by the locally very low nutritional content.
Figure 7.
Diagram of the life cycle of Trypanosoma cruzi.
Leishmania species are transmitted by sandflies, which infect the host with metacyclic promastigotes from the gut, by regurgitation during feeding (Fig. 8). In the mammalian host, these infected forms enter macrophages where they multiply as amastigotes in the phagolysosomes.
During the developmental cycles these parasites encounter highly different environments with different nutrients, different levels of oxygen supply and different temperatures. For example, T. brucei when living in blood encounters a rather constant, abundant supply of glucose. It generates ATP only by glycolysis because the mitochondrion is nearly completely repressed. In contrast, glucose is rarely present in the tsetse midgut. Therefore, amino acids, notably proline, become the major energy source, and they are oxidized in the then well-developed mitochondrion. Similarly, nutrients available to intracellular amastigotes of T. cruzi and Leishmania spp., present in the cytosol and phagolysosomes, respectively, are different from those in the blood and insect gut. Consequently, the metabolic profiles may differ in the different life-cycle stages of the various parasites, and it is expected that, at least where important metabolic reprogramming occurs, autophagy plays a major role in achieving this during transitions from one stage to the next.
Autophagy in Trypanosomatidae as inferred from bioinformatics studies of genome sequences. Genome database searches supplemented with more advanced analyses have revealed that only half of the autophagy-associated components characterized in S. cerevisiae are present in trypanosomatids.14,15,17,18,20 However, comparisons with other organisms show less dramatic differences since data suggest that a number of autophagy-related proteins e.g., Atg19, Atg21, Atg22, Atg23, Atg25, Atg28, Atg29, Atg30 and Cis1/Atg31 arose as innovations in the fungal lineage: There is no good evidence that they have orthologues outside fungi.6,14 Nevertheless, it remains likely that further lineage-specific innovations, as well as expansions outlined elsewhere, remain to be discovered. Until they are unraveled, it seems impossible to assess the true degree of similarity between the trypanosomatid autophagy machinery and the equipment of other species.
ATG8 has undergone an unusual evolution within the genus Leishmania.23 Bioinformatics analysis reveals that the L. major genome has 25 ORFs encoding ATG8-like proteins. They could be classified into four families, designated ATG8, ATG8A, ATG8B and ATG8C. The four families are also present in other Leishmania species, but with different copy numbers. The ATG8 protein is in all cases encoded by a single-copy gene. The members of the other three families are encoded by gene arrays. Experimental studies by Williams et al.23 discussed below, indicated different functions for the different families, although many questions remain to be answered about these functions and the reason for expansion of the ATG8 repertoire in this trypanosomatid genus. In contrast to the situation in Leishmania, the number of ATG8 genes found in T. brucei and T. cruzi is much more limited. T. brucei contains three isoforms of ATG8, designated ATG8.1, ATG8.2 and ATG8.3, while two isoforms, ATG8.1 and ATG8.2, have been detected in T. cruzi (discussed further below). The finding of multiple ATG8 homologues may reflect either functional redundancy and/or the ability of the proteins to carry out additional function(s).
Results of experimental studies. Trypanosoma brucei. The morphology of the different life-cycle stages of African trypanosomes has been studied in great detail by electron microscopy in the 1970s by Vickerman and colleagues.113,114 These studies provide the first indications that autophagy plays an important role during the transition from one differentiation stage to another. However, experimental evidence of autophagy in these organisms was only obtained in the last few years, after ATG homologues had been discovered and their function studied using specific antisera, inhibitors and reverse genetics. Rapamycin, the classical inhibitor of TOR, induces growth inhibition leading to cell death of bloodstream-form T. brucei with the formation of vacuoles stained with monodansylcadaverine that were proposed at the time to be autophagosomes.22 However, this effect was only seen at a high concentration (EC50 6.5 µM) of rapamycin, at least 10-fold higher than that required to induce autophagy in yeast. No apoptotis or necrosis was observed. In addition, electron microscopy suggested the formation of phagophores, autophagosomes and autolysosomes. Rapamycin also caused formation of densely packed membrane clusters associated with the kinetoplast near the flagellar pocket, which was also observed upon starvation, but not under several other stress conditions. These clusters were not seen in differentiating cells.
TOR is a kinase that regulates the balance between protein synthesis and degradation via autophagy. There are two different TOR-containing complexes with distinct functions: TOR complex 1 (TORC1) controls processes such as ribosome biogenesis, transcription, translation and induction of autophagy, while TORC2 controls spatial aspects of cell growth by actin cytoskeleton remodeling (review in ref. 115). Metazoans coordinate cell growth via a single TOR kinase (mTOR), while some single-cell organisms such as yeast have two TOR proteins, Tor1 and Tor2. Nonetheless, the different TOR complexes and their distinct functions are conserved in all eukaryotes studied. Like some other unicellular eukaryotes, trypanosomes contain two TOR kinases, TOR1 and TOR2; however, two additional TOR-related proteins (TOR-like 1 and TOR-like 2) have also been described.116 TOR1 and TOR2 participate in the complexes TORC1 and TORC2, respectively, which differ in function, subcellular localization and rapamycin sensitivity in trypanosomes.116 As in yeast, TORC1 and TORC2 are associated with the orthologues KOG1/raptor and AVO3/rictor. TOR1 is predominantly nuclear, whereas TOR2 is excluded from the nucleus and is associated with the ER and the mitochondrion. TORC1 is involved in regulating cell polarization and cytokinesis. In particular, rapamycin, with an EC50 of 152 nM severely reduces proliferation by inhibiting TORC2 formation. However, contrary to the situation in other eukaryotes, rapamycin, even at concentrations up to 1 µM, appears to have no effect on TORC1, since no cell cycle arrest in G1 or protein synthesis inhibition are detected. Moreover, a rapamycin-FKBP12 complex does not bind to the recombinant TOR1 rapamycin-binding domain, while it does to TOR2 in vitro and in vivo. Although trypanosome TORC1 is insensitive to rapamycin, it was shown by RNA interference that TOR1 knockdown triggers the appearance of autophagiclike vesicles. Its depletion also causes morphological changes such as abnormal appearance of the ER and formation of membranous whorls similar to those that occur in other eukaryotes upon TOR1 inhibition.117
Recently, punctate structures that were proposed to be autophagosomes, detected by expression of a fusion construct of yellow-fluorescent protein and ATG8.2, have been described for procyclic T. brucei cells exposed to ER stress.118 Furthermore, both ATG8.1 and ATG8.2 have been validated as autophagosome markers in both bloodstream and procyclic form T. brucei (Proto W and Mottram JC, unpublished).
Since TORC1 is entirely insensitive to rapamycin, it remains to be determined how the morphological effects observed after treatment with high concentrations of this compound are caused and if the observed structures are indicative of death by autophagy.
Another molecule that induces autophagy in African trypanosomes is dihydroxyacetone (DHA).19,119 Many cells can use DHA as an energy substrate because, after its phosphorylation by DHA kinase, it may enter the glycolytic pathway. Trypanosomes, however, which take up DHA via aquaglyceroporins, lack this kinase. As a consequence, DHA causes growth inhibition (at an EC50 of 1 mM) as a result of cell cycle arrest in the G2/M phase and morphological alterations (multivesicular bodies, vacuoles, characteristic changes of mitochondrial structure) typical of autophagy.
Also, some neuropeptides kill the mammal-infective form of T. brucei. Surprisingly parasite death does not occur by the classical antimicrobial peptide mechanisms, i.e., by forming pores or inducing membrane damage. Instead the neuropeptides are endocytosed and appear to induce autophagic cell death of the trypanosomes.120,121 The authors show that endocytosed peptides reach the lysosome whose integrity is subsequently disrupted. In addition, the peptides can be released from the late endosomes or lysosome into the cytosol, where they accumulate in various intracellular compartments. Interaction of peptides with glycosomes causes partial relocalization of the glycosomal enzymes, with the consequence that the cellular ATP is depleted. The failure of their energy metabolism results in cell death with involvement of autophagy, as suggested by ultrastructural morphological analysis, whereas no signs of necrosis or apoptosis are observed. These studies, which show the progressive formation and fusion of multiple lysosome-like vesicles and autophagic-like vacuoles, are corroborated by the results of immunostaining using mammalian anti-LC3 (ATG8) antibodies that are suggestive of the appearance of autophagosomes.
As mentioned above, T. brucei possesses three isoforms of ATG8 called ATG8.1, ATG8.2 and ATG8.3. ATG8.3 contains a unique insertion that is positioned near the interface in such a way that the binding properties of this protein might be different from those of other ATG8-like proteins. Moreover, some analyses suggest it is syntenic with an ATG8-like protein identified in L. major that has functionally been shown to act as an ATG12. ATG8.1 ends with a Gly residue and is therefore expected to be active without a need for processing by ATG4. The crystal structure of ATG8.2, with a C-terminal extension that should be processed, has been determined. It has the typical ubiquitin fold and its overall structure is very similar to that of its human homologue ATG8/LC3.122 T. brucei contains two ATG4 homologues, one of which (the closer relative of human ATG4) was modeled using available ATG4 crystal structures from other organisms. The model was docked onto the TbATG8 structure, and the complex compared with the known structure of the mammalian LC3-ATG4 complex. Despite the low sequence identity (30% between human and trypanosome ATG4) the overall structure is very similar and the catalytic C-D-H triad is also present in TbATG4, strongly suggesting conservation of function.122
Among the various unique features of Kinetoplastida is the possession of glycosomes.123 These organelles, which may be present in 10–100 copies per cell, dependent on the species and the respective developmental stage, belong to the peroxisome family but, unlike typical peroxisomes, contain the majority of the glycolytic enzymes, hence their name. In some of the Trypanosomatidae, such as the bloodstream form of T. brucei, glycolytic enzymes may contribute over 90% of the protein content of the organelles. Enzymes of some other important catabolic and anabolic pathways may also be present in glycosomes, such as those involved in gluconeogenesis, the pentose-phosphate pathway, β-oxidation of fatty acids, purine salvage and the biosynthesis of pyrimidines, ether lipids and squalenes, as well as typical peroxisomal enzymes responsible for peroxide metabolism. Dependent on the species, the importance of these different processes may vary with the environmental conditions encountered during the life cycle of the parasites. Indeed, glycosomes of African trypanosomes exhibit considerably different enzyme content among successive life-cycle stages. Consequently, an enhanced turnover of glycosomes during differentiation from one stage to another is expected. Pexophagy is invoked to be responsible for this turnover.18,21 Indeed, immunofluorescence microscopy studies reveal a small tendency of glycosomes to associate with the lysosome during differentiation from the slender to the stumpy bloodstream form: Colocalization is observed in 40% of the trypanosomes, whereas less than 10% of nondifferentiating cells display such colocalization. A fast and drastic increase in the association of the organelles is observed in all trypanosomes during their transition from stumpy to procyclic forms. Moreover, the lysosome shows a transient but dramatic increase in size during this latter differentiation process. Immunoelectron microscopy confirms the autophagic degradation of glycosomes and the appearance of glycosomal enzymes within lysosomes during the differentiation steps.21 Interestingly, the observed surrounding of glycosomes by the lysosome is suggestive of a process similar to micropexophagy, whereas the size increase of the lysosome may rather suggest macropexophagy involving the formation of autophagosomes. Indeed, bioinformatics analysis reveals putative orthologues of most of the Atg proteins specifically involved in micropexophagy in S. cerevisiae.18 In contrast, in Leishmania species, only macroautophagy has been experimentally observed so far (see below).16,17 Whether African trypanosomes and Leishmania spp. use different degradation pathways or if both pathways can function in all trypanosomatids but the induction of either depends on the conditions encountered, as observed for H. polymorpha and P. pastoris,124 remains to be determined.
Trypanosoma cruzi. Several papers have reported ultrastructural changes indicative of autophagy after treatment of T. cruzi parasites with toxic compounds or their exposure to adverse conditions. Protein kinase and PtdIns3K inhibitors like wortmannin, staurosporin and genistein led to growth inhibition of epimastigotes and caused various changes including the formation of autophagosome- and myelin-like structures. No effect was found on division of intracellular amastigotes or their differentiation to trypomastigotes.125 In epimastigotes, the lysophospholipid analogue edelfosine, alone or combined with the steroid inhibitor ketoconazole, induces morphological alterations, with changes in plasma membrane and lysosomes (also called reservosomes in this parasite), and promotes the appearance of autophagic structures and multinucleation.126 Other trypanocidal agents such as the dinitroanilines trifluralin and chloralin lead to the appearance of vacuoles containing damaged membranes.127 Also some natural products such as geranylgeraniol, propolis and naphthoquinones lead, in epimastigote and trypomastigote forms, to the appearance of concentric membrane structures in the cytosol, which are apparently associated with an autophagic process.127,128
Ultrastructural studies in epimastigotes and trypomastigotes treated with either one of three naphthoimidazoles N1, N2 and N3 derived from β-lapachone demonstrate that the mitochondrion, Golgi and reservosomes are the main targets.128,129 Frequently, treatment with these three compounds induces morphological autophagic characteristics in the parasites such as the occurrence of concentric membrane structures in the cytosol and an increase in the number of autophagosome-like bodies, observations reinforced by the strong increase of monodansylcadaverine labeling.130 The effect of the naphthoimidazoles on both forms of T. cruzi is blocked by E64 and calpain I inhibitor,129 suggesting the participation of calpain in the autophagic process as in mammalian cells.131 Pre-incubation with the autophagic inhibitors wortmannin and 3-methyladenine (3-MA) completely abolish the effects of N1, N2 and N3, pointing to autophagic cell death as a consequence of this treatment.130 In treated epimastigote forms, extensive ER profiles surround structures, especially reservosomes, proving a close contact between the membranes of both organelles (Fig. 9 and Table 1). This morphological finding supports the hypothesis that the ER provides membranes for PAS formation in T. cruzi, as recently shown for mammalian cells.33,34 The participation of reservosomes in T. cruzi autophagy deserves further investigation, as this organelle, a pre-lysosomal compartment present only in epimastigotes,132 may participate in autolysosome formation as lysosomes do in mammalian cells.
Figure 9.
Transmission electron microscopy analysis of Trypanosoma cruzi epimastigotes treated with naphthoimidazole. (A, B, D and E) The parasites showed well-developed endoplasmic reticulum profiles (black arrows) surrounding organelles, especially reservosomes (R), suggesting a close contact between both membranes (black arrowheads), and commonly loss of the integrity of these organelle structures (white arrowheads). (C) A cytoplasmic vesicle near to a reservosome was also observed, suggesting fusion with the organelle (white arrows). Scale bars = 0.5 µm.
Table 1.
Autophagic events in T. cruzi treated with naphthoimidazoles
| Epimastigotes | Trypomastigotes | |
| RE profiles close to organelles | + | + |
| Concentric membrane structures | + | + |
| ATG RNA detection | + | nd |
| Increase in number of autophagic vacuoles | + | + |
| Autophagy inhibition | + | + |
nd, not determined.
Subsequent studies unambiguously confirmed the occurrence of autophagy in T. cruzi. As mentioned above, homologues of about 20 yeast Atg proteins were identified. One of the steps where the bioinformatics analysis18 was confirmed by experimental data is vesicle expansion and completion.20 Although sequence length varies between the T. cruzi homologues and the corresponding yeast Atg protein, the protein domain composition remains conserved, supporting their proposed function. As has also been seen in some other trypanosomatids, T. cruzi may contain more than one protein where only a single one is found in yeast. Cases in point are ATG4-like and the ATG8-like proteins.20 T. cruzi contains two ATG4 and two ATG8 homologues, in contrast to the single-copy genes in yeasts. Like in T. brucei, one of these ATG8 homologues is possibly syntenic with L. major ATG12, but biochemical support for an ATG12 activity remains to be provided. Each of the ATG4 gene copies (TcATG4.1 and TcATG4.2) could, with similar efficiency, complement a yeast deletion strain and restore the Cvt and autophagy pathways, whereas only one of the ATG8 genes (TcATG8.1) is able to functionally reconstitute autophagy in the yeast deletion mutant, and the corresponding protein concentrates in large vesicles in epimastigotes after the cells are subjected to starvation.20 Mutation of the conserved Gly residue abolishes the localization of ATG8 into the vesicles, indicative of the need of ATG8-PE conjugation for association with the membrane. Only TcATG4.1 is able to efficiently process both ATG8.1 and ATG8.2 in vitro, suggesting that this protein is involved in autophagy, whereas the role of ATG4.2 remains to be established. Together, these results suggest that the ATG8 conjugation system in T. cruzi functions very similarly to that in yeasts and mammalian cells.
RT-PCR data indicate the participation of ATG3, ATG4, ATG7 and ATG8 in the autophagic process in epimastigotes treated with naphthoimidazoles,130 although the detailed pathway and proteins involved in the mechanisms of cell death require further investigation.
A major differentiation step in the life cycle of T. cruzi is the transition of epimastigotes into metacyclics triggered by nutritional stress within the rectum of the insect vector. This can be reconstituted in vitro, by starvation of cultured epimastigotes. Initially, the epimastigotes have access to sufficient nutrients due to the reservosome, an organelle unique to this stage. It is part of the lysosomal system and harbors endocytosed proteins and lipids, as well as secretory proteins synthesized by the parasite.133 Upon starvation, the proteins are degraded by cruzipain,20,134 the most abundant cysteine peptidase of the lysosome and reservosome and whose activity is critical for differentiation. The organelle then shrinks and disappears. Starved epimastigotes as well as cultured epimastigotes undergoing spontaneous differentiation to metacyclic trypomastigotes display structures related to autophagosomes that are highlighted by TcATG8.1 staining.20 The staining decreases considerably in metacyclics where, during differentiation, the ATG8 signal colocalizes with that of serine carboxypeptidase and decreases considerably in metacyclics, thus indicative of delivery of the autophagosome content to the reservosome/lysosome system.
Leishmania spp. The importance of protein turnover for differentiation of Leishmania has been known for several decades. Different life-cycle stages vary considerably in morphology, and the same holds true for its unusual lysosome. In promastigotes, the lysosome is present as a large single vesicular structure also known as ‘multivesicular tubule’ (MVT). It has, however, a low lytic capacity and a relatively high lumenal pH value. Upon differentiation of promastigotes into intracellular amastigotes the shape of the organelle changes into a large compartment with aspect, size and number dependent on the species. Moreover, its lytic capacity increases considerably. Initially this organelle was called a ‘megasome’ because of its large size. Megasomes may represent 5–15% of the total cell volume.135,136 Furthermore, Leishmania is known to have high peptidase activity, particularly of cysteine peptidases, that is upregulated during metacyclogenesis, the transformation of metacyclic promastigotes to amastigotes. The peptidases are involved in cellular reshaping and metabolic reprogramming during differentiation and also nutrient acquisition by the different life-cycle stages. Genome sequencing reveals a large arsenal of peptidases in L. major—at least 154 peptidases representing 1.8% of the genome—and a very similar repertoire of such enzymes in other Leishmania species (reviewed in ref. 137). Two main peptidases are the lysosomal cysteine peptidases CPA and CPB; the expression of these increases considerably during differentiation in parallel with the appearance of megasome-like structures. Leishmania, like all trypanosomatids, lack orthologues of the yeast major vacuolar aspartic peptidase Pep4 and serine peptidase Pbr1. There is evidence that their function is replaced by CPA and CPB, as inhibition of these enzymes or deletion of both genes results in promastigote cells that are more susceptible to nutrient withdrawal, fail to differentiate and do not degrade GFP-ATG8 labeled autophagosomes within the lysosome.17
Besteiro and coworkers also analyzed the phenotype of a L. major dominant-negative mutant of the yeast Vps4 orthologue, an ATPase known to be involved in the endosomal system and notably in the formation of late endosomes with a multivesicular aspect called multivesicular bodies (MVB). Indeed, as predicted, the mutant form of the VPS4 accumulated in vesicles of the endocytic pathway, and those cells are deficient in endosome sorting.16 Interestingly, autophagy and differentiation are also affected in these mutants. In wild-type cells, the GFP-ATG8 staining is most intense, and autophagosomal structures are most abundant, during differentiation of procyclic promastigotes to infective metacyclic promastigotes. In wild-type metacyclic parasites, the signal moves to the MVT-lysosome. However, in the mutant cells the number of autophagosomal structures that are stained remains high throughout. Furthermore, the level of ATG8—PE conjugate is consistently higher than in wild-type cells. These data suggest an inability of the autophagosomes to fuse with the endosomal/MVT-lysosomal compartment. They also underscore the importance of late endosomal function, and more specifically autophagy, for the transformation to the infective metacyclics. L. major contains two ATG4 genes, LmATG4.1 and LmATG4.2. A null mutant of the cysteine peptidase ATG4.2 is defective in the ability to differentiate, corroborating the earlier notion based on the results obtained with the VPS34 mutant that autophagy plays a role in this process.
As mentioned above, Leishmania species contain a large number of ATG8-like proteins, which can be classified into four families. The 25 ‘ATG8’ genes in L. major code for one ATG8, three ATG8A, eight ATG8B and 13 ATG8C proteins. Despite the low sequence identities with ATG8 homologues of other organisms, LmATG8 and representatives of each of the three other families are able to complement a S. cerevisiae atg8 null mutant, indicating that they can fulfill the Atg8 function in the Cvt pathway. However, a complete ATG8 physiological function can probably only be attributed to LmATG8, as it is involved in autophagosome formation during both differentiation and starvation. In contrast, the role of LmATG8A is probably only in starvation-induced autophagy as suggested by the observation that it forms puncta upon starvation. LmATG8B and LmATG8C are both located close to the flagellar pocket, but from the pattern of labeling of GFP-fusion products seem not to be involved in autophagy. In addition to the four ATG8 gene families, L. major has another ATG8-like gene that codes for a protein with a C-terminal extension beyond a scissile glycine that becomes exposed by ATG4 activity prior to conjugation to ATG5. Intriguingly, it has a 58-residue insertion somewhat similar to that seen in the yeast ATG12, raising the possibility of its having an ATG12-like function (see below).
L. major has two ATG4 genes, LmATG4.1 and LmATG4.2. LmATG4.1 exerts proteolytic activity towards ATG8, ATG8B and ATG8C, whereas LmATG4.2 shows only activity towards ATG8A. Searching the L. major genome, possible candidates are also found for ATG5, ATG10 and ATG12, although with low similarity with their orthologues in other organisms. These parasite genes are able to complement the respective yeast mutants, confirming their functional identity. However, the ATG12 gene complemented the respective mutant only when expressed without residues beyond the scissile glycine. In this heterologous expression system it is not processed by ATG4.1 or ATG4.2, suggesting that processing naturally in Leishmania requires an as yet to be identified peptidase. Furthermore, ATG12 localizes in part to ATG8-containing puncta in L. major promastigotes, suggesting the functionality of the ATG12-ATG5 conjugation pathway in this parasite. Using an in vitro assay, biochemical evidence has been obtained that ATG12 can be conjugated with ATG5 (Williams R, Mottram JC and Coombs GH, unpublished results). Interestingly, L. major ATG12 fails to cluster reliably with yeast and mammalian ATG12 sequences by phylogenetic analyses (neighbor-joining, minimum evolution and maximum parsimony trees calculated with MEGA-4;138 Daniel J. Rigden, data not shown) and bears a closer resemblance to ATG8 sequences, particularly in the N-terminal region. Thus, it appears possible that ATG12-like activity may have evolved more than once. The same phylogenetic analysis fails to consistently cluster any trypanosomal sequences with L. major ATG12, including those syntenic with it (see above), again highlighting the need to assess the roles of the latter experimentally.
Although Leishmania, like T. brucei, encounters different nutritional conditions during its life cycle, there are no conclusive data showing that the differentiation of promastigotes to amastigotes is accompanied with metabolic reprogramming involving an increased turnover of glycosomes by pexophagy, as is observed during differentiation of T. brucei. The size of amastigotes is reduced significantly, however, so it is likely that some glycosomes are digested.135,139 Pexophagy thus may be involved in adjusting the glycosome number, consistent with other observations on the role of autophagy in cellular remodeling. Gluconeogenesis rather than glycolysis is more important in amastigotes than in promastigotes and indeed proteomic analysis shows a two-fold decrease of the last three enzymes of the glycolytic pathway—phosphoglycerate mutase, enolase and pyruvate kinase, which are all present in the cytosol—during the late stage of differentiation. In contrast, the levels of glycolytic enzymes within the glycosomes are on average 1.4-fold increased.140 It seems probable that the altered metabolic fluxes result largely from the different nutrient availability and/or post-translational regulation of enzyme activity, rather than from a different enzyme network.141
Autophagy in Apicomplexa.
Apicomplexa: life cycle, morphology and metamorphosis. Apicomplexa form a large group of protists comprising approximately 5,000 known parasitic species, but this number is probably a gross underestimation. This group is characterized by an apical complex, a collection of organelles at the anterior end of the cell that serves to penetrate host cells. Apicomplexa are exclusively parasites of animals, both vertebrates—including humans, livestock and other mammals—and invertebrates. Apicomplexa are responsible for very serious human diseases like malaria and toxoplasmosis, as well as diseases of economically important livestock like theileriosis, babesiosis and coccidiosis. They are caused by species of the genera Plasmodium, Toxoplasma, Theileria, Babesia and Coccidia, respectively. In particular, malaria is a worldwide public health problem with 3.2 billion people at risk, an estimated 350–500 million new cases each year and over one million deaths annually. Apicomplexa have complex life cycles (Fig. 10), involving both asexual and sexual reproduction, but with considerable variation among the different subgroups. They can be transmitted to new hosts by insects, via feces or when a predator eats an infected prey. Typically, motile forms called sporozoites enter host cells within which the parasite divides to produce numerous merozoites. When the host cells burst, merozoites are released and infect new and sometimes different host cells, a process that can occur repeatedly. Some haploid asexual forms transform into sexually reproductive cells or gamonts. When haploid gamonts join in pairs to form gamontocysts, division occurs to produce multiple gametes. Pairs of gametes fuse to form zygotes, which by meiosis result in new sporozoites, so completing the life cycle.
Figure 10.
Diagram of the life cycle of Plasmodium spp.
In the next section, the role of autophagy in Plasmodium species will be discussed. Much less is known as yet about autophagy in other Apicomplexa genera. However, the role of host autophagy in controlling infections by Toxoplasma and Theileria is a topic of intense study and will be discussed in a later section of this review.
Plasmodium. Malaria sporozoites are transmitted from the mosquito vector to the mammalian host and first take up residence in the liver before initiating red blood cell infection (Fig. 10). Following penetration into hepatocytes, the parasite converts from an invasion-competent, motile, elongated sporozoite to a metabolically active, round trophozoite. Subsequent to this conversion, a mitotic phase begins, characterized by multiple fissions of nuclei, transforming the parasite into a giant syncytium (up to 10,000 nuclei). The end of the schizogony marks a phase of organelle biogenesis that is critical for erythrocyte invasion. Mature merozoites enclosed in a membrane, called a merosome, are released from hepatocytes and invade red blood cells: This erythrocyte infection is the cause of the pathology associated with malaria.142–144
The sporozoite is adeptly equipped for migration through tissues and penetration into hepatocytes. It possesses a robust membrane cytoskeleton essential for the maintenance of its shape and motility. The parasite body is delimited by a specialized cortical structure that is composed of the plasma membrane tightly associated with flattened membrane cisternae forming the inner membrane complex (IMC).145 The IMC is continuous along the entire length of the sporozoite. The process of sporozoite-to-trophozoite conversion is characterized by a spectacular change of the parasite shape and a major interior remodeling that is accompanied by the elimination of organelles unnecessary for the replication of the liver forms.146 The beginning of the metamorphosis is marked by the dismantling of the IMC, inducing the repositioning of the parasite nucleus to the middle of the parasite. As this median bulbous region expands, the two distal ends of the sporozoite gradually retract and disappear, leading to parasite sphericalization. The IMC is then reorganized into dense lamellar arrays within the cytoplasm and is expulsed by converting sporozoites in the parasitophorous vacuole (PV) wherein the parasite resides.
During metamorphosis, other organelles such as micronemes are also cleared from the sporozoite. Micronemes are abundant secretory vesicles that contain proteins such as the thrombospondin-related anonymous protein (TRAP), which facilitate parasite adhesion to hepatocytes.147,148 In converting parasites, micronemes are packaged into large compartments and partially discharged into the PV.146 Sporozoites maintained under axenic conditions undergo similar transformations as intravacuolar parasites, demonstrating that the signals and triggers of sporozoite differentiation are not host cell-dependent.146,149 At the completion of metamorphosis, mature trophozoites only retain organelles involved in biosyntheses, e.g., the ER, mitochondria and the three genomes (nuclear, mitochondria and apicoplast—i.e., the relic of a secondary endosymbiontic plastid; reviewed in ref. 150), that are necessary for the successive rounds of parasite divisions.
A critical role of autophagy for organelle turnover during the differentiation of parasitic protists that shuttle between an insect vector and mammalian host is clearly emerging.21 Scrutiny of the cytoplasmic content of converting sporozoites reveals the presence of novel double-membrane structures containing organelles such as micronemes (Fig. 11). These double-membrane compartments are morphologically similar to mammalian autophagosomes and, interestingly, the peak of their biogenesis temporally coincides with the packaging of micronemes within the parasite.146 These autophagosome-like profiles could then be indicative of functional autophagy in the malaria parasite. An autophagic activity is furthermore substantiated by treatment of parasites with the inhibitor 3-MA that induces a significant delay in the conversion process of sporozoites (Jayabalasingham B and Coppens I, unpublished data). 3-MA interferes with autophagy by blocking class III PtdIns3K activity.151,152 The Plasmodium orthologue of the main enzymatic component of the PtdIns3K complex and the target of 3-MA, VPS34, has been recently identified and characterized in blood-stage malaria parasites.152,153
Figure 11.
Presence of an autophagosomal structure in converting Plasmodium sporozoites. Electron microscopy of P. berghei sporozoites maintained in axenic condition for 3 h. The cytoplasm contains a double-membrane structure (arrow) sequestering micronemes (mi). Scale bar = 300 nm.
In addition to VPS34, Plasmodium genomes contain autophagy-related proteins. Of the 33 Atg proteins present in yeast, eight ATG putative orthologues have been clearly identified in the malaria parasites. These include: ATG1, ATG17 and ATG18 of the ATG9 cycling system, ATG12 of the ATG12 conjugation system, and all components of the ATG8 conjugation system comprising ATG3, ATG4, ATG7 and ATG8.14 The malaria parasite is therefore another example of a protist that apparently harbors a limited repertoire of autophagy-related genes. As reported for other protists, it remains also plausible that the set of ATG proteins expressed by Plasmodium define the minimum conserved cohort required for functional autophagy in this parasite.
The putative ATG orthologues of the human malaria parasite P. falciparum and the rodent malaria parasite P. berghei are 50–87% identical to each other. Among the components of the ATG8 conjugation system, ATG4 of P. falciparum (or PfATG4) shares 33% identity with yeast Atg4. An ATG4 homologue in P. berghei cannot be accurately analyzed because of incomplete annotation of this region of the genome. PfATG7 and ATG7 of P. berghei (PbATG7) show 38% and 36% identity to yeast Atg7, and PfATG3 and PbATG3 are 26% and 29% identical to yeast Atg3. PfATG8 and PbATG8 are the most similar to their yeast counterparts, sharing 40% and 45% identity, respectively. A putative orthologue of Atg12 is present in the P. falciparum genome, but like ATG4 cannot be identified in P. berghei. PfATG12 shares 32% identity with yeast Atg12.
The single P. berghei orthologues of ATG8 and ATG3 share close structural similarity with their yeast and mammalian counterparts (Jayabalasingham B and Coppens I, unpublished data). Cloned PbATG8 is 124 amino acids long and its sequence predicts two N-terminal α-helices tethered to an ubiquitin fold core. The prediction of the PbATG8 structure is supported by the conservation of amino acids involved in maintaining the tertiary structure of the protein. PbATG8 is predicted to have two hydrophobic patches as well as a number of basic residues on one face of the protein that form the scaffold for binding to ATG3, ATG4 or ATG7. While the surface-exposed residues located on one side of all ATG8 homologues are highly conserved, many of the surface-exposed residues located on the opposite face of the protein exhibit low conservation, even among homologues expressed in humans. It is possible that the reduced conservation of residues on one face reflects a reduction in evolutionary constraints, and it is tempting to hypothesize that this face mediates the interactions that are unique to different ATG8 homologues. Such potential binding partners remain to be identified for the ATG8 homologues expressed in Plasmodium species. To this point, PbATG8 and its orthologues in other Plasmodium species all share a unique inserted loop consisting of nine amino acids between α-helix 3 and β-sheet 3, suggesting that such binding partners may exist. A similar insert is also present in ATG8 homologues expressed in other protist species such as T. cruzi, T. gondii and Entamoeba species, though no function has yet been ascribed to these species-specific insertions. Finally, a functionally important residue conserved among all ATG8 homologues is a C-terminal Gly. In most homologues of ATG8, this Gly is followed by additional amino acids that are cleaved from the protein by ATG4. In this respect, the Plasmodium ATG8 sequences contain a C-terminal glycine that is not followed by additional residues, similarly to the T. gondii and one of the C. elegans ATG8 (CeLGG-2) homologues.154 Whether this implies that processing of PfATG8 by PfATG4 is not necessary is an interesting and open question.
Cloned PbATG3 is 286 residues long and the predicted sequence contains the catalytic cysteine as found in other ATG3 proteins. Two regions of interest have been identified in the yeast Atg3 homologue: the ‘handle region’ (HR), which mediates the binding between ATG3 and ATG8, and the ‘flexible region’ (FR), which is implicated in the enzyme's conjugation activity and binding to ATG7.155 Interestingly, the P. berghei orthologue shares greater structural similarity with the human and Arabidopsis thaliana orthologues, as all three either have a truncated HR or lack this region completely, and share an extension of the FR, which is absent in yeast Atg3. It is possible that the smaller HR in PbATG3 is functionally compensated by inserted residues present in the FR. Yeast Atg3 interacts with Atg8 via its N-terminus to form an intermediate that appears to be essential for the formation of the Atg8-PE moiety.156 The C-terminal HR of Atg3 mediates the interaction between Atg3 and Atg8.155 PbATG8 and PbATG3 also physically interact, as demonstrated in yeast two-hybrid assays (Jayabalasingham B and Coppens I, unpublished data). However, unlike the yeast Atg3-Atg8 interaction, the interaction between the P. berghei proteins is not dependent on any single region of ATG3. Indeed, PbATG8 is able to interact with the FR region of PbATG3, as well as with PbATG3 lacking either the FR region or the C-terminal region including the catalytic cysteine residue. Compared to yeast Atg3 in which the C terminus forms a long α-helix, the C terminus of PbATG3 is shorter and is not predicted to form such secondary structure.155 Unsurprisingly, PbATG8 cannot interact with yeast Atg3 and is unable to complement atg8Δ yeast probably due to these structural disparities between Plasmodium ATG3 and yeast Atg3. Thus, PbATG3 interacts with PbATG8 via several unique interfaces and this interaction more closely resembles that of the mammalian and plant homologues, which also have comparatively short C-terminal regions.
PbATG8 is expressed in Plasmodium blood and liver forms, and located in the peripheral membrane of large vesicles. This implies specific interaction of PbATG8 with membrane components of these organelles. In yeast and mammalian cells, nutrient deprivation stimulates Atg8 conjugation to a phospholipid of the phagophore to initiate the autophagy process. Lipidation has been visualized for many homologues of Atg8 by the presence of a second lower band in western blots after separation by SDS-PAGE in the presence of urea. However, only a single band corresponding to PbATG8 is detectable upon urea-SDS-PAGE separation (Jayabalasingham B and Coppens I, unpublished data). One possible explanation is that Plasmodium homologues of Atg8 are predominantly present in the parasite in either a conjugated or unconjugated state. Alternatively, PbATG8 may not be conjugated to a lipid moiety as reported for one of the two isoforms of ATG8 in T. cruzi, and GATE-16 and GABARAP, two mammalian homologues of Atg8.157 No other parasite-specific protein bands are detected in immunoblots against PbATG8, indicating that PbATG8 is not covalently linked to a protein like other ubiquitin-like molecules. When expressed in mammalian cells upon starvation conditions, Plasmodium ATG8 never associates with autophagosomes, but remains cytosolic. It is possible that the specific conformation of PbATG8 is responsible for the lack of interaction of this protein with the mammalian autophagy machinery, and that in general the interaction of PbATG8 and partners have evolved in a species-specific manner.
PbATG8-containing structures are prominently present in converting parasites, and some of them colocalize with TRAP-labeled structures (Jayabalasingham B and Coppens I, unpublished data). This suggests physical interactions between PbATG8-labeled compartments and micronemes and potentially implicates PbATG8 in facilitating microneme disposal. Interestingly, after sporozoite metamorphosis, PbATG8-labeled structures are still visible within the parasite and persist during the replication phase of development. In yeast, ATG8 is transcriptionally upregulated by starvation induction.158 By contrast, the PbATG8 transcripts are detected in sporozoites, hepatic stage parasites and mixed blood stage parasites, suggestive of constitutive expression of ATG8 by Plasmodium throughout the life cycle (Jayabalasingham B and Coppens I, unpublished data). PbATG3 transcription, however, is limited to sporozoites and mixed blood stages, and is below the detection level in intrahepatic parasites one day post-infection. This suggests that PbATG3 transcription is downregulated relative to PbATG8 during the hepatic phase of infection. Such a lack of PbATG3 expression suggests that autophagic activity is decreased as early as one day post-hepatocyte infection, and that PbATG8 may take on a second function during the anabolic phase of parasite growth in the liver.
Genetic ablation of Plasmodium ATG8 results in a lethal phenotype, indicating that ATG8 is essential for blood stage development. Other organisms including parasitic protists have several ATG8 homologues, permitting the deletion of one gene without deleterious consequences for the organism. For example, the human genome encodes three isotypes of the Atg8 homologue, LC3. By contrast, only a single Plasmodium ATG8 homologue has been identified. Such a duplication of ATG8 isotypes may reflect specific functions of each ATG8. Functional divergence of ATG8 homologues has been observed in mammals, in which LC3 and GATE-16/GABARAP subfamilies are both essential yet act differently in autophagosome biogenesis.159 However, even the function of LC3 does not seem to be limited to autophagy, as it is also able to bind fibronectin RNA enhancing its translation.160,161 Given the identification of only one ATG8 homologue in Plasmodium species it is possible that this single protein performs multiple functions within the parasite making its expression essential for parasite survival. Thus, in addition to a role in organelle degradation via autophagy, we hypothesize additional functions that could include a role in vesicular trafficking between organelles, as supported by the persistence of PbATG8-labeled vesicles during the anabolic phase of intrahepatic development.
Autophagy in Host-Parasitic Protist Interactions
Autophagy of host mammalian cells may play an important or even crucial role in the interaction with pathogenic protists, as it does with pathogenic viruses and bacteria. Autophagy may act to control the infection by microorganisms or the microorganisms may exploit the process to their advantage. The different roles of autophagy in host-pathogen interactions, and aspects of the mechanism of the interactions, have been studied in great detail for a variety of bacteria. A detailed description is beyond the scope of this paper about autophagy in protists, but can be found in a variety of authoritative reviews.162–166
Autophagy used by host cells to direct live parasites to lysosomes.
Macrophages play an important role in controlling infections by ingesting pathogens and trapping them in compartments such as phagosomes. These then fuse with a lysosome resulting into a phagolysosome, where the pathogen is digested. However, several pathogens, both bacteria (e.g., Mycobacterium tuberculosis) and protists (e.g., Leishmania, Toxoplasma, Theilera), are able to survive in macrophages. They achieve this by different strategies, such as inhibiting phagosome acidification, manipulating vesicular trafficking of the host cells and so avoiding fusion with lysosomes, egressing from phagosomes or creating a specialized compartment that remains isolated from the host endocytic network. T. gondii relies on the latter mechanism; the membrane of its PV is nonfusogenic due to its unique composition lacking host proteins. Nonetheless, macrophages infected with Toxoplasma can reroute the pathogen-containing compartment to lysosomes. Autophagy plays an important role in such an innate, cell-mediated immune defense mechanism.167,168 When CD40 of human or mouse macrophages infected with T. gondii is stimulated with (CD154+)CD4+ T cells or exposed to anti-CD4 antibodies, the nonfusogenic nature of the PV is reversed and the vacuole fuses with late endosomes and lysosomes. This fusion is dependent on autophagy, as indicated by observations that it is impaired by knockdown of Beclin 1 and treatment with 3-MA, an inhibitor of phagophore formation, whereas it is stimulated by rapamycin. Moreover, CD40 activation stimulates expression of LC3/Atg8 that localizes to the PV.168 Furthermore, bafilomycin A1 and LY294002, drugs that inhibit autophagosome formation and degradation of the content of autolysosomes, inhibit the fusion of the PV with lysosomes.
Autophagy used by host cells to eliminate dead parasites.
T. gondii not only infects macrophages, but also a variety of other cells such as fibroblasts, epithelial cells, endothelial cells, astrocytes and other microglial cells. Whereas the role of autophagy in parasite killing and elimination has been well established for macrophages, no evidence for the engulfment of the parasite or the PV in autophagosomes or autolysosomes has been found in other cells, such as astrocytes, where it has been sought. However, in these latter cells it has been observed that, after gamma interferon (IFNγ) stimulation, the parasites are degraded by another route that is also present in macrophages.169–173 This route involves the delivery of at least one of the p47 GTPases to the PV, followed by the disruption of first the PV membrane and subsequently the parasite's plasma membrane. Autophagy is then activated, resulting in the elimination of the damaged parasites. Treatment of infected cells with autophagy inhibitors leads to the accumulation of Toxoplasma debris in the cytosol. In this case, autophagy does not play a role in PV disruption or parasite killing, and indeed 3-MA and wortmannin have no effect on PV disposal.169 The process of clearance of dead parasites by autophagy seems necessary for host cell survival, since large amounts of parasite products accumulated in cells induce necrosis.169,172 In macrophages, the killing of parasites by the autophagic route, as described in the previous section, probably enables the cells to present pathogen antigens via the MHC class II pathway and thus to stimulate an adaptive immune response. It remains to be determined if autophagy of degraded parasite material in astrocytes also leads to antigen presentation and so would allow an intracerebral immune response to T. gondii.173 Intriguingly, in primary mouse macrophages IFNγ-induced recruitment of p47 GTPase to the PV is Atg5 dependent;171 no localization of the GTPase is observed in cells lacking this Atg protein, nor clearance of parasites. Yet, the IFNa-dependent nitric oxide production, necessary for parasite killing, is not affected. Since no classical hallmarks of autophagy are detected, it remains unknown how Atg5 is involved in causing this IFNγ-dependent cellular immunity against T. gondii.
Host cell autophagy exploited by parasites to stimulate their invasion or growth.
There are several examples where parasitic protists exploit the autophagy process of the host cell to their advantage. A first one is also from T. gondii, as observed when it grows in nonphagocytic cells, such as HeLa cells and primary fibroblasts.174 Upon infection of host cells and formation of the PV, the parasite undergoes a phase of rapid proliferation. This rapid growth is dependent on the host cell for the supply of nutrients, such as amino acids, lipids and purines. Interestingly, the PV is surrounded by host cell organelles such as endolysosomes, ER and mitochondria. Host lysosomes can even be in close contact with the PV and encroached in invaginations of the PV membrane, thereby allowing the parasite access to nutrients supplied by the host endocytic circuit.175 In general, autophagy contributes to increase the pool of nutrients derived from autolysosomes via the recycling of autophagy degradation products. Wang et al.174 showed that the host cell autophagy machinery is stimulated by Toxoplasma in order to exploit the nutritive function of autophagy and therefore boost its multiplication. Infection results in lipidation of LC3 and the accumulation of PtdIns(3)P or LC3-containing vesicles near the PV. Also Beclin 1 concentrates in the vicinity of the PV. All these effects are abolished in Atg5-deficient host cells, resulting in parasite growth arrest under conditions of low amino acid levels. The autophagic-dependent parasite growth correlates with an autophagy-dependent decrease of host cell mass. Interestingly, the stimulation of host autophagy by the parasite is independent of mTOR signaling, as suggested by the fact that no alteration in the phosphorylation state of the downstream effectors 4E-BP1 and S6K1 is observed. However, this process seems to be dependent on calcium signaling and abscisic acid, an inducer of calcium release from intracellular stores. In contrast, T. gondii stimulates mTOR-dependent host cell growth, as determined by rapamycin-sensitive phosphorylation of the ribosomal protein S6 and cell cycle progression.176
A second example of the exploitation of host autophagy by parasitic protists has been provided by studies of nonphagocytic cells infected with T. cruzi trypomastigotes.177 This parasite can enter its host cell via two routes: Lysosome-dependent and -independent entry pathways. The first route involves the calcium-dependent recruitment of host lysosomes to the site of parasite entry via the activation of PtdIns3Ks and the exploitation of lysosomal membranes by the parasite as sources of lipids to form its PV.178,179 Although nonlysosomal entry is quantitatively more frequent,180 the lysosome-dependent pathway is essential for establishment of a productive infection and intracellular proliferation.181 The alternate nonlysosomal and actin-independent entry pathway involves the formation of a tightly associated host cell plasma membrane-derived vacuole enriched in the lipid products of class I PtdIns3Ks.180 Using CHO cells transfected with GFP-LC3, Romano and coworkers showed that autophagosomes are also recruited to the entry site during T. cruzi invasion and are thus involved in formation of the PV as heterolysosomes.177 The low pH inside the PV may initiate the differentiation of trypomastigotes into amastigotes and activates the production of a pore-forming toxin, Tc-Tox, by trypomastigotes or intermediate forms allowing these forms to escape the PV.182 The completion of the differentiation process and proliferation of amastigotes occurs subsequently in the cytosol, followed by release from the cell. Induction of host cell autophagy by starvation or addition of rapamycin appears to favor the invasion process, but not the differentiation and proliferation of the parasites.177 The level of parasite infection in host cells starved for nutrients is decreased by classical inhibitors of autophagy such as wortmannin, 3-MA and vinblastine. Furthermore, in Atg5-deficient cells or in cells where Beclin 1 expression has been reduced by RNA interference, autophagosome formation, parasite entry and the association of the PV with the autolysosomal marker are reduced. Together, these results provide strong indications that T. cruzi exploits the autophagy process of its host cells to establish a productive infection.
A third example is illustrated by BALB/c macrophages infected with Leishmania amazonensis. Exogenously added IFNγ increases the intramacrophagic replication of L. amazonensis and this affect is attenuated by co-treating the cultures with either 3-MA or wortmannin.183 IFNγ treatment of infected cells stimulates the biogenesis of autophagosomal profiles, however, the amastigotes do not colocalize with these double-membrane vacuoles. Correlation between Leishmania replication and host autophagy activity is confirmed by induction of nutrient deprivation resulting in a boost of parasite growth. Starvation significantly increases the number of lipid bodies in infected BALB/c macrophages. Lipid bodies are involved in eicosanoid synthesis,184 and the prostanoid Prostaglandin E2 (PGE2) increases the intracellular load of Leishmania in macrophages.185 The levels of PGE2 are significantly augmented in infected BALB/c macrophages183 compared to uninfected macrophages, and starvation increases two-fold the PGE2 amounts produced by infected macrophages. Addition of the cyclooxygenase blocker indomethacin prevents the increase in parasite load induced by starvation. This suggests a role of PGE2 in macrophage responses to starvation-induced autophagy. Of interest, induction of autophagy that correlates with increased Leishmania load is selective for BALB/c and J774 macrophages but not C57BL/6 macrophages, suggesting the involvement of intrinsic host cell factors in the regulation of the outcome of infection following autophagy stimulation.
Suppression of host cell autophagy induction by parasites to avoid degradation.
Many intracellular pathogens can block their recognition by autophagosomes and/or the maturation of pathogen-containing autophagosomes into acidified autolysosomes. For example, viruses actively suppress initiation of the autophagy pathway, whereas certain bacterial virulence factors are necessary for bacterial evasion of autophagy. However, to date, there is no published evidence indicating that parasites block the induction of autophagy in infected cells. Nevertheless, unpublished work from the Langsley laboratory indicates that Theileria may be able to do so. Tropical Theileriosis and East Coast Fever are tickborn diseases of cattle caused by the parasitic protists Theileria annulata and T. parva, respectively. Both theilerioses display an enlargement of the lymph node draining the tick bite followed by a generalized lymphadenopathy associated with fever, anorexia and respiratory distress. Theileria sporozoites are injected with the tick saliva during a blood meal and they infect mononuclear cells. Their rapid development occurs in the absence of a PV membrane that characterizes the related Plasmodium and Toxoplasma parasites. Consequently, intracellular Theileria reside free in the host cell cytosol, closely associated with an array of host cell microtubules. In the case of T. parva, intracellular development to the multinucleated macroschizont only takes place in bovine T and B lymphocytes, whereas T. annulata transforms monocytes/macrophages and B cells (recently reviewed in ref. 186). The macroschizont stage is largely responsible for the pathology of Theileria infections, and multinucleated parasites induce leukocyte proliferation leading to clonal expansion of the parasitized cell population. Environmental signals trigger differentiation to the microschizont stage and upon merogony the host cell stops dividing. Released merozoites infect erythrocytes and differentiate into piroplasms that will eventually be taken up during a tick's blood meal. An efficacious immune response to East Coast fever involves cytotoxic CD8+ T cells, and additional CD4 responses are also thought to be necessary for protection against Tropical Theileriosis (reviewed in ref. 187). Very recently, a potent cysteine peptidase inhibitor of P. berghei PbICP, which is involved in sporozoite invasion and prevents hepatocyte cell death was identified. The inhibitor can potentially control parasites, as well as host cell-derived peptidases, which sheds some additional light on parasite-host cell interaction.188
Since autophagy plays a role in antigen presentation (reviewed in ref. 189), the hypothesis that intracellular Theileria parasites modify the autophagic response of infected leukocytes was tested. Theileria-infected lines that express Cherry-tagged LC3/Atg8 were made. Autophagy was induced by nutrient starvation of infected leukocytes and followed by the formation of large Cherry-LC3 aggregates. Confirmation that these corresponded to macroautophagosomes was provided by electron microscopy. Surprisingly, in the absence of nutrient starvation live Theileria parasites do not provoke Cherry-LC3 aggregates; i.e., the presence of intracellular Theileria appears to inhibit an autophagic response despite it being exposed (not enclosed with a PV) in the leukocyte cytosol. However, when parasites are killed with a specific drug called buparvacone they are readily recognized and digested by the autophagosome. As good CD4 and CD8 T cell responses to infection eliminate parasites, this suggests that Theileria might be deliberately inhibiting host cell autophagosome function to reduce peptide presentation to specific CD4 (by cross-presentation) and CD8 T cells, as a way of dampening the host's immune response and increasing its chances of survival. This exciting possibility is currently being tested in collaboration with I. Morrison (University of Edinburgh, UK).
Concluding remarks about autophagy in host-parasite interactions.
On the one hand, the original role of autophagy as an efficient antimicrobial defense mechanism has expanded to immunological processes that participate in both innate and adaptive immunity to neutralize infections. On the other hand, intracellular protist parasites have developed specialized anti-autophagy adaptations allowing them to prevent or block autophagic elimination. Many parasites have also built up multipronged strategies to utilize functions or components of autophagy to facilitate their own invasion of, and promote their replication rate in, mammalian cells. This concept of autophagy helping to ‘feed’ intracellular intruders is particularly relevant for parasites that reside in sequestered PV with limited access to host nutrients. However, observations on enhanced development of parasites upon autophagy induction are based upon in vitro studies and a “microbe-friendly” role of autophagy in parasite replication needs to be established in vivo. Returning to the point of origin, autophagy as a cell-autonomous defense of the host cell has met its adversarial counterpart in autophagy as a promicrobial process. Manifestly, autophagy works without respite to protect the master eukaryotic cell, often on both sides of the host-parasite relationship.
Discussion and Conclusions
Evolutionary aspects.
In this review, we addressed the occurrence, variability and evolution of autophagy in protists. However, the term ‘protists’ covers species of unicellular eukaryotes, which not only are very large in number, but also very broadly in diversity. In classical taxonomy they form one of the four kingdoms of eukaryotes, the other kingdoms being formed by the animals, plants and fungi, respectively. However, the increased availability of genome sequences for a large variety of organisms, in the last decade, and advances in molecular phylogenetics have allowed a reconsideration of this classical division. Today, most eukaryotes have been classified within one of six ‘suprakingdom-level’- or ‘super’-groups, each containing protists.48,49,60 One supergroup, the Opisthokonta, contains animals and fungi as well as certain protists, while another, the Archaeplastida, harbors plants and some groups of photosynthetic protists (e.g., Chlamydomonas). In the other supergroups (called Amoebozoa, Chromalveolata, Rhizaria and Excavata) only protist organisms are found. The presence of a nucleus, ER, Golgi, the occurrence of mitosis, and the possession of mitochondria or relic mitochondrial organelles in each of these major groups, indicates that the common ancestor of all extant eukaryotes was already a ‘typical’ eukaryotic cell. Also ATG genes have been found in protist representatives of the supergroups, examined thus far (there are no data available for the Rhizaria) as well as experimental indications that autophagy occurs in protists belonging to Amoebozoa, Chromalveolata, Archaeplastida and Excavata. For example, the trypanosomatids belong to the Excavata, the apicomplexans and ciliates to the Chromoalveolata, and Dictyostelium to the Amoebozoa (Fig. 2). Therefore, the process of macroautophagy must have been part of the repertoire of the common ancestor. Based on the distribution of ATGs in a taxonomically diverse range of protists, it may be concluded that this ancestor contained a core autophagy machinery that has been retained in all extant organisms in which autophagy can be found. The machinery has further developed with the acquisition of additional Atgs in opisthokonts (yeasts, mammals) and has been entirely lost in some protists, probably as a result of niche adaptation. In other protists the basic process has been simplified, by the probable loss of specific ATGs in some lineages or elaborated by paralogue expansion of other ATGs. It is equally possible that expansion of the core repertoire has occurred in other groups, but by recruitment of proteins unrelated to those developed in the opisthokont supergroup, and therefore not identified in the genomic searches performed with sequences of Atgs from experimentally better-characterized organisms like yeast. Only by future experimental cell biological and genetic research in more diverse unicellular taxa, similar to that performed in yeast, will the full autophagic process and the machinery involved be unraveled.
As mentioned, both ubiquitin-like proteins (Atg8, Atg12) and ubiquitin itself (with the involvement of p62 or NBR1) play key roles in the autophagy of ubiquitinated proteins. Following the recent discovery of ubiquitin-like proteins in bacteria190 it is tempting to speculate that they, too, may be involved in internal degradation mechanisms, but the differences in cell biology between prokaryotes and eukaryotes are so great that any such mechanisms would necessarily be very different relative to eukaryote autophagy.
Translational aspects.
A large number of the currently known protists are free-living organisms, but also a considerable number are pathogens of other eukaryotes, including human and domestic animals. Because of their importance for health and the economy, these pathogens have received attention in scientific research with an objective to explore the possibility of developing drugs against them. This interest for research of pathogenic protists also involved the study of the role of autophagy in these organisms and the identification of the autophagic machinery. Autophagy is essential for cellular differentiation of each of the three genera of human-infective trypanosomatid parasites and thus for the progress of the parasites though their life cycle. In addition, interference with the activity of individual proteins acting in different stages of autophagy pathways, either by reverse genetics of the use of inhibitors, affects in some cases not only the differentiation, but also the morphology, virulence or viability of the parasites. These observations, together with the generally low level of sequence conservation of ATGs and other proteins involved in autophagy in parasites, offer prospects for the development of new parasite-specific drugs.
Particularly, in the case of T. cruzi and Leishmania species, interference with the activity of proteins that are essential for differentiation may offer a possible chemotherapeutic strategy.191–195 After their introduction in the human host, these parasites have to undergo the transition from extracellular promastigotes or trypomastigotes to intracellular amastigotes, which are the stage responsible for the major pathogenic symptoms. The intracellular T. cruzi amastigotes, after several rounds of multiplication, differentiate again into trypomastigotes that burst out of the host cells to infect additional ones. In contrast, in the case of Leishmania, the parasites are released as amastigotes that initiate new macrophage infections.
Blocking of autophagy may prevent these differentiation steps and thus pathogenicity. For T. cruzi this has not yet been proven, contrary to Leishmania, where inactivation of autophagy-related gene products indeed prevents the differentiation of promastigotes.16 On the other hand, in T. brucei, the major morphological and metabolic changes, associated with upregulated autophagy, occur upon the transition from the human bloodstream form to the insect procyclic form and not when the parasite resides within the human host.21 Therefore, the effectiveness of autophagy inhibitors in treatment of sleeping sickness may be questionable. One could imagine, however, that African trypanosomes may try to overcome the deleterious effects caused by drugs by an autophagic response, and in that case combined administration with autophagy inhibitors may be effective.
One may wonder if treatment with autophagy inhibitors will be feasible for the initial phase of Chagas' disease and leishmaniasis because the period during which the insect-stage parasites will differentiate into the intracellular amastigotes, after the infection of human, is relatively short. However, at least in the case of T. cruzi, maintenance of infection involves a cyclic differentiation of amastigotes and trypomastigotes, suggesting that it could be targeted using inhibitors of autophagy.
In addition to interfering with the ATG proteins, it is possible to block autophagy also at a later stage, i.e., at the level of the autolysosome, the terminal step of the process. Using gene ablation or inhibition by chemical inhibitors it was demonstrated that the Leishmania mexicana cysteine peptidases CPA and CPB, which are orthologues of the mammalian lysosomal cysteine cathepsins, are important for both the autophagy and differentiation of the parasite. In addition, L. mexicana CPA/CPB-deficient mutants lack virulence in macrophages and mice.17 Similarly, chemically blocking the peptidases prevents amastigote replication and infectivity, although the effect on autophagy was not followed at that time.196 Moreover, similar results were obtained not only in Leishmania, but also in T. cruzi and other trypanosomatids,196–198 thereby setting the ground for possible therapeutic intervention.
An alternative chemotherapeutic strategy could be the induction of autophagic cell death. The feasibility of this approach has been shown for each of the two Trypanosoma species as discussed in the previous sections. For T. brucei, the example is the killing of parasites as a result of administration of DHA or neuropeptides,120,121 while for T. cruzi the same was achieved with lysophospholipid analogues and naphtoimidazoles.127,130 However, similar data regarding the induction of autophagic cell death of Leishmania parasites by administration of compounds are not available.
Apicomplexan parasites also undergo major morphological changes during the differentiation between the successive life-cycle stages. For Plasmodium, it has been discussed above that, during the sporozoite-to-trophozoite conversion, important interior remodeling occurs involving the elimination of many organelles. Morphological analysis, in conjunction with immuno-labeling with anti-ATG8, indicates that this conversion involves autophagy. Although the essentiality of autophagy for this differentiation has not yet been demonstrated by genetic means, the observed delay in stage conversion caused by the administration of the autophagy inhibitor 3-MA renders it very likely and suggest that specifically interfering with components of the parasite autophagy machinery might combat infection.
The development of inhibitors that interfere only with ATGs of parasitic protists and not with those of the host cells may be feasible because of the low level of sequence conservation of these proteins. This will require further studies on the structure of these ATGs, permitting the design and synthesis of selective inhibitors, or the production of recombinant ATGs and the development of sensitive binding assays to be used in high-throughput screening of compound libraries for the selection of inhibitors.
Alternative strategies may be attempted to interfere with specific host-parasite interactions. For example, stimulation of autophagy in macrophages infected with Toxoplasma parasites has been shown to promote the fusion of PVs with lysosomes.173 On the contrary, in cases where parasites (T. gondii, T. cruzi, L. amazonensis) exploit host autophagy to stimulate their invasion or growth, administration of inhibitors of the human autophagy process may be a means to control the infection. Equally, reversing this suppression may combat parasites such as Theileria, which appear to ablate host cell autophagy to avoid degradation. Of course, activation or inhibition of host autophagy is not selective and will also affect uninfected cells or tissues and thus risk causing side effects.
Autophagy has already received major attention in therapeutic intervention in human medicine—targeting autophagy may be beneficial as monotherapy or to sensitize cells to other conventional therapies (e.g., cancer). Research to explore the possibility of targeting autophagy in parasites is still at a very preliminary stage, but as reasoned above, the approach may be valuable for treatment of some of the parasite-associated diseases.
Acknowledgements
The authors gratefully acknowledge the following organizations: A.B., M.G.L. and P.A.M.M. the ‘Fonds de la Recherche Scientifique’ (FRS-FNRS) and associated foundations (FRSM and FRIA) and the Belgian Interuniversity Attraction Poles-Federal Office for Scientific, Technical and Cultural Affairs for support of their research on autophagy in trypanosomes; M.L.G. the Royal Society for an University Research Fellowship; G.L. the Wellcome Trust for a Special Initiative grant (075820/A/04/Z) for support for ‘An integrated approach for the development of sustainable methods to control tropical theileriosis’; J.C.M. and G.H.C. the Medical Research Council for support (G0700127); M.N. the MICINN for support (SAF2009-07587); B.T. and V.S. the Slovenian Research Agency for research grants (P1-0140, J1-9520, J3-2258); S.L.d.C. and R.M.B. the Brazilian agencies CNPq and Faperj; M.I.C. and P.R. the ANPCYT (PICT 2005 # 38420) and Comisión Nacional Salud Investiga in Argentina for research grants; B.T. and V.S. would like to thank Vito Turk for valuable discussions.
Abbreviations
- ATG
autophagy-related
- Cvt
cytoplasm-to-vacuole targeting
- EC50
effective concentration to cause 50% inhibition of growth
- ER
endoplasmic reticulum
- FR
flexible region
- GFP
green fluorescent protein
- HR
handle region
- IFNγ
gamma interferon
- IMC
inner membrane complex
- LECA
last eukaryotic common ancestor
- 3-MA
3-methyladenine
- MIPA
micropexophagic membrane apparatus
- MVB
multivesicular body
- MVT
multivesicular tube
- PAS
phagophore assembly site
- PCD
programmed cell death
- PE
phosphatidylethanolamine
- PGE2
prostaglandin E2
- PtdIns3K
phosphatidylinositol 3-kinase
- PtdIns(3)P
phosphatidylinositol 3-phosphate
- PMN
piecemeal microautophagy of the nucleus
- prApe1
precursor aminopeptidase
- PV
parasitophorous vacuole
- ROS
reactive oxygen species
- TOR
target of rapamycin
- TRAP
thrombospondin-related anonymous protein
Footnotes
Previously published online: www.landesbioscience.com/journals/autophagy/article/13310
Note
Slightly different systems are being used for abbreviations of proteins and their genes in mammals, yeasts and protists. Throughout this paper, we used for proteins and genes from protists the consensus nomenclature system for trypanosomatid protists: upper case for proteins, italicized upper case for genes.200
References
- 1.de Duve C. The lysosome turns fifty. Nat Cell Biol. 2005;7:847–849. doi: 10.1038/ncb0905-847. [DOI] [PubMed] [Google Scholar]
- 2.de Reuck AVS, Cameron MP. Ciba Foundation Symposium on Lysosomes. London: J.A. Chrurchill Ltd; 1963. [Google Scholar]
- 3.Klionsky DJ. Autophagy revisited: a conversation with Christian de Duve. Autophagy. 2008;4:740–743. doi: 10.4161/auto.6398. [DOI] [PubMed] [Google Scholar]
- 4.Klionsky DJ. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat Rev Mol Cell Biol. 2007;8:931–937. doi: 10.1038/nrm2245. [DOI] [PubMed] [Google Scholar]
- 5.Klionsky DJ, Cregg JM, Dunn WA, Jr, Emr SD, Sakai Y, Sandoval IV, et al. A unified nomenclature for yeast autophagy-related genes. Dev Cell. 2003;5:539–545. doi: 10.1016/s1534-5807(03)00296-x. [DOI] [PubMed] [Google Scholar]
- 6.Meijer WH, van der Klei IJ, Veenhuis M, Kiel JAKW. ATG genes involved in non-selective autophagy are conserved from yeast to man, but the selective Cvt and pexophagy pathways also require organism-specific genes. Autophagy. 2007;3:106–116. doi: 10.4161/auto.3595. [DOI] [PubMed] [Google Scholar]
- 7.Yang Z, Klionsky DJ. An overview of the molecular mechanism of autophagy. Curr Top Microbiol Immunol. 2009;335:1–32. doi: 10.1007/978-3-642-00302-8_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mehrpour M, Esclatine A, Beau I, Codogno P. Autophagy in health and disease. 1. Regulation and significance of autophagy: an overview. Am J Physiol Cell Physiol. 2010;298:776–785. doi: 10.1152/ajpcell.00507.2009. [DOI] [PubMed] [Google Scholar]
- 10.Kroemer G, Galluzzi L, Vandenabeele P, Abrams J, Alnemri ES, Baehrecke EH, et al. Nomenclature Committee on Cell Death 2009. Classification of cell death: recommendations of the Nomenclature Committee on Cell Death 2009. Cell Death Differ. 2009;16:3–11. doi: 10.1038/cdd.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yorimitsu T, Klionsky DJ. Autophagy: molecular machinery for self-eating. Cell Death Differ. 2005;12:1542–1552. doi: 10.1038/sj.cdd.4401765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bejarano E, Cuervo AM. Chaperone-mediated autophagy. Proc Am Thorac Soc. 2010;7:29–39. doi: 10.1513/pats.200909-102JS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fader CM, Sánchez DG, Mestre MB, Colombo MI. TI-VAMP/VAMP7 and VAMP3/cellubrevin: two v-SNARE proteins involved in specific steps of the autophagy/multivesicular body pathways. Biochim Biophys Acta. 2009;1793:1901–1916. doi: 10.1016/j.bbamcr.2009.09.011. [DOI] [PubMed] [Google Scholar]
- 14.Rigden DJ, Michels PAM, Ginger ML. Autophagy in protists: Examples of secondary loss, lineage-specific innovations and the conundrum of remodeling a single mitochondrion. Autophagy. 2009;5:784–794. doi: 10.4161/auto.8838. [DOI] [PubMed] [Google Scholar]
- 15.Kiel JA KW. Autophagy in unicellular eukaryotes. Philos Trans R Soc Lond B Biol Sci. 2010;365:819–830. doi: 10.1098/rstb.2009.0237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Besteiro S, Williams RA, Morrison LS, Coombs GH, Mottram JC. Endosome sorting and autophagy are essential for differentiation and virulence of Leishmania major. J Biol Chem. 2006;281:11384–11396. doi: 10.1074/jbc.M512307200. [DOI] [PubMed] [Google Scholar]
- 17.Williams RA, Tetley L, Mottram JC, Coombs GH. Cysteine peptidases CPA and CPB are vital for autophagy and differentiation in Leishmania mexicana. Mol Microbiol. 2006;61:655–674. doi: 10.1111/j.1365-2958.2006.05274.x. [DOI] [PubMed] [Google Scholar]
- 18.Herman M, Gillies S, Michels PA, Rigden DJ. Autophagy and related processes in trypanosomatids: insights from genomic and bioinformatic analyses. Autophagy. 2006;2:107–118. doi: 10.4161/auto.2.2.2369. [DOI] [PubMed] [Google Scholar]
- 19.Uzcátegui NL, Denninger V, Merkel P, Schoenfeld C, Figarella K, Duszenko M. Dihydroxyacetone induced autophagy in African trypanosomes. Autophagy. 2007;3:626–629. doi: 10.4161/auto.4907. [DOI] [PubMed] [Google Scholar]
- 20.Alvarez VE, Kosec G, Sant'Anna C, Turk V, Cazzulo JJ, Turk B. Autophagy is involved in nutritional stress response and differentiation in Trypanosoma cruzi. J Biol Chem. 2008;283:3454–3464. doi: 10.1074/jbc.M708474200. [DOI] [PubMed] [Google Scholar]
- 21.Herman M, Pérez-Morga D, Schtickzelle N, Michels PA. Turnover of glycosomes during life-cycle differentiation of Trypanosoma brucei. Autophagy. 2008;4:294–308. doi: 10.4161/auto.5443. [DOI] [PubMed] [Google Scholar]
- 22.Denninger V, Koopmann R, Muhammad K, Barth T, Bassarak B, Sch∞nfeld C, et al. Kinetoplastida: model organisms for simple autophagic pathways? Methods Enzymol. 2008;451:373–408. doi: 10.1016/S0076-6879(08)03225-4. [DOI] [PubMed] [Google Scholar]
- 23.Williams RA, Woods KL, Juliano L, Mottram JC, Coombs GH. Characterization of unusual families of ATG8-like proteins and ATG12 in the protozoan parasite Leishmania major. Autophagy. 2009;5:159–172. doi: 10.4161/auto.5.2.7328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klionsky DJ. The molecular machinery of autophagy: unanswered questions. J Cell Sci. 2005;118:7–18. doi: 10.1242/jcs.01620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suzuki K, Ohsumi Y. Molecular machinery of autophagosome formation in yeast, Saccharomyces cerevisiae. FEBS Lett. 2007;581:2156–2161. doi: 10.1016/j.febslet.2007.01.096. [DOI] [PubMed] [Google Scholar]
- 26.Díaz-Troya S, Pérez-Pérez ME, Florencio FJ, Crespo JL. The role of TOR in autophagy regulation from yeast to plants and mammals. Autophagy. 2008;4:851–865. doi: 10.4161/auto.6555. [DOI] [PubMed] [Google Scholar]
- 27.Jung CH, Ro SH, Cao J, Otto NM, Kim DH. mTOR regulation of autophagy. FEBS Lett. 2010;584:1287–1295. doi: 10.1016/j.febslet.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manjithaya R, Nazarko TY, Farré JC, Subramani S. Molecular mechanism and physiological role of pexophagy. FEBS Lett. 2010;584:1367–1373. doi: 10.1016/j.febslet.2010.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ano Y, Hattori T, Kato N, Sakai Y. Intracellular ATP correlates with mode of pexophagy in Pichia pastoris. Biosci Biotechnol Biochem. 2005;69:1527–1533. doi: 10.1271/bbb.69.1527. [DOI] [PubMed] [Google Scholar]
- 30.Okamoto K, Kondo-Okamoto N, Ohsumi Y. Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev Cell. 2009;17:87–97. doi: 10.1016/j.devcel.2009.06.013. [DOI] [PubMed] [Google Scholar]
- 31.Kanki T, Wang K, Cao Y, Baba M, Klionsky DJ. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev Cell. 2009;17:98–109. doi: 10.1016/j.devcel.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kanki T, Wang K, Baba M, Bartholomew CR, Lynch-Day MA, Du Z, et al. A genomic screen for yeast mutants defective in selective mitochondria autophagy. Mol Biol Cell. 2009;20:4730–4738. doi: 10.1091/mbc.E09-03-0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hayashi-Nishino M, Fujita N, Noda T, Yamaguchi A, Yoshimori T, Yamamoto A. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat Cell Biol. 2009;11:1433–1437. doi: 10.1038/ncb1991. [DOI] [PubMed] [Google Scholar]
- 34.Ylä-Anttila P, Vihinen H, Jokitalo E, Eskelinen EL. 3D tomography reveals connections between the phagophore and endoplasmic reticulum. Autophagy. 2009;5:1180–1185. doi: 10.4161/auto.5.8.10274. [DOI] [PubMed] [Google Scholar]
- 35.Hailey DW, Rambold AS, Satpute-Krishnan PP, Mitra K, Sougrat R, Kim PK, Lippincott-Schwartz J. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell. 2010;141:656–667. doi: 10.1016/j.cell.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Geng J, Klionsky DJ. The Atg8 and Atg12 ubiquitin-like conjugation systems in macroautophagy. ‘Protein modifications: beyond the usual suspects’ review series. EMBO Rep. 2008;9:859–864. doi: 10.1038/embor.2008.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kirkin V, McEwan DG, Novak I, Dikic I. A role for ubiquitin in selective autophagy. Mol Cell. 2009;34:259–269. doi: 10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- 38.Kim PK, Hailey DW, Mullen RT, Lippincott-Schwartz J. Ubiquitin signals autophagic degradation of cytosolic proteins and peroxisomes. Proc Natl Acad Sci USA. 2008;105:20567–20574. doi: 10.1073/pnas.0810611105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSIBLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Altschul SF, Wootton JC, Gertz EM, Agarwala R, Morgulis A, Schaffer AA, Yu YK. Protein database searches using compositionally adjusted substitution matrices. FEBS J. 2005;272:5101–5109. doi: 10.1111/j.1742-4658.2005.04945.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Patthy L. Protein Evolution. Oxford: UK: Blackwell; 2008. [Google Scholar]
- 42.Addou S, Rentzsch R, Lee D, Orengo CA. Domain-based and family-specific sequence identity thresholds increase the levels of reliable protein function transfer. J Mol Biol. 2009;387:416–430. doi: 10.1016/j.jmb.2008.12.045. [DOI] [PubMed] [Google Scholar]
- 43.Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, et al. The COG database: An updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Soding J. Protein homology detection by HMMHMM comparison. Bioinformatics. 2005;21:951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- 45.Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, et al. The pfam protein families database. Nucleic Acids Res. 2010;38:211–222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marchler-Bauer A, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, et al. CDD: Specific functional annotation with the conserved domain database. Nucleic Acids Res. 2009;37:205–210. doi: 10.1093/nar/gkn845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Turk B, Turk V. Lysosomes as ‘suicide bags’ in cell death: myth or reality? J Biol Chem. 2009;284:21783–21787. doi: 10.1074/jbc.R109.023820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Simpson AG, Roger AJ. The real ‘kingdoms’ of eukaryotes. Curr Biol. 2004;14:693–696. doi: 10.1016/j.cub.2004.08.038. [DOI] [PubMed] [Google Scholar]
- 49.Roger AJ, Simpson AG. Evolution: revisiting the root of the eukaryote tree. Curr Biol. 2009;19:165–167. doi: 10.1016/j.cub.2008.12.032. [DOI] [PubMed] [Google Scholar]
- 50.Embley TM, Martin W. Eukaryotic evolution, changes and challenges. Nature. 2006;440:623–630. doi: 10.1038/nature04546. [DOI] [PubMed] [Google Scholar]
- 51.Rogozin IB, Basu MK, Csürös M, Koonin EV. Analysis of rare genomic changes does not support the unikontbikont phylogeny and suggests cyanobacterial symbiosis as the point of primary radiation of eukaryotes. Genome Biol Evol. 2009;2009:99–113. doi: 10.1093/gbe/evp011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cavalier-Smith T. Kingdoms Protozoa and Chromista and the eozoan root of the eukaryotic tree. Biol Lett. 2010;6:342–345. doi: 10.1098/rsbl.2009.0948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Katinka MD, Duprat S, Cornillot E, Méténier G, Thomarat F, Prensier G, et al. Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature. 2001;414:450–453. doi: 10.1038/35106579. [DOI] [PubMed] [Google Scholar]
- 54.Matsuzaki M, Misumi O, Shin-I T, Maruyama S, Takahara M, Miyagishima SY, et al. Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature. 2004;428:653–657. doi: 10.1038/nature02398. [DOI] [PubMed] [Google Scholar]
- 55.Morrison HG, McArthur AG, Gillin FD, Aley SB, Adam RD, Olsen GJ, et al. Genomic minimalism in the early diverging intestinal parasite Giardia lamblia. Science. 2007;317:1921–1926. doi: 10.1126/science.1143837. [DOI] [PubMed] [Google Scholar]
- 56.Tovar J, León-Avila G, Sánchez LB, Sutak R, Tachezy J, van der Giezen M, et al. Mitochondrial remnant organelles of Giardia function in iron-sulphur protein maturation. Nature. 2003;426:172–176. doi: 10.1038/nature01945. [DOI] [PubMed] [Google Scholar]
- 57.Abodeely M, DuBois KN, Hehl A, Stefanic S, Sajid M, DeSouza W, et al. A contiguous compartment functions as endoplasmic reticulum and endosome/lysosome in Giardia lamblia. Eukaryot Cell. 2009;8:1665–1676. doi: 10.1128/EC.00123-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Marti M, Regös A, Li Y, Schraner EM, Wild P, Müller N, et al. An ancestral secretory apparatus in the protozoan parasite Giardia intestinalis. J Biol Chem. 2003;278:24837–24848. doi: 10.1074/jbc.M302082200. [DOI] [PubMed] [Google Scholar]
- 59.Roger AJ, Hug LA. The origin and diversification of eukaryotes: problems with molecular phylogenetics and molecular clock estimation. Philos Trans R Soc Lond B Biol Sci. 2006;361:1039–1054. doi: 10.1098/rstb.2006.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hampl V, Hug L, Leigh JW, Dacks JB, Lang BF, Simpson AG, Roger AJ. Phylogenomic analyses support the monophyly of Excavata and resolve relationships among eukaryotic “supergroups”. Proc Natl Acad Sci USA. 2009;106:3859–3864. doi: 10.1073/pnas.0807880106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Benchimol M. Hydrogenosome autophagy: an ultrastructural and cytochemical study. Biol Cell. 1999;91:165–174. doi: 10.1016/s0248-4900(99)80039-2. [DOI] [PubMed] [Google Scholar]
- 62.Kaneda Y, Torii M, Tanaka T, Aikawa M. In vitro effects of berberine sulphate on the growth and structure of Entamoeba histolytica, Giardia lamblia and Trichomonas vaginalis. Ann Trop Med Parasitol. 1991;85:417–425. doi: 10.1080/00034983.1991.11812586. [DOI] [PubMed] [Google Scholar]
- 63.Carlton JM, Hirt RP, Silva JC, Delcher AL, Schatz M, Zhao Q, et al. Draft genome sequence of the sexually transmitted pathogen Trichomonas vaginalis. Science. 2007;315:207–212. doi: 10.1126/science.1132894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Aury JM, Jaillon O, Duret L, Noel B, Jubin C, Porcel BM, et al. Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature. 2006;444:171–178. doi: 10.1038/nature05230. [DOI] [PubMed] [Google Scholar]
- 65.Sehring IM, Reiner C, Mansfeld J, Plattner H, Kissmehl R. A broad spectrum of actin paralogs in Paramecium tetraurelia cells display differential localization and function. J Cell Sci. 2007;120:177–190. doi: 10.1242/jcs.03313. [DOI] [PubMed] [Google Scholar]
- 66.Wassmer T, Kissmehl R, Cohen J, Plattner H. Seventeen a-subunit isoforms of Paramecium V-ATPase provide high specialization in localization and function. Mol Biol Cell. 2006;17:917–930. doi: 10.1091/mbc.E05-06-0511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kirkin V, Lamark T, Sou YS, Bjorkoy G, Nunn JL, Bruun JA, et al. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell. 2009;33:505–516. doi: 10.1016/j.molcel.2009.01.020. [DOI] [PubMed] [Google Scholar]
- 68.Okamoto K, Kondo-Okamoto N, Ohsumi Y. Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev Cell. 2009;17:87–97. doi: 10.1016/j.devcel.2009.06.013. [DOI] [PubMed] [Google Scholar]
- 69.Calvo-Garrido J, Escalante R. Autophagy dysfunction and ubiquitin-positive protein aggregates in Dictyostelium cells lacking Vmp1. Autophagy. 2010;6:100–109. doi: 10.4161/auto.6.1.10697. [DOI] [PubMed] [Google Scholar]
- 70.Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 71.Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 72.Pawlowski J, Burki F. Untangling the phylogeny of amoeboid protists. J Eukaryot Microbiol. 2009;56:16–25. doi: 10.1111/j.1550-7408.2008.00379.x. [DOI] [PubMed] [Google Scholar]
- 73.Schaap P, Winckler T, Nelson M, Alvarez-Curto E, Elgie B, Hagiwara H, et al. Molecular phylogeny and evolution of morphology in the social amoebas. Science. 2006;314:661–663. doi: 10.1126/science.1130670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hilbi H, Weber SS, Ragaz C, Nyfeler Y, Urwyler S. Environmental predators as models for bacterial pathogenesis. Environ Microbiol. 2007;9:563–575. doi: 10.1111/j.1462-2920.2007.01238.x. [DOI] [PubMed] [Google Scholar]
- 75.Calvo-Garrido J, Carilla-Latorre S, Escalante R. Vacuole membrane protein 1, autophagy and much more. Autophagy. 2008;4:835–837. doi: 10.4161/auto.6574. [DOI] [PubMed] [Google Scholar]
- 76.Calvo-Garrido J, Carilla-Latorre S, Lazaro-Dieguez F, Egea G, Escalante R. Vacuole membrane protein 1 is an endoplasmic reticulum protein required for organelle biogenesis, protein secretion and development. Mol Biol Cell. 2008;19:3442–3453. doi: 10.1091/mbc.E08-01-0075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mizushima N, Kuma A, Kobayashi Y, Yamamoto A, Matsubae M, Takao T, et al. Mouse Apg16L, a novel WD-repeat protein, targets to the autophagic isolation membrane with the Apg12-Apg5 conjugate. J Cell Sci. 2003;116:1679–1688. doi: 10.1242/jcs.00381. [DOI] [PubMed] [Google Scholar]
- 78.Tekinay T, Wu MY, Otto GP, Anderson OR, Kessin RH. Function of the Dictyostelium discoideum Atg1 kinase during autophagy and development. Eukaryot Cell. 2006;5:1797–1806. doi: 10.1128/EC.00342-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Otto GP, Wu MY, Kazgan N, Anderson OR, Kessin RH. Macroautophagy is required for multicellular development of the social amoeba Dictyostelium discoideum. J Biol Chem. 2003;278:17636–17645. doi: 10.1074/jbc.M212467200. [DOI] [PubMed] [Google Scholar]
- 80.Giusti C, Tresse E, Luciani MF, Golstein P. Autophagic cell death: analysis in Dictyostelium. Biochim Biophys Acta. 2009;1793:1422–1431. doi: 10.1016/j.bbamcr.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 81.Lam D, Kosta A, Luciani MF, Golstein P. The inositol 1,4,5-trisphosphate receptor is required to signal autophagic cell death. Mol Biol Cell. 2008;19:691–700. doi: 10.1091/mbc.E07-08-0823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tresse E, Kosta A, Giusti C, Luciani MF, Golstein P. A UDP-glucose derivative is required for vacuolar autophagic cell death. Autophagy. 2008;4:680–691. doi: 10.4161/auto.6084. [DOI] [PubMed] [Google Scholar]
- 83.Otto GP, Wu MY, Kazgan N, Anderson OR, Kessin RH. Dictyostelium macroautophagy mutants vary in the severity of their developmental defects. J Biol Chem. 2004;279:15621–15629. doi: 10.1074/jbc.M311139200. [DOI] [PubMed] [Google Scholar]
- 84.Tung SM, Unal C, Ley A, Pena C, Tunggal B, Noegel AA, et al. Loss of Dictyostelium ATG9 results in a pleiotropic phenotype affecting growth, development, phagocytosis and clearance and replication of Legionella pneumophila. Cell Microbiol. 2010;12:765–780. doi: 10.1111/j.1462-5822.2010.01432.x. [DOI] [PubMed] [Google Scholar]
- 85.Ogura K, Shirakawa M, Barnes TM, Hekimi S, Ohshima Y. The UNC-14 protein required for axonal elongation and guidance in Caenorhabditis elegans interacts with the serine/threonine kinase UNC-51. Genes Dev. 1997;11:1801–1811. doi: 10.1101/gad.11.14.1801. [DOI] [PubMed] [Google Scholar]
- 86.Reggiori F, Tucker KA, Stromhaug PE, Klionsky DJ. The Atg1-Atg13 complex regulates Atg9 and Atg23 retrieval transport from the pre-autophagosomal structure. Dev Cell. 2004;6:79–90. doi: 10.1016/s1534-5807(03)00402-7. [DOI] [PubMed] [Google Scholar]
- 87.Jia K, Thomas C, Akbar M, Sun Q, Adams-Huet B, Gilpin C, Levine B. Autophagy genes protect against Salmonella typhimurium infection and mediate insulin signaling-regulated pathogen resistance. Proc Natl Acad Sci USA. 2009;106:14564–14569. doi: 10.1073/pnas.0813319106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kobayashi T, Endoh H. Caspase-like activity in programmed nuclear death during conjugation of Tetrahymena thermophila. Cell Death Differ. 2003;10:634–640. doi: 10.1038/sj.cdd.4401216. [DOI] [PubMed] [Google Scholar]
- 89.McArdle EW, Bergquist BL, Ehret CF. Structural changes in Tetrahymena rostrata during induced encystment. J Protozool. 1980;27:388–397. [Google Scholar]
- 90.Martin NC, Goodenough UW. Gametic differentiation in Chlamydomonas reinhardtii. I. Production of gametes and their fine structure. J Cell Biol. 1975;67:587–605. doi: 10.1083/jcb.67.3.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang ZT, Ullrich N, Joo S, Waffenschmidt S, Goodenough U. Algal lipid bodies: stress induction, purification and biochemical characterization in wild-type and starchless Chlamydomonas reinhardtii. Eukaryot Cell. 2009;8:1856–1868. doi: 10.1128/EC.00272-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Affenzeller MJ, Darehshouri A, Andosch A, Lutz C, Lutz-Meindl U. PCD and autophagy in the unicellular green alga Micrasterias denticulata. Autophagy. 2009;5:854–855. doi: 10.4161/auto.8791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Affenzeller MJ, Darehshouri A, Andosch A, Lutz C, Lutz-Meindl U. Salt stress-induced cell death in the unicellular green alga Micrasterias denticulata. J Exp Bot. 2009;60:939–954. doi: 10.1093/jxb/ern348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fenchel T. The life history of Flabellula baltica Smirnov (Gymnamoebae, Rhizopoda): adaptations to a spatially and temporally heterogeneous environment. Protist. 2010;161:279–287. doi: 10.1016/j.protis.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 95.Inwood W, Yoshihara C, Zalpuri R, Kim KS, Kustu S. The ultrastructure of a Chlamydomonas reinhardtii mutant strain lacking phytoene synthase resembles that of a colorless alga. Mol Plant. 2008;1:925–937. doi: 10.1093/mp/ssn046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wloga D, Strzyzewska-Jowko I, Gaertig J, Jerka-Dziadosz M. Septins stabilize mitochondria in Tetrahymena thermophila. Eukaryot Cell. 2008;7:1373–1386. doi: 10.1128/EC.00085-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Morais P, Lamas J, Sanmartín ML, Orallo F, Leiro J. Resveratrol induces mitochondrial alterations, autophagy and a cryptobiosis-like state in scuticociliates. Protist. 2009;160:552–564. doi: 10.1016/j.protis.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 98.Martin NC, Chiang KS, Goodenough UW. Turnover of chloroplast and cytoplasmic ribosomes during gametogenesis in Chlamydomonas reinhardi. Dev Biol. 1976;5:190–201. doi: 10.1016/0012-1606(76)90137-8. [DOI] [PubMed] [Google Scholar]
- 99.Diaz-Troya S, Perez-Perez ME, Florencio FJ, Crespo JL. The role of TOR in autophagy regulation from yeast to plants and mammals. Autophagy. 2008;4:851–865. doi: 10.4161/auto.6555. [DOI] [PubMed] [Google Scholar]
- 100.Parker MS, Mock T, Armbrust EV. Genomic insights into marine microalgae. Annu Rev Genet. 2008;42:619–645. doi: 10.1146/annurev.genet.42.110807.091417. [DOI] [PubMed] [Google Scholar]
- 101.Franklin DJ, Brussaard CPD, Berges JA. What is the role and nature of programmed cell death in phytoplankton ecology? Eur J Phycol. 2006;41:1–14. [Google Scholar]
- 102.Duszenko M, Figarella K, Macleod ET, Welburn SC. Death of a trypanosome: a selfish altruism. Trends Parasitol. 2006;22:536–542. doi: 10.1016/j.pt.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 103.Golstein P, Aubry L, Levraud JP. Cell-death alternative model organisms: why and which? Nat Rev Mol Cell Biol. 2003;4:798–807. doi: 10.1038/nrm1224. [DOI] [PubMed] [Google Scholar]
- 104.Pérez-Pérez ME, Florencio FJ, Crespo JL. Inhibition of target of rapamycin signaling and stress activate autophagy in Chlamydomonas reinhardtii. Plant Physiol. 2010;152:1874–1888. doi: 10.1104/pp.109.152520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Crespo JL, Diaz-Troya S, Florencio FJ. Inhibition of target of rapamycin signaling by rapamycin in the unicellular green alga Chlamydomonas reinhardtii. Plant Physiol. 2005;139:1736–1749. doi: 10.1104/pp.105.070847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Diaz-Troya S, Florencio FJ, Crespo JL. Target of rapamycin and LST8 proteins associate with membranes from the endoplasmic reticulum in the unicellular green alga Chlamydomonas reinhardtii. Eukaryot Cell. 2008;7:212–222. doi: 10.1128/EC.00361-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Huang K, Diener DR, Mitchell A, Pazour GJ, Witman GB, Rosenbaum JL. Function and dynamics of PKD2 in Chlamydomonas reinhardtii flagella. J Cell Biol. 2007;179:501–514. doi: 10.1083/jcb.200704069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mostov KE. mTOR is out of control in polycystic kidney disease. Proc Natl Acad Sci USA. 2006;103:5247–5248. doi: 10.1073/pnas.0601352103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Shillingford JM, Murcia NS, Larson CH, Low SH, Hedgepeth R, Brown N, et al. The mTOR pathway is regulated by polycystin-1, and its inhibition reverses renal cystogenesis in polycystic kidney disease. Proc Natl Acad Sci USA. 2006;103:5466–5471. doi: 10.1073/pnas.0509694103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Picazarri K, Nakada-Tsukui K, Nozaki T. Autophagy during proliferation and encystation in the protozoan parasite Entamoeba invadens. Infect Immun. 2008;76:278–288. doi: 10.1128/IAI.00636-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Moon EK, Chung DI, Hong YC, Kong HH. Autophagy protein 8 mediating autophagosome in encysting Acanthamoeba. Mol Biochem Parasitol. 2009;168:43–48. doi: 10.1016/j.molbiopara.2009.06.005. [DOI] [PubMed] [Google Scholar]
- 112.Lukes J, Hashimi H, Zíková A. Unexplained complexity of the mitochondrial genome and transcriptome in kinetoplastid flagellates. Curr Genet. 2005;48:277–299. doi: 10.1007/s00294-005-0027-0. [DOI] [PubMed] [Google Scholar]
- 113.Brown RC, Evans DA, Vickerman K. Developmental changes in ultrastructure and physiology of Trypanosoma brucei. Trans R Soc Trop Med Hyg. 1972;66:336–337. doi: 10.1016/0035-9203(72)90215-5. [DOI] [PubMed] [Google Scholar]
- 114.Vickerman K, Tetley L. Recent ultrastructural studies on trypanosomes. Ann Soc Belg Med Trop. 1977;57:441–457. [PubMed] [Google Scholar]
- 115.Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 116.Barquilla A, Crespo JL, Navarro M. Rapamycin inhibits trypanosome cell growth by preventing TOR complex 2 formation. Proc Natl Acad Sci USA. 2008;105:14579–14584. doi: 10.1073/pnas.0802668105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Barquilla A, Navarro M. Trypanosome TOR as a major regulator of cell growth and autophagy. Autophagy. 2009;5:256–258. doi: 10.4161/auto.5.2.7591. [DOI] [PubMed] [Google Scholar]
- 118.Goldshmidt H, Matas D, Kabi A, Carmi S, Hope R, Michaeli S. Persistent ER stress induces the spliced leader RNA silencing pathway (SLS), leading to programmed cell death in Trypanosoma brucei. PLoS Pathog. 2010;6:1000731. doi: 10.1371/journal.ppat.1000731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Uzcátegui NL, Carmona-Gutiérrez D, Denninger V, Schoenfeld C, Lang F, Figarella K, Duszenko M. Antiproliferative effect of dihydroxyacetone on Trypanosoma brucei bloodstream forms: cell cycle progression, subcellular alterations and cell death. Antimicrob Agents Chemother. 2007;51:3960–3968. doi: 10.1128/AAC.00423-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Delgado M, Anderson P, Garcia-Salcedo JA, Caro M, Gonzalez-Rey E. Neuropeptides kill African trypanosomes by targeting intracellular compartments and inducing autophagic-like cell death. Cell Death Differ. 2009;16:406–416. doi: 10.1038/cdd.2008.161. [DOI] [PubMed] [Google Scholar]
- 121.Campos-Salinas J, Gonzalez-Rey E. Autophagy and neuropeptides at the crossroad for parasites: to survive or to die? Autophagy. 2009;5:551–554. doi: 10.4161/auto.5.4.8365. [DOI] [PubMed] [Google Scholar]
- 122.Koopmann R, Muhammad K, Perbandt M, Betzel C, Duszenko M. Trypanosoma brucei ATG8: structural insights into autophagic-like mechanisms in protozoa. Autophagy. 2009;5:1085–1091. doi: 10.4161/auto.5.8.9611. [DOI] [PubMed] [Google Scholar]
- 123.Michels PA, Bringaud F, Herman M, Hannaert V. Metabolic functions of glycosomes in trypanosomatids. Biochim Biophys Acta. 2006;1763:1463–1477. doi: 10.1016/j.bbamcr.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 124.Bellu AR, Kiel JAKW. Selective degradation of peroxisomes in yeasts. Microsc Res Tech. 2003;61:161–170. doi: 10.1002/jemt.10325. [DOI] [PubMed] [Google Scholar]
- 125.Braga MV, de Souza W. Effects of protein kinase and phosphatidylinositol-3 kinase inhibitors on growth and ultrastructure of Trypanosoma cruzi. FEMS Microbiol Lett. 2006;256:209–216. doi: 10.1111/j.1574-6968.2006.00125.x. [DOI] [PubMed] [Google Scholar]
- 126.Santa-Rita RM, Lira R, Barbosa HS, Urbina JA, de Castro SL. Anti-proliferative synergy of lysophospholipid analogues and ketoconazole against Trypanosoma cruzi (Kinetoplastida: Trypanosomatidae): cellular and ultrastructural analysis. J Antimicrob Chemother. 2005;55:780–784. doi: 10.1093/jac/dki087. [DOI] [PubMed] [Google Scholar]
- 127.Menna-Barreto RFS, Salomão K, Dantas AP, Santa-Rita RM, Soares MJ, Barbosa HS, De Castro SL. Different cell death pathways induced by drugs in Trypanosoma cruzi: an ultrastructural study. Micron. 2009;40:157–168. doi: 10.1016/j.micron.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 128.Menna-Barreto RFS, Henriques-Pons A, Pinto AV, Morgado-Diaz JA, Soares MJ, De Castro SL. Effect of a β-lapachone-derived naphthoimidazole on Trypanosoma cruzi: identification of target organelles. J Antimicrob Chemother. 2005;56:1034–1041. doi: 10.1093/jac/dki403. [DOI] [PubMed] [Google Scholar]
- 129.Menna-Barreto RF, Corrêa JR, Pinto AV, Soares MJ, de Castro SL. Mitochondrial disruption and DNA fragmentation in Trypanosoma cruzi induced by naphthoimidazoles synthesized from β-lapachone. Parasitol Res. 2007;101:895–905. doi: 10.1007/s00436-007-0556-1. [DOI] [PubMed] [Google Scholar]
- 130.Menna-Barreto RFS, Corrêa JR, Cascabulho CM, Fernandes MC, Pinto AV, Soares MJ, De Castro SL. Naphthoimidazoles promote different death phenotypes in Trypanosoma cruzi. Parasitology. 2009;136:499–510. doi: 10.1017/S0031182009005745. [DOI] [PubMed] [Google Scholar]
- 131.Codogno P, Meijer AJ. Atg5: more than an autophagy factor. Nat Cell Biol. 2006;8:1045–1047. doi: 10.1038/ncb1006-1045. [DOI] [PubMed] [Google Scholar]
- 132.Soares MJ. The reservosome of Trypanosoma cruzi epimastigotes: an organelle of the endocytic pathway with a role on metacyclogenesis. Mem Inst. Oswaldo Cruz. 1999;94:139–141. doi: 10.1590/s0074-02761999000700015. [DOI] [PubMed] [Google Scholar]
- 133.Sant'Anna C, Nakayasu ES, Pereira MG, Lourenço D, de Souza W, Almeida IC, Cunha-E-Silva NL. Subcellular proteomics of Trypanosoma cruzi reservosomes. Proteomics. 2009;9:1782–1794. doi: 10.1002/pmic.200800730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Cazzulo JJ, Stoka V, Turk V. Cruzipain, the major cysteine proteinase from the protozoan parasite Trypanosoma cruzi. Biol Chem. 1997;378:1–10. doi: 10.1515/bchm.1997.378.1.1. [DOI] [PubMed] [Google Scholar]
- 135.Coombs GH, Tetley L, Moss VA, Vickerman K. Three-dimensional structure of the Leishmania amastigote as revealed by computer-aided reconstruction from serial sections. Parasitology. 1986;92:13–23. doi: 10.1017/s0031182000063411. [DOI] [PubMed] [Google Scholar]
- 136.Waller RF, McConville MJ. Developmental changes in lysosome morphology and function Leishmania parasites. Int J Parasitol. 2002;32:1435–1445. doi: 10.1016/s0020-7519(02)00140-6. [DOI] [PubMed] [Google Scholar]
- 137.Besteiro S, Williams RA, Coombs GH, Mottram JC. Protein turnover and differentiation in Leishmania. Int J Parasitol. 2007;37:1063–1075. doi: 10.1016/j.ijpara.2007.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kumar S, Nei M, Dudley J, Tamura K. MEGA: A biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform. 2008;9:299–306. doi: 10.1093/bib/bbn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Hart DT, Opperdoes FR. The occurrence of glycosomes (microbodies) in the promastigote stage of four major Leishmania species. Mol Biochem Parasitol. 1984;13:159–172. doi: 10.1016/0166-6851(84)90110-5. [DOI] [PubMed] [Google Scholar]
- 140.Rosenzweig D, Smith D, Opperdoes F, Stern S, Olafson RW, Zilberstein D. Retooling Leishmania metabolism: from sand fly gut to human macrophage. FASEB J. 2008;22:590–602. doi: 10.1096/fj.07-9254com. [DOI] [PubMed] [Google Scholar]
- 141.Saunders EC, Souza DP, Naderer T, Sernee MF, Ralton JE, Doyle MA, et al. Central carbon metabolism of Leishmania parasites. Parasitology. 2010;137:1303–1313. doi: 10.1017/S0031182010000077. [DOI] [PubMed] [Google Scholar]
- 142.Frevert U, Engelmann S, Zougbede S, Stange J, Ng B, Matuschewski K, et al. Intravital observation of Plasmodium berghei sporozoite infection of the liver. PLoS Biol. 2005;3:192.. doi: 10.1371/journal.pbio.0030192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Amino R, Thiberge S, Martin B, Celli S, Shorte S, Frischknecht F, Ménard R. Quantitative imaging of Plasmodium transmission from mosquito to mammal. Nat Med. 2006;12:220–224. doi: 10.1038/nm1350. [DOI] [PubMed] [Google Scholar]
- 144.Sturm A, Amino R, van de Sand C, Regen T, Retzlaff S, Rennenberg A, et al. Manipulation of host hepatocytes by the malaria parasite for delivery into liver sinusoids. Science. 2006;313:1287–1290. doi: 10.1126/science.1129720. [DOI] [PubMed] [Google Scholar]
- 145.Morrissette NS, Sibley LD. Cytoskeleton of apicomplexan parasites. Microbiol Mol Biol Rev. 2002;66:21–38. doi: 10.1128/MMBR.66.1.21-38.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Jayabalasingham B, Bano N, Coppens I. Metamorphosis of the malaria parasite in the liver is associated with organelle clearance. Cell Res. 2010;20:1043–1059. doi: 10.1038/cr.2010.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Robson KJ, Frevert U, Reckmann I, Cowan G, Beier J, Scragg IG, et al. Thrombospondin-related adhesive protein (TRAP) of Plasmodium falciparum: expression during sporozoite ontogeny and binding to human hepatocytes. EMBO J. 1995;14:3883–3894. doi: 10.1002/j.1460-2075.1995.tb00060.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Sinnis P, Sim BK. Cell invasion by the vertebrate stages of Plasmodium. Trends Microbiol. 1997;5:52–58. doi: 10.1016/s0966-842x(97)84657-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kaiser K, Camargo, Kappe SH. Transformation of sporozoites into early exoerythrocytic malaria parasites does not require host cells. J Exp Med. 2003;197:1045–1050. doi: 10.1084/jem.20022100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Lim L, McFadden GI. The evolution, metabolism and functions of the apicoplast. Philos Trans R Soc Lond B Biol Sci. 2010;365:749–763. doi: 10.1098/rstb.2009.0273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Seglen PO. Gordon PBb 3-Methyladenine: specific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes. Proc Natl Acad Sci USA. 1982;79:1889–1892. doi: 10.1073/pnas.79.6.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Petiot A, Ogier-Denis E, Blommaart EF, Meijer AJ, Codogno P. Distinct classes of phosphatidylinositol 3'-kinases are involved in signaling pathways that control macroautophagy in HT-29 cells. J Biol Chem. 2000;275:992–998. doi: 10.1074/jbc.275.2.992. [DOI] [PubMed] [Google Scholar]
- 153.Vaid A, Ranjan R, Smythe WA, Hoppe HC, Sharma P. PfPI3K, a phosphatidylinositol-3 kinase from Plasmodium falciparum, is exported to the host erythrocyte and is involved in hemoglobin trafficking. Blood. 2010;115:2500–2507. doi: 10.1182/blood-2009-08-238972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Alberti A, Michelet X, Djeddi A, Legouis R. The autophagosomal protein LGG-2 acts synergistically with LGG-1 in dauer formation and longevity in C. elegans. Autophagy. 2010;6:622–633. doi: 10.4161/auto.6.5.12252. [DOI] [PubMed] [Google Scholar]
- 155.Yamada Y, Suzuki NN, Hanada T, Ichimura Y, Kumeta H, Fujioka Y, et al. The crystal structure of Atg3, an autophagy-related ubiquitin carrier protein (E2) enzyme that mediates Atg8 lipidation. J Biol Chem. 2007;282:8036–8043. doi: 10.1074/jbc.M611473200. [DOI] [PubMed] [Google Scholar]
- 156.Hanada T, Satomi Y, Takao T, Ohsumi Y. The amino-terminal region of Atg3 is essential for association with phosphatidylethanolamine in Atg8 lipidation. FEBS Lett. 2009;583:1078–1083. doi: 10.1016/j.febslet.2009.03.009. [DOI] [PubMed] [Google Scholar]
- 157.Kabeya Y, Mizushima N, Yamamoto A, Oshitani-Okamoto S, Ohsumi Y, Yoshimori T. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J Cell Sci. 2004;117:2805–2812. doi: 10.1242/jcs.01131. [DOI] [PubMed] [Google Scholar]
- 158.Kirisako T, Baba M, Ishihara N, Miyazawa K, Ohsumi M, Yoshimori T. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J Cell Biol. 1999;147:435–446. doi: 10.1083/jcb.147.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Weidberg H, Shvets E, Shpilka T, Shimron F, Shinder V, Elazar Z. LC3 and GATE-16/GABARAP subfamilies are both essential yet act differently in autophagosome biogenesis. EMBO J. 2010;29:1792–1802. doi: 10.1038/emboj.2010.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Zhou B, Boudreau N, Coulber C, Hammarback J, Rabinovitch M. Microtubule-associated protein 1 light chain 3 is a fibronectin mRNA-binding protein linked to mRNA translation in lamb vascular smooth muscle cells. J Clin Invest. 1997;100:3070–3082. doi: 10.1172/JCI119862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Cann GM, Guignabert C, Ying L, Deshpande N, Bekker JM, Wang L, et al. Developmental expression of LC3α and β absence of fibronectin or autophagy phenotype in LC3β knockout mice. Dev Dyn. 2008;237:187–195. doi: 10.1002/dvdy.21392. [DOI] [PubMed] [Google Scholar]
- 162.Swanson MS. Autophagy: eating for good health. J Immunol. 2006;177:4945–4951. doi: 10.4049/jimmunol.177.8.4945. [DOI] [PubMed] [Google Scholar]
- 163.Colombo MI. Autophagy: a pathogen driven process. IUBMB Life. 2007;59:238–242. doi: 10.1080/15216540701230503. [DOI] [PubMed] [Google Scholar]
- 164.Huang J, Klionsky DJ. Autophagy and human disease. Cell Cycle. 2007;6:1837–1849. doi: 10.4161/cc.6.15.4511. [DOI] [PubMed] [Google Scholar]
- 165.Deretic V. Autophagy in infection. Curr Opin Cell Biol. 2009;22:252–262. doi: 10.1016/j.ceb.2009.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Deretic V, Levine B. Autophagy, immunity and microbial adaptations. Cell Host Microbe. 2009;5:527–549. doi: 10.1016/j.chom.2009.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Andrade RM, Wessendarp M, Gubbels MJ, Striepen B, Subauste CS. CD40 induces macrophage anti-Toxoplasma gondii activity by triggering autophagy-dependent fusion of pathogen-containing vacuoles and lysosomes. J Clin Invest. 2006;116:2366–2377. doi: 10.1172/JCI28796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Subauste CS. CD40, autophagy and Toxoplasma gondii. Mem Inst Oswaldo Cruz. 2009;104:267–272. doi: 10.1590/s0074-02762009000200020. [DOI] [PubMed] [Google Scholar]
- 169.Melzer T, Duffy A, Weiss LM, Halonen SK. The gamma interferon (IFNγ)-inducible GTP-binding protein IGTP is necessary for Toxoplasma vacuolar disruption and induces parasite egression in IFNγ-stimulated astrocytes. Infect Immun. 2008;76:4883–4894. doi: 10.1128/IAI.01288-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Ling YM, Shaw MH, Ayala C, Coppens I, Taylor GA, Ferguson DJ, Yap GS. Vacuolar and plasma membrane stripping and autophagic elimination of Toxoplasma gondii in primed effector macrophages. J Exp Med. 2006;203:2063–2071. doi: 10.1084/jem.20061318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Zhao Z, Fux B, Goodwin M, Dunay IR, Strong D, Miller BC, et al. Autophagosome-independent essential function for the autophagy protein Atg5 in cellular immunity to intracellular pathogens. Cell Host Microbe. 2008;4:458–469. doi: 10.1016/j.chom.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Zhao YO, Khaminets A, Hunn JP, Howard JC. Disruption of the Toxoplasma gondii parasitophorous vacuole by IFNgamma-inducible immunity-related GTPases (IRG proteins) triggers necrotic cell death. PLoS Pathog. 2009;5:1000288. doi: 10.1371/journal.ppat.1000288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Halonen SK. Role of autophagy in the host defense against Toxoplasma gondii in astrocytes. Autophagy. 2009;5:268–269. doi: 10.4161/auto.5.2.7637. [DOI] [PubMed] [Google Scholar]
- 174.Wang Y, Weiss LM, Orlofsky A. Host cell autophagy is induced by Toxoplasma gondii and contributes to parasite growth. J Biol Chem. 2009;284:1694–1701. doi: 10.1074/jbc.M807890200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Coppens I, Dunn JD, Romano JD, Pypaert M, Zhang H, Boothroyd JC, Joiner KA. Toxoplasma gondii sequesters lysosomes from mammalian hosts in the vacuolar space. Cell. 2006;125:261–274. doi: 10.1016/j.cell.2006.01.056. [DOI] [PubMed] [Google Scholar]
- 176.Wang Y, Weiss LM, Orlofsky A. Intracellular parasitism with Toxoplasma gondii stimulates mammalian-target-of-rapamycin-dependent host cell growth despite impaired signalling to S6K1 and 4E-BP1. Cell Microbiol. 2009;11:983–1000. doi: 10.1111/j.1462-5822.2009.01305.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Romano PS, Arboit MA, Vázquez CL, Colombo MI. The autophagic pathway is a key component in the lysosomal dependent entry of Trypanosoma cruzi into the host cell. Autophagy. 2009;5:6–18. doi: 10.4161/auto.5.1.7160. [DOI] [PubMed] [Google Scholar]
- 178.Tardieux I, Webster P, Ravesloot J, Boron W, Lunn JA, Heuser JE, Andrews NW. Lysosome recruitment and fusion are early events required for trypanosome invasion of mammalian cells. Cell. 1992;71:1117–1130. doi: 10.1016/s0092-8674(05)80061-3. [DOI] [PubMed] [Google Scholar]
- 179.Rodríguez A, Samoff E, Rioult MG, Chung A, Andrews NW. Host cell invasion by trypanosomes requires lysosomes and microtubule/kinesin-mediated transport. J Cell Biol. 1996;134:349–362. doi: 10.1083/jcb.134.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Woolsey AM, Sunwoo L, Petersen CA, Brachmann SM, Cantley LC, Burleigh BA. Novel PI 3-kinase-dependent mechanisms of trypanosome invasion and vacuole maturation. J Cell Sci. 2003;116:3611–3622. doi: 10.1242/jcs.00666. [DOI] [PubMed] [Google Scholar]
- 181.Andrade LO, Andrews NW. The Trypanosoma cruzi-host-cell interplay: location, invasion, retention. Nat Rev Microbiol. 2005;3:819–823. doi: 10.1038/nrmicro1249. [DOI] [PubMed] [Google Scholar]
- 182.Ley V, Robbins ES, Nussenzweig V, Andrews NW. The exit of Trypanosoma cruzi from the phagosome is inhibited by raising the pH of acidic compartments. J Exp Med. 1990;171:401–413. doi: 10.1084/jem.171.2.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Pinheiro RO, Nunes MP, Pinheiro CS, D'Avila H, Bozza PT, Takiya CM, et al. Induction of autophagy correlates with increased parasite load of Leishmania amazonensis in BALB/c but not C57BL/6 macrophages. Microbes Infect. 2009;11:181–190. doi: 10.1016/j.micinf.2008.11.006. [DOI] [PubMed] [Google Scholar]
- 184.Bozza PT, Melo RC, Bandeira-Melo C. Leukocyte lipid bodies regulation and function: contribution to allergy and host defense. Pharmacol Ther. 2007;113:30–49. doi: 10.1016/j.pharmthera.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 185.Guimarães ET, Santos LA. Ribeiro dos Santos R, Teixeira MM, dos Santos WL, Soares MB, Role of interleukin-4 and prostaglandin E2 in Leishmania amazonensis infection of BALB/c mice. Microbes Infect. 2006;8:1219–1226. doi: 10.1016/j.micinf.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 186.Chaussepied M, Langsley G. Theileria-induced leukocyte transformation: an example of oncogene addiction? In: Kaufmann SHE, Rouse BT, Sacks DL, editors. The Immune Response to Infection. Washington DC: ASM Press; 2010. In press. [Google Scholar]
- 187.Morrison WI. Progress towards understanding the immunobiology of Theileria parasites. Parasitology. 2009;136:1415–1426. doi: 10.1017/S0031182009990916. [DOI] [PubMed] [Google Scholar]
- 188.Rennenberg A, Lehmann C, Heitmann A, Witt T, Hansen G, Nagarajan K, et al. Exoerythrocytic Plasmodium parasites secrete a cysteine protease inhibitor involved in sporozoite invasion and capable of blocking cell death of host hepatocytes. PLoS Pathog. 2010;6:1000825. doi: 10.1371/journal.ppat.1000825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Münz C. Antigen processing via autophagy—not only for MHC class II presentation anymore? Curr Opin Immunol. 2010;22:89–93. doi: 10.1016/j.coi.2010.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Pearce MJ, Mintseris J, Ferreyra J, Gygi SP, Darwin KH. Ubiquitin-like protein involved in the proteasome pathway of Mycobacterium tuberculosis. Science. 2008;322:1104–1107. doi: 10.1126/science.1163885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Cazzulo JJ, Stoka V, Turk V. The major cysteine proteinase of Trypanosoma cruzi: a valid target for chemotherapy of Chagas disease. Curr Pharm Des. 2001;7:1143–1156. doi: 10.2174/1381612013397528. [DOI] [PubMed] [Google Scholar]
- 192.Docampo R, Moreno SN. Current chemotherapy of human African trypanosomiasis. Parasitol Res. 2003;90:10–13. doi: 10.1007/s00436-002-0752-y. [DOI] [PubMed] [Google Scholar]
- 193.Steverding D, Tyler KM. Novel antitrypanosomal agents. Expert Opin Investig Drugs. 2005;14:939–955. doi: 10.1517/13543784.14.8.939. [DOI] [PubMed] [Google Scholar]
- 194.Alvarez VE, Kosec G, Sant Anna C, Turk V, Cazzulo JJ, Turk B. Blocking autophagy to prevent parasite differentiation: a possible new strategy for fighting parasitic infections? Autophagy. 2008;4:361–363. doi: 10.4161/auto.5592. [DOI] [PubMed] [Google Scholar]
- 195.Duschak VG, Couto AS. Cruzipain, the major cysteine protease of Trypanosoma cruzi: a sulfated glycoprotein antigen as relevant candidate for vaccine development and drug target. A review. Curr Med Chem. 2009;16:3174–3202. doi: 10.2174/092986709788802971. [DOI] [PubMed] [Google Scholar]
- 196.Selzer PM, Pingel S, Hsieh I, Ugele B, Chan VJ, Engel JC, et al. Cysteine protease inhibitors as chemotherapy: lessons from a parasite target. Proc Natl Acad Sci USA. 1999;96:11015–11022. doi: 10.1073/pnas.96.20.11015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Franke de Cazzulo BM, Martínez J, North MJ, Coombs GH, Cazzulo JJ. Effects of proteinase inhibitors on the growth and differentiation of Trypanosoma cruzi. FEMS Microbiol Lett. 1994;124:81–86. doi: 10.1111/j.1574-6968.1994.tb07265.x. [DOI] [PubMed] [Google Scholar]
- 198.McKerrow JH, Engel JC, Caffrey CR. Cysteine protease inhibitors as chemotherapy for parasitic infections. Bioorg Med Chem. 1999;7:639–644. doi: 10.1016/s0968-0896(99)00008-5. [DOI] [PubMed] [Google Scholar]
- 199.Stechmann A, Cavalier-Smith T. The root of the eukaryote tree pinpointed. Curr Biol. 2003;13:665–666. doi: 10.1016/s0960-9822(03)00602-x. [DOI] [PubMed] [Google Scholar]
- 200.Clayton C, Adams M, Almeida R, Baltz T, Barrett M, Bastien P, et al. Genetic nomenclature for Trypanosoma and Leishmania. Mol Biochem Parasitol. 1998;97:221–224. doi: 10.1016/s0166-6851(98)00115-7. [DOI] [PubMed] [Google Scholar]